Abstract
The life cycles of human papillomaviruses (HPVs) are intimately linked to the differentiation program of infected stratified epithelia, with both viral gene expression and replication being maintained at low levels in undifferentiated basal cells and increased upon host cell differentiation. We recently identified, in HPV-16, a negative regulatory element between the epithelial-cell-specific enhancer and the E6 promoter that is capable of silencing E6 promoter activity, and we termed this element a papillomavirus silencing motif (PSM) and the unknown cellular factor that bound to it PSM binding protein (PSM-BP). Here we show that the homologous genomic segments of six other distantly related genital HPV types contain a PSM that binds PSM-BP and is capable of repressing transcription. Conservation of the PSM suggests that it is indispensable for the HPV life cycle. Purification, electrophoretic mobility shift assay experiments, and the use of specific antibodies proved that the cellular factor PSM-BP is identical to a previously described transcriptional repressor, the CCAAT displacement protein (CDP), also referred to as the human Cut protein (Cut). CDP/Cut repression of HPV-16 may stem from the modification of specifically positioned nucleosomes, as suggested by transcriptional stimulation under the influence of the histone deacetylase inhibitor trichostatin A. CDP/Cut is an important developmental regulator in several different tissues. It was recently shown that CDP/Cut is expressed in basal epithelial cells but not in differentiated primary keratinocytes. This suggests the possibility that repression by PSM couples HPV transcription to the stratification of epithelia. In each of the studied HPV types, the two CDP/Cut binding sites of PSM overlap with the known or presumed binding sites of the replication initiator protein E1. Transfection of CDP/Cut expression vectors into cells that support HPV-16 or HPV-31 replication leads to the elimination of viral episomes. Similarly, two PSM-like motifs overlapping the E1 binding site of bovine papillomavirus type 1 bind CDP/Cut, and CDP/Cut overexpression reduces the copy number of episomally replicating BPV-1 genomes in mouse fibroblasts. CDP/Cut appears to be a master regulator of HPV transcription and replication during epithelial differentiation, and PSMs are important cis-responsive targets of this repressor.
Human papillomaviruses (HPVs) infect mucosal and cutaneous epithelia and cause benign and malignant lesions, such as common warts, genital warts, and cervical cancer (29, 67). Transcription of HPVs is regulated in a complex manner in response to the identity of the infected epithelial cell type, the differentiation state of the stratified epithelium, the physiological state of the host, and the episomal or chromosomally integrated state of the viral genome. Most of the cis-responsive elements that mediate the activity of cellular and viral factors on HPV gene regulation are located in the long control region (LCR), which lies between the 3′ terminus of the L1 capsid gene and the start of the E6 oncogene. With a size of 850 bp in HPV-16 and a similar length in other genital HPVs, the LCR takes up about 11% of the 7.9-kb circular DNA genome. When one compares remotely related HPV types, one finds that their genes are clearly homologous but their LCRs have diverged so much that they cannot be unequivocally aligned and their nucleotide sequences lack long, similar segments. However, when one compares the LCRs of more closely related HPV types, such as the large group of genital HPVs, one finds many short similar cis-responsive elements, which are activated by a defined subset of the cellular transcription factors of the infected cell. This conservation of sequence and function suggests that these particular mechanisms are so appropriate for the biology of these HPV types that they could not undergo evolutionary changes (13, 44). Here we study such an element that is conserved among genital HPVs, and for the first time we describe an element that represses both transcription and replication, most probably in response to the differentiation of stratified epithelia.
The central part of the LCR of genital HPVs functions as an epithelial-cell-specific transcriptional enhancer (14, 23, 50), and the 3′ part contains the replication origin and the E6 promoter (12, 16, 19, 31, 44, 59). We have recently detected a novel transcriptional silencer, which we called a papillomavirus silencing motif (PSM), in this 3′ part of the LCR of HPV-16 (45). The PSM is physically separate from and functionally independent of another repressor domain, 50 to 100 bp further in the 5′ direction, which consists of five binding sites for the transcription factor YY1 (9, 38, 46). We have defined the PSM as a 25-bp segment, containing two direct repeats of the sequence 5′-TAYAATAAT-3′ spaced by 7 nucleotides consisting of the sequence 5′-ACTAAA*C-3′, where A* is position 1 of the genomic map of HPV-16. Each of the two TAYAATAAT repeats binds the same factor, which we called the PSM binding protein (PSM-BP). Binding of a PSM-BP dimer to the two flanking repeats occurred cooperatively and led to synergistic transcriptional repression.
In this study we obtained evidence that all genital HPV types and bovine papillomavirus type 1 (BPV-1) contain very similar sequence motifs, and we observed identical binding properties for PSM-BP in the eight papillomavirus types that we examined. All of these motifs are located in a similar position between the enhancer and the E6 promoter, and they overlap with documented or presumed binding sites for the replication initiator protein E1. We identified PSM-BP as the previously described protein CCAAT displacement protein (CDP/Cut), and we show that CDP/Cut represses the transcription and replication of genital HPVs. CDP/Cut is a known repressor of numerous cellular genes that are developmentally regulated in a variety of cell types (8, 34). CDP/Cut binds to and represses these cell-type-specific promoters in developmental stages when these specific genes are not required. Tissue-specific activation of these promoters results upon terminal differentiation when CDP/Cut availability decreases and promoter binding is lost. A similar phenomenon has been reported for stratified epithelia (1). Consequently, it is likely that CDP/Cut is a general repressor of transcription and replication of genital HPVs and a key regulatory device in coupling these two functions to the differentiation program of the infected stratified epithelium.
MATERIALS AND METHODS
Plasmid constructs.
All constructs used in functional assays were based on the chloramphenicol acetyltransferase (CAT) reporter plasmid pBLCAT3dH/N, a modified version of pBLCAT3 (51). The derivative p80 (46) contains HPV-16 promoter sequences from nucleotides 16 to 80 cloned into the BglII and XhoI sites of pBLCAT3dH/N and contains a conserved Sp1 and two E2 binding sites and a TATA box, typical for all genital HPVs (61). The construct p80SV was created by cloning the EcoRI fragment containing the simian virus 40 (SV40) enhancer from the oligonucleotide vector (64) into pBluescript SK(+) (Stratagene). This fragment was then excised with HindIII and BamHI and cloned into p80. Double-stranded DNA oligonucleotides containing HPV-16 sequences (genomic position 7883-15), either wild type or mutated (m), and with complementary BamHI and BglII ends, were cloned into the BamHI site of p80SV to give the constructs p80SV16 or p80SV16m respectively. Similar double-stranded oligonucleotides (see Fig. 2) containing homologous sequences flanking the E1 binding sites of other HPVs (HPV-2, HPV-11, HPV-18, HPV-31, HPV-33, and HPV-45) were cloned using an identical strategy to generate the plasmids p80SV2, p80SV11, p80SV18, p80SV31, p80SV33, and p80SV45. The structures of all constructs were confirmed by determination of their nucleotide sequence. The structures of pBR322-HPV-31 (22, 27), used in replication, and of the CDP expression vector pMT2-CDP and the parental vector pMT2 (42) have been published.
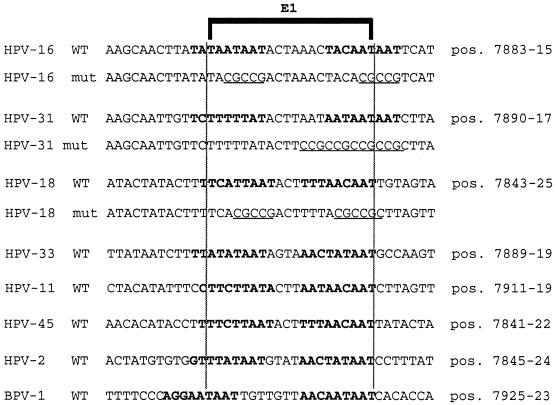
FIG. 2.
Alignments of presumed and known binding sites for the replication factor E1 in seven genital HPV types and in BPV-1 coincide with binding sites for the transcriptional repressor PSM-BP (or CDP/Cut, respectively). The alignment of E1 binding sites was done as in a study by Holt and Wilson (26) extended by a sequence comparison (44). A bracket indicates the presumed E1 consensus binding sequence. Bold letters identify the PSM of HPV-16 and similarities in six other HPV types. Since CDP/Cut binding sites do not show a strict consensus sequence, with multiple 5′-TAAT-3′ 5′-CAAT-3′ motifs at variable positions and orientations being the hallmark of the generally AT-rich sequences, the identification of CDP/Cut targets in these PSMs is conjectural. The mutations (underlined) were aimed at eliminating all of the 5′-TAAT-3′ and most of the 5′-CAAT-3′ elements. All of these sequences overlap with the position 1 of the circular genomic map. The exact genomic position is shown on the right. The sequences shown represent oligonucleotides that were examined by EMSA and cloned in the form of BamHI-BglII fragments into the CAT expression vector p80SV for functional tests. Oligonucleotides that were inserted in the sense orientation in this vector were identified by DNA sequencing.
