Abstract
While the ecological role that Trichodesmium sp. play in nitrogen fixation has been widely studied, little information is available on potential specialized metabolites that are associated with blooms and standing stock Trichodesmium colonies. While a collection of biological material from a T. thiebautii bloom event from North Padre Island, Texas, in 2014 indicated that this species was a prolific producer of chlorinated specialized metabolites, additional spatial and temporal resolution was needed. We have completed these metabolite comparison studies, detailed in the current report, utilizing LC-MS/MS-based molecular networking to visualize and annotate the specialized metabolite composition of these Trichodesmium blooms and colonies in the Gulf of Mexico (GoM) and other waters. Our results showed that T. thiebautii blooms and colonies found in the GoM have a remarkably consistent specialized metabolome. Additionally, we isolated and characterized one new macrocyclic compound from T. thiebautii, trichothilone A (1), which was also detected in three independent cultures of T. erythraeum. Genome mining identified genes predicted to synthesize certain functional groups in the T. thiebautii metabolites. These results provoke intriguing questions of how these specialized metabolites affect Trichodesmium ecophysiology, symbioses with marine invertebrates, and niche development in the global oligotrophic ocean.
Keywords: cyanobacterial blooms, mass spectrometry, molecular networking, specialized metabolites, Trichodesmium
Short abstract
A multiyear study of environmental samples and cultivated samples of Trichodesmium species showed that T. thiebautii produces dozens of specialized metabolites and these molecules are widespread in environmental samples.
Introduction
As climate change results in both increased global temperatures and CO2 concentrations in ocean waters, species of the cyanobacterial genus Trichodesmium are projected to expand their already substantial, oligotrophic ocean range as the subtropics move poleward.1−3 An increase in oceanic and coastal growth of the periodically bloom-forming Trichodesmium will raise both the direct and indirect exposure risk to humans from potential toxins produced by this genus. While the ecological role of Trichodesmium as nitrogen fixers is well studied, the specialized metabolism of the genus along with the impact of potential toxins on human health and animal health is unknown. The latter is of increasing importance because of the aforementioned range and abundance expansions predicted for Trichodesmium.
Many aspects of Trichodesmium’s toxicity have been somewhat enigmatic and inconsistent. Multiple classes of metabolites and toxins have been identified from Trichodesmium blooms and environmental collections such as saxitoxin, microcystin, trichamide, and palytoxin.4−8 Previous Trichodesmium toxicity studies performed using homogenized cells, filtrates, aging cells, and crude extracts of Trichodesmium thiebautii have shown toxicity to copepod grazers, while those from Trichodesmium erythraeum did not show the same levels of toxicity.9,10 However, extracts of T. erythraeum samples obtained from coastal India showed toxicity to shrimp and multiple human cell lines.11 This previous work has not included metabolite analysis of repeated field collections, biosynthetic gene cluster information, and culture confirmations, which has made interpreting toxicity studies challenging.
To address this gap, we completed an integrative, systematic study of the metabolites produced by Trichodesmium species in the environment and from isolates in culture. Our previous work characterized T. thiebautii metabolites that represented new analogs from existing cytotoxic compound classes (e.g., smenamides C, D, and E)12 and new classes of cytotoxic metabolites (e.g., the trichophycins, tricholides, and trichothiazoles), which had not been described previously.13−23
In the current report, we detailed the results of continued field studies in 2017, 2019, and 2021 in the GoM to provide additional temporal and spatial resolution of specialized metabolite production. The culmination of these studies has shown that these metabolites can be detected in T. thiebautii colonies year after year and throughout the GoM. We have used genetic tools to determine the Trichodesmium species present in these collections and untargeted MS/MS-based molecular networking to provide a chemical inventory of colony and bloom metabolites. It appears clear from our data that T. thiebautii in the oligotrophic GoM is a prolific producer of specialized metabolites, many of which possess nanomolar cytotoxicity against human cells (certain smenamides and smenothiazoles).17,18 We report a new metabolite in this work, trichothilone A (1), which was confirmed in multiple Trichodesmium laboratory cultures. Additionally, we identified key genetic architecture in the T. thiebautii H94 genome that is consistent with the generation of key functional groups in many of the metabolites we have characterized, i.e., the chlorovinylidene group and terminal vinyl chloride present in the trichophycins and smenolactones, the trichotoxins, the smenamides, the conulothiazoles, and trichothiazole A.12−16,19−23 This study provokes intriguing questions with respect to the full chemical potential of the members of this genus.
Materials and Methods
Collection of Cyanobacteria, Genetic Analysis, and Extraction Procedures
Trichodesmium biomass was collected from the Gulf of Mexico near North Padre Island, Texas, in 2014. As described previously, the dominant organism was identified as Trichodesmium sp., and the material was shipped to our laboratory.13,24 The biomass (14 g dry weight) was repeatedly extracted with a 2:1 mixture of CH2Cl2:CH3OH, and the crude extract was concentrated to an oil under reduced pressure. Collections were also made from the GoM in 2017, 2019, and 2021. Select samples were preserved in RNAlater and transferred to the laboratory. DNA isolation, PCR amplification of 16S rRNA genes, and phylogenetic analysis followed the procedures of McManus and co-workers exactly.23 16S rRNA sequences have been deposited in NCBI GenBank (GoM2017 accession no. OR661266; GoM2019-11 accession no. OR665426). Additionally, samples were acquired from the TriCoLim 2018 expedition25 and from culture collections (Table S1). The biomass from each of these sites was extracted in 2:1 CH2Cl2:CH3OH, followed by a 1:1 CH2Cl2:CH3OH, and 100% CH3OH. The resulting crude extracts were separately concentrated to oils under reduced pressure. Small aliquots of the resulting crude extracts were passed over a 100 mg C18 SPE column to prepare for LC-MS/MS analysis. Trichothilone A (1) was isolated from 2014 and 2019 extracts, and information on the isolation and physical characteristics of the molecule can be found in the Supporting Information.
LC-MS/MS-Based Molecular Networking
The resulting extracts described above were analyzed on a liquid chromatography system, a Dionex UltiMate 3000 HPLC, coupled to a high-resolution electrospray mass spectrometer, a Thermo LTQ Orbitrap XL system with postacquisition analysis performed at the GNPS Web site.26 All raw mass spectrometry files and .mzXML files can be found in the Center for Computational Mass Spectrometry MassIVE repository under number MSV000093069. For LC-MS/MS methodological details, please refer to the Supporting Information.
Cytotoxicity Assays
Assays were carried out as previously described using neuro-2A cells.13 Compound 1 was dissolved in DMSO (1% v/v) and added to the cells in the range of 100 to 0.1 μM in order to generate EC50 curves against both cell lines. Four technical replicates were prepared for each concentration, and each assay was performed in triplicate. Doxorubicin was used as a positive control, and DMSO (1% v/v) was used as a negative control.
