Abstract
Ethnopharmacological relevance:
The last three decades have witnessed a surge in popularity and consumption of herbal products. An unintended consequence of such popularity is that chronic consumption of these products can often modulate the functions of various proteins involved in drug disposition and may, in turn, impose risks for herb-drug interactions (HDIs), leading to serious adverse health outcomes. Identifying plants that may give rise to clinically relevant HDIs is essential, and proactive dissemination of such research outcomes is necessary for researchers, clinicians, and average consumers.
Aim of the study:
The main objective of this study was to evaluate the HDI potential of plants commonly used as ingredients in many herbal products, including BDS.
Materials and methods:
The dried material of 123 plants selected from the NCNPR repository was extracted with 95% ethanol. The extracts were screened for agonistic effects on nuclear receptors (PXR and AhR) by reporter gene assays in PXR-transfected HepG2 and AhR-reporter cells. For cytochrome P450 enzyme (CYP) inhibition studies, CYP450 baculosomes were incubated with enzyme-specific probe substrates by varying concentrations of extracts. The inhibitory effect on the efflux transporter P-glycoprotein (P-gp) was investigated via rhodamine (Rh-123) uptake assay in P-gp overexpressing MDR1-MDCK cells.
Results:
Out of 123 plants, 16 increased transcriptional activity of human PXR up to 4 to 7-fold at 60 μg/mL, while 18 plants were able to increase AhR activity up to 10 to 40-fold at 30 μg/mL. Thirteen plants inhibited the activity of CYP3A4, while 10 plants inhibited CYP1A2 activity with IC50 values in the range of 1.3–10 μg/mL. Eighteen plants (at 50 μg/mL) increased intracellular accumulation of Rh-123 (>150%) in MDR1-MDCK cells. Additionally, other plants tested in this study were able to activate PXR, AhR, or both to lesser extents, and several inhibited the catalytic activity of CYPs at higher concentrations (IC50 >10 μg/mL).
Conclusions:
The results indicate that prolonged or excessive consumption of herbal preparations rich in such plants (presented in Figs. 1a, 2a, 3a, 4a, and 5a) may pose a risk for CYP- and P-gp-mediated HDIs, leading to unwanted side effects due to the altered pharmacokinetics of concomitantly ingested medications.
Keywords: Herbal medicine, Herb-drug interactions, Pregnane X receptor, Aryl hydrocarbon receptor, Cytochrome P450s, P-glycoprotein
1. Introduction
For millennia, humans have relied upon plants to help satisfy many of their primary medicine needs. In many ancient and modern cultures, plants are an integral part of herbal medicines, viz., Ayurveda, Yunani, Traditional Chinese Medicine (TCM), and Complementary and Alternative Medicine (CAM), and have been used to treat many ailments. Today, however, medicinal herbs are more often formulated and marketed as botanical dietary supplements (BDS) that people consume as a source of essential nutrients and for the maintenance of a healthy lifestyle (Husain et al., 2021a). The ongoing COVID-19 pandemic marked a milestone in this trend and enormously increased the popularity, sale, and use of these products (Ahmad et al., 2021). In the United States (U. S.), sale of BDS surpassed $11 billion in 2020, and historically, for the first time, this sector achieved two-digit growth (17.3%) (Smith et al., 2021). Most herbal products are available over-the-counter (OTC), and consumers can purchase them without a prescription. It has been noticed that people consume BDS under the misconstrued notion that “they have no side effects because they are of natural origin” (Raizner and Cooke, 2019). While generally recognized as safe, certain BDS ingredients may pose specific health risks if not taken as directed. Moreover, most consumers are not aware of potential BDS-induced interactions with conventional medications and, as a result, often fail to inform their primary health care providers of such concomitant usage. Interestingly, trained physicians also lack sufficient understanding of BDS and their potential for herb-drug interactions (HDIs) (Fasinu et al., 2012).
Cytochrome P450 enzymes (CYPs) are expressed in multiple organs, although predominantly in the liver and intestine, where they are actively involved in the metabolism and detoxification of xenobiotics and endobiotics (Goodwin et al., 2002; McDonnell and Dang, 2013). In humans, CYPs are expressed constitutively, but they may be inducible, specifically those involved in xenobiotic metabolism (Zanger and Schwab, 2013). CYP induction, in large part, is controlled via activated transcription factors. Pregnane X receptor (PXR) and aryl hydrocarbon receptor (AhR) are two such transcriptional factors belonging to the class of ligand-activated nuclear receptors (NRs). They activate a set of overlapping genes involved in the metabolism and transport of xenobiotics (including drugs) and endogenous substances (Goodwin et al., 2002; Kamenickova et al., 2013). Recent studies have established that medicinal herbs and their finished products, like BDS, are a rich source of active phytochemicals, which may affect the transcriptional activity of NRs and downstream targets, including CYPs and drug transporters (Husain et al., 2021b; Gurley, 2012). Hence, the chronic and indiscriminate consumption of BDS could pose a risk for HDIs by altering the pharmacokinetics or pharmacodynamics of co-administrated conventional pharmaceutical drugs (Husain et al., 2021b; Haron et al., 2022a). As a result, HDIs can be classified as either pharmacokinetic- or pharmacodynamic-mediated interactions (Gurley, 2020). Often, HDIs are clinically insignificant and do not require any special medical care, but sometimes they may become clinically relevant and require medical intervention (Borse et al., 2019). In the recent past, the transcriptional regulation of hepatic drug metabolism has been extensively studied for Hypericum perforatum (St. John’s wort). This herb is used to treat mild depression that can upregulate the expression of several important CYPs (e.g., CYP2C9, CYP2C19, CYP3A4) and efflux transporters (i.e., ATP-binding cassette [ABC] transporters like P-glycoprotein [P-gp]) involved in human drug disposition. As a result, H. perforatum can decrease the bioavailability of numerous conventional drugs that are substrates for CYPs and P-gp (e.g., warfarin, cyclosporine, digoxin, indinavir, irinotecan, midazolam, and others) leading to reduced efficacy (Moore et al., 2000; Markowitz et al., 2003; Gurley et al., 2012). In contrast, grapefruit juice inhibits the catalytic activity of intestinal CYP3A4, thereby altering the pharmacokinetics and ultimately the pharmacodynamic profiles of prescription medications like cyclosporine and felodipine by inhibiting their presystemic metabolism and subsequently enhancing their oral bioavailability (Bailey et al., 2013). The HDIs caused by other medicinally important herbs such as Hydrastis canadensis (goldenseal), Glycyrrhiza glabra (licorice), and Camellia sinensis (green tea) are also well documented (Gurley et al., 2012; Chen et al., 2012; Husain et al., 2021a). However, the HDI potential of many other commonly traded medicinal plants is largely unknown. At present, the most clinically relevant HDIs appear to have a pharmacokinetic mechanism (Gurley, 2012); therefore, it is imperative to identify botanicals that pose risks for pharmacokinetic-mediated HDIs by screening them through a battery of in vitro methods that can gauge their modulatory effects on drug-metabolizing enzymes and transporters. However, in vitro predictions may not always translate into in vivo realities (Gurley, 2012). Hence, the in vivo relevancy of in vitro results always requires clinical confirmation.
Despite their widespread use in traditional medicines, limited information is available on the safety and adverse effects of medicinal plants, especially their HDI potential. Moreover, to avoid the untoward consequences of clinically significant HDIs, basic information on their mechanism(s) is essential. In the present report, 123 medicinal plants belonging to 55 families, often used in herbal medicines, were selected and their inductive effects on PXR- and AhR-mediated transcription of human drug-metabolizing enzymes and transporters were evaluated using reporter gene assays. Our reasoning for targeting botanical-mediated activation of PXR and AhR is that the most prevalent and clinically significant HDIs recognized to date involve the induction of human CYPs and efflux transporters (Gurley et al., 2018). Accordingly, the expression of two key CYP isoforms, CYP1A2 and CYP3A4, are not only regulated by AhR and PXR, respectively, but are involved in many HDIs. In addition, the inhibitory effects of these plants on the activity of CYP3A4 and CYP1A2 as well as the human efflux transporter, P-gp, were investigated using fluorescence-based high-throughput screening assays. This, in turn, enabled us to evaluate the degree to which both induction and/or inhibition mechanisms influence the trend and magnitude of any predicted HDIs for these plants. The findings of this report will be a valuable addition to our current knowledge of HDIs and will aid herbalists, physicians, researchers, and regulatory personnel working in herbal medicine and related areas. The detailed information related to the plants selected for screening is given in Table S1 along with the references for their traditional use in authentic ethnobotanical databases.
2. Materials and Methods
2.1. Biochemicals and reagents
DMEM-F12, HEPES, trypsin EDTA, penicillin-streptomycin, and sodium pyruvate were purchased from GIBCO BRL, Invitrogen Corp., USA. Fetal Bovine Serum (FBS) was purchased from Hyclone Lab Inc. USA. Omeprazole, rifampicin, doxorubicin, camptothecin, MTT, DMSO, phosphate buffer saline (PBS), Hank’s buffer, Bradford reagent, and rhodamine-123 were from Sigma Chem. Co., USA. Aryl Hydrocarbon Receptor (AhR) reporter cells were procured from Indigo Biosciences, USA. Vivid cytochrome P450s screening kits were procured from Thermo Fisher Scientific, USA. All other reagents and solvents were of analytical grade and procured from Fisher Scientific, USA.
2.2. Preparation of extracts
The authenticated dried plant material from the repository of NCNPR (10 g) was subjected to extraction in 95% ethanol using an automated Accelerated Solvent Extractor (Dionex™ ASE™ 350, Thermo Scientific). The extract was dried completely by evaporation under reduced pressure. The stock solution of each extract (20 mg/mL) was prepared by dissolving in dimethyl sulfoxide (DMSO) and stored at 4 °C.
2.3. Cell culture
Human hepatocellular carcinoma (HepG2) and multidrug resistance protein 1-Madin-Darby canine kidney (MDR1-MDCK) cell lines were obtained from the American Type Culture Collection (ATCC), USA. The HepG2 cells were cultured in DMEM/F12 medium. MDR1-MDCK cells were cultured in high glucose DMEM/F12 medium supplemented with 1% non-essential amino acids, 1% L-glutamine, and 0.02 μM colchicine. Additionally, both culture media were supplemented with heat-inactivated FBS (10%), 2.4 g/L sodium bicarbonate, 100 μg/mL streptomycin, and 100 units/mL penicillin. Cells were grown at 37 °C in an environment of 5% CO2 and 98% relative humidity.
2.3.1. Cell viability assay
The viability of HepG2 cells was measured using the MTT assay as described previously (Husain et al., 2017; Sharma and Husain, 2017). In brief, exponentially growing cells (1.2 × 104 to 1.5 × 104 per 200 μL) were seeded in 96 well plates and allowed to grow overnight. Cells were treated with different concentrations of plant extracts (60, 20 and 6.66 μg/mL) and positive control (doxorubicin; 10, 5 and 2.5 μM) serially diluted in serum free medium. After incubation for 24 h, 10 μL of MTT dye (5 mg/mL in PBS) was added to each well, and plates were additionally incubated for 4 h. Further, media was aspirated, and cells were washed with 100 μL of phosphate buffer saline. The formazan blue crystals formed by viable cells were dissolved in 150 μL of DMSO, and the absorbance was measured at 580 nm on a Bio-Tek, Synergy HT Multi-Mode, plate reader. The % decrease in viability of sample-treated cells was calculated compared to vehicle-treated cells.
2.4. Luciferase reporter gene assay for PXR activation
The pSG5-hPXR expression vector was generously provided by Dr. Steven Kliewer, University of Texas Southwestern Medical Center, USA (Kliewer et al., 1998). The reporter plasmid PCR-5 [CYP3A4-PXR response element (PXRE)-LUC containing the proximal 0/−362 and distal 7208/7797 PXRE regions fused upstream of luciferase] was a kind gift from Dr. Christopher Liddle, University of Sydney, Australia (Goodwin et al., 1999). The PXR activation potential of extracts was determined in transiently transfected HepG2 cells, as described earlier (Husain et al., 2021b). Briefly, 24 h old HepG2 cells having 70–80% confluency were trypsinized and suspended in 0.4 mL of transfection medium (DMEM/F12) containing 1% FBS and no antibiotics. Subsequently, pSG5-hPXR (25 μg) and PCR-5 (25 μg) plasmid DNA were added to the cell suspension. To attach the plasmid to the cell membrane, transfection cuvettes were incubated for 5 min at room temperature. Transfection was performed by electroporation at 180 V, one pulse for 70 msec. Further, cells were incubated at room temperature for 8 min, and floating dead cells were removed. The transfected cells were seeded in 96-well plates at a density of 50,000 cells/200 μL per well. After 24 h, when cells attain >90% confluency, test samples of extracts serially diluted in serum free medium (extract: 60, 20, and 6.66 μg/mL) and positive control (rifampicin: 30, 10, and 3.33 μM) were added. After 24 h of incubation, the culture medium from the plates was aspirated, and 40 μL of luciferase reagent (Promega Corporation, USA) was added to each well. Luminescence was measured using a Spectramax M5 plate reader (Molecular Devices, USA), and a fold increase in luciferase activity in sample treated cells was calculated compared to the vehicle (0.3% DMSO) treated cells.