Cell culture and gene expression assays.
C33A, an HPV-free cell line derived from a cervical carcinoma, was cultured in Dulbecco's Modified Eagle's medium containing 10% fetal calf serum, plated onto 10-cm culture dishes, and transfected at 50 to 70% confluency with Lipofectin reagent (GIBCO-BRL). For each transfection, 30 μl of Lipofectin was mixed with 5 μg of DNA in 1 ml of medium and left at room temperature for 15 min before being added to 9 ml of medium. After 18 to 24 h, the medium containing Lipofectin was replaced with 10 ml of Lipofectin-free medium, and the cells were incubated for a further 24 h before harvesting.
Chloramphenicol acetyltransferase (CAT) assays were performed as modified by our laboratory (11). CAT activities were reported as picomoles of chloramphenicol acetylated per minute per milligram of protein extract and were measured by quantification of radioactive spots on thin-layer chromatograms. Each value represents the average of three to six independent transfections with at least two different DNA preparations.
The cell line Cho 4.15 (E1/E2) was a gift from M. Ustav and I. Ilves. ID13, a mouse fibroblast line derived from c127 with episomally replicating copies of the BPV-1, has been maintained in our laboratory for more than 10 years and was originally obtained from P. M. Howley (32a).
For stimulation by trichostatin A (TSA), cells were electroporated with vector DNA and incubated in growth medium for 24 h before the addition of TSA in ethanolic solution to a final concentration of 100 μg/ml; luciferase activity was determined after another 24 h.
Electrophoretic mobility shift assay (EMSA).
A 50-ng portion of annealed oligonucleotide was labelled with [32P]dATP and [32P]dCTP nucleotides by using Klenow polymerase. Approximately 250 pg of purified labelled probe with an activity of approximately 20,000 cpm was used in a previously described standard reaction (46). Samples were run at 200 V for 2 h on a 4% polyacrylamide gel containing 0.25× Tris-borate-EDTA buffer (TBE). The gels were transferred onto blotting paper, dried for 1 h, and exposed to autoradiographic film. For supershift experiments, 1 μl (1.5 μg) of anti-CDP antiserum (42) or the same amount of a preimmune serum was added to the reaction mixture, radiolabelled probe was added, and the mixture was incubated on ice for 10 min.
Purification of proteins by column chromatography.
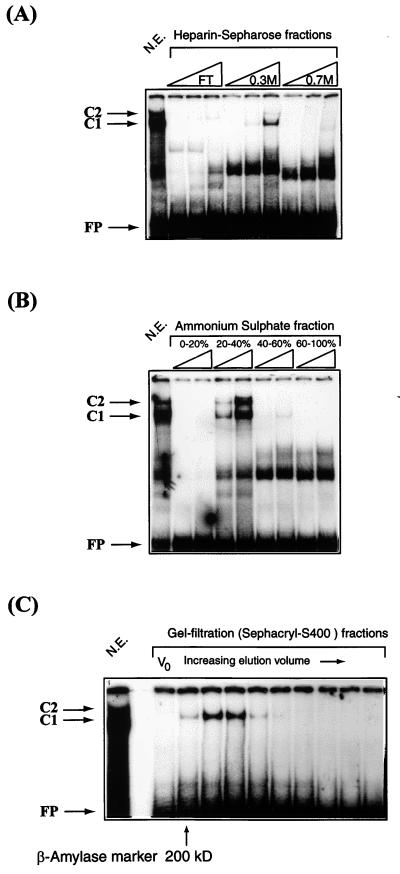
To enrich the PSM-BP, we either prepared nuclear extract from HeLa cells ourselves (17) or purchased it from the Computer Cell Culture Center (Brussels, Belgium). During fractionation, binding of proteins to an HPV-16 oligonucleotide with the two PSMs was monitored in EMSAs. As a first step toward purification, 10 ml of nuclear extract corresponding to a protein content of 150 mg was applied to a heparin-Sepharose column (Pharmacia), previously equilibrated with 5 column volumes of buffer D (0.1 M KCl, 20 mM HEPES [pH 7.9], 20% [vol/vol] glycerol, 0.2 mM EDTA, 1 mM dithiothreitol, 0.5 mM phenylmethylsulfonyl fluoride). After loading of nuclear extract, the column was washed with 2 volumes of buffer D and bound proteins were eluted stepwise with increasing concentrations of KCl in buffer D. PSM-BP EMSA activity was observed in the 0.3 M fraction. This fraction was dialyzed against buffer D and fractionated by ammonium sulfate precipitation. Binding activity was found in the 20 to 40% ammonium sulfate precipitate after overnight dialysis against buffer D. Then 200-μl volumes of active fractions were loaded onto a Sephacryl S400 column (Pharmacia) equilibrated with buffer D. The EMSA activity eluted with a molecular mass of around 200 kDa by comparison with a β-amylase size marker.
In vivo replication assays.
To support the replication of HPV genomes or chimeric HPV constructs, we used the cell line Cho4.15, which expresses the BPV-1 E1 and E2 proteins from stably transfected intrachromosomal E1 and E2 genes (48). The cells were transfected by electroporation of 3 μg either of pBluescript KS harboring a 500-bp XhoI-HindIII fragment of the LCR of HPV-16 with the enhancer, the replication origin, and the E6 promoter or of HPV-31 genomes free of plasmid sequences. The latter were obtained by separating them from vector sequences of the pBR322-HPV-31 construct by EcoRI digestion and religation of the 8 kb viral DNA (27). DNAs were mixed with the cells and subjected to 250 V and 960 μF with a Bio-Rad gene pulser. The transfected cells were cultivated in 10-cm plates and harvested after 48 h. The cells were lysed by an alkali lysis method (37) and centrifuged, and the low-molecular-weight DNA was precipitated by addition of 0.7 volume of isopropanol to the supernatant. The pellets were dissolved in 200 μl of a buffer containing 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, 10 mM EDTA, 0.2% sodium dodecyl sulfate (SDS), and 100 μg of proteinase K per ml and incubated for 2 h at 37°C. After treatment with phenol-chloroform-isoamyl alcohol and ethanol precipitation, the pelleted DNA was washed with 70% ethanol and dissolved in TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). To linearize the DNA, the samples were digested with 10 U of EcoRI for HPV-16 and 10 U of HpaI for HPV-31 and concomitantly with 10 U of DpnI to cut the originally transfected, bacterially replicated, and therefore methylated DNA. The restriction digests were incubated for 5 h at 37°C, and the DNA was purified by standard procedures, run on 0.8% agarose gels, and blotted onto a Hybond-N nylon membrane. After the membrane was baked at 80°C for 2 h, 32P-end-labelled probes derived from HPV-16, HPV-31, and BPV-1, respectively, were hybridized against the blotted DNA. The membrane was washed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–10% SDS for 20 min at 65°C and autoradiographed.
RESULTS
Sequences resembling the HPV-16 PSM are present between the enhancers and promoters of all genital HPV types and bind the same cellular factor.
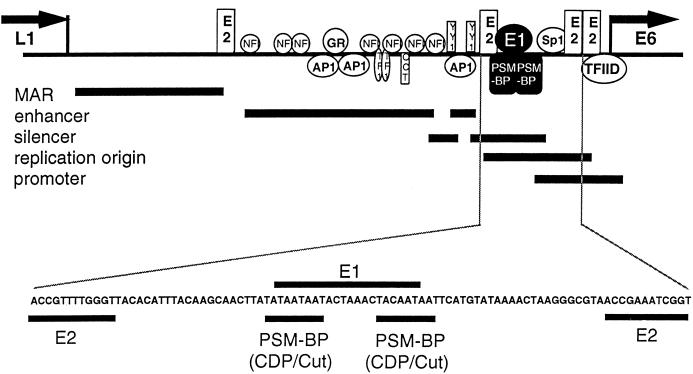
All genital HPV types appear to have an epithelial-cell-specific enhancer (14, 18, 23, 24, 32, 50) that is flanked by two E2 binding sites, roughly 150 and 550 bp upstream of the E6 promoter (for overviews, see references 44 and 50). The E6 promoter is modulated by an Sp1 site and two further E2 binding sites (16, 59). The 60-bp region upstream of the Sp1 site toward the enhancer is very AT rich in all genital HPVs (41). A 25-bp segment within this AT-rich segment of HPV-16 represses epithelial-cell enhancer-dependent transcription from the E6 promoter (45). We refer to this segment as PSM and to the protein that binds and functionally activates a direct repeat of the sequence 5′-TAYAATAAT-3′, within this segment, as PSM-BP. The 60-bp AT-rich region and its two flanking E2 binding sites seem to be the replication origin of all papillomaviruses, as was shown for BPV-1 (26, 39, 40, 61, 62), HPV-11 (31, 49), HPV-16 (20), HPV-18 (57), and HPV-31 (21, 22, 27). Replication is initiated by the E2 protein bound to either of the two flanking binding sites (22, 35, 54, 57) directing the E1 replication initiation protein to an AT-rich 18-mer binding site (26; for an alignment, see reference 44). Figure 1 summarizes these elements of HPV-16, and shows that the E1 binding site overlaps with the PSM.