Genomic Analysis
The T. erythraeum IMS101 assembled genome sequence was retrieved from the European Nucleotide Archive (accession #CP000393). The T. thiebautii H94 genome and a group of field sample metagenome-assembled genome (MAG) assemblies were retrieved from NCBI’s SRA (BioProject PRJNA828267).25
Results
Morphological Analysis and DNA Sequencing of Organism Collections
Our previous work from an initial collection of T. thiebautii biomass near North Padre Island, Texas, in 2014 provided the first evidence that there was a Trichodesmium species capable of prolific specialized metabolite production in the GoM. Mining this biomass resulted in the characterization of 24 new molecules,12−16,19−23 including 1 presented in the current report. Additionally, seven chlorinated metabolites were detected from the 2014 collection via analysis of LC-MS/MS molecular networks. These metabolites were originally isolated and characterized from the sponge Smenospongia aurea but putatively considered as the products of associated cyanobacteria due to the chlorovinylidene functional group present in these metabolites.17,18,20−22
The primary purpose of the current work was to determine if metabolite composition of these T. thiebautii colonies in the GoM was consistent over time and geographic location. To accomplish this, a second collection of Trichodesmium colonies was made in the GoM (27.00° N; 92.00° W) in 2017 (GoM2017, Table S1). Additionally in 2019, aboard the R/V Oregon II (National Oceanic and Atmospheric Administration), we visited several stations in the GoM and collected Trichodesmium colonies and biomass (GoM 2019-2-4, 6, 11, Table S1 and Figure S1). Collections from 2017 and 2019 were identified as predominantly Trichodesmium sp. by examining filaments and puff and tuft colonies (Figure S2). Phylogenetic analysis of partial sequences of the 16S rRNA gene supported identification of samples as T. thiebautii (GoM2017, GoM2019-11), clustering with the T. thiebautii identified in the 2014 collection (Figure 2B). During the 2019 research cruise, we collected bulk biomass from a surface accumulation (GoM2019-6) and were also able to harvest individual tuft and puff colonies from certain stations, sequestering individual colonies with a sterile loop and preserving them for laboratory analysis (e.g., GoM2019-11) (Figure S3). More field samples were collected from the GoM in 2021 (GoM2021-2-4, Table S1), and three cultivated samples of T. erythraeum IMS101 were examined—two separate cultures obtained from the Bigelow Laboratory for Ocean Sciences (IMS101) and another from the University of Southern California Trichodesmium Culture Collection (USCTCC) (T. erythraeum ST8) (Table S1). Finally, we examined legacy samples from a 2018 TriCoLim research expedition (AT39-05) in the Caribbean and Atlantic Ocean (TriCoLim St 3, 4, 6, 8, 13, 15, 16, 17, 20) (Table S1), which expanded the geographic coverage of metabolite composition of Trichodesmium colonies outside of the GoM. Collections and cultures used in this study are summarized in Table S1.
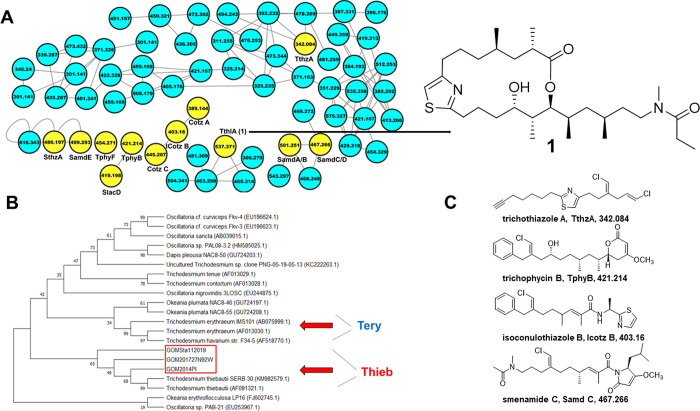
Figure 2.
(A) T. thiebautii specialized metabolites in “picked colonies”. Partial LC-MS/MS-based molecular network of extract from GoM2019-11. Colonies were hand-picked with a sterile loop from bulk collection, and an extract was generated. An arrow points from node m/z 537 (ThlA) to the trichothilone A (1) structure. Yellow nodes in panel (A) indicate that these metabolites were previously characterized by our group (cf. Figure S4). The full network can be found at http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=9b6cefbe17c44ecf897caec9aba38031 (picked colonies–total number of nodes 241). (B) 16S rRNA phylogenetic tree aligning Trichodesmium species from this study (red box—PI2014, GoM 2017, and GoM2019-11). All samples clustered with T. thiebautii. The tree was created using the Maximum Likelihood method and the Tamura-Nei model. The bootstrap consensus tree is inferred from 1000 replicates, and the percentage of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to branches. Analysis was conducted in MEGA. GenBank accession numbers of sequences are noted in parentheses. Red arrows show clusters for T. erythraeum (Tery) and T. thiebautii (Thieb). (C) Representative structures that have been characterized from Trichodesmium collections previously and were found in the “picked colonies” molecular network with their names, abbreviations, and m/z values below the structures.
LC-MS/MS-Based Molecular Networking: Spatial and Temporal Resolution of Trichodesmium Metabolites
We utilized LC-MS/MS paired with molecular networking to compare the metabolite composition of Trichodesmium collections over time and geographic area in the GoM. The previously characterized metabolites from the 2014 collection served as a “screening library” for specialized metabolites (Figure S4). Collections from 2017 and 2019 showed remarkable consistency with respect to metabolite content when compared to the previous 2014 analysis. Several of the previously characterized metabolites from the 2014 collection were identified in samples from 2017 (21 metabolites annotated) and 2019 (18 metabolites annotated). These include many of the trichophycins, smenamides, tricholides, trichothiazoles, conulothiazoles, smenothiazole A, and trichothilone A (1) (Figure 1A).
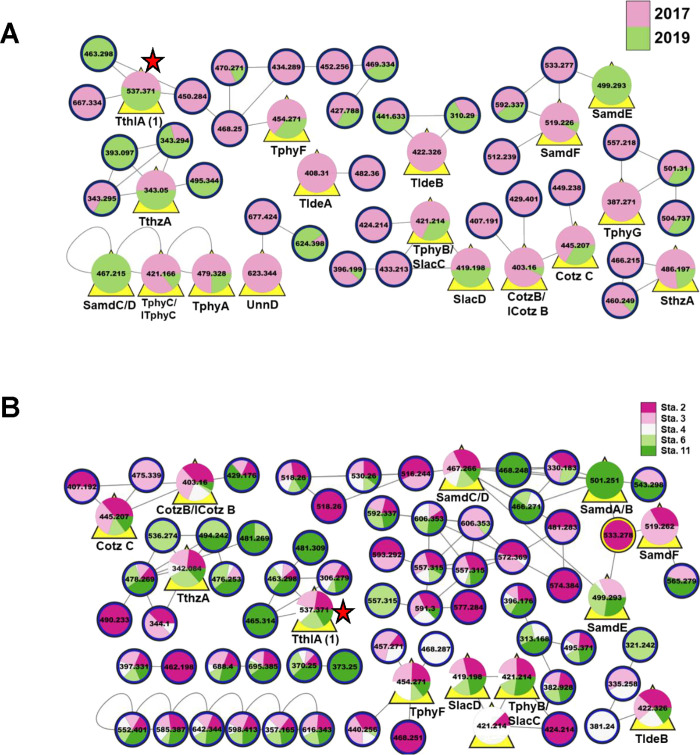
Figure 1.
T. thiebautii specialized metabolites were found year after year and across the GoM. (A) Partial LC-MS/MS-based molecular network of extracts from collection GoM2017 (pink) and GoM2019-6 (green). (B) T. thiebautii specialized metabolites found across the GoM in 2019. Partial LC-MS/MS-based molecular network of extracts from 2019 collections (stations 2–4, 6, and 11). In all panels, nodes are designated by their precursor masses. In panels (A) and (B), yellow triangles behind the node indicate that these metabolites were previously characterized by our group (cf. Figure S4). In panel (B), “pie slices” designate stations where the metabolite was detected. Red stars show the position of the node for trichothilone A (1) (m/z 537). Cotz, conulothiazole; ICotz, isoconulothiazole; ITphy, isotrichophycin; Sam, smenamide; Slac, smenolactone; Sthz, smenothiazole; Tlde, tricholide; Tphy, trichophycin; Tthl, trichothilone; Tthz, trichothiazole; Unn, unnarmicin. The full networks can be found at http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task = 27cc1b6493804e0283a08d5d056d015d (temporal–total number of nodes in full network 1465), http://gnps.ucsd.edu/ProteoSAFe/status.jsp?task=4b5821e25366435fae44da433db8819e (spatial—total number of nodes in full network 538).