2.5. Aryl hydrocarbon receptor (AhR) reporter gene assay
According to the manufacturer’s instructions, activation of AhR was evaluated using human AhR reporter cells (Indigo Biosciences). In brief, AhR reporter cells were recovered from the vial using a pre-warmed cell recovery medium. To disperse cell aggregates, the tube of reporter cells was gently inverted several times. The homogenous cell suspension (200 μL) was dispensed into each well of the 96-well white flat bottom plate. The plate was incubated for 5 h in a 5% CO2 incubator at 37 °C and 95% humidity to facilitate the cell attachment. After the incubation, media was aspirated, 200 μL of screening medium containing serially diluted test samples of extracts (30, 10, and 3.33 μg/mL) and positive control (Me-Bio: 1000, 500, and 250 nM) were added. Plates were again incubated for 24 h in the same culture environment. Following incubation, the culture medium was aspirated, and 100 μL of luciferase detection reagent was added to each well. Plates were allowed to rest for 5 min at room temperature, and luminescence was measured using a Spectramax M5 plate reader (Molecular Devices, USA). The fold increase in luciferase activity of treated wells was calculated compared to vehicle-treated (0.3% DMSO) cells.
2.6. Cytochrome P450 inhibition assay
CYP inhibition assays were performed to investigate the inhibitory potential of various plant extracts on the catalytic activity of human CYP3A4 and CYP1A2. For this, CYP450 baculosomes were used provided with Vivid CYP450 screening kits. In brief, stock solutions of extracts and specific positive controls were serially diluted in methanol, and reactions were performed in a 96-well plate according to the supplier’s instructions. Briefly, 40 μL (2.5X) of a serially diluted test sample and 50 μL master premix were added to 96 well plates. To allow sufficient interaction of the test sample with the enzyme, plates were incubated for 10 min at room temperature. Following incubation, 10 μL (10X) of enzyme-specific substrates were added, and the plates were vortexed for 30 s. Fluorescence was measured on the Spectramax M5 plate reader (Molecular Devices, USA) at specified excitation and emission wavelengths for each substrate as recommended by the kit’s protocols. Ketoconazole (10 μM) and α-naphthoflavone (10 μM) were used as positive controls for CYP3A4, and CYP1A2 isozymes, respectively. IC50 values were obtained from concentration-response curves generated by plotting percent inhibition versus tested concentrations.
2.7. Rhodamine-123 uptake assay for P-gp inhibition
P-gp inhibition was determined in P-gp-overexpressing MDR1-MDCK cells by Rh-123 uptake assay (Kotwal et al., 2020). In brief, cells in the logarithmic growth phase with 70–80% confluency were harvested and seeded at a density of 2 × 104 per well of a 96-well plate. Cells were allowed to grow for 24 h in a CO2 incubator. After the incubation, the media was aspirated, 150 μL Hanks salt solution was added, and the plate was again incubated for 40 min. Further, Hanks salt solution was aspirated, and phenol-free DMEM/F12 medium (no serum) containing 10 mM of Rh-123 and various concentrations of test samples of extracts (50, 25, and 12.5 μg/mL) and positive control (cyclosporin 10, 5, and 2.5 μM) were added and incubated for 90 min. The cells were washed three times with ice-cold PBS, and 200 μL of lysis buffer (0.1% Triton X 100 and 0.2 N NaOH) was added. The plate was kept on a shaker for 1 h at room temperature, and 100 μL of lysate was used to measure the Rh-123 fluorescence at 485/529 nm. Further, 10 μL of lysate was taken in a separate 96-well plate, and 190 μL Bradford reagent was added. The plate was shaken for 5–10 min, and absorbance was measured at 595 nm. Each sample was normalized by dividing the fluorescence of each sample by the total protein present in the lysate.
2.8. Statistical analysis
Results are expressed as mean ± SD of at least two experiments. Analysis of variance (ANOVA) was used to determine the significant difference between untreated and treated groups. The Bonferroni multi-comparison test was used for group comparisons using GraphPad Prism Software Version 7 (San Diego, CA, USA). The value of p < 0.05 was considered to be statistically significant.
3. Results
3.1. Effect of plant extracts on the cell viability
The effect on the viability of HepG2 cells was determined after exposing the cells to increasing concentrations of extracts for 24 h. It was noticed that >70% of cells remain viable after 24 h of exposure. In contrast, upon exposure to 10 μM doxorubicin, the cell viability of HepG2 cells was decreased to 56%. These results suggest that plant extracts have minimal effect on cell viability up to the highest concentrations tested in various experiments (data not shown).
3.2. Effects of plant extracts on transcriptional activity of human PXR
To evaluate the transactivation of PXR by selected medicinal plants, reporter gene assays were carried out in transiently transfected hepatic cells (HepG2). Based on fold activation of PXR, tested plants were divided into four groups: 1) potent activators (fold activation of 4 and above); 2) strong activators (3- to <4-fold); 3) mild (or moderate) activators (2- to <3-fold); 4) weak (<2-fold) or non-activators. Results as summarized in Table 1, showed that out of 123 extracts, 16 (A. rugosa, A. graveolens, A. lancea, C. annuum, C. longa, C. rotundus, H. serrata, P. olacoides, P. lobata, P. montana, S. chinensis, S. aromaticum, T. terrestris, T. diffusa and V. myrtillus) increased transcriptional activity of PXR up to 4 – 7-fold at 60 μg/mL (Fig. 1a). These plants were grouped as potent activators. The extract of P. nigrum showed >4-fold activation of PXR at a lower concentration (6.66 μg/mL) (Fig. 1a). Therefore, P. nigrum was also included in the potent PXR activators. Subsequently, the extracts of 22 plants, including A. milliefolium, A. sinensis, A. uva-ursi, A. macrocephala, C. angustifolia, C. asiatica, C. aurantium, C. forskohlii, E. japonica, G. cambogia, G. mangostana, H. canadensis, H. perforatum, B. aquifolium, N. sativa, R. vomitoria, S. cordifolia, U. fulva, V. macrocarpon, V. officinalis, Z. officinale and Z. jujuba increased the transcriptional activity of PXR in the range of 3- to <4-fold, at concentrations of 60 μg/mL except G. mangostana. These plants were characterized as strong activators (Fig. 1b). Interestingly, the extract prepared from G. mangostana showed >3-fold increase in PXR’s transcriptional activity at lower concentrations (20 and 6.66 μg/mL). Hence, this plant was included among the strong activators (Fig. 1b). The PXR activation effects of C. forskohlii, C. longa, G. cambogia, H. canadensis, H. perforatum, P. nigrum, V. officinalis and Z. officinale are in agreement with the data reported previously (Haron et al., 2022b). Another 48 plants were found to activate PXR between 2- to <3-fold at the highest tested concentration (60 μg/mL), but lower concentrations were not as effective. Thus, these plants were termed as mild activators (Fig. 1c). In contrast, the remaining 37 plants had weak or no agonistic effects on PXR (0 to <2-fold). These plants were categorized as weak activators or non-activators, and the data are presented in the supplementary information (Table S2).
Table 1.
Summary of screening results.
| Plant Name | PXR Activation | AhR Activation | CYP3A4 Inhibition | CYP1A2 Inhibition | P-gp Inhibition |
|---|---|---|---|---|---|
|
| |||||
| Acacia rigidula [gum] | M | – | S | W | S |
| Achillea millefolium L. [flower] | S | – | W | W | – |
| Actaea racemosa L. [root] | – | M | – | – | – |
| Agastache rugosa Kuntze [mixed parts] | P | S | – | W | – |
| Aloe vera (L.) Burm.f. [leaf] | M | P | M | M | – |
| Andrographis paniculata (Burm.f.) Wall. [mixed parts] | – | – | W | – | M |
| Angelica sinensis (Oliv.) Diels [root] | S | S | S | M | – |
| Apium graveolens L. [seed] | P | S | W | M | – |
| Arctostaphylos uva-ursi (L.) Spreng. [fruit] | M | M | – | – | M |
| Arctostaphylos uva-ursi (L.) Spreng. [leaf] | S | – | – | – | – |
| Astragalus membranaceus [root] | – | M | W | – | M |
| Artemisia annua L. [mixed parts] | M | M | – | – | – |
| Atractylodes lancea DC. [root] | P | M | W | W | – |
| Atractylodes macrocephala Koidz. [root] | S | – | W | – | – |
| Atropa belladonna L. [leaf] | M | – | W | – | – |
| Avena sativa L. [fruit coat] | – | – | – | – | – |
| Avena sativa L. [seed] | – | – | W | S | S |
| Berberis aquifolium Pursh [mixed part] | S | S | M | – | – |
| Boswellia serrata Roxb. [gum] | – | – | M | – | – |
| Camellia sinensis (L.) Kuntze [leaf] | – | – | – | – | M |
| Capsicum annuum L. [fruit] | P | S | M | S | M |
| Capsicum chinense Jacq. [fruit] | – | – | W | – | – |
| Capsicum frutescens L. [fruit] | M | – | W | W | S |
| Carthamus tinctorius L. [flower] | M | M | W | – | – |
