Abstract
The poly(A) tail of influenza virus mRNA is synthesized by reiterative copying of a U track near the 5′ end of the virion RNA (vRNA) template by the viral RNA polymerase. We have engineered a novel influenza A/WSN/33 virus which contains a neuraminidase (NA) vRNA with its U track mutated into an A track. Instead of synthesizing poly(A)-tailed NA mRNA, this novel virus synthesizes poly(U)-tailed NA mRNA. In infected cells, most poly(U)-tailed NA mRNA was retained in the nucleus, while most control polyadenylated NA mRNA was transported to the cytoplasm. These results suggest that the poly(A) tail is important for efficient nuclear export of NA mRNA. The mutant virus produced a reduced amount of NA and showed an attenuated phenotype, suggesting that poly(A) signal mutants of this type might be useful as potential live attenuated virus vaccines. In addition, this virus mutant might provide a useful model to further elucidate the basic mechanisms of mRNA nuclear export.
mRNAs are synthesized in the cell nucleus by RNA polymerase II as precursors which undergo a series of RNA-processing events during maturation. These processing events include (i) the addition of a 7-methylguanosine and 2′-O methylation to form a cap at the 5′ end, (ii) the removal of intron sequences by splicing, and (iii) the generation of a mature 3′ end by endonucleolytic cleavage and polyadenylation (25). Most of the fully processed eukaryotic mRNAs have poly(A) tails at their 3′ ends. Eukaryotic mRNA precursors are cleaved 10 to 30 bases downstream of the highly conserved polyadenylation signal sequence, AAUAAA (50), followed by poly(A) synthesis (3). However, some viruses, such as influenza virus, use an entirely different mechanism for adding a poly(A) tail to their mRNAs (46).
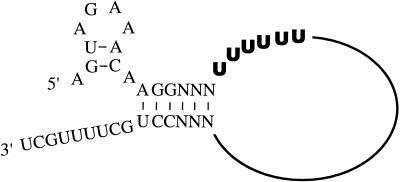
The genome of influenza A virus is composed of eight negative-strand RNA segments, which are packaged into virions as ribonucleoprotein (RNP) complexes (38). In infected cells, the virion RNA (vRNA) is transcribed into mRNA and replicated into cRNA in the cell nuclei (19, 22). cRNA is a full-length copy of vRNA and acts as a template for vRNA synthesis. Viral RNA replication is initiated by primer-independent transcription (18). By contrast, mRNA synthesis is initiated by a capped primer which is derived from host cellular mRNAs (44). For mRNA synthesis, the influenza virus RNA polymerase has to bind to the 5′ and 3′ ends of the vRNA both for cap utilization (58) and for transcription initiation (9). In mRNA transcription, it is proposed that the RNA polymerase remains bound to the 5′ end sequence throughout chain elongation (9, 45, 48, 58). At the end of transcription, the polymerase is unable to copy the site to which it is bound and pauses on a track of uridine residues near the 5′ end of vRNA (Fig. 1) (27, 53). Instead of transcribing the 5′ end of the vRNA, the RNA polymerase reiteratively copies the U track and polyadenylates the mRNA transcript by polymerase slippage (46). RNP isolated from viral particles can synthesize polyadenylated transcripts in vitro, demonstrating that polyadenylation of influenza virus mRNA is a host-independent process (43).
FIG. 1.
Model of vRNA with the wild-type conserved terminal sequences. The proposed RNA hook model of the vRNA template (49) is shown. The U6 track (the polyadenylation site) is in boldface.
Although the mechanism of RNA nuclear export is not well understood, it is clear that different classes of proteins are involved in exporting different classes of RNAs (21, 23, 32, 34). For mRNA nuclear export, several lines of evidence suggest that heterogeneous nuclear RNA-binding proteins (hnRNP) are involved in this process (5). Some of these proteins (e.g., hnRNP A1) were shown to bind to mRNA and shuttle between nucleus and cytoplasm (41). In addition, several mRNA maturation processes, such as the addition of the 5′ cap, the removal of introns by splicing, and the generation of the mature 3′ end, are known to influence mRNA export (6, 16, 20, 26), suggesting that fully processed mRNAs are targets for mRNA export. Interestingly, several viral proteins, such as the Rev protein of human immunodeficiency virus type 1 (29) and nonstructural protein 1 (NS1) of influenza A virus (1, 11, 30, 51, 52), facilitate viral mRNA export selectively.
Recently, by replacing the U track with an A track at the polyadenylation site of model vRNA templates, we demonstrated that the influenza virus polymerase was able to synthesize polyuridylated mRNA in vitro and in vivo (46). Here, we extended these studies by engineering a novel influenza virus which synthesizes a neuraminidase (NA) mRNA with a poly(U) tail. Interestingly, the majority of poly(U)-tailed NA mRNA was found to be retained in the nuclei of infected cells. As a result, the mutant virus produced reduced amounts of NA and showed an attenuated phenotype, suggesting a new method for generating live attenuated vaccines. These results also support the idea that the poly(A) tail is important for influenza virus mRNA nuclear export. Thus, this novel virus might be a useful model for further elucidating the basic mechanisms of mRNA nuclear export.