Examining by geographic location using the 2019 samples, we again annotated many of the previously characterized metabolites from 2014 including trichophycins B and F, smenolactones C and D, smenamides A-F, isoconulothiazole B and conulothiazole C, trichothiazole A, tricholide B, and 1. There were also over 25 metabolites that remain unidentified but were detected from all five stations (Figure 1B).
We next analyzed the sample GoM2019-11, composed of “picked” colonies (dominated by puff morphology). These colonies were collected via net tow, washed in a sieve, and transferred one by one to a preservation vial using a sterile loop. This sample does have associated bacteria in it, but it is devoid of other phytoplankton and loosely associated ectocommensals, which was verified by microscopy. Many of the previously characterized metabolites were identified in this sample as well (2A,C). These results point to the existence of a core metabolome possessed by resident T. thiebautii colonies in the GoM, which has never been documented previously. This species produces hundreds of small organic metabolites, many of which are hallmarked by the incorporation of at least one halogen atom, typically chlorine (Figure S4). Following examination of all networks, one unidentified metabolite was prioritized for isolation and structure elucidation—m/z 537. This molecule was found in samples from 2014, 2017, all 2019 samples, and in the “picked colonies” sample. Additionally, ion counts obtained from LC-MS/MS indicated that this molecule was abundant in all samples. In contrast to most T. thiebautii metabolites, mass spectrometry analysis indicated that 1 did not contain any halogen atoms. Following the metabolite composition analysis, HPLC-DAD- and LC-MS-guided isolations were performed.
Structure Characterization of 1
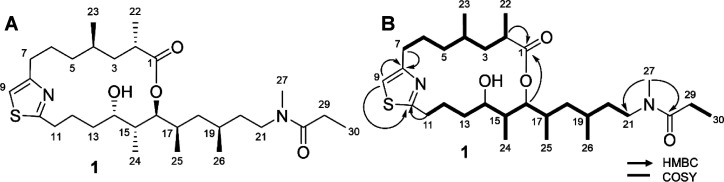
HPLC-DAD and mass spectrometry-guided isolation resulted in the isolation of an optically active pale yellow oil (1) (Figure 3A). HRESIMS analysis of 1 gave a protonated molecule [M + H]+ of m/z 537.3725 suggesting a molecular formula of C30H52N2O4S and a requirement of six degrees of unsaturation (Figure S5). Examination of 1D and 2D NMR data (Figures S6–S11) were used to establish the planar structure of the molecule. Certain 1H NMR resonances, most notably the N-methyl singlet, were split into two signals in a ratio of about 1:1. This suggested two conformers at the amide bond, which are detailed in Figure S12. The text discusses only the Z-conformer. This phenomenon has been described for other metabolites from Trichodesmium that contain an N-methyl amide functionality.12 Inspection of the COSY spectrum and 1H–1H spin systems allowed for the assignment of three partial structures: the first from H-2 to H2-7 including the 1,3-dimethyl system from C-1 to C-5, and a second from H2-11 to H2-21 including a 1,3,5-trimethyl system from C-14 to C-20 (Figure 3B). A third partial structure consisted of a single COSY correlation between H3-30 and H2-29. HMBC correlations between H2-7 and C-8 (δC 156.0) and H2-11 and C-10 (δC 169.1) along with the H-9 resonance (δH 7.01) established a thiazole functionality that connected the first two partial structures. An N-methyl signal (δH 2.88) showed HMBC correlations to C-21 (δC 44.5) and C-28 (δC 172.1) and connected partial structure two to an N-methylpropanamide functionality. HMBC correlations of H-2 (δH 2.44) and H-16 (δH 4.81) with C-1 (δC 175.5) established the ester of a macrolactone and satisfied the final degree of unsaturation characterizing a hybrid polyketide–peptide metabolite that was named trichothilone A (1) (Table 1).
Figure 3.
Structure of trichothilone A (1) (left) and key 2D NMR correlations (right).
Table 1. NMR Data for Trichothilone A (1)a in DMSO-d6 (500 MHz for 1H NMR, 125 MHz for 13C NMR).
| position | δC, type | δH (J in Hz) | HMBC | COSY |
|---|---|---|---|---|
| 1 | 175.5, C | |||
| 2 | 35.5, CH | 2.44, ovlpb | 1, 3, 22 | 22, 3a, 3b |
| 3a | 40.0, CH2 | 1.32, m | 2, 4, 22, 23 | 2, 4 |
| 3b | 1.16, m | 2, 4, 22, 23 | 2, 4 | |
| 4 | 29.1, CH | 1.42, ovlp | 3, 5, 6, 23 | 3b, 5, 23 |
| 5 | 34.9, CH2 | 1.06, ovlp | 6, 23 | 4, 6a, 6b |
| 6a | 25.0, CH2 | 1.70, m | 5, 7a, 7b | |
| 6b | 1.59, m | 5, 7a | ||
| 7a | 29.9, CH2 | 2.73, m | 6, 8 | 6b |
| 7b | 2.49, m | 6a | ||
| 8 | 156.0, C | |||
| 9 | 112.8, CH | 7.01, s | 8, 10 | |
| 10 | 169.1, C | |||
| 11a | 31.2, CH2 | 2.96, ddd (14.9, 6.7, 3.8) | 10, 13 | 12 |
| 11b | 2.77, m | 10, 13 | 12 | |
| 12 | 26.0, CH2 | 1.75, m | 11a, 11b, 13a, 13b | |
| 13a | 32.5, CH2 | 1.40, ovlp | 12, 14 | |
| 13b | 1.13, ovlp | 12, 14 | ||
| 14 | 68.9, CH | 3.18, m | 13a, 13b, 15, OH | |
| OH | 4.19, br | 14 | ||
| 15 | 38.8, CH | 1.51, p (7.0) | 16, 24 | 16, 24 |
| 16 | 76.7, CH | 4.81, ddd (9.5, 7.2, 2.5) | 1, 25 | 15, 17 |
| 17 | 30.5, CH | 1.80, m | ||
| 16, 18, 25 | ||||
| 18 | 41.0, CH2 | 1.01, m | 17, 19, 25, 26 | 17, 19 |
| 19 | 26.9, CH | 1.42, ovlp | 18, 26 | |
| 20a | 34.2, CH2 | 1.38, ovlp | 19, 26 | 19, 21 |
| 20b | 1.19, ovlp | 19, 21, 26 | 19, 21 | |
| 21 | 44.5, CH2 | 3.25, t (7.4) | 19, 20, 28 | 20a, 20b |
| 22 | 16.4, CH3 | 1.01, d (6.8) | 1, 2, 3 | 2 |
| 23 | 19.9, CH3 | 0.81, d (6.8) | 3, 4, 5 | 4 |
| 24 | 9.4, CH3 | 0.71, ovlp | 14, 15 | 15 |
| 25 | 12.3, CH3 | 0.73, ovlp | 16, 17, 18 | 17 |
| 26 | 19.3, CH3 | 0.80, d (6.8) | 18, 19, 20 | 19 |
| 27 | 34.3, CH3 | 2.88, s | 21, 28 | |
| 28 | 172.1, C | |||
| 29 | 25.8, CH2 | 2.24, m | 28, 30 | 30 |
| 30 | 9.2, CH3 | 0.94, t (7.4) | 28, 29 | 29 |
Z-conformer chemical shift values.
Overlapping signals.