| Cassia angustifolia [leaf] | S | P | S | W | – |
| Centella asiatica (L.) Urb. [mixed parts] | S | M | – | – | – |
| Chrysanthemum indicum L. [flower] | M | M | W | W | M |
| Citrus aurantium L. [fruit] | S | S | W | W | – |
| Citrus limon (L.) Osbeck [fruit] | – | S | – | – | – |
| Citrus paradisi [fruit] | – | – | – | – | – |
| Citrus reticulata [fruit] | – | M | – | – | – |
| Citrus sinensis [leaf] | – | – | W | – | – |
| Citrus sinensis [fruit] | – | M | – | – | – |
| Cnidium monnieri Cusson [seed] | M | P | S | S | S |
| Coffea arabica L. [seed] | – | – | M | – | – |
| Coleus forskohlii Briq. [stem] | S | M | W | W | – |
| Crataegus monogyna Jacq. [fruit] | M | – | – | – | – |
| Crataegus monogyna Jacq. [mixed parts] | M | – | W | W | – |
| Crataegus laevigata (Poir.) DC. | – | – | – | – | – |
| Crataegus pinnatifida Bunge [fruit] | – | M | – | – | – |
| Curcuma longa L. [rhizome] | P | P | S | S | S |
| Cyanotis vaga (Lour.) Schult.f. [mixed parts] | M | S | M | W | – |
| Cynara cardunculus L. [leaf] | M | M | S | W | M |
| Cyperus rotundus L. [root] | P | P | M | W | S |
| Dendrobium nobile Lindl. [stem] | M | P | S | S | S |
| Echinacea angustifolia DC [root] | M | M | S | – | M |
| Epimedium brevicornu Maxim. [mixed parts] | M | P | M | – | – |
| Eriobotrya japonica (Thunb.) Lindl. [leaf] | S | – | M | W | M |
| Eurycoma longifolia Jack [root] | M | M | – | – | – |
| Euterpe oleracea Mart. [fruit] | M | – | W | – | – |
| Foeniculum vulgare Mill. [seed] | M | M | – | – | – |
| Forsythia suspensa Vahl [fruit] | M | – | W | – | – |
| Garcinia cambogia [fruit] | S | P | W | – | S |
| Garcinia mangostana L. [fruit] | S | P | W | W | M |
| Ginkgo biloba L. [leaf] | M | M | S | – | S |
| Gymnema sylvestre (Retz.) R.Br. ex Sm. [leaf] | – | M | W | – | – |
| Hoodia gordonii (Masson) Sweet [mixed parts] | – | – | W | – | – |
| Huperzia serrata (Thunb.) Trevis. [mixed parts] | P | S | M | – | S |
| Hydrastis canadensis L. [leaf] | S | P | S | W | – |
| Hydrastis canadensis L. [root] | – | S | – | S | – |
| Hypericum perforatum L. [mixed parts] | S | S | M | – | – |
| Hex paraguariensis A.St.-Hil. [leaf] | M | M | W | – | – |
| Hex vomitoria Aiton [fruit] | M | – | W | W | – |
| Hex vomitoria Aiton [leaf] | – | M | – | – | – |
| Lycium barbarum L. [fruit] | – | M | – | – | – |
| Lonicera japonica Thunb. [flower bud] | – | M | – | – | – |
| Medicago sativa L. [mixed parts] | – | M | W | – | – |
| Moringa oleifera Lam. [aerial parts] | M | M | W | – | – |
| Mucuna pruriens (L.) DC. [mixed parts] | M | M | W | – | – |
| Mucuna pruriens (L.) DC. [seed] | – | – | M | – | – |
| Murraya koenigii (L.) Spreng. [leaf] | – | M | M | M | S |
| Nelumbo nucifera Gaertn. [flower] | M | – | W | – | – |
| Nigella sativa L. [seed] | S | – | W | M | – |
| Olea europaea L. [leaf] | – | – | W | – | – |
| Panax ginseng C.A.Mey. [root] | – | – | – | – | – |
| Paullinia cupana Kunth [seed] | – | – | W | W | – |
| Pausinystalia johimbe (K.Schum.) Pierre [bark] | M | P | W | – | S |
| Petasites vulgaris [root] | M | – | M | W | S |
| Pfaffia paniculata [root] | M | M | – | – | – |
| Phyllanthus emblica L. [fruit] | – | – | – | W | – |
| Piper nigrum L. [fruit] | P | P | S | S | – |
| Plantago asiatica L. [seed] | – | – | W | W | – |
| Polygonum cuspidatum Willd. ex Spreng. [root] | – | – | M | S | – |
| Polygonum multiflorum [mixed parts] | M | – | – | – | – |
| Ptychopetalum olacoides Benth. [root] | P | P | W | W | S |
| Pueraria lobata [root] | P | S | M | W | – |
| Pueraria montana (Lour.) Merr. [root] | P | – | – | – | – |
| Rauvolfia vomitoria Wennberg [root] | S | P | W | M | S |
| Rhamnus frangula [bark] | M | – | W | M | M |
| Rhamnus purshiana (DC.) A.Gray [bark] | M | – | – | W | – |
| Rhodiola rosea L. [root] | M | P | W | W | S |
| Rosmarinus officinalis L. [leaf] | M | S | M | M | M |
| Salix alba L. [bark] | – | M | M | W | M |
| Sambucus nigra L. [fruit] | M | M | M | W | M |
| Sceletium tortuosum (L.) N.E.Br. [leaf] | M | S | M | – | – |
| Schisandra chinensis (Turcz.) Baill. [fruit] | P | – | W | – | – |
| Scutellaria baicalensis Georgi [root] | M | S | W | S | – |
| Serenoa repens (W.Bartram) Small [fruit] | – | – | M | W | – |
| Sida cordifolia L. [mixed parts] | S | S | W | – | – |
| Sida rhombifolia L. [mixed parts] | M | M | W | – | – |
| Syzygium aromaticum (L.) Merr. [flower] | P | P | M | M | – |
| Taraxacum officinale F.H.Wigg. [leaf] | M | – | M | W | – |
| Terminalia arjuna Wight & Arn. [bark] | – | – | M | W | – |
| Terminalia bellirica (Gaertn.) Roxb. [fruit] | – | M | M | S | S |
| Terminalia chebula Retz. [fruit] | M | – | W | M | – |
| Theobroma cacao L. [seed] | – | – | – | – | – |
| Tribulus terrestris L. [fruit] | – | M | W | M | – |
| Tribulus terrestris L. [mixed parts] | P | P | W | – | – |
| Trifolium pratense L. [flower] | M | – | – | – | – |
| Turnera diffusa Willd. ex Schult. [leaf] | P | P | S | M | – |
| Turnera diffusa Willd. ex Schult. [mixed parts] | M | M | M | W | S |
| Ulmus fulva [bark] | S | – | W | W | – |
| Urtica dioica L. [leaf] | M | M | W | – | – |
| Vaccinium macrocarpon Aiton [fruit] | S | – | – | – | – |
| Vaccinium myrtillus L. [fruit] | P | M | W | W | – |
| Valeriana edulis Nutt. [root] | M | M | W | – | – |
| Valeriana officinalis L. [root] | S | M | – | – | – |
| Viburnum opulus L. [bark] | M | M | W | W | – |
| Vitis vinifera L. [fruit] | – | – | – | – | – |
| Vitis vinifera L. [seed] | M | M | – | – | – |
| Withania somnifera (L.) Dunal [leaf] | M | – | M | – | – |
| Withania somnifera (L.) Dunal [root] | M | S | W | – | – |
| Xanthium sibiricum [fruit] | – | – | M | – | – |
| Zingiber officinale Roscoe [rhizome] | S | M | S | W | – |
| Z. jujuba [seed] | S | – | W | M | M |
M = mild; P = potent; S = strong; W = weak; - = no activation or no inhibition.
Fig. 1.
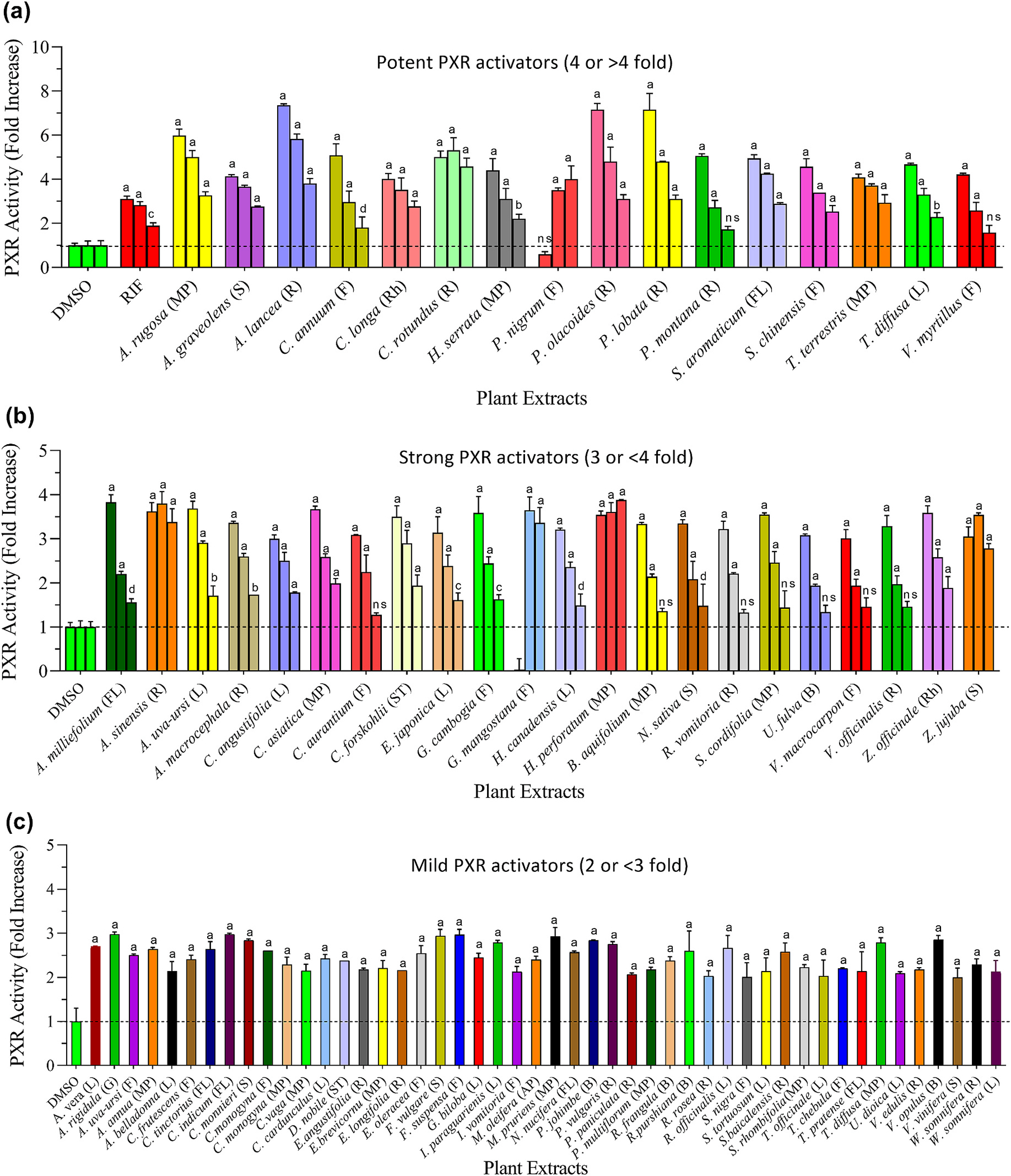
PXR activation by plant extracts commonly used in herbal medicine and other related products. Transiently transfected HepG2 cells were treated with various concentrations of extract (60, 20, and 6.66 μg/mL), positive control [rifampicin (RIF): 30, 10, and 3.33 μM], or vehicle control (0.3% DMSO) for 24 h. Following the incubation, luciferase activity was measured. Based on fold increase in luciferase activity, tested plants were categorized as follows: (a) Potent activators (PXR activation up to 4 or >4 fold); (b) Strong activators (PXR activation up to 3 to <4 fold); (c) Mild activators (PXR activation up to 2 to <3 fold). Weak or non-activator plants have been presented in the supplementary data (Table S2). Statistical significance is shown compared to DMSO control. ap < 0.0001; bp < 0.001; cp < 0.01, dp < 0.05 and ns = not significant. The abbreviations are representing as L = leaves, S = seed, R = root, F = fruit, ST = stem, B = bark, G = gum, FL = flower, SC = seed coat, Rh = rhizome, AP = aerial part, and MP = mixed part.
3.3. Effect of plant extracts on transcriptional activity of human AhR
Human AhR reporter cells were used to examine the effect of selected plant extracts on the AhR’s transcriptional activity. Depending upon AhR activation potential, all tested plants were mainly divided into four groups: 1) potent activators (fold activation of ≥10); 2) strong activators (>5 to <10-fold); 3) mild or moderate activators (>2 to <5-fold); 4) weak or non-activators (<2-fold). Results are presented in Fig. 2 and Table 1. As shown in Fig. 2a, extracts of 18 plants, including A. vera, C. angustifolia, C. monnieri, C. longa, C. rotundus, D. nobile, E. brevicornum, G. cambogia, G. mangostana, H. canadensis, P. johimbe, P. nigrum, P. olacoides, R. vomitoria, R. rosea, S. aromaticum, T. terrestris and T. diffusa showed strong agonistic effects and increased AhR activity >10-fold. These plants were considered potent activators. Subsequently, 17 plants (A. rugosa, A. sinensis, A. graveolens, C. annuum, C. aurantium, C. limon, C. vaga, H. serrata, H. canadensis, H. perforatum, B. aquifolium, P. lobata, R. officinalis, S. tortuosum, S. baicalensis, S. cordifolia, and W. somnifera) were able to increase AhR activity between >5 and <10-fold and these were termed strong activators (shown in Fig. 2b). Further, 41 plants were able to enhance the AhR activity between >2 and <5-fold at the highest tested concentration of 30 μg/mL (Fig. 2c), but at lower concentrations (10 and 3.33 μg/mL) had no influence (data not shown). Interestingly, the extracts of the remaining 47 plants did not affect AhR activation, and they were characterized as non-activators (Table S3). The positive control, Me-Bio (at 1 μM), induced >70-fold increase in AhR’s transcriptional activity over the solvent control under similar experimental conditions. The AhR activation effects of A. vera, C. longa, G. cambogia, H. canadensis, P. johimbe, P. nigrum R. rosea, H. canadensis, H. perforatum and W. somnifera are in agreement with previously reported data (Haron et al., 2022b).
Fig. 2.