MATERIALS AND METHODS
Viruses.
Influenza A/WSN/HK virus, a reassortant virus containing an NA segment from influenza A/HK/8/68 virus and all other segments from influenza A/WSN/33 virus, was grown in embryonic chicken eggs. Influenza X-31 virus, a reassortant virus of influenza A/HK/8/68 virus and influenza A/PR/8/34 virus was supplied commercially.
Plasmid transfection and CAT assay.
Plasmids pGT-h-PB1, pGT-h-PB2, pGT-h-PA, and pGT-h-NP, which express the influenza PB1, PB2, PA, and NP proteins, respectively, and pPOLI-CAT-RT were generously provided by P. Palese (42, 46). One microgram of each plasmid was transfected into 293 cells with 25 μl of DOTAP transfection reagent (Boehringer Mannheim) as instructed by the manufacturer. Mutated versions of the pPOLI-CAT-RT plasmid were made by an inverse-PCR technique with Pfu DNA polymerase, and the mutated sequences were confirmed by DNA sequencing. At about 60 h posttransfection, cell extracts were harvested and tested for chloramphenicol acetyltransferase (CAT) activity as described previously (15). Fifty microliters of undiluted or diluted cell extracts was incubated in 75-μl reaction mixtures containing 0.08 μCi of [14C]chloramphenicol, 1 mM acetyl-coenzyme A, and 250 mM Tris-HCl (pH 7.5). After incubation at 37°C for 2 h, reaction products were extracted by ethyl acetate and then separated by thin-layer chromatography. Acetylated products were detected by autoradiography and analyzed in a phosphorimager.
Preparation of influenza A virus RNP and RNP transfection.
RNA polymerase and nucleoprotein were isolated from influenza A virus, strain X-31, as described previously (55). Briefly, the virus was disrupted with a mixture containing Triton X-100, NP-40, and lysolecithin. RNP was separated by glycerol step gradient centrifugation. Fractions enriched in RNP were treated with micrococcal nuclease and then used, after the addition of EGTA, in RNP transfection. Plasmid pT3NAM1 (12), which encodes the full-length wild-type NA vRNA segment of influenza A/WSN/33, was generously provided by P. Palese. Mutated versions of the plasmid were made by an inverse-PCR technique with Pfu DNA polymerase, and the mutated sequences were confirmed by DNA sequencing. RNP transfections were conducted as described previously (7). Briefly, BpuAI-linearized pT3NAM1 was transcribed by T3 RNA polymerase at 37°C for 30 min in the presence of micrococcal nuclease-treated RNP. The reconstituted RNP complexes were transfected with DEAE-dextran into Madin-Darby bovine kidney (MDBK) cells infected with influenza A/WSN/HK helper virus. After a 1-day incubation, medium from transfected cells was checked for the presence of transfectant viruses by plaque assay on MDBK cells.
Virus purification.
Transfectant viruses were plaque purified three times in MDBK cells covered with agar overlay medium. Purified viruses were then grown in MDBK cells and purified through a 30% sucrose cushion, followed by purification on a 30 to 60% sucrose gradient.
Sequencing of the 5′ and 3′ ends of the NA gene.
cDNA clones of the 5′ end of the NA gene segment from transfectant viruses were obtained by rapid amplification of 5′ DNA ends (5′ RACE). NA vRNA from the purified transfectant viruses was reverse transcribed by using primer 5′-GGGTGTCCTTCGACCAAAAC-3′ (complementary to nucleotides [nt] 879 to 898 of the NA vRNA) and SuperScript II reverse transcriptase (RT; Gibco BRL). The reverse-transcribed product was tailed with a poly(C) sequence at its 3′ end by terminal deoxynucleotidyl transferase. The poly(C)-tailed cDNAs were amplified by PCR with primer 5′-TGGACTAGTGGGAGCATCAT-3′ (complementary to nt 1280 to 1299 of the NA vRNA), and a 5′ RACE-abridged anchor primer as instructed by the manufacturer (Gibco BRL). PCR products were then cloned into pGEM-T vector (Promega) and sequenced. To synthesize cDNA of the 3′-end sequence of the NA gene segment, vRNAs from purified viruses were first polyadenylated by poly(A) polymerase (Gibco BRL). The polyadenylated vRNA was reverse transcribed by using primer 5′-GCGCTCTAGAATTCTTTTTTTTTTTTTTTTAGC-3′. cDNA was amplified by PCR with the primer for reverse transcription and primer 5′-GTTGAGTCCTGCCCAGCAACAACT-3′ corresponding to nt 1197 to 1220 of the NA vRNA. PCR products were then cloned into pGEM-T and sequenced.