Relative Configuration of 1
Relative configuration of the four stereogenic centers, C-14 to C-17, was determined as 14S*,15R*,16S*,17R* using the Murata’s approach,27 which is based on 1H–1H and 1H–13C scalar couplings and NOE effects, as detailed in Figure S13. The 1H–13C scalar couplings were directly measured using a HECADE-HSQC experiment (Figure S13)28 or estimated as “large” or “small” from the ratio of the relative magnitudes of their cross peak measured in the HMBC spectrum with respect to a common proton, an intense peak indicating a large 2,3JCH, a weak or missing peak a small 2,3JCH.29 Further support to the configuration of the segments C-14 to C-17 of 1 was provided by Kishi’s method for the relative configuration of contiguous propionate units.30,31 Comparison of 13C NMR signals in 1 recorded in three different solvents (CDCl3, CD3OD, and DMSO-d6) to Kishi’s 13C NMR database determined the relative configuration as α,α,β,β for this segment (cf. Table S2 and Figure S14), fully confirming the stereochemical assignment based on Murata’s method.
Determination of the relative configuration at C-2 and C-4 in 1 with respect to the C-14/C-17 segment was achieved using quantum-mechanical computational chemistry, namely, DFT prediction of 1H and 13C NMR chemical shifts (DFT-NMR).32 Considering the structural complexity of compound 1, the truncated model compound 1m was used for the calculations, in which the flexible side chain of the natural compound is replaced by an isopropyl group. This strongly reduced the number of low-energy conformations of the molecule, while it did not significantly affect the chemical shifts of the region of the molecule under study.33 The four possible stereoisomers of 1m at C-2 and C-4 (RR-1m, RS-1m, SR-1m, SS-1m) were considered in the calculations (Figure S15). Conformational analysis of the four 4m stereoisomers was performed using the program Pcmodel 10.0 (GMMX algorithm, MMFF94 force field),34 which generated 35 through 66 conformers for each stereoisomer within 4 kcal/mol from the lowest-energy conformer (see SI Experimental Procedures for details).
All the conformers were optimized by DFT minimization using the Gaussian program at the B3LYP/6-31G(d,p)/SMD level,35 and their 1H and 13C chemical shifts were calculated at the mPW1PW91/6-311+G(d,p)/PCM level. Average chemical shifts over all the conformers were then calculated for each of RR-1m, RS-1m, SR-1m, and SS-1m based on the Boltzmann distribution and compared with the experimental values measured for 1. Comparison of the root-mean-square deviations (RMSD) of 13C and 1H chemical shifts immediately showed that SR-1m fit experimental data much better than the other three stereoisomers (RR-1m: 2.32 and 0.150 ppm; RS-1m: 2.24 and 0.145 ppm; SR-1m: 1.98 and 0.122 ppm; SS-1m: 2.46 and 0.149 ppm, respectively). The DP4+ statistical analysis of computational results fully confirmed this,36 in that it assigned 100.00% probability to SR-1m being the correct stereoisomer (Figure S16). The anti orientation of the methyl groups at CH3-22 and CH3-23 of 1 determined above was further supported by the Δ(Ha–Hb) value (0.16 ppm) of the methylene protons attached to C-3, which is consistent with the trend shown for 1,3-anti dimethyl groups in the macrocycle myxovirescin (anti = 0.20 ppm; syn = 0.70 ppm).37 Therefore, the relative configuration of 1 was extended to 2S*,4R*,14S*,15R*,16S*,17R* and only configuration at C-19 remained to be determined.
An attempt to determine the relative configuration at C-19 using DFT-NMR was unsuccessful because the predicted chemical shifts of the two possible epimers showed similar agreement with experimental data, with DP4+ probability <80% and therefore not conclusive (data not shown). Moreover, the Δ(Ha–Hb) value of the methylene protons attached to C-18, which was close to 0, could not be used for determination of relative configuration of the 1,3-dimethyl system CH3-25/CH3-26 because the method is unreliable when the methyl branches are adjacent to a lactone, which is the case for this system in 1.37 To overcome this limitation, we generated the acyclic methyl ester of 1, characterized this derivative (2) using HRMS and 1HNMR (Figures S17 and S18), and reanalyzed this compound to determine the chemical shifts of the intervening methylene unit in the 1,3-dimethyl system. The Δ(Ha–Hb) value of the methylene protons attached to C-18 of 2 (Δ(Ha – Hb) = 0.43) strongly supported a syn relative configuration for CH3-25 and CH3-26 (Figure S14).37 In addition, the Δ(Ha–Hb) measured for 2 was almost identical to the value (0.40 ppm) measured for the methylene protons attached to C-5 of the 4-bromophenacyl derivative of hemibourgeanic acid (3),38 thus ruling out the possibility that the stereogenic centers C-16 and C-15 could affect the reliability of the method (Figure S19). These data defined the full relative configuration of 1 as 2S*,4R*,14S*,15R*,16S*,17R*,19S*.
Absolute Configuration Analysis of 1
A single secondary alcohol in 1 (attached to C-14) allowed us to generate Mosher esters at that position. We analyzed the S and R esters of 1 via 1H NMR, COSY, and TOCSY. A positive Δ(δHS–δHR) value for H-14 and negative Δ(δHS–δHR) values for H-9 and H-11 supported a 14S configuration (Figure S20). Relaying stereochemical assignments based on the previously determined relative configuration supported a 2S,4R,14S,15R,16S,17R,19S configuration of 1.
Specialized Metabolites in Cultures and from Collections in the Wider Caribbean and Atlantic Ocean
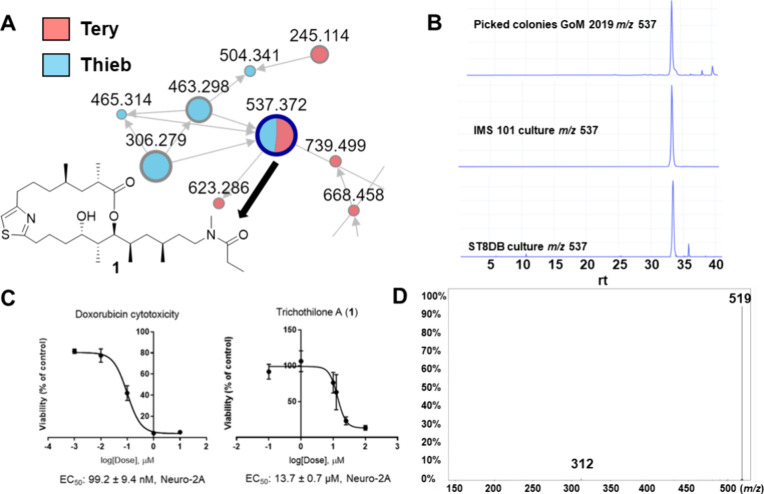
Trichothilone A (1) was detected in both the cultivated T. erythraeum IMS101 samples and the T. erythraeum ST8 sample with matches to retention time, high-resolution mass spectrometry measurement, and MS/MS fragmentation pattern (Figure 4A,B,D). An examination of the cell mass and media of a second IMS101 culture also showed the presence of trichothilone A (1) (Figure S21), and its presence in the media may be related to its ecological relevance. Examining collections from the GoM collected aboard NOAA’s R/V Oregon II in 2021 (revisiting stations from 2019) and samples from the TriCoLim expedition,25 we continued to identify the presence of metabolites from our 2014 library by matching MS/MS fragmentation patterns. The GoM samples showed the presence of certain trichophycins, smenamides, and other molecules (Table S3). We detected trichothiazole A in extracts from TriCoLim stations 3, 8, 16, and 17 (Figure S22) and trichophycin F in 3, 6, and 17 (Figure S23). We also detected smenamide A/B from station 3 extracts (Figure S24).
Figure 4.
(A) Molecular network of extracts from GoM2019-11 and IMS101 and ST8 showing the trichothilone A (1) node present in both species. (B) Extracted ion chromatogram (XIC) shows detection of 1 in T. erythraeum IMS101 and ST8 extracts. (C) Cytotoxicity of 1 compared to the positive control doxorubicin against neuro-2A murine neuroblastoma cells. (D) MS/MS spectrum of trichothilone A (1).