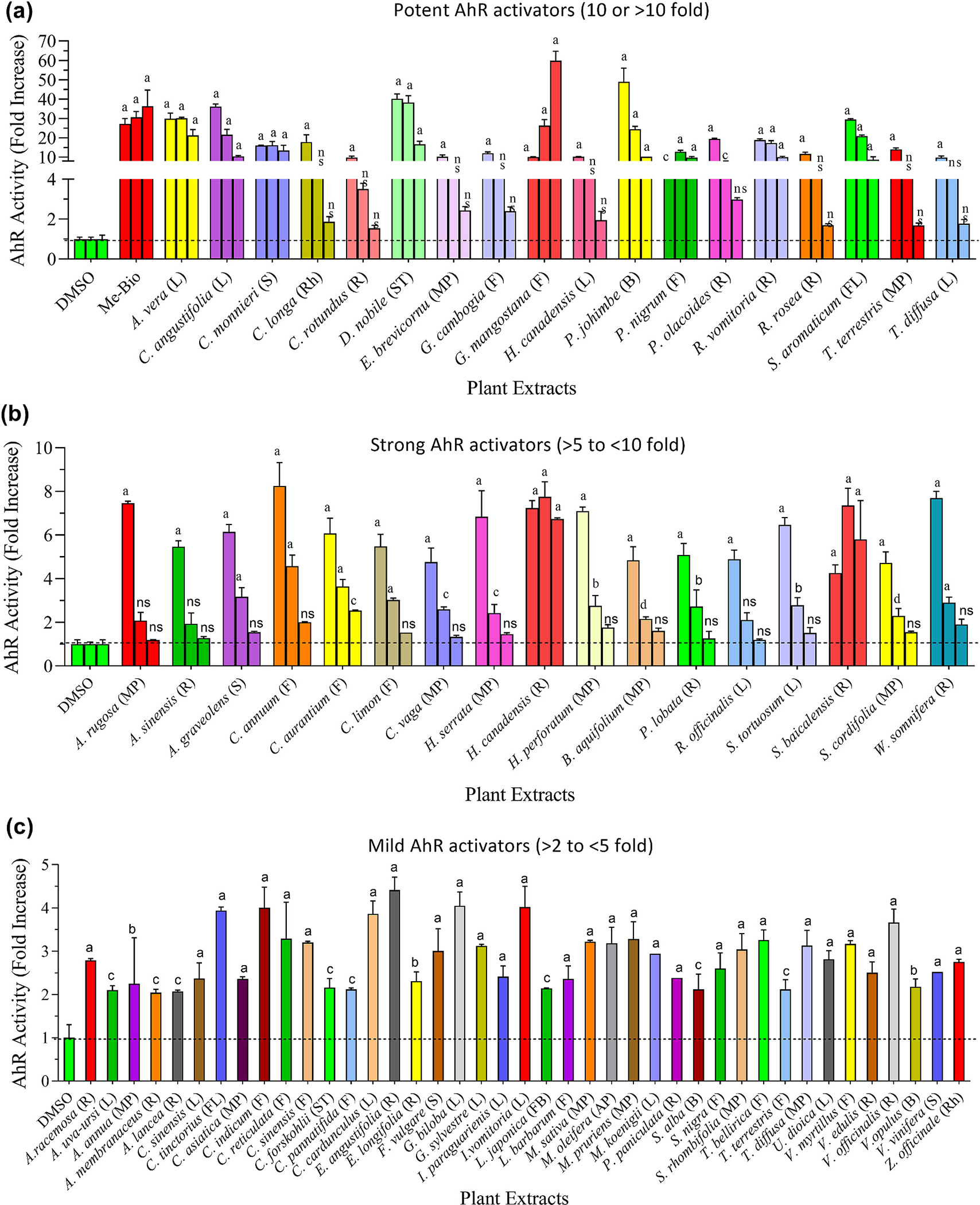
Effects of various medicinal plant extracts on the activity of hAhR. AhR reporter cells were treated with different concentrations of extract (30, 10, and 3.33 μg/mL), positive control (Me-bio: 1000, 500, and 250 nM), or vehicle control (DMSO) for 24 h. Following incubation, luciferase activity was measured. Based upon fold increase in luciferase activity, tested plants were categorized as follows: (a) Potent activators (≥10-fold); (b) Strong activators (>5 – <10-fold); (c) Mild activators (>2 – <5-fold). Weak (<2-fold) or non-activators are presented in the supplementary data (Table S3). L, S, R and MP represent leaf, stem, root and mixed parts, respectively. Statistical significance is shown compared to DMSO control. ap < 0.0001; bp < 0.001; cp < 0.01, dp < 0.05 and ns = not significant. The abbreviations are representing as L = leaves, S = seed, R = root, F = fruit, ST = stem, B = bark, G = gum, FL = flower, FB = flower bud, SC = seed coat, Rh = rhizome, AP = aerial part, and MP = mixed part.
3.4. Effect of plant extracts on the catalytic activity of CYP3A4
To evaluate CYP3A4 inhibition, plant extracts at various concentrations were incubated with CYP3A4-baculosomes, and enzyme activity was measured (Fig. 3). Results are summarized in Table 1. Based on IC50 values, plants were divided into four groups: 1) Strong inhibitors: this group of plants inhibited the catalytic activity of tested CYPs with IC50 values between 1.3 and 10 μg/mL, as shown in Fig. 3a. Out of 123 plants screened, 13 extracts originated from A. rigidula, A. sinensis, C. angustifolia, C. monnieri, C. longa, C. cardunculus, D. nobile, E. angustifolia, G. biloba, H. canadensis, P. nigrum, T. diffusa, and Z. officinale showed a strong inhibition for the catalytic activity of CYP3A4. 2) Mild inhibitors: this group of plants inhibited the catalytic activity of tested CYPs with IC50 values between >10 and 20 μg/mL, as shown in Fig. 3b. The extracts of 28 plants representing A. vera, B. serrata, C. annuum, C. arabica, C. vaga, C. rotundus, E. brevicornum, E. japonica, H. serrata, H. perforatum, B. aquifolium, M. pruriens, M. koenigii, P. vulgaris, P. cuspidatum, P. lobata, R. officinalis, S. alba, S. nigra, S. tortuosum, S. repens, S. aromaticum, T. officinale, T. arjuna, T. bellirica, T. diffusa, W. somnifera, and X. sibiricum were considered as mild inhibitors. 3) Weak inhibitors: this group of plants inhibited CYPs with IC50 values between >20 and 50 μg/mL. The description of this group of 51 candidates is shown in Fig. 3c. 4) Non-inhibitors: this group of plants showed no inhibitory effect on tested CYPs. A total of 31 plants were identified as non-inhibitors and are presented in the supplementary information (Table S4). Ketoconazole (KZN), a well-known CYP3A4 inhibitor with an IC50 of 0.02 μM under similar experimental conditions, was used as a positive control.
Fig. 3.
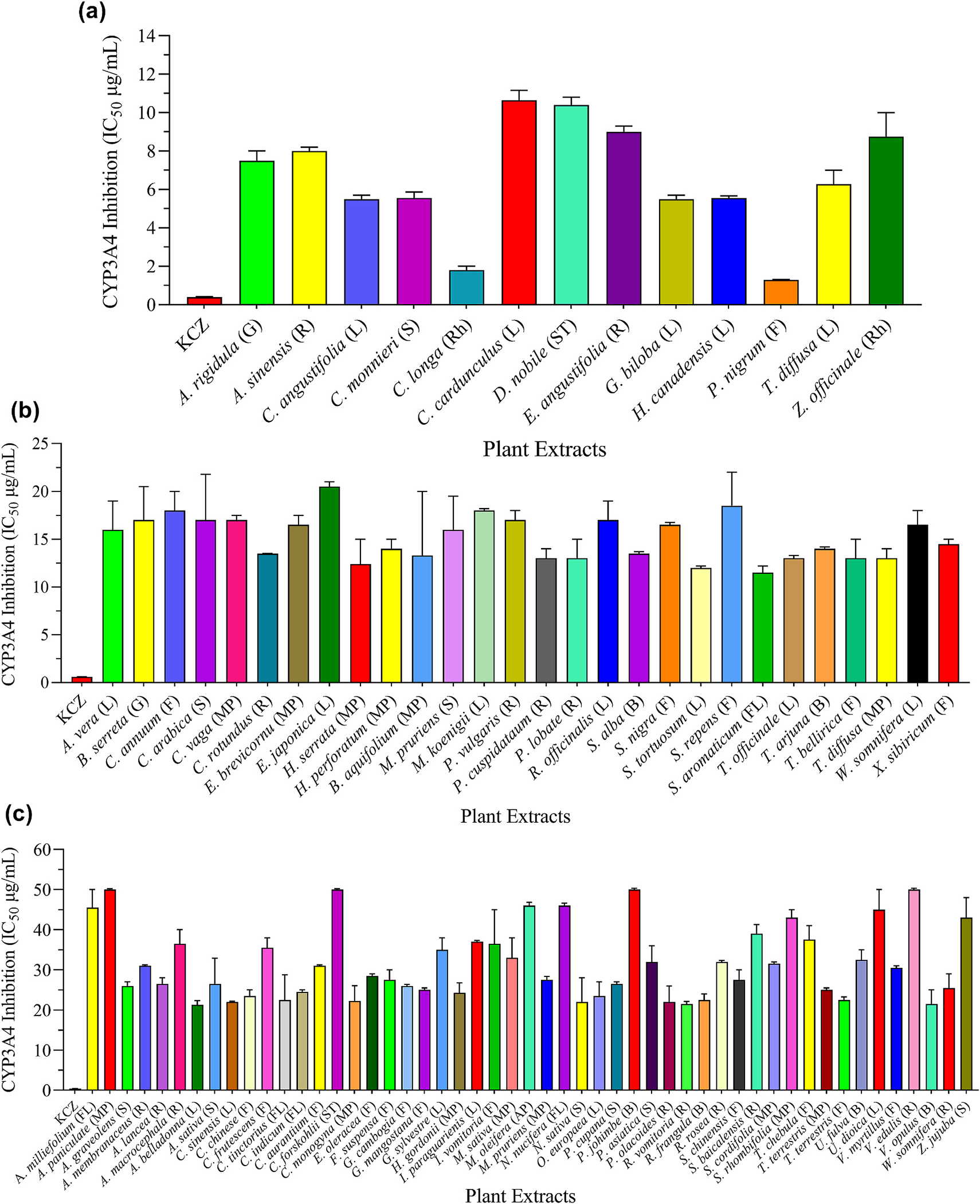
Inhibitory effects of medicinal plants on CYP3A4 activity. The CYP3A4-baculosomes were incubated with enzyme-specific substrate and increasing concentrations of the extracts. Ketoconazole (KCZ) was used as a positive control. After incubation, the reaction was stopped, and fluorescence was measured. The percent inhibition was calculated, and results were presented in terms of IC50 values. Based on IC50 values, tested plants were categorized as follows: (a) Strong inhibitors (IC50 values of 1.3–~10 μg/mL); (b) Mild inhibitors (IC50 values of >10–~20 μg/mL; (c) Weak inhibitors (IC50 values in the range of >20–~50 μg/mL). Non-inhibitors are presented in the supplementary materials (Table S4). The abbreviations are representing as L = leaves, S = seed, R = root, F = fruit, G = gum, ST = stem, B = bark, FL = flower, SC = seed coat, Rh = rhizome, AP = aerial part, and MP = mixed part.
3.5. Effect of plant extracts on the catalytic activity of CYP1A2
To examine CYP1A2 inhibition, designated baculosomes and Vivid EOMCC substrate were incubated with various concentrations of plant extracts, and fluorescence was monitored in a kinetic mode. The percent inhibition in enzyme activity was calculated, and data (IC50 values) were presented in Fig. 4. The results are also summarized in Table 1. As shown in Figs. 4a 10 plants (A. sativa, C. annuum, C. monnieri, C. longa, D. nobile, H. canadensis, P. nigrum, P. cuspidatum, S. baicalensis, and T. bellirica) strongly hampered the catalytic activity of CYP1A2 with IC50 values between 1.3 and ~10 μg/mL. These 10 plants were designated as strong CYP1A2 inhibitors. As shown in Figs. 4b 13 plants, including A. vera, A. sinensis, A. graveolens, M. koenigii, N. sativa, R. vomitoria, R. frangula, R. officinalis, S. aromaticum, T. diffusa, T. chebula, T. terrestris, and Z. jujube inhibited CYP1A2 with IC50 values ranging between >10 and ~20 μg/mL. These plants were considered mild inhibitors. However, 35 plants were deemed weak inhibitors with IC50 values between >20 and ~50 μg/mL (Fig. 4c). Interestingly, the remaining 65 plants demonstrated no noticeable inhibitory effects on CYP1A2 activity. These plants were considered non-inhibitors and are shown in Table S5. α-Naphthoflavone (α-NAP), a known inhibitor of the CYP1A2 isoform, was included as a positive control, which inhibited CYP1A2 with an IC50 of 0.06 μM. Altogether, the CYP inhibition data indicated that plants such as C. monnieri, C. longa, D. nobile, H. canadensis, and P. nigrum demonstrated strong inhibition of both CYP isoforms (3A4 and 1A2) and may pose a potential risk for HDIs.