FIG. 1.
Location of the PSM, the overlapping binding sites of the replication protein E1, and other major binding sites for transcriptional regulators in the LCR of HPV-16. The identification of the nuclear MAR is described in reference 60, the enhancer and its elements are described in references 4, 11, 13, 14, 23, and 44, the silencer elements are described in references 38, 45, and 46, the replication origin is described in reference 15, and the promoter elements are described in references 16 and 59. Abbreviations: L1, L1 gene; E6, E6 gene; NFI, nuclear factor 1; GR, glucocorticoid receptor; AP1, activator protein 1; TF1, transcription enhancer factor 1; oct, octamer binding factor 1.
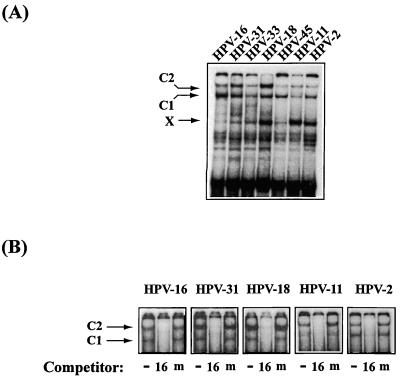
An examination of this genomic region of many genital HPV types reveals sequences bearing similarity to the two 5′-TAYAATAAT-3′ repeats of the HPV-16 PSM (41). To determine whether these sequences could bind the same cellular factor, PSM-BP, as HPV-16, we analyzed sequences from a number of genital HPVs, namely, those from HPV-2, HPV-11, HPV-16, HPV-18, HPV-31, HPV-33, and HPV-45 (Fig. 2). Figure 3A depicts the results of an EMSA in which double-stranded oligonucleotides containing the sequences shown in Fig. 2 were radiolabelled and used as probes for HeLa nuclear extract. The characteristic low-mobility complexes (denoted C1 and C2) observed with the HPV-16 probe were seen with all six other HPV sequences. Based on a mutational analysis of HPV-16, we had proposed that C1 may represent a monomer of PSM-BP complexed with one 5′-TAYAATAAT-3′ submotif and that C2 may represent a PSM-BP dimer, which binds efficiently to the 5′-TAYAATAAT-3′ repeat within PSM but may form inefficiently on singular binding sites. It cannot be excluded, however, that C2 represents a heterodimer. Several fast-migrating bands most probably stem from nonspecifically binding proteins, while one additional complex, X, is barely visible in HPV-16 and some other types but appears as a strong band in HPV-18, HPV-11, and HPV-2. The nature of the protein that gives rise to complex X is not known (see below).
FIG. 3.
The cellular factor PSM-BP binds to the homologous genomic region of seven different HPV types. (A) EMSA studies with probes containing PSM-like sequences (Fig. 2) including the origin of replication of HPV-16, HPV-31, HPV-33, HPV-18, HPV-45, HPV-11, and HPV-2 give rise to the same slow-migrating complexes C1 and C2 previously seen with the HPV-16 PSM. The nature of complex X, which is barely visible in HPV-16 but strong in several other HPV types, is not known. (B) The C1 and C2 complexes of HPV-31, HPV-18, HPV-11, and HPV-2 are competitively inhibited by an HPV-16 probe containing a wild-type PSM (labelled 16 and consisting of the sequence 5′-CTAAACTACAATAATTCAT-3′) but not by one containing mutations within the PSM that inhibit PSM-BP binding (labelled m and consisting of the sequence 5′-CTAAACTACACGCCGTCAT-3′). These results suggest that the PSM-BP complexes C1 and C2 that form with the sequences of these four HPV types are the same complexes previously described for HPV-16 (45).
Five of these seven HPV types were studied further in competition experiments, and Fig. 3B shows that formation of the C1 and C2 complexes could be successfully inhibited by an oligonucleotide with the wild-type HPV-16 5′-TACAATAAT-3′ sequence (labelled 16) but not by a mutant HPV-16 sequence, 5′-TACACGCCG-3′, that fails to bind PSM-BP (labelled m). We conclude from this that the seven studied HPV types (and, as sequence inspection suggests, probably all genital HPVs) have motifs with similar PSM-BP binding capabilities.
PSMs from different genital HPV types function as transcriptional repressors.
We published recently the finding that the contiguous LCR of many different HPV types has a dramatically lower transcriptional activity than chimeric constructs in which the enhancers of the same viruses are cloned in front of a canonical Sp1-activated HPV E6 promoter (50). It became clear to us that these two groups of constructs differed from one another by the absence in the latter group of the 60-bp AT-rich segments containing potential PSM elements. This strongly suggests the presence of a silencer in these AT-rich segments of each of these HPV types. To verify this and to confirm that the PSM is the element responsible for silencing the contiguous LCRs, we decided to compare the potential PSMs of different HPV types in standardized test constructs.
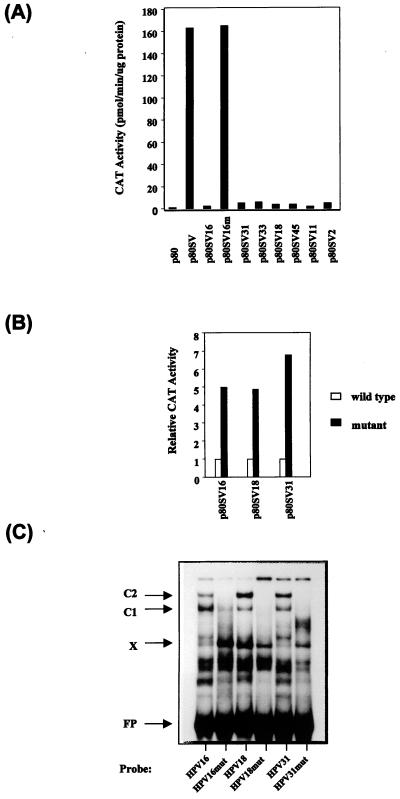
We had previously observed that the HPV-16 PSM had similar silencing activity whether it was positioned between the homologous HPV-16 enhancer and promoter or when it was tested in the context of heterologous elements such as the SV40 enhancer (45). To investigate whether the potential PSMs from the HPV-2, HPV-11, HPV-18, HPV-31, HPV-33, and HPV-45 would indeed have a silencing activity similar to that of HPV-16, we cloned the sequences used in the EMSA experiments into the p80SV CAT reporter plasmid between the SV40 enhancer and the HPV-16 E6 promoter. Figure 4A shows the outcome of CAT assays with extracts of C33A cells transiently transfected with these six constructs and control plasmids. As seen in the figure, the SV40 enhancer strongly activates the HPV-16 promoter in construct p80SV compared with the promoter activity alone for construct p80. This activity is almost completely repressed by the presence of the HPV-16 PSM in p80SV16 but not by a mutant sequence of the PSM in p80SV16m (Fig. 2). The repressing effect of the sequences of all of the six other HPV types is similar to that of HPV-16, identifying them as functional PSMs.
FIG. 4.
The PSM-like elements of seven different HPV types function as strong transcriptional repressors. (A) As previously described (45), the HPV-16 sequences containing a functional PSM repress the activity of the heterologous SV40 enhancer (p80SV16) while corresponding sequences containing mutations within the 5′-TAYAATAAT-3′ motifs fail to do so (p80SV16m). Likewise, the PSM-like sequences of six other HPV types (HPV-31, HPV-33, HPV-18, HPV-45, HPV-11, and HPV-2) repress CAT expression comparably. (B) Mutation of the PSM sequences of HPV-16, HPV-18, and HPV-31 (Fig. 2) releases the repression of the SV40 enhancer–HPV-16 promoter chimera. (C) When tested in EMSA in form of free oligonucleotides, the same mutations as functionally tested in the experiment in panel B fail to give rise to the characteristic C1 and C2 complexes of PSM-BP. The nature of complex X, which is barely visible with the HPV-16 wild type but strong in several other HPV types and in the HPV-16 mutant, is not known. The fastest-migrating complex above the free probe (FP), which is visible with wild-type sequences but absent with mutants, is likely to be a degradation product of the C1/C2 protein.