Cytotoxicity of 1
Trichothilone A (1) showed modest cytotoxicity against neuro-2A cells with an IC50 value of 13.7 μM (Figure 4C). Due to the limited amount remaining following chemical degradation and derivative formation experiments, we were not able to investigate other bioactivities that the molecule might possess. The structural similarity of 1 to the thuggacins A and B, macrolactone antibiotics which showed biological activity against Mycobacterium tuberculosis,39 provides optimism that additional testing of trichothilone A (1) could uncover potent therapeutically relevant biological activity.
Genomic Analysis of T. thiebautii H94, T. erythraeum IMS101, and Field Metagenomes
We examined several genome and MAGs that were publicly available,25 searching for genetic architecture consistent with the unique functional groups (chlorovinylidene group and terminal vinyl chloride) present in T. thiebautii metabolites (although these functional groups were not in 1) and genetic architecture consistent with putative PKS-NRPS-derived metabolite, e.g., tricholides A and B and 1. We also examined genomes and MAGs from T. thiebautii and T. erythraeum for putative full biosynthetic gene clusters (BGCs) that might include 1. Specifically, this included the genomes of T. thiebautii H94, T. erythraeum IMS101, and the metagenome assemblies obtained from samples collected during the R/V Atlantis TriCoLim cruise,25 having some colocation with the archived samples from the cruise that we analyzed chemically during this work. Analysis of PKS clusters present in the T. thiebautii H94 genome using the antiSMASH platform40 and the known cluster BLAST function identified a module in the T. thiebautii H94 genome, which showed similarity to modules in the curacin A pathway.41 Curacin A possesses a terminal alkene functionality and the biochemistry surrounding its construction, and the activity of the decarboxylating thioesterase has been experimentally shown.42 Additional mining and annotation using DELTA-BLAST43 showed the presence of the following domains in the T. thiebautii cluster of interest: ketosynthase (KS), acyltransferase (AT), ketoreductase (KR), acyl carrier protein (ACP), sulfotransferase (ST), and a thioesterase domain (TE). These domains are consistent with those that form the terminal alkene in curacin A.41 Furthermore, adjacent to this module was a gene that BLAST searching annotated as an Fe(II)-dependent halogenase. This partial cluster was similar to the genes predicted to form the terminal vinyl chloride in many of the trichophycins and trichothiazole (Figure S25A). A second partial gene cluster showed remarkable consistency with the genetic architecture necessary to form the chlorovinylidene group. An HCS cassette was present in this cluster consisting of an HMG-CoA synthase and two enoyl CoA hydratases and ketosynthase, acyltransferase, and three ACP domains with additional enoyl reductase and ketosynthase domains (Figure S25B). The HCS cassette has been observed in gene clusters that produce products with the chlorovinylidene moiety, such as the jamaicamide biosynthetic pathway.44 While a specific halogenase was not identified in this partial cluster, the elements are present for the vinylidene group observed in nearly all the metabolites isolated from T. thiebautii collections (Figure S4). The HMG-CoA synthase was also identified in MAGs Bin1_Station19 and Bin1_Station18, which phylogenomics analysis supported as T. thiebautii.25 We predict that a PKS-NRPS system that generates 1 and one of the NRPS modules would have a cyclization domain to create the thiazole. Additionally, we predict that this gene cluster would have several methyltransferases including an N-methyl transferase. However, no cluster was consistent with these predictions in any genome examined and we could not identify the putative gene cluster that creates trichothilone A (1).
Discussion
Trichodesmium Species as Toxin and Specialized Metabolite Producers
With the occurrence of cyanobacterial blooms increasing concurrent with climatological changes, there is a need to determine the ecological role of specialized metabolites and how they affect speciation, ecotoxicology, and range expansion. The rarity of Trichodesmium cultures stems from an inability for most laboratories to obtain hand samples and to scale up cultures of Trichodesmium for the biomass amounts that can lead to the isolation of metabolites. This has necessitated that many toxin and specialized metabolite isolation efforts come from environmental collections. While many classes of metabolites have been detected from these blooms, it is difficult to attribute production of these metabolites, including 1, to Trichodesmium without analysis of axenic cultures. However, the widespread presence of 1 in environmental collections and its detection in three independent unialgal cultures of Trichodesmium supports its cyanobacterial production, and its presence in the media may indicate an important ecological role. The T. thiebautii-dominated blooms and collected colonies showed intriguing profiles with respect to their metabolite composition. Continuing longitudinal and geographic studies will aid in understanding the metabolite profiles of this group and potentially lead to understanding the ecological role of these metabolites. The widespread presence of trichothiazole, trichothilone A (1), and other metabolites presumes that they likely have an important physiological or ecological role. Additionally, the widespread presence of smenamides A/B and smenothiazole A warrant a reinvestigation of Trichodesmium toxicity, as these metabolites showed cytotoxic effects to human cells at nanomolar concentrations.17,18 While the trichophycins, trichothiazoles, tricholides, and other metabolites we have isolated from T. thiebautii have shown toxicity to human cells generally at low micromolar levels,13−19 it is important to consider the additive or synergistic effect of toxin mixtures. We have established a demonstrated analytical capability to show that certain specialized metabolites are found in colonies in the GoM and the wider Caribbean and Atlantic Ocean, but Trichodesmium is found around the globe with notable repeated blooms in the Pacific and Indian Oceans.45 It would be most interesting to continue analysis in collections from these areas using our established methods to provide further foundational studies on Trichodesmium-specialized metabolism. The ability to accumulate more material from yearly blooms will enhance the potential for 1 and other metabolites to be comprehensively evaluated for their potential therapeutic benefit and ecological function. Furthermore, there remain hundreds of uncharacterized metabolites as shown in the various molecular networks we have generated. We are only scratching the surface of the molecular potential of Trichodesmium.
Connecting Biosynthetic Pathways to Trichodesmium-Specialized Metabolites
Field samples are more accurately defined in a clade format dominated by one of the two major groups of Trichodesmium (T. erythraeum or T. thiebautii), and recent work has shown that there are four clades of N2-fixing Trichodesmium (T. erythraeum A and B, T. thiebautii A and B).25 Until there is an unequivocal biosynthetic identification of complete gene clusters in the T. thiebautii genomes, and the lack of these clusters in strains of T. erythraeum, no definitive answer can be given on species chemotaxonomy and metabologenomics. There is also previously published evidence that indicates environment populations are composed of mixed species.46 Defining species assemblages in collections will be necessary to understand community composition and how this may affect specialized metabolite production. Furthermore, while the cultivated sample of T. erythraeum only showed the presence of one of the specialized metabolites we have characterized (1), the culture conditions may not be conducive to metabolite production, or we do not understand the environmental triggers necessary for metabolite expression. While dozens of molecules have been characterized from Trichodesmium by our group, they have similar carbon scaffolds and most are analogues of each other. We speculate that the biosynthesis of these products in Trichodesmium follows that deciphered for the vatiamides in which a unique combinatorial nonlinear hybrid PKS-NRPS system generated a series of analogues adding to molecular diversity.47
Holistic molecular approaches must be taken to define species assemblages, genomic analysis to define biosynthetic architecture, and culture and field experiments to understand potential abiotic and biotic triggers that control metabolite production. Additionally, as the bulk of these samples were from field collections, there are many variables such as the impact of the bacterial and microbial community and the sample volumes collected vary widely from a few colonies to bulk collections. Instituting water volume measures, colony counts, and more molecular community data will aid in better detection and more comparable data across samples. Untargeted LC-MS/MS networking has shown utility in the discovery of new cyanobacterial toxins and metabolites previously,48 and it was well tailored for the present study. Recent work has shown that along the West Florida Shelf, there is a coastal vs open ocean separation for T. erythraeum and T. thiebautii, respectively.49 It would be most interesting to investigate if specialized metabolism plays a role in this niche differentiation. The discovery of a nondiazotrophic Trichodesmium species50 provokes additional questions with respect to comparative metabolomics studies and the potential role of specialized metabolites in nitrogen fixation. Understanding how potential abiotic stressors regulate specialized metabolite and protein production will be key in determining the ecological role of the molecules we have discovered.51Trichodesmium’s iron uptake is well studied, and the impact of specialized metabolites on iron bioavailability in the phycosphere52 should be an area of further study to fully understand the ecophysiological role of these specialized metabolites.