Fig. 4.
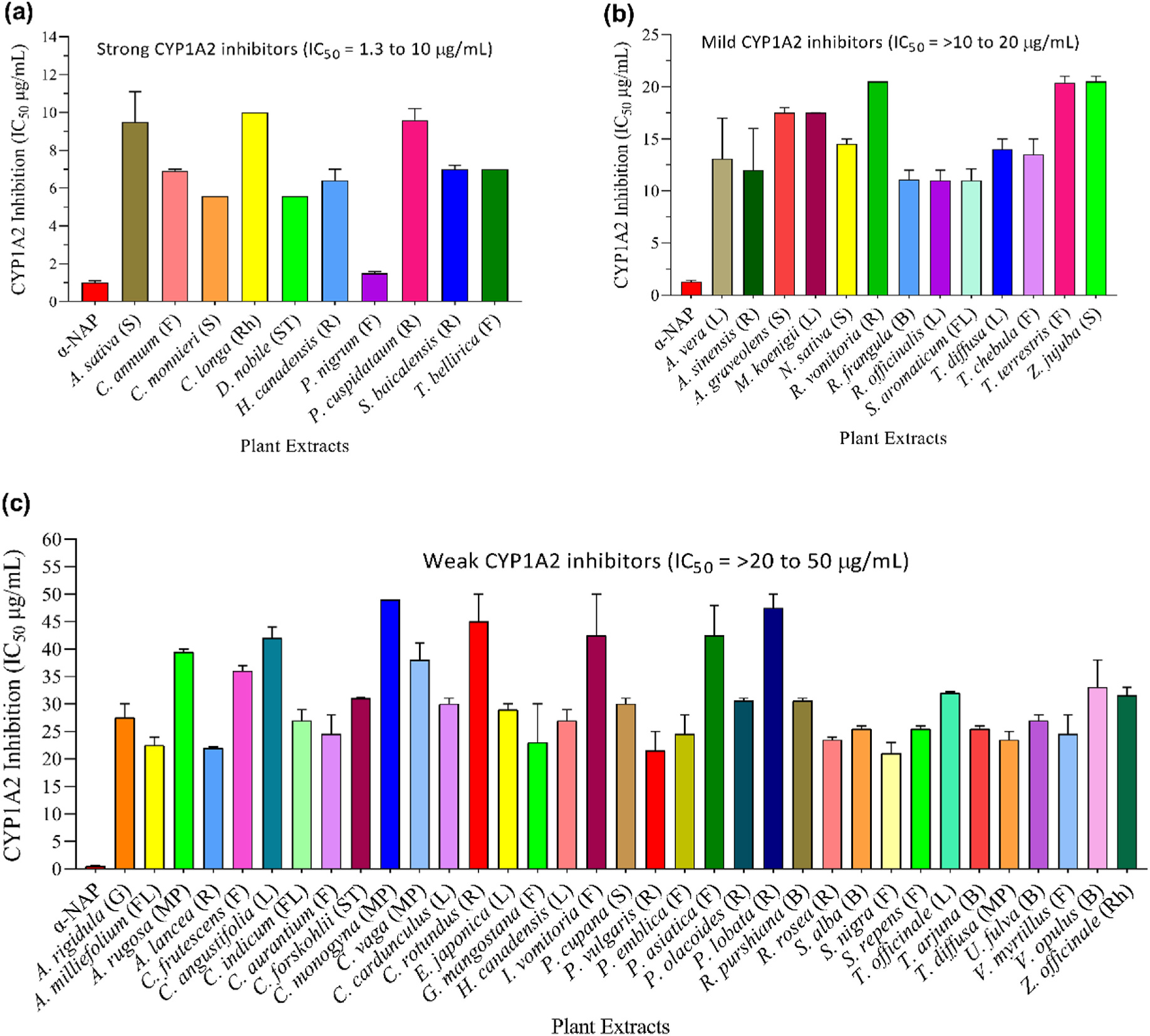
Inhibitory effects of medicinal plants on CYP1A2 activity. The CYP1A2-baculosomes were incubated with the enzyme-specific substrate, increasing concentrations of extracts or positive control α-naphthoflavone (α-NAP). Following the incubation, the reaction was stopped, and fluorescence was measured. The inhibitory effect was represented in terms of IC50 values. Based on the IC50 values, all tested plants were categorized as follows: (a) Strong inhibitors (IC50 values of 1.3–~10 μg/mL); (b) Mild inhibitors (IC50 values of >10–~20 μg/mL); (c) Weak inhibitors (IC50 values of >20–~50 μg/mL). Data for non-inhibitors is presented in the supplementary materials (Table S5). The abbreviations are representing as L = leaves, S = seed; R = root; F = fruit, ST = stem, B = bark, G = gum, FL = flower, SC = Seed coat; Rh = rhizome, and MP = mixed part.
3.6. Effects of plant extracts on P-glycoprotein (P-gp)
Inhibition of P-gp by select medicinal plants was investigated using Rh-123 uptake-based efflux assay in P-gp overexpressing MDR1-MDCK cells. To rule out possible false-positives due to the toxic effects of extracts, all samples were normalized by dividing the fluorescence by total protein. The results are summarized in Table 1. Based on the results the extracts were divided into three groups. 1) Strong inhibitors: extracts in this group strongly attenuated the function of P-gp, and Rh-123 uptake was increased in the range of 150–490%. 2) Mild inhibitors: extracts in this group moderately inhibited the P-gp activity with Rh-123 uptake increase in the range of 120–150%. 3) Non-inhibitors: this group produced little to no inhibition of P-gp activity, and Rh-123 uptake remained almost equal to vehicle control (100%). As shown in Fig. 5a, 18 plants (A. rigidula, A. sativa, C. frutescens, C. longa, C. monnieri, C. rotundus, D. nobile, G. cambogia, G. biloba, H. serrata, M. koenigii, P. johimbe, P. vulgaris, P. olacoides, R. vomitoria, R. rosea, T. bellirica, and T. diffusa) significantly inhibited the P-gp mediated efflux of Rh-123 at the highest tested concentration of 50 μg/mL. These plants were grouped as strong P-gp inhibitors. As shown in Fig. 5b, extracts of 15 plants (A. paniculata, A. uva-ursi, A. membranaceus, C. sinensis, C. annuum, C. indicum, C. cardunculus, E. angustifolia, E. japonica, G. mangostana, R. frangula, R. officinalis, S. alba, S. nigra, and Z. jujube) increased the intracellular accumulation of Rh-123 to lesser extents (120% to <150%), in comparison to vehicle control (100%). These plants were collectively considered mild P-gp inhibitors (Fig. 5b). On the contrary, the remaining 90 plants showed no significant increase in Rh-123 accumulation (Table S6), which suggests these plants have no inhibitory effect on this specific efflux pump. Cyclosporine, a first-generation P-gp inhibitor, was used as a positive control in all efflux assays, which exhibited dose-dependent P-gp inhibition. These results (summarized in Table 1) revealed that some medicinal herbs, categorized as strong inhibitors, may impede P-gp-mediated efflux of drugs in primary tissues such as the kidney, liver, and intestine. However, properly designed clinical studies will be needed to confirm the clinical relevance of this conjecture.
Fig. 5.
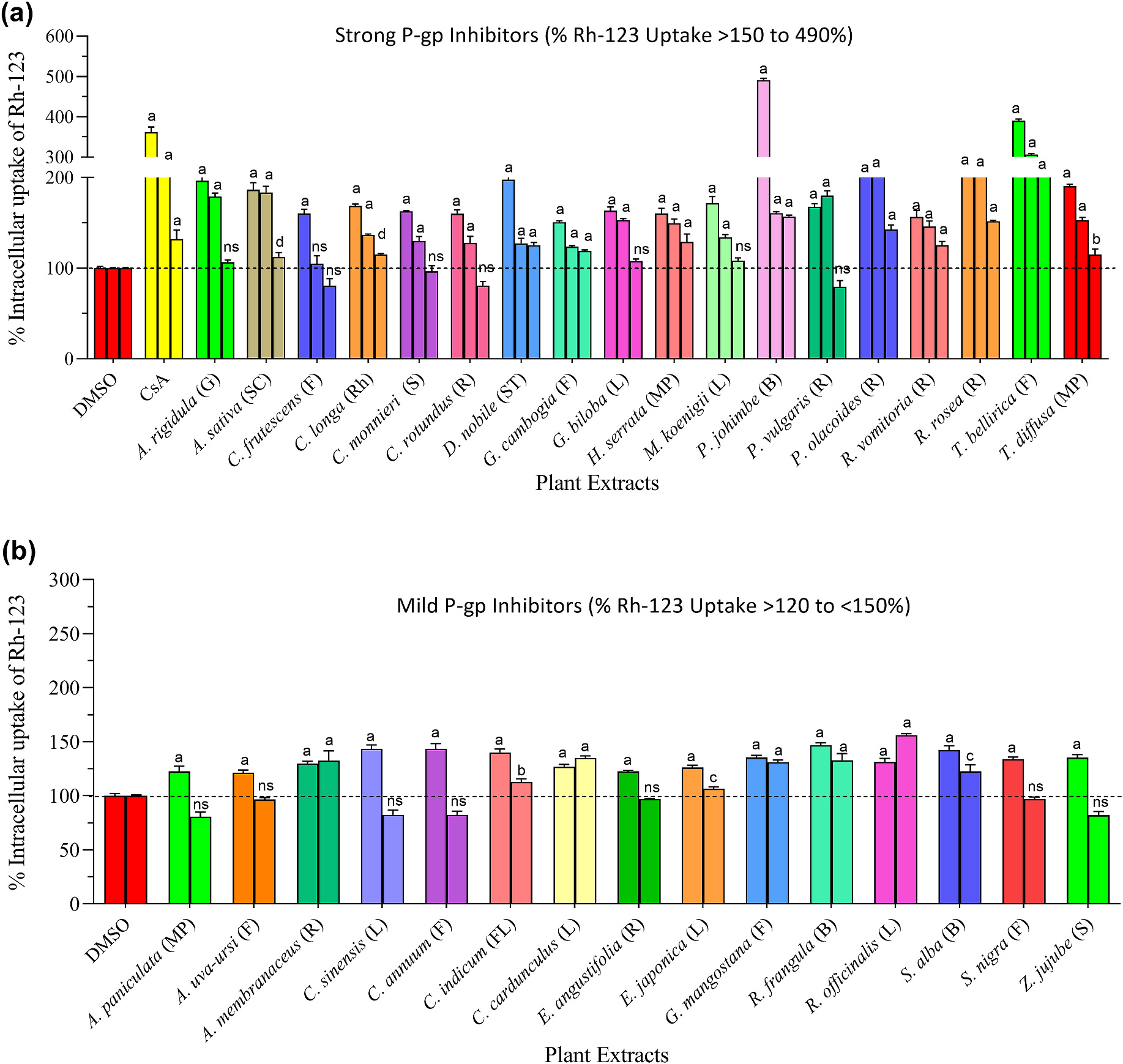
P-gp inhibition by medicinal plants. The P-gp overexpressing MDR1-MDCK cells were incubated with increasing concentrations of plant extracts (50, 25, and 12.5 μg/mL) or positive control [cyclosporine A (CsA): 10 μM)] for 90 min. Following incubation, cells were washed with ice-cold PBS, lysed with lysis buffer, and Rh-123 uptake was measured at 485/529 nm. Based on % Rh-123 uptake, results were categorized into three groups: (a) Strong inhibitors; the extracts in this group showed significant P-gp inhibition and increased Rh-123 uptake in the range of >150–490%. (b) Mild inhibitors; the extracts in this group exhibited P-gp inhibition, but the magnitude of inhibition was limited (>120% to <150%). The extracts of 90 plants had no significant P-gp inhibition (<120%). These plants were grouped as non-inhibitors and are presented in the supplementary data (Table S6). Statistical significance is shown compared to DMSO control. ap < 0.0001; bp < 0.001; cp < 0.01, dp < 0.05 and ns = not significant. The abbreviations are representing as L = leaves, S = seed; R = root; F = fruit, ST = stem, B = bark, G = gum, FL = flower, SC = Seed coat; Rh = rhizome, and MP = mixed part.
4. Discussion
Overview.