We had observed that the deletion of one of the two 5′-TAYAATAAT-3′ repeats in HPV-16 led to a partial release of repression. Here we have examined the functional effect of multiple point mutations that altered most of the 5′-TAAT-3′ and 5′-CAAT-3′ motifs within the PSMs. Figure 4B shows that these mutations of the PSMs of HPV-16, HPV-18, and HPV-31 (Fig. 2) released the repressor activity of the PSMs. The wild-type and mutant HPV-16 vectors used in the experiments in Fig. 4A and B are identical, but the two panels were derived from separate transfections. Figure 4C shows that these mutations no longer allow the formation of complexes C1 and C2 on the free oligonucleotides. The faster-migrating complex X is not affected by these mutations in HPV-18 and HPV-31 but increases in amount in HPV-16. We do not believe that the factor giving rise to complex X has any stimulatory effect on the transcription of these constructs, since deletion of the whole HPV-16 PSM segment (including the binding site for X) (45) has the same effect as these point mutations that increase complex X formation. Together, these results suggest that the functional organization of the PSMs of different HPV types is conserved.
The cellular factor PSM-BP is identical to the differentiation-specific transcriptional repressor CDP/Cut.
Since our findings had confirmed the PSM-BP as an important transcriptional regulator of genital HPVs with a binding site overlapping the replication origin, we were interested in identifying and characterizing this factor. Figure 5 documents our attempts to purify PSM-BP from crude HeLa nuclear extract. The first step involved a heparin-Sepharose ion-exchange column. After binding of HeLa nuclear extract, maximal PSM-BP activity eluted in the 0.3 M KCl fraction (Fig. 5A). A second enrichment was achieved by precipitation of the proteins in this fraction by using a fractionated ammonium sulfate precipitation and screening by EMSA for PSM-BP activity. Maximal PSM-BP activity was observed in the 20 to 40% fraction (Fig. 5B). This fraction was further purified on a Sephacryl-S400 column by gel filtration (Fig. 5C). A series of molecular mass markers was used to provide an indication of the size of PSM-BP, and it can be seen from Fig. 5C that it elutes close to the position of the β-amylase protein, which has a molecular mass of approximately 200 kDa.
FIG. 5.
The purification profile of PSM-BP, as monitored by EMSA, is similar to that of the differentiation-specific factor CDP/Cut. (A) Ion-exchange chromatography purification of HeLa nuclear extracts on a heparin-Sepharose column demonstrates that PSM-BP binding activity is maximal in the 0.3 M KCl fraction. FT, flowthrough. (B) Ammonium sulfate precipitation of PSM-BP-enriched fractions of the heparin-Sepharose chromatography defines PSM-BP activity in the 20 to 40% fraction. (C) A gel filtration experiment, with the 20 to 40% ammonium sulfate fraction from the heparin-Sepharose column, indicates that PSM-BP is a very large protein of approximately 180 kDa (as defined by the β-amylase molecular mass marker). V0, void volume. The purification profile of PSM-BP presented here is consistent with that described for the differentiation-specific transcriptional repressor CDP/Cut (42). FP, free probe.
We considered the possibility that PSM-BP is a previously known transcription factor. Only a few repressors have been characterized so far from human and mammalian cells, and one of them, the CCAAT displacement protein (CDP/Cut), is a large protein with a size similar to that observed for PSM-BP. CDP/Cut consists of 1,505 amino acid residues, with a calculated molecular mass of 165 kDa, and migrates in SDS-polyacrylamide gel electrophoresis with an apparent molecular mass of 180 to 190 kDa. Moreover, CDP/Cut demonstrates a purification profile similar to that of PSM-BP (42) and has a propensity for binding AT-rich sequences (3).
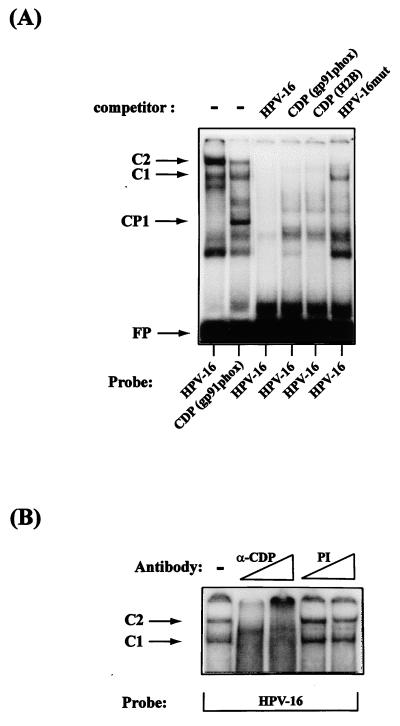
To determine whether PSM-BP and CDP/Cut were the same protein, we carried out a series of EMSA studies including competition experiments with different regulatory sequences that have previously been shown to interact with CDP/Cut. Figure 6A shows that sequences from the phagocyte-specific cytochrome heavy-chain gene promoter, a well-characterized binding site of CDP/Cut, give rise to complexes with identical mobilities to the HPV-16 PSM-BP complexes C1 and C2. The fact that the C2 complex is weaker than C1 in the gp91-phox site may stem from the fact that formation of a potential dimer occurs less efficiently on this singular binding site than on the dimeric binding sites of HPV-16. In competition experiments, the gp91-phox probe and another well-known CDP/Cut binding site from a Psammechinus miliaris histone H2B promoter (8) competitively inhibit the formation of C1 and C2 on the HPV-16 probe. Further evidence suggesting that PSM-BP is CDP/Cut is presented in Fig. 6B, where a specific anti-CDP/Cut polyclonal antibody results in the loss of C1 and C2 complexes from the HPV-16 probe while a preimmune serum fails to prevent C1 and C2 formation. From the combination of these data, we conclude that PSM-BP is identical to CDP/Cut.
FIG. 6.
The repressor PSM-BP of HPV transcription is identical to CDP/Cut. (A) An EMSA shows similarity of the complexes C1 and C2 generated by an HPV-16 PSM oligonucleotide and an oligonucleotide representing a known CDP/Cut binding site of the phagocyte-specific cytochrome heavy-chain gene gp91-phox. As outlined in reference 45, we interpret the higher intensity of C2 in the left lane as a reflection of a dimer binding the two repeats of PSM, while only a partially dimeric CDP/Cut factor population binds the singular binding site of gp91-phox. The strong complex with higher mobility in the gp91-phox lane reflects the binding of the CCAAT binding protein CP1, and the fast-migrating complex in both lanes is a possible CDP/Cut degradation product. The next three lanes show competition for complex formation by a homologous probe (lane HPV-16) and probes containing CDP/Cut binding sequences from the gp91-phox gene and the P. miliaris histone H2B gene. By contrast, a probe containing a mutated PSM only slightly interferes with C1 and C2 formation (right lane). CP1 identifies the complex formed on the gp91-phox promoter with the CCAAT binding protein. Bands that migrated faster than CP1 are degradation products of CDP/Cut. FP, free probe (B) An anti-CDP-specific antibody (α-CDP) abolishes the formation of HPV-16 PSM-BP complexes C1 and C2. This effect is specific and is not seen upon the addition of a preimmune serum (PI) to the EMSA reaction.
Repression of HPV-16 transcription by CDP/Cut involves a histone-mediated mechanism.
CDP/Cut represses transcription by two alternative mechanisms, displacement of activators (8, 36, 63) and binding of the histone deacetylase HDAC1, whose activity changes nucleosomes such that it becomes difficult for the transcriptional machinery to access the DNA (33). For genital HPVs, this means either that the PSMs may bind an activator that is displaced by CDP/Cut or that HDAC1 bound to CDP/Cut may modify two nucleosomes that overlap (at least in HPV-16 and HPV-18) with the enhancer and the E6 promoter (56) such that these elements become inaccessible to transcription factors. We considered the second possibility more likely, since PSM deletion mutants of HPV-16, which would also delete the binding site of a potential activator, are strongly upregulated (45).
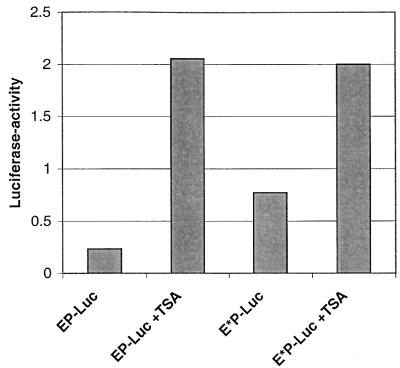
To examine this possibility, we transiently transfected HeLa cells with vectors containing the natural (EP-Luc) and mutated (E*P-Luc) CDP/Cut binding sites (Fig. 2) of HPV-16 and treated the cells with the HDAC1 inhibitor TSA. These luciferase test vectors contained the HPV-16 enhancer and promoter based on a cassette system used in the context of the CAT reporter gene (45). This analysis is complicated by the fact that the HPV-16 silencer contains five binding sites for the YY1 factor, which is, just like CDP/Cut, also known to interact with a histone deacetylase (65). Figure 7 shows the outcome of this experiment. Expression of luciferase from EP-Luc, containing two binding sites for CDP/Cut and five sites for YY1, is stimulated by TSA 9-fold, probably due to the combined effect of TSA on the histone deacetylase complexed with both factors. Mutation of the two CDP/Cut sites in E*P-Luc leads to a threefold increase of the uninduced level and, through a further 2.5-fold stimulation by TSA, to a total expression similar to that of the TSA stimulated EP-Luc. In the absence of CDP/Cut binding sites, this stimulation should stem from interference with the YY1-associated histone deacetylase. From this we conclude that the ninefold TSA stimulation of EP-Luc contains two components, namely, the 2.5-fold stimulation still detectable with E*P-Luc due to YY1, and a further 3- to 4-fold stimulation due to the CDP/Cut associated HDAC1. This amount should be identical to the difference between the uninduced EP-Luc and E*P-Luc vector. These observations indicate that CDP/Cut bound to PSM represses by modifying the nucleosomes established on the HPV LCR.