Acknowledgments
We profusely thank Dr. Simon J. Geist at Texas A&M, Corpus Christi, for assistance with collections. We gratefully acknowledge the crew of the Oregon II and NOAA with special thanks to Andrew Millett and Glenn Zapfe. The Table of Contents Graphic and Figure S1 were created in part with Biorender.com (agreement #XL26T0KI8K and # WU26T0KYIN, respectively).
Glossary
Abbreviations
- BGC
biosynthetic gene cluster
- DAD
diode array detector
- DFT
density functional theory
- ESI
electrospray ionization
- GNPS
Global Natural Product Social Molecular Networking
- GoM
Gulf of Mexico
- HRMS
high-resolution mass spectrometry
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- MAG
metagenome-assembled genome
- MTPA-Cl
α-methoxy-α-(trifluoromethyl)phenylacetyl chloride
- NMR
nuclear magnetic resonance
- PKS-NRPS
polyketide synthase-nonribosomal peptide synthetase
- USCTCC
University of Southern California Trichodesmium Culture Collection
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.est.3c10739.
Author Contributions
◆ C.W.V. and L.G. contributed equally.
Author Contributions
M.J.B. and A.M. conceived the study and performed analysis. C.W.V., L.G., K.M.M., R.D.K., A.M.K., and S.S. performed data acquisition and analysis. E.A.W., N.A.H., M.A.S., P.V.Z., and P.D.R.M. provided key samples and data interpretation. The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript. C.W.V. and L.G. contributed equally.
Acquisition of certain data in this publication was made possible by the use of equipment and services available through grant number P20GM103430. We also gratefully acknowledge support from NSF-2125191 to M.A.S. and E.A.W. Research reported in this article was supported in part by the National Institute of Environmental Health Sciences of the National Institutes of Health under Award Number R21ES033758 (M.J.B.) and the American Society of Pharmacognosy Starter Grant (M.J.B).
The authors declare no competing financial interest.
Supplementary Material
References
- Hutchins D. A.; Fu F.-X.; Webb E. A.; Walworth N.; Tagliabue A. Taxon-Specific Response of Marine Nitrogen Fixers to Elevated Carbon Dioxide Concentrations. Nature Geosci 2013, 6 (9), 790–795. 10.1038/ngeo1858. [DOI] [Google Scholar]
- Hutchins D. A.; Walworth N. G.; Webb E. A.; Saito M. A.; Moran D.; McIlvin M. R.; Gale J.; Fu F.-X. Irreversibly Increased Nitrogen Fixation in Trichodesmium Experimentally Adapted to Elevated Carbon Dioxide. Nat. Commun. 2015, 6 (1), 8155. 10.1038/ncomms9155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boatman T. G.; Lawson T.; Geider R. J. A Key Marine Diazotroph in a Changing Ocean: The Interacting Effects of Temperature, CO2 and Light on the Growth of Trichodesmium erythraeum IMS101. PLoS One 2017, 12 (1), e0168796 10.1371/journal.pone.0168796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacilotto Detoni A. M.; Fonseca Costa L. D.; Pacheco L. A.; Yunes J. S. Toxic Trichodesmium Bloom Occurrence in the Southwestern South Atlantic Ocean. Toxicon 2016, 110, 51–55. 10.1016/j.toxicon.2015.12.003. [DOI] [PubMed] [Google Scholar]
- Shunmugam S.; Gayathri M.; Prasannabalaji N.; Thajuddin N.; Muralitharan G. Unraveling the Presence of Multi-Class Toxins from Trichodesmium Bloom in the Gulf of Mannar Region of the Bay of Bengal. Toxicon 2017, 135, 43–50. 10.1016/j.toxicon.2017.06.003. [DOI] [PubMed] [Google Scholar]
- Ramos A.; Martel A.; Codd G.; Soler E.; Coca J.; Redondo A.; Morrison L.; Metcalf J.; Ojeda A.; Suárez S.; Petit M. Bloom of the Marine Diazotrophic Cyanobacterium Trichodesmium erythraeum in the Northwest African Upwelling. Mar. Ecol.: Prog. Ser. 2005, 301, 303–305. 10.3354/meps301303. [DOI] [Google Scholar]
- Kerbrat A. S.; Amzil Z.; Pawlowiez R.; Golubic S.; Sibat M.; Darius H. T.; Chinain M.; Laurent D. First Evidence of palytoxin and 42-Hydroxy-palytoxin in the Marine Cyanobacterium Trichodesmium. Marine Drugs 2011, 9 (4), 543–560. 10.3390/md9040543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudek S.; Haygood M. G.; Youssef D. T. A.; Schmidt E. W. Structure of trichamide, a Cyclic Peptide from the Bloom-Forming Cyanobacterium Trichodesmium erythraeum, Predicted from the Genome Sequence. Appl. Environ. Microbiol. 2006, 72 (6), 4382–4387. 10.1128/AEM.00380-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawser S. P.; O’Neil J. M.; Roman M. R.; Codd G. A. Toxicity of Blooms of the Cyanobacterium Trichodesmium to Zooplankton. J. Appl. Phycol 1992, 4 (1), 79–86. 10.1007/BF00003963. [DOI] [Google Scholar]
- Guo C.; Tester P. A. Toxic Effect of the Bloom-Forming Trichodesmium sp. (Cyanophyta) to the copepod Acartia tonsa. Nat. Toxins 1994, 2 (4), 222–227. 10.1002/nt.2620020411. [DOI] [PubMed] [Google Scholar]
- Narayana S.; Chitra J.; Tapase S. R.; Thamke V.; Karthick P.; Ramesh Ch.; Murthy K. N.; Ramasamy M.; Kodam K. M.; Mohanraju R. Toxicity Studies of Trichodesmium erythraeum (Ehrenberg, 1830) Bloom Extracts, from Phoenix Bay, Port Blair, Andamans. Harmful Algae 2014, 40, 34–39. 10.1016/j.hal.2014.10.003. [DOI] [Google Scholar]
- Via C. W.; Glukhov E.; Costa S.; Zimba P. V.; Moeller P. D. R.; Gerwick W. H.; Bertin M. J. The Metabolome of a Cyanobacterial Bloom Visualized by MS/MS-Based Molecular Networking Reveals New Neurotoxic Smenamide Analogs (C, D, and E). Front. Chem. 2018, 316. 10.3389/fchem.2018.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin M. J.; Wahome P. G.; Zimba P. V.; He H.; Moeller P. D. R. Trichophycin A, a Cytotoxic Linear Polyketide Isolated from a Trichodesmium thiebautii Bloom. Mar. Drugs 2017, 15, 10. 10.3390/md15010010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin M. J.; Saurí J.; Liu Y.; Via C. W.; Roduit A. F.; Williamson R. T. Trichophycins B-F, Chlorovinylidene-containing Polyketides Isolated from a Cyanobacterial Bloom. J. Org. Chem. 2018, 83, 13256–13266. 10.1021/acs.joc.8b02070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin M. J.; Roduit A. F.; Sun J.; Alves G. E.; Via C. W.; Gonzalez M. A.; Zimba P. V.; Moeller P. D. R. tricholides A and B and Unnarmicin D: New Hybrid PKS-NRPS Macrocycles Isolated from an Environmental Collection of Trichodesmium thiebautii. Mar. Drugs 2017, 15, 206. 10.3390/md15070206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belisle R. S.; Via C. W.; Schock T. B.; Villareal T. A.; Zimba P. V.; Beauchesne K. R.; Moeller P. D. R.; Bertin M. J. Trichothiazole A, a Dichlorinated Polyketide Containing an Embedded thiazole Isolated from Trichodesmium Blooms. Tetrahedron Lett. 2017, 58, 4066–4068. 10.1016/j.tetlet.