As a testament to medicinal plants’ popularity, a significant segment of US population regularly ingests BDS. This is understandable given that >50 K different dietary supplement products are currently available on the US market (Dietary Supplement Label Database, 2017). Recent surveys indicate that more than half of US adults above the age of 20 have utilized some type of dietary supplement within 30 days. Furthermore, it has also been noticed that, with increasing age, consumers tend to ingest multiple (sometimes four or more) dietary supplements (Mishra et al., 2021). The popularity of herbals can be more clearly understood by examining the SPINS® report of 2020 (Smith et al., 2021). As per this report, the top-10 selling herbs were Sambucus nigra (elderberry), Marrubium vulgare (horehound), Vaccinium macrocarpon (cranberry), Curcuma longa (turmeric), Malus spp. (apple cider vinegar), Zingiber officinale (ginger), Echinacea spp. (echinacea), Allium sativum (garlic), Trigonella foenum-graecum (fenugreek), and Triticum aestivum/Hordeum vulgare (wheatgrass/barley grass). In addition, recent studies suggest that patients with chronic or serious diseases are more prone to use herbal products (Peltzer and Pengpid, 2019). COVID-19 is a relevant example. During the pandemic, consumers used various herbal remedies to alleviate common symptoms associated with viral infections or improve overall immune function. Additionally, clinicians and government health care agencies also issued recommendations for their safe use (Ahmad et al., 2021). Agencies like the US Food and Drug Administration (FDA) regulate herbal dietary supplements, but not as rigorously as that for conventional medications. They do, however, constantly surveille supplement marketing practices and send warning letters to manufacturers for unproven medical claims (FDA, 2021a). The FDA may also release press notices for consumers related to the limited or restricted use of particular herbal products (FDA, 2017, 2021b). Still, the average consumer is often unaware of the undesirable side effects of overconsumption of herbal products. This is important because several clinical reports have shown that chronic intake of certain botanicals may pose health risks (Zanger and Schwab, 2013).
PXR Modulation.
PXR (hPXR, NR1I2) is a ligand-activated transcriptional factor significantly expressed in organs actively involved in the xenobiotic transformation. Structurally, PXR has a ligand-binding domain (LBD) at its C-terminus and DNA binding domain (DBD) at the N-terminus. Upon activation, the DBD binds to the regulatory DNA sequence of response elements, whereas the LBD has a dual function: one to bind ligand, the other to provide an interface for HDIs (Ihunnah et al., 2011). Medicinal herbs and related products like BDS are rich sources of diverse phytochemicals (e.g., alkaloids, polyphenols, flavonoids, coumarins, tannins, terpenoids, stilbenes, etc.). At the cellular level, phytochemicals perform dual functions: 1) they can act as agonists by binding to the ligand-binding domain of PXR, where they can upregulate the transcriptional activity of downstream genes, especially CYP3A4, CYP2B, CYP2C, glutathione S-transferases, sulfotransferases, UGTs, and drug transporters (OATP2, MDR1, MRP2, and MRP3) (Chang et al., 2007; Goldstein et al., 2013); or 2) they can act as antagonists, either by functioning as repressors of PXR transcription or binding to active sites of post-translationally matured CYPs to inhibit their catalytic activities. At this point, either function could be detrimental as they form the basis for HDIs.
As per the Food and Agriculture Organization (FAO) (2002), worldwide, >50,000 plants are being used for medicinal purposes (Schippmann et al., 2002). Moreover, only a few medicinal plants such as Hypericum perforatum (hyperforin), Commiphora mukul (E- and Z-guggulsterone), Piper methysticum (kavalactones; desmethoxyangonin and dihydromethysticin), Coleus forskohlii (forskolin and 1,9-dideoxyforskolin), and Ginkgo biloba (ginkgolide A and B) are recognized as efficient PXR agonists and pose risks for HDIs when taken concomitantly with prescription medications (Staudinger et al., 2006). Still, many more plants may potentially modulate the transcriptional activity of PXR. Thus, identifying such candidates is extremely important and requires systematic investigation in a high throughput manner. This study used transfected HepG2 cells as a cellular model coupled with hydro-ethanolic extracts of 123 medicinal herbs to screen for PXR activity using a luciferase assay. We were able to detect 16 plants, namely A. rugosa, A. graveolens, A. lancea, C. annuum, C. longa, C. rotundus, H. serrata, P. olacoides, P. lobata, P. montana, S. chinensis, S. aromaticum, T. terrestris, T. diffusa and V. myrtillus, and P. nigrum (Fig. 1a) which augmented the transcriptional activity of PXR from 4- to 7-fold. In this model, these plants were considered potent PXR agonists. Another 23 plants were also strong activators of PXR (3- to <4-fold increase). Among both groups, certain plants like C. forskohlii, H. perforatum, and C. rotundus are already recognized as efficient PXR agonists, but to the best of our knowledge, adequate information is unavailable for plants like C. annum, A. rugosa, A. graveolens, P. lobata, P. montana, and T. diffusa. Therefore, our findings could be an important step forward in identifying new plant-based PXR activators. CYP3A4 and CYP2B6 are involved in the metabolism of 50–60% and 8% of drugs, nutraceuticals, and herbal molecules, respectively (Hernandez et al., 2009; Hedrich et al., 2016), and downstream induction of detoxifying enzymes and transporters can increase the metabolism and clearance of xenobiotics, an action that may have either positive or negative physiological consequences. For example, limited consumption of these plants (Fig. 1a and 1b) and their formulated products could provide protective effects against potential toxicants in healthy individuals. On the other hand, phytochemical-mediated activation of PXR and its targeted downstream gene products could markedly increase life-saving drugs’ metabolism and clearance, rendering them less efficacious. Accordingly, the plants shown in Fig. 1a, and 1b (and products containing them) may pose a risk for induction-mediated HDIs. However, further studies are required at the expression level (e.g., CYP and ABC transporters) to understand the physiological consequences of PXR activation and their clinical relevancy.
AhR Modulation.
AhR is an inducible ligand-dependent transcriptional factor belonging to the basic helix-loop-helix family and structurally has a Per-Arnt-Sim (PAS) domain that binds to various endogenous and exogenous ligands. In a non-active state, AhR resides in the cytoplasm with chaperone proteins as an AHR-protein complex (Zhu et al., 2019). On activation, it translocates into the nucleus, releases chaperones, and heterodimerizes with its nuclear protein partner ARNT (AhR nuclear translocator). Upon ligand binding, the ligand-AhR-ARNT complex attaches to a specific DNA recognition site and triggers the transcription of responsive genes, including CYP1A2, CYP1A1, CYP1B1, glutathione-S-transferase, UDP-glucuronosyl transferase, NADPH quinone reductase, and others (Jeuken et al., 2003; Larigot et al., 2018). Among the CYP1 family, isoforms 1A2, 1A1, and 1B1 are very important as they metabolize several exogenous and endogenous compounds to biologically active metabolites (Wang and Zhou, 2009). Two common examples are arachidonic and eicosapentaenoic acids, both polyunsaturated fatty acids metabolized by CYP1A1, whose products play key roles in vital physiological activities (Schwarz et al., 2004). The active role of these isoforms is also reported in the metabolic activation of dietary compounds that possess chemoprevention activities. On the contrary, isoforms 1A2, 1A1, and 1B1 catalyze the metabolism and activation of procarcinogenic xenobiotics (e.g., 7,12-dimethyl benzanthracene [DMBA] and 2,3,7,8-tetrachlorodibenzo-p-dioxin [TCDD]) to carcinogenic reactive intermediates, such as polycyclic aromatic hydrocarbons and other environmental pollutants, which significantly contribute to carcinogenesis (Schwarz et al., 2004; Choi and Kim, 2008). A regulatory role of AhR in overexpression of CYP1 isoforms, including 1A2, A1, and B1, has been suggested (Chang et al., 2007). Hence, constant exposure to dietary products which can induce chronic activation of AhR could be more problematic and perhaps life-threatening. The agonistic effect of dietary supplements, food products, and medicinal herbs for AhR has been reported by several investigators, but most of these studies focused on individual dietary phytochemicals such as quercetin, quercetin glycosides, rutin, kaempferol, ginkgolides, bilobalide, taxifolin, curcumin, etc. (Choi and Kim, 2008; Jin et al., 2018). In contrast, very few reports appear in the public domain on AhR agonism of crude extracts prepared from medicinal herbs (Jeuken et al., 2003; Weiss, 2018; Husain et al., 2021b). As part of our quest to ensure botanical safety, extracts of 123 medicinally important plants were primarily screened to probe their agonism using AhR reporter cells. Our findings demonstrated that 18 plant extracts increased AhR transcription by > 10- <48-fold and were considered potent agonists. Seventeen plants increased AhR activity between >5 and <10-fold, and these plants were considered strong agonists. In contrast, 41 plant extracts were identified as less potent (2- to <5-fold increase), and 47 plant extracts had no impact on AhR activity. Accordingly, these plants were considered weak or non-agonists. Based on these findings, it is suggested that the herbs depicted in Fig. 2a and 2b and Table 1 possess sufficient potential to induce AhR activation. Thus, these plants’ regular and indiscriminate consumption and/or their associated BDS may be able to transcriptionally activate relevant downstream genes. Therefore, AhR-mediated overexpression of CYP1 isoforms (1A1, 1A2, and 1B1) can greatly influence xenobiotic metabolism and may even increase the untoward deleterious effects if consumed chronically. However, further molecular and clinical studies are necessary to establish the clinical relevancy of these extracts’ AhR-mediated effects.
Inhibition of CYP3A4 and 1A2.
Around 15 different CYP isoforms are involved in the metabolism of drugs and xenobiotics. CYP3A4 accounts for ~28% of the total hepatic CYP pool and is actively involved in the metabolism of almost half of all currently prescribed drugs. Numerous drugs of natural (e.g., berberine, colchicine, cyclosporine, ergotamine, quinidine, vinblastine) and synthetic (e.g., atorvastatin, itraconazole, midazolam, nifedipine, saquinavir) origin are recognized as substrates of CYP3A4 (Zanger and Schwab, 2013). CYP1A2, another important isoform, is mainly expressed in the liver and represents ~4–16% of the total hepatic CYP pool. Studies suggest that 9% of currently prescribed drugs (e.g., theophylline, caffeine, clozapine, olanzapine, tizanidine, duloxetine, ramelteon) are principal substrates for CYP1A2 (Zanger and Schwab, 2013). It is well known that phytochemicals significantly contribute to the medicinal and nutritional value of food, herbals, and BDS. What is not as readily appreciated is that certain phytochemicals may also act as substrates for various CYP isoforms to inhibit (reversibly or irreversibly) their catalytic activity (Husain et al., 2021c). Some phytochemicals appear to be capable of both inducing nuclear receptor activation and inhibiting CYP activity. This dual divergent phenomenon has been observed for rhapontigenin (Rheum undulatum), furanocoumarins (grapefruit juice), resveratrol (red wine), gomicin C (Schisandra fruit), rutaecarpine and limonin (Evodia rutaecarpa), methylenedioxyphenyl lignans (Piper cubeba), kaempferol glycosides (Zingiber aromaticum), and serpentine (Catharanthus roseus) (Subehan et al., 2006). Recently, several clinical cases have described patients that experienced unwanted toxic effects of co-administrated drugs in conjunction with the overconsumption of select herbals or BDS (Rubin et al., 2019; Revol et al., 2020).
As an initial screening tool for possible HDIs, invitro CYP inhibition studies offer several practical advantages. The first is that multiple botanicals and/or purified phytochemicals can be evaluated simultaneously using well-established systems (e.g., microsomal systems, supersomes, transfected cell lines, human hepatocyte cultures, etc.). The second is mechanistic information associated with reversible inhibition, time-dependent inhibition, etc. Third, and perhaps most appealing, is that in vitro inhibition studies are relatively inexpensive to perform (Gurley, 2012).