FIG. 7.
Repression of HPV-16 transcription by CDP/Cut involves a histone-mediated mechanism. Luciferase expression in HeLa cells transiently transfected with vectors containing the natural (EP-Luc) and mutated (E*P-Luc) CDP/Cut binding sites of HPV-16 is stimulated by treatment with the HDAC1 inhibitor TSA.
Overexpression of the CDP/Cut cDNA represses replication of HPV-16, HPV-31, and BPV-1.
The presumed PSMs of all genital HPVs are found in the same genomic region, overlapping with position 1 of the nucleotide sequence of the respective genome (Fig. 2). As determined for a number of HPV types (20, 21, 27, 31, 49, 57) and presumed for a number of others, this region represents the binding sites of the replication factor E1 (for an alignment of HPV replication origins, see reference 44). It is therefore distinctly possible that E1 and CDP/Cut sites overlap, and it presents the interesting possibility that CDP/Cut also regulates HPV replication.
To test this possibility, we studied the effect of overexpressing CDP/Cut on HPV replication in vivo. Toward this end, we used both the HPV-16 replication origin present within the complete LCR cloned into a bacterial plasmid and the complete recircularized and plasmid-free HPV-31 genome. As recipient cells, we used the cell line Cho4.15, which has been engineered to express the BPV-1 proteins E1 and E2 under the influence of heterologous promoters from intrachromosomally recombined genes. These proteins are known to activate heterologous HPV replication origins (15, 48).
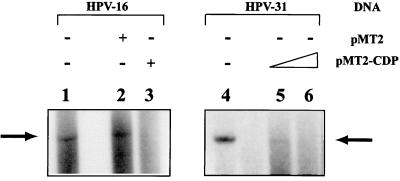
Figure 8 shows that the HPV-16 LCR construct was indeed able to replicate in these cells (lane 1). Cotransfection of the empty expression vector pMT2 (42) had no effect on HPV-16 replication (lane 2). By contrast, coexpression of a pMT2-CDP construct that contains the full-length CDP/Cut cDNA abolished HPV-16 replication (lane 3). This effect was not limited to HPV-16, since replication of HPV-31 was also repressed in a dose-dependent manner by the overexpression of CDP/Cut (compare lanes 5 and 6 with lane 4). These data and the intriguing overlap of the E1 and the CDP/Cut binding sites suggest that CDP/Cut can repress HPV replication and that CDP/Cut simultaneously regulates both transcription and replication.
FIG. 8.
CDP/Cut inhibits transient HPV replication in vivo. Cho cells stably expressing E1 and E2 proteins were transfected with pBluescript SK(+) carrying the HPV-16 LCR or with recircularized HPV-31 DNA. Total cellular DNA was prepared after 48 h, digested with DpnI to remove the input bacterially replicated DNA, linearized with EcoRI (HPV-16) or HpaI (HPV-31), respectively, blotted, and hybridized with an LCR fragment of HPV-16 (lanes 1 to 3) or of HPV-31 (lanes 4 to 6). The arrow denotes the position of the linearized replication products, which have molecular sizes of 4 kb in pBluescript SK(+) HPV-16LCR (arrow in the left panel) and 7.9 kb in the linearized HPV-31 genome (arrow in the right panel). Cotransfection of the CDP expression vector pMT2-CDP strongly represses replication for both HPV types (compare lane 1 with lane 3, and lanes 4 with lanes 5 and 6. The expression vector pMT2 without CDP sequences does not interfere with the replication of HPV-16 (lane 2).
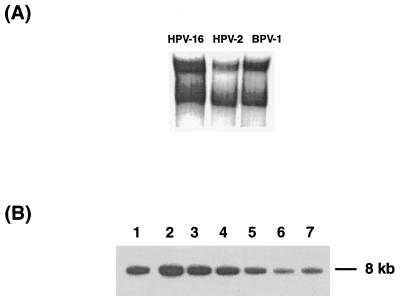
As an alternative to this interpretation, one could suspect that CDP/Cut influences the expression of E1 and E2 from the heterologous promoters used in Cho4.15 with the downstream consequence of repression of replication. To show that CDP/Cut represses replication independently from such a potentially artifactual system and to extend our study to a papillomavirus type unrelated to the genital HPVs, we also examined the replication of BPV-1. Figure 2 shows that this virus contains potential CDP/Cut binding sites that overlap with the binding site of the E1 protein in a manner similar to the arrangement of sequences in genital HPVs. EMSA results in Fig. 9A show that these homologous sequences indeed lead to band shifts indistinguishable from those with HPV-16 and HPV-2 sequences. To study the effect of CDP/Cut on the replication of BPV-1, we transfected ID13 fibroblasts, a mouse c127 cell-derived line with about 100 stably episomally replicating BPV-1 genomes, with the pMT2-CDP expression vector. Figure 9B shows stepwise decreasing concentrations of BPV-1 genomes after application of 0.5, 5, and 12 μg of pMT2-CDP (slots 2 plus 3, 4 plus 5, and 6 plus 7, respectively) in comparison with cells transfected only with the empty expression vector (slot 1). These data prove that CDP/Cut interferes efficiently with papillomavirus replication, although this experiment does not distinguish between an interference with E1 function, as suggested by us, and a potential repression of the homologous E1 and E2 promoters of BPV-1 with the downstream consequence of repression of replication.
FIG. 9.
(A) An oligonucleotide representing the replication origin and presumed CDP/Cut binding sites of BPV-1 generates bandshifts that are indistinguishable from those of HPV-16 and HPV-2. (B) Transfection of ID13 cells, which contain about 100 episomally replicating copies of BPV-1, with increasing amounts of the CDP/Cut expression vector pMT2-CDP, decreases the copy number of BPV-1. The cells were electroporated with 4 μg of the empty vector (slot 1), 0.5 (slots 2 plus 3), 4 (slots 4 plus 5), and 12 μg (slots 6 plus 7) of pMT2-CDP and grown for 48 h. Then DNA was harvested, cleaved with HindIII, and processed with a radioactive probe for BPV-1 DNA. The arrow points to the 8-kb band generated by the linearized BPV-1 genome.
DISCUSSION
The repressor CDP/Cut binds a nucleotide sequence that is conserved among many HPV types.
We have reported here five lines of evidence suggesting that PSM-BP is identical to the transcription factor CDP/Cut: (i) PSM-BP and CDP/Cut are both transcriptional repressors with a propensity for binding AT-rich sequences, (ii) PSM-BP and CDP/Cut have identical purification profiles and similar molecular weight, (iii) the known CDP/Cut binding site on gp91-phox gives rise to complexes in EMSA studies that have identical mobilities to the HPV-16 PSM-BP C1 and C2 complexes, (iv) formation of the HPV-16 C1 and C2 complexes on the PSM can be competitively inhibited by two known CDP binding sites, and (v) a specific anti-CDP/Cut antibody abolishes the formation of the C1 and C2 complexes in EMSA experiments with the HPV-16 probe. Collectively, these data leave little doubt that the transcriptional repressor termed PSM-BP in our previous study is, in fact, CDP/Cut.
CDP was originally described from a study of a histone H2B gene from the sea urchin P. miliaris. This gene is selectively expressed in sperm cells but repressed in embryonic cells. Activation of the H2B promoter in sperm cells depends on the complex formation between a CCAAT motif and the CCAAT binding factor, CP1. However, formation of this complex is abrogated in embryonic cells by CDP, which binds sequences overlapping the CCAAT motif, thus blocking DNA binding by CP1 (8). CDP was consequently named to reflect this initial observation. Later, it was found that CDP can repress by two distinct mechanisms, factor displacement (8, 53) and the function of a silencer domain without displacing an activating transcription factor (36, 63). The recent finding that CDP/Cut associates with the histone deacetylase HDAC1 provides a potential explanation for this second activity, namely, structural alteration of nucleosomes (33). Our observation of the stimulation of HPV-16 transcription by TSA suggests that this mechanism is active in genital HPVs.