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta R.; Irollo E.; Della Sala G.; Pirozzi G.; Mangoni A.; Costantino V. smenamides A and B, Chlorinated Peptide/Polyketide Hybrids Containing a Dolapyrrolidinone Unit from the Caribbean Sponge Smenospongia aurea. Evaluation of Their Role as Leads in Antitumor Drug Research. Marine Drugs 2013, 11 (11), 4451–4463. 10.3390/md11114451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G.; Teta R.; Miceli R.; Ceccarelli L.; Della Sala G.; Camerlingo R.; Irollo E.; Mangoni A.; Pirozzi G.; Costantino V. Isolation and Assessment of the in Vitro Anti-Tumor Activity of smenothiazole A and B, Chlorinated thiazole-Containing Peptide/Polyketides from the Caribbean Sponge. Smenospongia aurea. Marine Drugs 2015, 13 (1), 444–459. 10.3390/md13010444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertin M. J.; Zimba P. V.; He H.; Moeller P. D. R. Structure Revision of Trichotoxin, a Chlorinated Polyketide Isolated from a Trichodesmium thiebautii Bloom. Tetrahedron Lett. 2016, 57, 5864–5867. 10.1016/j.tetlet.2016.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caso A.; Esposito G.; Della Sala G.; Pawlik J. R.; Teta R.; Mangoni A.; Costantino V. Fast Detection of Two Smenamide Family Members Using Molecular Networking. Marine Drugs 2019, 17 (11), 618. 10.3390/md17110618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito G.; Della Sala G.; Teta R.; Caso A.; Bourguet-Kondracki M.; Pawlik J. R.; Mangoni A.; Costantino V. Chlorinated thiazole-Containing Polyketide-Peptides from the Caribbean Sponge Smenospongia conulosa: Structure Elucidation on Microgram Scale. Eur. J. Org. Chem. 2016, 2016 (16), 2871–2875. 10.1002/ejoc.201600370. [DOI] [Google Scholar]
- Teta R.; Della Sala G.; Esposito G.; Via C. W.; Mazzoccoli C.; Piccoli C.; Bertin M. J.; Costantino V.; Mangoni A. A Joint Molecular Networking Study of a Smenospongia Sponge and a Cyanobacterial Bloom Revealed New Antiproliferative Chlorinated Polyketides. Org. Chem. Front. 2019, 6, 1762–1774. 10.1039/C9QO00074G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus K. M.; Kirk R. D.; Via C. W.; Lotti J. S.; Roduit A. F.; Teta R.; Scarpato S.; Mangoni A.; Bertin M. J. Isolation of Isotrichophycin C and Trichophycins G–I from a Collection of Trichodesmium thiebautii. J. Nat. Prod. 2020, 83 (9), 2664–2671. 10.1021/acs.jnatprod.0c00550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komárek J; Anagnostidis K. Cyanoprokarota Part 2: Oscillatoriales, Freshwater Flora of Central Europe; Spektrum Akademischer Verlag: Heidelberg, Germany, 2007. pp 1- 759. [Google Scholar]
- Webb E. A.; Held N. A.; Zhao Y.; Graham E. D.; Conover A. E.; Semones J.; Lee M. D.; Feng Y.; Fu F.; Saito M. A.; Hutchins D. A. Importance of Mobile Genetic Element Immunity in Numerically Abundant Trichodesmium Clades. ISME Commun. 2023, 3 (1), 15. 10.1038/s43705-023-00214-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M.; Carver J. J.; Phelan V. V.; Sanchez L. M.; Garg N.; Peng Y.; Nguyen D. D.; Watrous J.; Kapono C. A.; Luzzatto-Knaan T.; Porto C.; Bouslimani A.; Melnik A. V.; Meehan M. J.; Liu W.-T.; Crüsemann M.; Boudreau P. D.; Esquenazi E.; Sandoval-Calderón M.; Kersten R. D.; Pace L. A.; Quinn R. A.; Duncan K. R.; Hsu C.-C.; Floros D. J.; Gavilan R. G.; Kleigrewe K.; Northen T.; Dutton R. J.; Parrot D.; Carlson E. E.; Aigle B.; Michelsen C. F.; Jelsbak L.; Sohlenkamp C.; Pevzner P.; Edlund A.; McLean J.; Piel J.; Murphy B. T.; Gerwick L.; Liaw C.-C.; Yang Y.-L.; Humpf H.-U.; Maansson M.; Keyzers R. A.; Sims A. C.; Johnson A. R.; Sidebottom A. M.; Sedio B. E.; Klitgaard A.; Larson C. B.; Boya P. C. A.; Torres-Mendoza D.; Gonzalez D. J.; Silva D. B.; Marques L. M.; Demarque D. P.; Pociute E.; O’Neill E. C.; Briand E.; Helfrich E. J. N.; Granatosky E. A.; Glukhov E.; Ryffel F.; Houson H.; Mohimani H.; Kharbush J. J.; Zeng Y.; Vorholt J. A.; Kurita K. L.; Charusanti P.; McPhail K. L.; Nielsen K. F.; Vuong L.; Elfeki M.; Traxler M. F.; Engene N.; Koyama N.; Vining O. B.; Baric R.; Silva R. R.; Mascuch S. J.; Tomasi S.; Jenkins S.; Macherla V.; Hoffman T.; Agarwal V.; Williams P. G.; Dai J.; Neupane R.; Gurr J.; Rodríguez A. M. C.; Lamsa A.; Zhang C.; Dorrestein K.; Duggan B. M.; Almaliti J.; Allard P.-M.; Phapale P.; Nothias L.-F.; Alexandrov T.; Litaudon M.; Wolfender J.-L.; Kyle J. E.; Metz T. O.; Peryea T.; Nguyen D.-T.; VanLeer D.; Shinn P.; Jadhav A.; Müller R.; Waters K. M.; Shi W.; Liu X.; Zhang L.; Knight R.; Jensen P. R.; Palsson B. Ø.; Pogliano K.; Linington R. G.; Gutiérrez M.; Lopes N. P.; Gerwick W. H.; Moore B. S.; Dorrestein P. C.; Bandeira N. Sharing and Community Curation of Mass Spectrometry Data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34 (8), 828–837. 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumori N.; Kaneno D.; Murata M.; Nakamura H.; Tachibana K. Stereochemical Determination of Acyclic Structures Based on Carbon–Proton Spin-Coupling Constants. A Method of Configuration Analysis for Natural Products. J. Org. Chem. 1999, 64 (3), 866–876. 10.1021/jo981810k. [DOI] [PubMed] [Google Scholar]
- Koźmiński W.; Nanz D. J. Magn. Reson. 2000, 142 (2), 294–299. 10.1006/jmre.1999.1939. [DOI] [PubMed] [Google Scholar]
- Ciminiello P.; Dell’Aversano C.; Dello Iacovo E.; Fattorusso E.; Forino M.; Grauso L.; Tartaglione L. Eur. J. Chem. 2012, 18 (52), 16836–16843. 10.1002/chem.201201357. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y.; Lee J.; Tezuka K.; Kishi Y. Toward Creation of a Universal NMR Database for the Stereochemical Assignment of Acyclic Compounds: The Case of Two Contiguous Propionate Units. Org. Lett. 1999, 1 (13), 2177–2180. 10.1021/ol9903786. [DOI] [PubMed] [Google Scholar]
- Lee J.; Kobayashi Y.; Tezuka K.; Kishi Y. Toward Creation of a Universal NMR Database for the Stereochemical Assignment of Acyclic Compounds: Proof of Concept. Org. Lett. 1999, 1 (13), 2181–2184. 10.1021/ol990379y. [DOI] [PubMed] [Google Scholar]
- Grauso L.; Li Y.; Scarpato S.; Cacciola N. A.; De Cicco P.; Zidorn C.; Mangoni A. A Cytotoxic Heterodimeric Cyclic Diarylheptanoid with a Rearranged Benzene Ring from the Seagrass Zostera Marina. J. Nat. Prod. 2022, 85 (10), 2468–2473. 10.1021/acs.jnatprod.2c00796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpato S.; Teta R.; De Cicco P.; Borrelli F.; Pawlik J. R.; Costantino V.; Mangoni A. Molecular Networking Revealed Unique UV-Absorbing Phospholipids: Favilipids from the Marine Sponge Clathria faviformis. Mar. Drugs 2023, 21 (2), 58. 10.3390/md21020058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert K. E.Pcmodel (version 10.0); Serena Software: Bloomington, IN, 2013.