In the present investigation, we used fluorescence-based throughput screening assays to examine the inhibitory effects of 123 selected medicinal plants on human CYP3A4 and 1A2. Our results showed that extracts derived from 13 plants, namely A. rigidula, A. sinensis, C. angustifolia, C. monnieri, C. longa, C. cardunculus, D. nobile, E. angustifolia, G. biloba, H. canadensis, P. nigrum, T. diffusa, and Z. officinale, strongly hampered the catalytic activity of CYP3A4 in a cell-free system and exhibited IC50 values in the range of 1.3–~10 μg/mL (Fig. 3a). In addition, 10 plants (A. sativa, C. annuum, C. monnieri, C. longa, D. nobile, H. canadensis, P. nigrum, P. cuspidatum, S. baicalensis, and T. bellirica) inhibited CYP1A2 activity with IC50 values in the same range (1.3–~10 μg/mL) (Fig. 4a). Interestingly, four plants (C. monnieri, C. longa, D. nobile, and P. nigrum) inhibited the catalytic activity of both tested isoforms. Likewise, extracts of H. canadensis leaf and root inhibited CYP3A4 and CYP1A2 with IC50 values < 10 μg/mL. We also observed that 29 extracts impaired CYP3A4 activity with IC50 values in the range of >10–20 μg/mL, while 13 others similarly inhibited CY1A2 (Figs. 3b and 4b). The CYP inhibitory activity of Z. officinale (Revol et al., 2020), C. cardunculus (Campos et al., 2018), and P. nigrum (Rezaee et al., 2014) have already been reported in clinical studies. These observations suggest that prolonged exposure to, or overconsumption of, these plants or their affiliated BDS with drugs exhibiting narrow therapeutic indices that are substrates for either of these two isoforms may give rise to HDIs.
Modulation of P-gp.
P-gp is a transmembrane glycoprotein that belongs to the adenosine triphosphate-binding cassette (ABC) transporters superfamily. It is highly expressed in the apical membrane of pharmacologically important epithelial barriers such as kidney, liver, intestine, blood-brain barrier, etc. P-gp uses energy generated from ATP hydrolysis to regulate the absorption and efflux of various exogenous substrates across cell membranes (Li et al., 2013; Cao et al., 2020). Studies suggest that the phytochemicals of co-administered herbs sometimes act as modulators of P-gp activity (Kim et al., 2018; Fasinu et al., 2012). As inhibitors, herbals may reduce P-gp-mediated drug efflux, which may lead to enhanced bioavailability. Conversely, enhanced efflux can markedly reduce drug oral bioavailability or increase hepatic biliary clearance as inducers. Hence, both conditions raise the risk of P-gp mediated HDIs (Mannel, 2004).
Botanical modulation of P-gp expression and activity has been demonstrated both in vitro and in the clinic, with St. John’s wort and grapefruit juice being two well-recognized examples of induction (Gurley et al., 2008) and inhibition (Hermann et al., 2002), respectively. We used P-gp-overexpressing MDR1-MDCK cells to gauge inhibition of efflux transporter activity by selected herbals using Rh-123 uptake assay. Our findings indicated that of the 123 plants screened, 18 (A. rigidula, A. sativa, C. frutescens, C. longa, C. monnieri, C. rotundus, D. nobile, G. cambogia, G. biloba, H. serrata, M. koenigii, P. johimbe, P. vulgaris, P. olacoides, R. vomitoria, R. rosea, T. bellirica, and T. diffusa) increased intracellular Rh-123 up to ~150%–490%, compared to vehicle control (100%). We designated these plants as strong P-gp inhibitors (Fig. 5a, Table 1). Seven of these plants (e.g., C. frutescens, C. longa, C. monnieri, G. biloba, H. serrata, P. johimbe, and R. rosea), have been reported earlier by others (Hellum and Nilsen, 2008; Eyal et al., 2010; Cao et al., 2020; Ganesan et al., 2021), while the remaining 11 plants represent potentially new candidates as P-gp inhibitors. We also noticed that several other plants (Fig. 5b, Table 1) exhibited P-gp inhibition by <50%. These plants were considered as weak/mild inhibitors. Collectively, our initial P-gp inhibition observations indicated that the regular and overconsumption of plants presented in Fig. 5a, or their associated BDS, may induce P-gp-facilitated HDIs, although further in vivo research is warranted to confirm the clinical relevancy of these findings.
5. Conclusion
Ethanolic extracts of 123 medicinal plants were examined for PXR and AhR induction activities. Among them, 19 plants considerably increased the transcriptional activity of PXR (4- to 7-fold), and 19 plants increased the transcriptional activity of AhR (10- to 103-fold). The extracts of 18 plants dose-dependently increased the intracellular uptake of Rh-123 up to >150–490% in P-gp overexpressing cells. CYP inhibition analysis showed that 13 plants inhibited the catalytic activity of CYP3A4 at concentrations that may be physiologically relevant (IC50, 1.3–~10 μg/mL). Likewise, 10 plants could also inhibit CYP1A2 activity with similar IC50 values. Interestingly, 4 plant extracts (e.g., C. monnieri, C. longa, D. nobile, and P. nigrum) were able to inhibit both tested CYP isoforms with IC50 values in the range of 1.3–~10 μg/mL.
Hence, the present study’s findings indicate concomitant and overconsumption of certain herbal plants (seen in Figs. 1a, 2a and 3a, 4a, 5a and Table 1) and their related products (e.g., BDS, beverages, functional foods, etc.) may pose a risk for HDIs. However, more detailed preclinical and clinical studies are required to identify the primary phytoconstituent(s) responsible for these plants, their pharmacokinetic behavior, and the clinical relevance of any suspected HDIs.
Supplementary Material
Funding
This research was supported in part by “Science-Based Authentication of Botanical Ingredients” funded by the Center for Food Safety and Applied Nutrition, US Food and Drug Administration grant number 5U01FD004246, and “Discovery & Development of Natural Products for Pharmaceutical & Agricultural Applications” funded by the United States Department of Agriculture, Agricultural Research Service, Specific Cooperative Agreement No. 58-6060-6-015.
Abbreviations:
- TCM
Traditional Chinese Medicine
- CAM
Complementary and Alternative Medicine
- BDS
Botanical dietary supplements
- CYP
Cytochrome P450
- OTC
Over-the-counter
- EC50
Effective concentration for half-maximal response
- US-FDA
U.S. Food and Drug Administration
- HDI
Herb-drug interaction
- LBD
Ligand binding domain
- DBD
DNA binding domain
- FAO
Food and Agriculture Organization
- P-gp
P-glycoprotein
- PBS
Phosphate buffered saline
- PXR
Pregnane Xenobiotic Receptor
- AhR
Aryl Hydrocarbon Receptor
- PAS
Per-Arnt-Sim
- DMBA
7,12-Dimethyl benzanthracene
- TCDD
2,3,7,8-Tetrachlorodibenzo-p-dioxin
- MDR1
MDCK Multidrug Resistance gene-1-Madin-Darby canine kidney
- ATCC
American Type Culture Collection
Footnotes
Declaration of competing interest
The authors declare that they have no conflict of interest.
CRediT authorship contribution statement
Islam Husain: Investigation, Formal analysis, Writing – original draft. Olivia R. Dale: Data curation, Visualization, Investigation. Katherine Martin: Data curation, Visualization, Investigation. Bill J. Gurley: Writing – review & editing. Sebastian J. Adams: Formal analysis. Bharathi Avula: Conceptualization, Methodology. Amar G. Chittiboyina: Conceptualization, Methodology, Funding acquisition. Ikhlas A. Khan: Conceptualization, Methodology, Supervision, Funding acquisition. Shabana I. Khan: Conceptualization, Methodology, Project administration, Supervision, Writing – review & editing, All authors read and approved the final version of the manuscript.
Appendix B. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jep.2022.115822.
Data availability
No data was used for the research described in the article.
References
- Ahmad S, Zahiruddin S, Parveen B, Basist P, Parveen A, Gaurav, Parveen R, Ahmad M, 2021. Indian medicinal plants and formulations and their potential against COVID-19 preclinical and clinical research. Front. Pharmacol. 2 11, 578970 10.3389/fphar.2020.578970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey DG, Dresser G, Arnold JM, 2013. Grapefruit-medication interactions: forbidden fruit or avoidable consequences? CMAJ (Can. Med. Assoc. J.) 185 (4), 309–316. 10.1503/cmaj.120951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borse SP, Singh DP, Nivsarkar M, 2019. Understanding the relevance of herb-drug interaction studies with special focus on interplays: a prerequisite for integrative medicine. Porto Biomed J 4 (2), e15. 10.1016/j.pbj.0000000000000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos MG, Machado J, Costa ML, Lino S, Correia F, Maltez F, 2018. Case report: severe hematological, muscle and liver toxicity caused by drugs and artichoke infusion interaction in an elderly polymedicated patient. Curr. Drug Saf. 13 (1), 44–50. 10.2174/1574886312666170912163746. [DOI] [PubMed] [Google Scholar]
- Cao Y, Shi Y, Cai Y, Hong Z, Chai Y, 2020. The effects of traditional Chinese medicine on p-glycoprotein-mediated multidrug resistance and approaches for studying the herb P-glycoprotein interactions. Drug Metab. Dispos. 48 (10), 972–979. 10.1124/dmd.120.000050. [DOI] [PubMed] [Google Scholar]
- Chang JT, Chang H, Chen PH, Lin SL, Lin P, 2007. Requirement of aryl hydrocarbon receptor overexpression for CYP1B1 up-regulation and cell growth in human lung adenocarcinomas. Clin. Cancer Res. 13 (1), 38–45. 10.1158/1078-0432.CCR-06-1166. [DOI] [PubMed] [Google Scholar]
- Chen XW, Sneed KB, Pan SY, Cao C, Kanwar JR, Chew H, Zhou SF, 2012. Herb-drug interactions and mechanistic and clinical considerations. Curr. Drug Metabol. 13 (5), 640–651. 10.2174/1389200211209050640. [DOI] [PubMed] [Google Scholar]
- Choi EJ, Kim T, 2008. Daidzein modulates induction of hepatic CYP1A1, 1B1, and AhR by 7,12-dimethylbenz[a]anthracene in mice. Arch Pharm. Res. (Seoul) 31 (9), 1115–1119. 10.1007/s12272-001-1277-3. [DOI] [PubMed] [Google Scholar]
- Dietary Supplement Label Database, 2017. Office of Dietary Supplements. National Institutes of Health, USA. [Google Scholar]
- Eyal S, Ke B, Muzi M, Link JM, Mankoff DA, Collier AC, Unadkat JD, 2010. Regional P-glycoprotein activity and inhibition at the human blood-brain barrier as imaged by positron emission tomography. Clin. Pharmacol. Ther. 87 (5), 579–585. 10.1038/clpt.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FDA, 2017. https://www.fda.gov/consumers/consumer-updates/black-licorice-trick-or-treat.
- FDA, 2021a. https://www.fda.gov/food/cfsan-constituent-updates/fda-sends-warning-letters-10-companies-illegally-selling-dietary-supplements-claiming-treat.
- FDA, 2021b. https://www.fda.gov/news-events/press-announcements/fda-warns-15-companies-illegally-selling-various-products-containing-cannabidiol-agency-details.