CDP was found to be a homologue of the Drosophila Cut homeodomain protein, which plays a role in the determination of cell fate in various tissues (reference 42 and references therein); it is now frequently referred to by the combined abbreviation CDP/Cut. As in Drosophila, CDP is believed to be a general repressor of developmentally regulated genes in mammals. Examples include regulation of the cytochrome heavy-chain gene gp91-phox during myeloid differentiation (34, 53) and expression of the immunoglobulin heavy chain during B-cell differentiation (63).
CDP/Cut recognizes defined binding sites, as judged, for example, by footprint analysis, but it does not specifically recognize CCAAT boxes or any other clearly recognizable and conserved sequence motif. Natural and artificial CDP/Cut binding sites are AT rich and often contain the sequences 5′-TAAT-3′ and 5′-CAAT-3′ repeated directly or invertedly with variable spacing (3). Lack of specificity is in part explained by the finding that the CDP/Cut contains four DNA binding domains with different but overlapping sequence specificity (6, 25). Multiple 5′-TAAT-3′ and 5′-CAAT-3′ motifs occur in each of the PSMs of the seven genital HPV types studied here.
PSM coupling of HPV transcription and epithelial differentiation.
The activity of HPV enhancers and promoters depends upon the cooperation between more than 10 different factors (for a review, see reference 44). The specificity of HPVs for epithelial cells is apparently dependent upon only a subset of these factors, with most data available from studies of NF-I (4), AP-1 (60), and Sp1 (5). These three factors are each derived from multigene families, with the epithelial-cell specificity originating from the fact that different members of these gene families differ in function and are expressed in a cell-type-specific manner. While these three factors appear to bind the LCRs of all genital HPV types (44, 50), another factor, skn-1, expressed exclusively in epithelial cells, has so far been found to activate only HPV-1 and HPV-18 gene expression (2, 66). Consequently, regulation by skn-1 does not provide an explanation for the general phenomenon of epithelial-cell specificity of genital HPV enhancers.
It is probably the principal function of these three transcription factors (NFI, AP-1, and Sp1) to activate HPV gene expression selectively in an epithelial environment, leaving HPV genomes dormant in nonepithelial cells. However, other factors may regulate transcription in a differentiation-dependent manner during stratification. Differential gene expression in the different layers of epithelia is well documented for both cellular (30) and HPV (28, 55) genes. The sequences contained within the E6 promoter may represent one way by which HPVs modulate early transcription, since the relative concentration between the activator Sp1 and its antagonist Sp3 changes during epithelial differentiation (5).
Recent publications have provided new and exciting data suggesting that CDP/Cut may represent another and quantitatively more important factor that couples HPV transcription to epithelial-cell differentiation (1, 47). This is because the activity of CDP/Cut strongly decreases during epithelial-cell differentiation and correlates with increased transcription from three different early promoters of HPV-6. These findings suggest that stratified epithelia are yet another example where CDP/Cut influences the developmental regulation of gene expression. The modulation of the HPV-6 E6 promoter observed by Roman and colleagues (1, 47) is probably derived from the HPV-6 PSM, since the promoter fragment studied by these authors includes this motif. Together, these findings, along with the results presented here, suggest that CDP/Cut, by binding the conserved PSM of HPVs, may provide an important regulatory link between epithelial differentiation and HPV early gene expression.
Roman and colleagues also reported that other regions of the HPV-6 genome, namely, the 5′ part of the LCR, the E6 gene, and the E7 gene, bind CDP/Cut (1, 47), resulting in the repression of the E6 promoter, the E7 promoter (which is absent from HPV-16 and most other genital HPVs), and the E1 promoter. In our ongoing research on the nature of nuclear matrix attachment regions (MARs) of HPVs (58), we have found that HPV-16 MARs also contain clusters of CDP/Cut binding sites (W. Stünkel, M. J. O'Connor, and H. U. Bernard, unpublished results). The MARs of HPV-16 are located in the same genomic positions as the CDP/Cut binding regions of HPV-6 (1, 47) outside the HPV-6 E6 promoter fragment. The CDP/Cut binding regions of HPV-6 are, by sequence analysis, likely to be MARs (58). CDP/Cut has been proposed to be attached the nuclear matrix (7), suggesting some functional relationship between MARs and clusters of CDP/Cut binding sites. While details of these mechanisms await further research, it is apparent that in addition to the important role played by the PSM of each HPV type, there are, in the form of MARs, conserved silencing elements within HPV genomes with CDP/Cut binding sites. The PSM and MARs together may form a complex network of cooperations among one another and with other cellular components to achieve synchronization of HPV gene regulation and epithelial differentiation.
CDP/Cut represses papillomavirus replication and may couple this process to epithelial-cell differentiation.
HPV genomes are much more abundant in supraepithelial cells than in basal cells (43), and their efficiency of replication apparently depends on epithelial-cell differentiation (10). There are a number of observations suggesting that CDP/Cut binding to the PSM of HPVs could be partly or wholly responsible for the differentiation-specific regulation of HPV replication. These include (i) the overlapping of the PSM (and therefore the CDP/Cut binding sites) with the HPV E1 binding site, (ii) the repression of HPV-16 and HPV-31 replication by exogenously expressed CDP/Cut protein, and (iii) as described above, the differentiation-specific expression of the CDP/Cut protein itself.
While we have provided the first examples of how CDP/Cut can repress papillomavirus replication in vivo, it is not clear exactly what mechanism is involved. One obvious possibility is that CDP/Cut prevents E1 from binding to its cognate sites within the origin. However, E1 binding to sequences within the origin is a complex affair in which active E1 complexes depend upon the correct multimerization of E1 proteins (52). Moreover, E1 has the potential to bind to alternative sites within the AT-rich sequences that include the origin, although binding to these sites might not necessarily give rise to active E1 complexes (A. Stenlund, personal communication). Thus, CDP/Cut binding to the PSM might not prevent the binding of E1 to the origin of replication but might prevent the formation of an active E1 complex. Another nonexclusive mechanism could be invoked that involves the association of histone deacetylase activity with CDP/Cut (33). In such a mechanism, deacetylation of the histones of nucleosomes positioned in the vicinity of the replication origin (56) could result in a chromatin structure that prevents active replication. The need for nucleosomal alterations during the initiation of papillomavirus replication has recently been highlighted by the finding that E1 interacts with components of the Swi/Snf complex (32b). Future studies should provide insight into these possible mechanisms.
In summary, we have provided evidence that the PSM originally identified in HPV-16 is conserved in all genital HPVs tested. The factor that binds to the PSM has now been identified as the differentiation-specific repressor CDP/Cut. What is more, the conservation of positioning within the origin of replication does not appear to be a coincidence, and we provide evidence that in addition to transcriptional repression, CDP can repress HPV replication in vivo. Thus, through one regulatory motif, both HPV early-gene expression and replication may be coupled to the differentiation program of the host stratified epithelia.
ACKNOWLEDGMENTS
M.J.O. and W.S. contributed equally to this work.
We thank S. H. Orkin and E. Neufeld for plasmids pMT2 and pMT2-CDP and anti-CDP antiserum, W. G. Hubert and L. A. Laimins for pBR322-HPV-31, and M. Ustav and I. Ilves for the cell line Cho 4.15 (E1/E2).