- Gaussian 16, Revision C.01; Gaussian Inc.: Wallingford CT, USA.
- Grimblat N.; Zanardi M. M.; Sarotti A. M. Beyond DP4: An Improved Probability for the Stereochemical Assignment of Isomeric Compounds Using Quantum Chemical Calculations of NMR Shifts. J. Org. Chem. 2015, 80 (24), 12526–12534. 10.1021/acs.joc.5b02396. [DOI] [PubMed] [Google Scholar]
- Schmidt Y.; Lehr K.; Colas L.; Breit B. Assignment of relative Configuration of Desoxypropionates by 1H NMR Spectroscopy: Method Development, Proof of Principle by Asymmetric Total Synthesis of Xylarinic Acid A and Applications. Chem.—Eur. J. 2012, 18, 7071–7081. 10.1002/chem.201103988. [DOI] [PubMed] [Google Scholar]
- Bodo B.; Trowitzsch-Kienast W.; Schomburg D. Absolute Configuration of Bourgeanic Acid: X-Ray Crystal Structure of a 4-Bromophenacyl Derivative of Hemibourgeanic Acid. Tetrahedron Lett. 1986, 27 (7), 847–848. 10.1016/S0040-4039(00)84117-9. [DOI] [Google Scholar]
- Steinmetz H.; Irschik H.; Kunze B.; Reichenbach H.; Höfle G.; Jansen R. thuggacins, Macrolide Antibiotics Active against Mycobacterium Tuberculosis: Isolation from Myxobacteria, Structure Elucidation, Conformation Analysis and Biosynthesis. Chem.—Eur. J. 2007, 13 (20), 5822–5832. 10.1002/chem.200700269. [DOI] [PubMed] [Google Scholar]
- Blin K.; Shaw S.; Kloosterman A. M.; Charlop-Powers Z.; van Wezel G. P.; Medema M. H.; Weber T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49 (W1), W29–W35. 10.1093/nar/gkab335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z.; Sitachitta N.; Rossi J. V.; Roberts M. A.; Flatt P. M.; Jia J.; Sherman D. H.; Gerwick W. H. Biosynthetic Pathway and Gene Cluster Analysis of Curacin A, an Antitubulin Natural Product from the Tropical Marine Cyanobacterium Lyngbya majuscula. J. Nat. Prod. 2004, 67 (8), 1356–1367. 10.1021/np0499261. [DOI] [PubMed] [Google Scholar]
- Gehret J. J.; Gu L.; Gerwick W. H.; Wipf P.; Sherman D. H.; Smith J. L. Terminal alkene Formation by the thioesterase of Curacin A Biosynthesis. J. Biol. Chem. 2011, 286 (16), 14445–14454. 10.1074/jbc.M110.214635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boratyn G. M.; Schäffer A. A.; Agarwala R.; Altschul S. F.; Lipman D. J.; Madden T. L. Domain Enhanced Lookup Time Accelerated BLAST. Biol. Direct 2012, 7 (1), 12. 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards D. J.; Marquez B. L.; Nogle L. M.; McPhail K.; Goeger D. E.; Roberts M. A.; Gerwick W. H. Structure and Biosynthesis of the Jamaicamides, New Mixed Polyketide-Peptide Neurotoxins from the Marine Cyanobacterium Lyngbya majuscula. Chem. Biol. 2004, 11 (6), 817–833. 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- Bergman B.; Sandh G.; Lin S.; Larsson J.; Carpenter E. J. Trichodesmium – a widespread marine cyanobacterium with unusual nitrogen fixation properties. FEMS Microbiol. Rev. 2013, 37, 286–302. 10.1111/j.1574-6976.2012.00352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hynes A. M.; Webb E. A.; Doney S. C.; Waterbury J. B. Comparison of Cultured Trichodesmium (Cyanophyceae) with Species Characterized from the Field. Journal of Phycology 2012, 48 (1), 196–210. 10.1111/j.1529-8817.2011.01096.x. [DOI] [PubMed] [Google Scholar]
- Moss N. A.; Seiler G.; Leão T. F.; Castro-Falcón G.; Gerwick L.; Hughes C. C.; Gerwick W. H. Nature’s Combinatorial Biosynthesis Produces vatiamides A–F.. Angew. Chem., Int. Ed. 2019, 58 (27), 9027–9031. 10.1002/anie.201902571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teta R.; Della Sala G.; Glukhov E.; Gerwick L.; Gerwick W. H.; Mangoni A.; Costantino V. Combined LC–MS/MS and Molecular Networking Approach Reveals New Cyanotoxins from the 2014 Cyanobacterial Bloom in Green Lake, Seattle. Environ. Sci. Technol. 2015, 49 (24), 14301–14310. 10.1021/acs.est.5b04415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Confesor K. A.; Selden C. R.; Powell K. E.; Donahue L. A.; Mellett T.; Caprara S.; Knapp A. N.; Buck K. N.; Chappell P. D. Defining the Realized Niche of the Two Major Clades of Trichodesmium: A Study on the West Florida Shelf. Front. Mar. Sci. 2022, 9, 821655 10.3389/fmars.2022.821655. [DOI] [Google Scholar]
- Delmont T. O. Discovery of nondiazotrophic Trichodesmium species abundant and widespread in the open ocean. Proc. Natl. Acad. Sci. U. S. A. 2021, 118 (46), e2112355118 10.1073/pnas.2112355118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held N. A.; Webb E. A.; McIlvin M. M.; Hutchins D. A.; Cohen N. R.; Moran D. M.; Kunde K.; Lohan M. C.; Mahaffey C.; Woodward E. M. S.; Saito M. A. Co-Occurrence of Fe and P Stress in Natural Populations of the Marine Diazotroph Trichodesmium. Biogeosciences 2020, 17 (9), 2537–2551. 10.5194/bg-17-2537-2020. [DOI] [Google Scholar]
- Liu F.; Tan Q.-G.; Weiss D.; Crémazy A.; Fortin C.; Campbell P. G. C. Unravelling Metal Speciation in the Microenvironment Surrounding Phytoplankton Cells to Improve Predictions of Metal Bioavailability. Environ. Sci. Technol. 2020, 54 (13), 8177–8185. 10.1021/acs.est.9b07773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.