- Fasinu PS, Bouic PJ, Rosenkranz B, 2012. An overview of the evidence and mechanisms of herb-drug interactions. Front. Pharmacol. 30 3, 69. 10.3389/fphar.2012.00069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan M, Kanimozhi G, Pradhapsingh B, Khan HA, Alhomida AS, Ekhzaimy A, Brindha GR, Prasad NR, 2021. Phytochemicals reverse P-glycoprotein mediated multidrug resistance via signal transduction pathways. Biomed. Pharmacother. 139, 111632 10.1016/j.biopha.2021.111632. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Rivlin N, Shoshana OY, Ezra O, Madar S, Goldfinger N, Rotter V, 2013. Chemotherapeutic agents induce the expression and activity of their clearing enzyme CYP3A4 by activating p53. Carcinogenesis 34 (1), 190–198. 10.1093/carcin/bgs318. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Hodgson E, Liddle C, 1999. The orphan human pregnane X receptor mediates the transcriptional activation of CYP3A4 by rifampicin through a distal enhancer module. Mol. Pharmacol. 56 (6), 1329–1339. 10.1124/mol.56.6.1329. [DOI] [PubMed] [Google Scholar]
- Goodwin B, Redinbo MR, Kliewer SA, 2002. Regulation of cyp3a gene transcription by the pregnane x receptor. Annu. Rev. Pharmacol. Toxicol. 42, 1–23. 10.1146/annurev.pharmtox.42.111901.111051. [DOI] [PubMed] [Google Scholar]
- Gurley BJ, 2012. Pharmacokinetic herb-drug interactions (part 1): origins, mechanisms, and the impact of botanical dietary supplements. Planta Med. 78 (13), 1478–1489. 10.1055/s-0031-1298273. [DOI] [PubMed] [Google Scholar]
- Gurley BJ, 2020. Clinically-relevant Herb-Drug Interactions: Current Status and Practical Considerations. In: Spagnuolo Paul A. (Ed.), In Nutraceuticals and Human Health: the Food-To-Supplement Paradigm. The Royal Society of Chemistry, London, 2020. [Google Scholar]
- Gurley BJ, Swain A, Williams DK, Barone G, Battu SK, 2008. Gauging the clinical significance of P-glycoprotein-mediated herb-drug interactions: Comparative effects of St. John’s wort, Echinacea, clarithromycin, and rifampin on digoxin pharmacokinetics. Mol. Nutr. Food Res. 772–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley BJ, Fifer KE, Gardner Z, 2012. Pharmacokinetic Herb-Drug Interactions (Part 2): Drug Interactions Involving Popular Botanical Dietary Supplements and Their Clinical Relevance. In: Planta Med, vol. 78, pp. 1490–1514. 10.1055/s-0031-129833113. [DOI] [PubMed] [Google Scholar]
- Gurley BJ, Yates CR, Markowitz JS, 2018. Not intended to diagnose, treat, cure or prevent any disease.” 25 Years of botanical dietary supplement research and the lessons learned. Clin. Pharmacol. Ther. 104 (3), 470–483. 10.1002/cpt.1131. [DOI] [PubMed] [Google Scholar]
- Haron MH, Avula B, Ali Z, Chittiboyina AG, Khan IA, Li J, Wang V, Wu C, Khan SI, 2022a. Assessment of herb-drug interaction potential of five common species of licorice and their phytochemical constituents. J. Diet. Suppl. 18, 1–20. 10.1080/19390211.2022.2050875. [DOI] [PubMed] [Google Scholar]
- Haron MH, Dale O, Martin K, Avula B, Chittiboyina AG, Khan IA, Gurley BJ, Khan SI, 2022b. Evaluation of the herb-drug interaction potential of commonly used botanicals on the US market with regard to PXR- and AhR-mediated influences on CYP3A4 and CYP1A2. J. Diet. Suppl. 10.1080/19390211.2022.2110351. [DOI] [PubMed] [Google Scholar]
- Hedrich WD, Hassan HE, Wang H, 2016. Insights into CYP2B6-mediated drug-drug interactions. Acta Pharm. Sin. B. 6 (5), 413–425. 10.1016/j.apsb.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellum BH, Nilsen OG, 2008. In vitro inhibition of CYP3A4 metabolism and P-glycoprotein-mediated transport by trade herbal products. Basic Clin. Pharmacol. Toxicol. 102 (5), 466–475. 10.1111/j.1742-7843.2008.00227.x. [DOI] [PubMed] [Google Scholar]
- Hermann M, Asberg A, Reubsaet JL, Sather S, Berg KJ, Christensen H, 2002. Intake of grapefruit juice alters the metabolic pattern of cyclosporin A in renal transplant recipients. Int. J. Clin. Pharm. Ther. 40 (10), 451–456. 10.5414/cpp40451. [DOI] [PubMed] [Google Scholar]
- Hernandez JP, Mota LC, Baldwin WS, 2009. Activation of CAR and PXR by dietary, environmental and occupational chemicals alters drug metabolism, intermediary metabolism, and cell proliferation. Curr. Pharmacogenomics Personalized Med. (CPPM) 7 (2), 81–105. 10.2174/187569209788654005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain I, Bala K, Wani A, Makhdoomi U, Malik F, Sharma A, 2017. Arginase purified from endophytic Pseudomonas aeruginosa IH2: induce apoptosis through both cell cycle arrest and MMP loss in human leukemic HL-60 cells. Chem. Biol. Interact. 274, 35–49. 10.1016/j.cbi.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Husain I, Bala K, Khan IA, Khan SI, 2021a. A review on phytochemicals, pharmacological activities, drug interactions, and associated toxicities of licorice (Glycyrrhiza sp.). Food Frontiers. 10.1002/fft2.110. [DOI] [Google Scholar]
- Husain I, Dale OR, Manda V, Ali Z, Gurley BJ, Chittiboyina AG, Khan IA, Khan SI, 2021b. Bulbine natalensis (currently Bulbine latifolia) and select bulbine knipholones modulate the activity of AhR, CYP1A2, CYP2B6, and P-gp. Planta Med. 88, 975–984. 10.1055/a-1557-2113. [DOI] [PubMed] [Google Scholar]
- Husain I, Manda V, Alhusban M, Dale OR, Bae JY, Avula B, Gurley BJ, Chittiboyina AG, Khan IA, Khan SI, 2021c. Modulation of CYP3A4 and CYP2C9 activity by Bulbine natalensis and its constituents: an assessment of HDI risk of B. natalensis containing supplements. Phytomedicine 81, 153416. 10.1016/j.phymed.2020.153416. [DOI] [PubMed] [Google Scholar]
- Ihunnah CA, Jiang M, Xie W, 2011. Nuclear receptor PXR, transcriptional circuits and metabolic relevance. Biochim. Biophys. Acta 1812 (8), 956–963. 10.1016/j.bbadis.2011.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeuken A, Keser BJ, Khan E, Brouwer A, Koeman J, Denison MS, 2003. Activation of the AhR receptor by extracts of dietary herbal supplements, vegetables, and fruits. J. Agric. Food Chem. 51 (18), 5478–5487. 10.1021/jf030252u. [DOI] [PubMed] [Google Scholar]
- Jin UH, Park H, Li X, Davidson LA, Allred C, Patil B, Jayaprakasha G, Orr AA, Mao L, Chapkin RS, Jayaraman A, Tamamis P, Safe S, 2018. Structure-dependent modulation of aryl hydrocarbon receptor-mediated activities by flavonoids. Toxicol. Sci. 164 (1), 205–217. 10.1093/toxsci/kfy075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamenickova A, Anzenbacherova E, Pavek P, Soshilov AA, Denison MS, Anzenbacher P, Dvorak Z, 2013. Pelargonidin activates the AhR and induces CYP1A1 in primary human hepatocytes and human cancer cell lines HepG2 and LS174T. Toxicol. Lett. 218 (3), 253–259. 10.1016/j.toxlet.2013.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Shin S, Yoo SD, Shin BS, 2018. Effects of phytochemical P-glycoprotein modulators on the pharmacokinetics and tissue distribution of doxorubicin in mice. Molecules 23 (2), 349. 10.3390/molecules23020349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA, McKee DD, Oliver BB, Willson TM, Zetterstrom RH, Perlmann T, Lehmann JM, 1998. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 92 (1), 73–82. 10.1016/S0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kotwal P, Dogra A, Sharma A, Bhatt S, Gour A, Sharma S, Wazir P, Singh PP, Kumar A, Nandi U, 2020. Effect of natural phenolics on pharmacokinetic modulation of bedaquiline in rat to assess the likelihood of potential food-drug interaction. J. Agric. Food Chem. 68 (5), 1257–1265. 10.1021/acs.jafc.9b06529. [DOI] [PubMed] [Google Scholar]
- Larigot L, Juricek L, Dairou J, Coumoul X, 2018. AhR signaling pathways and regulatory functions. Biochim Open 7, 1–9. 10.1016/j.biopen.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen PZ, Yue QX, Li JQ, Chu RA, Zhang W, Wang H, 2013. Pungent ginger components modulate human cytochrome P450 enzymes in vitro. Acta Pharmacol. Sin. 34 (9), 1237–1242. 10.1038/aps.2013.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannel M, 2004. Drug interactions with St John’s Wort: mechanisms and clinical implications. Drug Saf. 27 (11), 773–797. 10.2165/00002018-200427110-00003. [DOI] [PubMed] [Google Scholar]
- Markowitz JS, Donovan JL, DeVane CL, Taylor RM, Ruan Y, Wang JS, Chavin KD, 2003. Effect of St John’s wort on drug metabolism by induction of cytochrome P450 3A4 enzyme. JAMA 290 (11), 1500–1504. 10.1001/jama.290.11.1500. [DOI] [PubMed] [Google Scholar]
- McDonnell AM, Dang CH, 2013. Basic review of the cytochrome p450 system. J. Adv. Pract. Oncol. 4 (4), 263–268. 10.6004/jadpro.2013.4.4.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra S, Stierman B, Gahche JJ, Potischman N, 2021. Dietary supplement use among adults: United States, 2017–2018. NCHS Data Brief (399), 1–8. 10.15620/cdc:101131. [DOI] [PubMed] [Google Scholar]
- Moore LB, Goodwin B, Jones SA, Wisely GB, Serabjit-Singh CJ, Willson TM, Collins JL, Kliewer SA, 2000. St. John’s wort induces hepatic drug metabolism through activation of the pregnane X receptor. Proc. Natl. Acad. Sci. USA 97 (13), 7500–7502. 10.1073/pnas.130155097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltzer K, Pengpid S, 2019. The use of herbal medicines among chronic disease patients in Thailand: a cross-sectional survey. J. Multidiscip. Healthc. 22 (12), 573–582. 10.2147/JMDH.S212953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizner AE, Cooke JP, 2019. Dietary supplements: facts and fallacies. Methodist Debakey Cardiovasc J 15 (3), 169–170. 10.14797/mdcj-15-3-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revol B, Gautier-Veyret E, Arrive C, Fouilhe Sam-Lai N, McLeer-Florin A, Pluchart H, Pinsolle J, Toffart AC, 2020. Pharmacokinetic herb-drug interaction between ginger and crizotinib. Br. J. Clin. Pharmacol. 86 (9), 1892–1893. 10.1111/bcp.13862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rezaee MM, Kazemi S, Kazemi MT, Gharooee S, Yazdani E, Gharooee H, Shiran MR, Moghadamnia AA, 2014. The effect of piperine on midazolam plasma concentration in healthy volunteers, a research on the CYP3A-involving metabolism. Daru 7 (1), 8. 10.1186/2008-2231-22-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin D, Patel V, Dietrich E, 2019. Effects of oral ginger supplementation on the INR. Case Rep. Med 11, 8784029. 10.1155/2019/8784029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippmann U, Leaman DJ, Cunningham AB, 2002. Impact of Cultivation and Gathering of Medicinal Plants on Biodiversity: Global Trends and Issues. FAO, Rome Italy. [Google Scholar]
- Schwarz D, Kisselev P, Ericksen SS, Szklarz GD, Chernogolov A, Honeck H, Schunck WH, Roots I, 2004. Arachidonic and eicosapentaenoic acid metabolism by human CYP1A1: highly stereoselective formation of 17(R),18(S)-epoxyeicosatetraenoic acid. Biochem. Pharmacol. 67 (8), 1445–1457. 10.1016/j.bcp.2003.12.023. [DOI] [PubMed] [Google Scholar]
- Sharma A, Husain I, 2017. Evaluation of antitumor activity of glutaminase-free periplasmic asparaginase from indigenous bacterial isolates as candidates for cancer therapy. Proc. Natl. Acad. Sci. India B Biol. Sci. 87, 997–1004. 10.1007/s40011-015-0681-z. [DOI] [Google Scholar]
- Smith T, Majid F, Eckl V, Reynolds CM, 2021. Herbal supplement sales in US increase by record-breaking 17.3% in 2020. HerbalGram 131, 52–65. [Google Scholar]
- Staudinger JL, Ding X, Lichti K, 2006. Pregnane X receptor and natural products: beyond drug-drug interactions. Expet Opin. Drug Metabol. Toxicol. 2 (6), 847–857. 10.1517/17425255.2.6.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subehan T Usia, Iwata H, Kadota S, Tezuka Y, 2006. Mechanism-based inhibition of CYP3A4 and CYP2D6 by Indonesian medicinal plants. J. Ethnopharmacol. 105 (3), 449–455. 10.1016/j.jep.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Wang B, Zhou SF, 2009. Synthetic and natural compounds that interact with human cytochrome P450 1A2 and implications in drug development. Curr. Med. Chem. 16 (31), 4066–4218. 10.2174/092986709789378198. [DOI] [PubMed] [Google Scholar]
- Weiss J, 2018. Herb-drug interaction potential of anti-borreliae effective extracts from Uncaria tomentosa (Samento) and Otoba parvifolia (Banderol) assessed in vitro. Molecules 24 (1), 137. 10.3390/molecules24010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger UM, Schwab M, 2013. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol. Ther. 138 (1), 103–141. 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- Zhu K, Meng Q, Zhang Z, Yi T, He Y, Zheng J, Lei W, 2019. Aryl hydrocarbon receptor pathway: role, regulation and intervention in atherosclerosis therapy (Review). Mol. Med. Rep. 20 (6), 4763–4773. 10.3892/mmr.2019.10748. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No data was used for the research described in the article.