REFERENCES
- 1.Ai W, Toussaint E, Roman A. CCAAT displacement protein binds to and negatively regulates human papillomavirus type 6 E6, E7, and E1 promoters. J Virol. 1999;73:4220–4229. doi: 10.1128/jvi.73.5.4220-4229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson B, Hariri A, Pittelkow M R, Rosenfeld M G. Characterization of Skin-1a/i POU domain factors and linkage to papillomavirus gene expression. J Biol Chem. 1997;272:15905–15913. doi: 10.1074/jbc.272.25.15905. [DOI] [PubMed] [Google Scholar]
- 3.Andres V, Chiara M D, Mahdavi V. A new bipartite DNA-binding domain: cooperative interaction between the cut repeat and homeo domain of the cut homeo proteins. Genes Dev. 1994;8:245–257. doi: 10.1101/gad.8.2.245. [DOI] [PubMed] [Google Scholar]
- 4.Apt D, Liu Y, Bernard H U. Cloning and functional analysis of spliced isoforms of human nuclear factor I-X: interference with transcriptional activation by NFI/CTF in a cell type specific manner. Nucleic Acids Res. 1994;22:3825–3833. doi: 10.1093/nar/22.19.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apt D, Watts R M, Suske G, Bernard H U. High Sp1/Sp3 rations in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology. 1996;224:281–291. doi: 10.1006/viro.1996.0530. [DOI] [PubMed] [Google Scholar]
- 6.Aufiero B, Neufeld E J, Orkin S H. Sequence-specific DNA binding of individual cut repeat of the human CCAAT displacement/cut homeodomain protein. Proc Natl Acad Sci USA. 1994;91:7757–7761. doi: 10.1073/pnas.91.16.7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Banan M, Rojas I C, Lee W H, King H L, Harriss J V, Kobayashi R, Webb C F, Gottlieb P D. Interaction of the nuclear matrix associated region (MAR)-binding proteins, SATB1 and CDP/Cux, with a MAR element (L2a) in an upstream regulatory region of the mouse CD8a gene. J Biol Chem. 1997;272:18440–18452. doi: 10.1074/jbc.272.29.18440. [DOI] [PubMed] [Google Scholar]
- 8.Barberis A, Superti-Furga G, Busslinger M. Mutually exclusive interaction of the CCAAT-binding factor and of a displacement protein with overlapping sequences of a histone gene promoter. Cell. 1987;50:347–359. doi: 10.1016/0092-8674(87)90489-2. [DOI] [PubMed] [Google Scholar]
- 9.Bauknecht T, Angel P, Royer H D, zur Hausen H. Identification of a negative regulatory domain in the human papillomavirus type 18 promoter: interaction with the transcriptional repressor YY1. EMBO J. 1992;11:4607–4617. doi: 10.1002/j.1460-2075.1992.tb05563.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bedell M A, Hudson J B, Golub T R, Turyk M E, Hosken M, Wilbanks G D, Laimins L A. Amplification of human papillomavirus genomes in vitro is dependent on epithelial differentiation. J Virol. 1991;65:2254–2260. doi: 10.1128/jvi.65.5.2254-2260.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan W K, Klock G, Bernard H U. Progesterone and glucocorticoid response elements occur in the long control regions of several human papillomaviruses involved in anogenital neoplasia. J Virol. 1989;63:3261–3269. doi: 10.1128/jvi.63.8.3261-3269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiang C M, Ustav M, Stenlund A, Ho T F, Broker T R, Chow L. Viral E1 and E2 proteins support replication of homologous and heterologous papillomaviral origins. Proc Natl Acad Sci USA. 1992;89:5799–5803. doi: 10.1073/pnas.89.13.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chong T, Apt D, Gloss B, Isa M, Bernard H U. The enhancer of human papillomavirus-16: Binding sites for the ubiquitous transcription factors oct-1, NFA, TEF-2, NFI, and AP1 participate in the epithelial specific transcription. J Virol. 1991;65:5933–5943. doi: 10.1128/jvi.65.11.5933-5943.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cripe T P, Haugen T H, Turk J P, Tabatabai F, Schmid P G, Dürst M, Gissmann L, Roman A, Turek L P. Transcriptional regulation of the human papillomavirus-16 E6-E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans-activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 1987;6:3745–3753. doi: 10.1002/j.1460-2075.1987.tb02709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Del Vecchio A M, Romanczuk H, Howley P M, Baker C C. Transient replication of human papillomavirus DNAs. J Virol. 1992;66:5949–5948. doi: 10.1128/jvi.66.10.5949-5958.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Demeret C, Desaintes C, Yaniv M, Thierry F. Different mechanisms contribute to the E2-mediated transcriptional repression of human papillomavirus type 18 viral oncogenes. J Virol. 1997;71:9343–9349. doi: 10.1128/jvi.71.12.9343-9349.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dignam J D, Lebowitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dollard S C, Broker T R, Chow L T. Regulation of the human papillomavirus type 11 E6 promoter by viral and host transcription factors in primary human keratinocytes. J Virol. 1993;67:1721–1726. doi: 10.1128/jvi.67.3.1721-1726.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong G, Broker T R, Chow L T. Human papillomavirus type 11 E2 proteins repress the homologous E6 promoter by interfering with the binding of host transcription factors to adjacent elements. J Virol. 1994;68:1115–1127. doi: 10.1128/jvi.68.2.1115-1127.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flores E R, Lambert P F. Evidence for a switch in the mode of human papillomavirus type 16 DNA replication during the viral life cycle. J Virol. 1997;71:7167–7179. doi: 10.1128/jvi.71.10.7167-7179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frattini M G, Laimins L A. Binding of the human papillomavirus E1 origin-recognition protein is regulated through complex formation with the E2 enhancer-binding protein. Proc Natl Acad Sci USA. 1994;91:12398–12402. doi: 10.1073/pnas.91.26.12398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frattini M G, Lim H B, Laimins L A. In vitro synthesis of oncogenic human papillomaviruses requires episomal genomes for differentiation-dependent late expression. Proc Natl Acad Sci USA. 1996;93:3062–3067. doi: 10.1073/pnas.93.7.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gloss B, Bernard H U, Seedorf K, Klock G. The upstream regulatory region of the human papillomavirus-16 contains and E2 protein independent enhancer which is specific for cervical carcinoma cells and regulated by glucocorticoid hormones. EMBO J. 1987;6:3735–3743. doi: 10.1002/j.1460-2075.1987.tb02708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gloss B, Chong T, Bernard H U. Numerous nuclear proteins bind the long control region of human papillomavirus type 16: a subset of 6 of 23 DNase I-protected segments coincides with the location of the cell-type-specific enhancer. J Virol. 1989;63:1142–1152. doi: 10.1128/jvi.63.3.1142-1152.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harada R, Berube G, Tamplin O J, Denis-Larose C, Nepveu A. DNA-binding specificity of the cut repeats from the human cut-like protein. Mol Cell Biol. 1995;15:129–140. doi: 10.1128/mcb.15.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt S E, Wilson V G. Mutational analysis of the 18-base-pair inverted repeat element at the bovine papillomavirus origin of replication: identification of critical sequences for E1 binding and in vivo replication. J Virol. 1995;69:6525–6532. doi: 10.1128/jvi.69.10.6525-6532.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hubert W G, Kanaya T, Laimins L A. DNA replication of human papillomavirus type 31 is modulated by elements of the upstream regulatory region that lie 5′ of the minimal origin. J Virol. 1999;73:1835–1845. doi: 10.1128/jvi.73.3.1835-1845.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iftner T, Oft M, Bohm S, Wilczynski S P, Pfister H. Transcription of the E6 and E7 genes of human papillomavirus type 6 in anogenital condylomata is restricted to undifferentiated cell layers of the epithelium. J Virol. 1992;66:4639–4646. doi: 10.1128/jvi.66.8.4639-4646.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.International Agency for Research on Cancer. Human papillomaviruses. IARC Monogr Eval Carcinog Risks Hum. 1995;64:35–86. [PMC free article] [PubMed] [Google Scholar]
- 30.Kopan R, Fuchs E. A new look into an old problem: keratins as tools to investigate determination, morphogenesis, and differentiation in skin. Genes Dev. 1989;3:1–15. doi: 10.1101/gad.3.1.1. [DOI] [PubMed] [Google Scholar]
- 31.Kuo S R, Liu J S, Broker T R, Chow L T. Cell-free replication of the human papillomavirus DNA with homologous viral E1 and E2 proteins and human cell extracts. J Biol Chem. 1994;269:24058–24065. [PubMed] [Google Scholar]
- 32.Kyo S, Tam A, Laimins L A. Transcriptional activity of human papillomavirus type 31b enhancer is regulated through synergistic interaction of AP-1 with two novel cellular factors. Virology. 1995;211:184–197. doi: 10.1006/viro.1995.1390. [DOI] [PubMed] [Google Scholar]
- 32a.Law M F, Lowy D R, Dvoretzky I, Howley P M. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc Natl Acad Sci USA. 1981;78:2727–2731. doi: 10.1073/pnas.78.5.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32b.Lee D, Sohn H, Kalpana G V, Choe J. Interaction of E1 and hSNF5 proteins stimulates replication of human papillomavirus DNA. Nature. 1999;399:487–491. doi: 10.1038/20966. [DOI] [PubMed] [Google Scholar]
- 33.Li S, Moy L, Pittman N, Shue G, Aufiero B, Neufeld E J, LeLeiko N S, Walsh M J. Transcriptional repression of the cystic fibrosis transmembrane conductance regulator gene, mediated by CCAAT displacement protein/cut homolog, is associated with histone deacetylation. J Biol Chem. 1999;274:7803–7815. doi: 10.1074/jbc.274.12.7803. [DOI] [PubMed] [Google Scholar]
- 34.Lievens P M, Donady J J, Tufarelli C, Neufeld E J. Repressor activity of CCAAT displacement protein in HL-60 myeloid leukemia cells. J Biol Chem. 1995;270:12745–12750. doi: 10.1074/jbc.270.21.12745. [DOI] [PubMed] [Google Scholar]
- 35.Lu J Z, Sun Y N, Rose R C, Bonnez W, McCance D J. Two E2 binding sites (E2BS) alone or one E2BS plus an A/T-rich region are minimal requirements for the replication of the human papillomavirus type 11 origin. J Virol. 1993;67:7131–7139. doi: 10.1128/jvi.67.12.7131-7139.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mailly F, Berube G, Harada R, Mao P L, Phillips S, Nepveu A. The human cut homeodomain protein can repress gene expression by two distinct mechanisms: active repression and competition for binding site occupancy. Mol Cell Biol. 1996;16:5346–5357. doi: 10.1128/mcb.16.10.5346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. p. 90. [Google Scholar]
- 38.May M, Dong X P, Beyer-Finkler E, Stubenrauch F, Fuchs P G, Pfister H. The E6/E7 promoter of extrachromosomal HPV16 DNA in cervical cancers escapes from cellular repression by mutation of target sequences for YY1. EMBO J. 1994;13:1460–1466. doi: 10.1002/j.1460-2075.1994.tb06400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mendoza R, Gandhi L, Botchan M R. E1 recognition sequences in the bovine papillomavirus type 1 origin of DNA replication: interaction between half sites of the inverted repeats. J Virol. 1995;69:3789–3798. doi: 10.1128/jvi.69.6.3789-3798.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohr I J, Clark R, Sun S, Androphy E J, MacPherson P, Botchan M R. Targeting the E1 replication protein to the papillomavirus origin of replication by complex formation with the E2 transactivator. Science. 1990;250:1694–1699. doi: 10.1126/science.2176744. [DOI] [PubMed] [Google Scholar]
- 41.Myers G, Bernard H U, Delius H, Favre M, Icenogle J, van Ranst M, Wheeler C. Human papillomaviruses. A compilation and analysis of nucleic acid and amino acid sequences, sect. II-LCR. Los Alamos, N.M: Los Alamos National Laboratory; 1994. pp. 14–16. [Google Scholar]
- 42.Neufeld E J, Skalnick D G, Lievens P M J, Orkin S H. Human CCAAT displacement protein is homologous to the Drosophila homeoprotein, cut. Nat Genet. 1992;1:50–55. doi: 10.1038/ng0492-50. [DOI] [PubMed] [Google Scholar]
- 43.Nuovo G J, MacConnell P, Forde A, Delvenne P. Detection of human papillomavirus DNA in formalin-fixed tissues by in situ hybridization after amplification by polymerase chain reaction. Am J Pathol. 1991;139:847–854. [PMC free article] [PubMed] [Google Scholar]
- 44.O'Connor M, Chan S Y, Bernard H U. Transcription factor binding sites in the long control regions of genital HPVs, part III-A. In: Myers G, Bernard H U, Delius H, Baker C, Icenogle J, Halpern A, Wheeler C, editors. Human papillomaviruses 1995 compendium. Los Alamos, N.M. USA: Los Alamos National Laboratory; 1995. pp. 21–40. [Google Scholar]
- 45.O'Connor M J, Stünkel W, Zimmermann H, Koh C H, Bernard H U. A novel YY1-independent silencer represses the activity of the human papillomavirus type 16 enhancer. J Virol. 1998;72:10083–10092. doi: 10.1128/jvi.72.12.10083-10092.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Connor M J, Tan S H, Tan C H, Bernard H U. YY1 represses human papillomavirus type 16 transcription by quenching AP-1 activity. J Virol. 1996;70:6529–6539. doi: 10.1128/jvi.70.10.6529-6539.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pattison S, Skalnik D G, Roman A. CCAAT displacement protein, a regulator of differentiation-specific gene expression, binds a negative regulatory element within the 5′ end of the human papillomavirus type 6 long control region. J Virol. 1997;71:2013–2022. doi: 10.1128/jvi.71.3.2013-2022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Piirsoo M, Ustav E, Mandel T, Stenlund A, Ustav M. Cis and trans requirements for stable episomal maintenance of the BPV-1 replicator. EMBO J. 1996;15:1–11. [PMC free article] [PubMed] [Google Scholar]
- 49.Russell J, Botchan M R. cis-acting components of human papillomavirus (HPV) DNA replication: linker substitution analysis of the HPV type 11 origin. J Virol. 1995;69:651–660. doi: 10.1128/jvi.69.2.651-660.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sailaja G, Watts R M, Bernard H U. Many different papillomaviruses have low transcriptional activity in spite of strong epithelial specific enhancers. J Gen Virol. 1999;80:1715–1724. doi: 10.1099/0022-1317-80-7-1715. [DOI] [PubMed] [Google Scholar]
- 51.Schule R, Muller M, Otsuka-Murakami H, Renkawitz R. Cooperativity of the glucocorticoid receptor and the CACCC-box binding factor. Nature. 1988;332:87–90. doi: 10.1038/332087a0. [DOI] [PubMed] [Google Scholar]
- 52.Sedman J, Stenlund A. The initiator protein E1 binds to the bovine papillomavirus origin of replication as a trimeric ring-like structure. EMBO J. 1996;15:5085–5092. [PMC free article] [PubMed] [Google Scholar]
- 53.Skalnik D G, Strauss E C, Orkin S H. CCAAT displacement protein as a repressor of the myelomonocytic-specific gp91-phox gene promoter. J Biol Chem. 1991;266:16736–16744. [PubMed] [Google Scholar]
- 54.Spalholz B A, McBride A A, Sarafi T, Quintero J. Binding of bovine papillomavirus E1 to the origin is not sufficient for DNA replication. Virology. 1993;193:201–212. doi: 10.1006/viro.1993.1116. [DOI] [PubMed] [Google Scholar]
- 55.Stoler M H, Wolinsky S M, Whitbeck A, Broker T R, Chow L T. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology. 1989;172:331–340. doi: 10.1016/0042-6822(89)90135-9. [DOI] [PubMed] [Google Scholar]
- 56.Stünkel W, Bernard H U. The chromatin structure of the long control region of human papillomavirus type 16 represses viral oncoprotein expression. J Virol. 1999;73:1918–1930. doi: 10.1128/jvi.73.3.1918-1930.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sverdrup F, Khan S A. Two E2 binding sites alone are sufficient to function as the minimal origin of replication of human papillomavirus type 18 DNA. J Virol. 1995;69:1319–1323. doi: 10.1128/jvi.69.2.1319-1323.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tan S H, Bartsch D, Schwarz E, Bernard H U. Nuclear matrix attachment regions of human papillomavirus type 16 point toward conservation of these genomic elements in all genital papillomaviruses. J Virol. 1998;72:3610–3622. doi: 10.1128/jvi.72.5.3610-3622.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tan S H, Leong L E C, Walker P, Bernard H U. The human papillomavirus type 16 E2 transcription factor binds with low cooperativity to two flanking sites and represses the E6 promoter through displacement of Sp1 and TFIID. J Virol. 1994;68:6411–6420. doi: 10.1128/jvi.68.10.6411-6420.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thierry F, Spyrou G, Yaniv M, Howley P M. Two AP1 sites binding JunB are essential for human papillomavirus type 18 transcription in keratinocytes. J Virol. 1992;66:3740–3748. doi: 10.1128/jvi.66.6.3740-3748.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ustav E, Ustav M, Szymanski P, Stenlund A. The bovine papillomavirus origin of replication requires a binding site for the E2 transcriptional activator. Proc Natl Acad Sci USA. 1993;90:898–902. doi: 10.1073/pnas.90.3.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ustav M, Ustav E, Szymanski P, Stenlund A. Identification of the origin of replication of bovine papillomavirus and characterization of the viral origin recognition factor E1. EMBO J. 1991;10:4321–4329. doi: 10.1002/j.1460-2075.1991.tb05010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z, Goldstein A, Zong R T, Lin D, Neufeld E J, Scheuermann R H, Tucker P W. Cux/CDP homeoprotein is a component of NF-mNR and represses the immunoglobulin heavy chain intronic enhancer by antagonizing the bright transcription enhancer. Mol Cell Biol. 1999;19:284–298. doi: 10.1128/mcb.19.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Westin G, Gerster T, Muller M M, Schaffner G, Schaffner W. OVEC, a versatile system to study transcription in mammalian cells and cell-free extracts. Nucleic Acids Res. 1987;15:6787–6798. doi: 10.1093/nar/15.17.6787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang W M, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci USA. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yukawa K, Butz K, Yasui T, Kikutani H, Hoppe-Seyler F. Regulation of human papillomavirus transcription by the differentiation-dependent epithelial factor Epoc-1/skn-1a. J Virol. 1996;70:10–16. doi: 10.1128/jvi.70.1.10-16.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.zur Hausen H. Papillomavirus infections—a major cause of human cancers. Biochim Biophys Acta. 1996;1288:F55–F78. doi: 10.1016/0304-419x(96)00020-0. [DOI] [PubMed] [Google Scholar]