NA activity assay.
The NA activity was determined as described before (10). Briefly, purified viruses (∼3 μg) were incubated in 100 μl of 150 mM phosphate buffer (pH 6.0) containing 1 mM CaCl2 and 50 nmol of 2′-(4-methylumbelliferyl)–α-d-N-acetylneuraminic acid (Sigma) for 10 min at 37°C. Reactions were then stopped by adding 2 ml of 0.5 M glycine-NaOH (pH 10.4), and the released 4-methylumbelliferone was assayed by using a spectrofluorometer (Shimadzu; RF-540). A 0.1 mM solution of 4-methylumbelliferone (Sigma) was used as a standard control.
Immunofluorescence microscopy.
MDBK cells grown on glass cover slides were washed with phosphate-buffered saline (PBS) and then infected with viruses at a multiplicity of infection (MOI) of about 0.5. Infected cells were harvested at 10 h postinfection, (p.i.), washed twice in PBS, and fixed in 4% paraformaldehyde in PBS. Fixed cells were permeabilized with 0.5% Triton X-100 in PBS for 15 min, followed by two washes in PBS. Cells were then incubated with a 100-fold-diluted mouse monoclonal anti-NA antibody (H17-L17-5; a generous gift from Walter Gerhard) overnight at 4°C. After two washes in PBS with 0.01% Triton X-100, cells were incubated with a horse anti-mouse antibody conjugated with fluorescein (Vector; 1:200 dilution) for 1 h at room temperature. Samples were washed twice in PBS containing 0.01% Triton X-100 and then mounted in antibleach medium containing 4′,6-diamidino-2-phenylindole (DAPI) for DNA staining (Vector). Samples were examined on a Zeiss Axiophot microscope.
RT-PCR assays for mRNA and vRNA from infected cells.
MDBK cells infected with viruses at an MOI of 2 were harvested at 12 h p.i. Total RNA was isolated with Trizol (Gibco BRL) as instructed by the manufacturer. For detecting the NA vRNA, about 1.5 μg of total RNA was first reverse transcribed by 25 U of Moloney murine leukemia virus (Mo-MuLV) RT (Promega) at 42°C for 30 min in the presence of NA vRNA-specific primer 5′-GGGTGTCCTTCGACCAAAAC-3′ (complementary to nt 879 to 898 of the NA vRNA). The reverse-transcribed products were then amplified by Taq polymerase with primers 5′-TGGACTAGTGGGAGCATCAT-3′ (complementary to nt 1280 to 1299 of the NA vRNA) and 5′ GGACTAGTAGTAGTAGAAACAAGG-3′ (underlined nucleotides correspond to the first 13 nt of the 5′ end of NA vRNA).
To detect the homopolymeric tails of NA mRNA, RT-PCR was performed as described before (48). About 1.5 μg of total RNA was first reverse transcribed by 25 U of Mo-MuLV RT (Promega) at 42°C for 30 min in the presence of either a 5′ GC-clamped T20 primer [5′-GCCCCGGGATCCT20-3′; specific for poly(A)-tailed mRNA] or a 5′ GC-clamped A20 primer [5′-GCCCCGGGATCCA20-3′; specific for poly(U)-tailed mRNA]. The reverse-transcribed products were then amplified by Taq polymerase with 5′-TGGACTAGTGGGAGCATCAT-3′ (complementary to nt 1280 to 1299 of the NA gene) and the corresponding primer specific for the homopolymeric tail. All PCR products were then cloned into pGEM-T and sequenced.
Primer extension assay.
Total RNA from cells infected with viruses at an MOI of 2 was isolated with Trizol (Gibco BRL) at the time points indicated in Fig. 5 and 6. Primer extension assays were performed as described before (10). About 0.2 μg of viral RNA or 5 μg of total RNA was reverse transcribed by 200 U of SuperScript II RT (Gibco BRL) for 45 min at 42°C in the presence of the corresponding 32P-labelled primer. Reactions were terminated by adding an equal volume of formamide loading dye. Products were then heated to 99°C for 2 min and analyzed on 5% polyacrylamide–7 M urea gels. To quantify the yield of the reverse-transcribed products, gels were dried and analyzed in a phosphorimager.
FIG. 5.
Primer extension assay for NA-specific vRNA. (A) NA-specific vRNA levels in cells infected with the A6 mutant (A6) and the wild-type virus (WT). Total RNAs from infected cells were isolated at the indicated time points p.i. The expected products for NA (259 nt) and NS (196 nt) vRNA are indicated. (B) NA-specific vRNA levels in purified A6 mutant and wild-type virus. The expected products for NA (259 nt) and NS (196 nt) vRNA are indicated. (C) NA vRNA-to-NS vRNA ratio in cells infected with the A6 mutant (A6) or wild-type virus (WT).
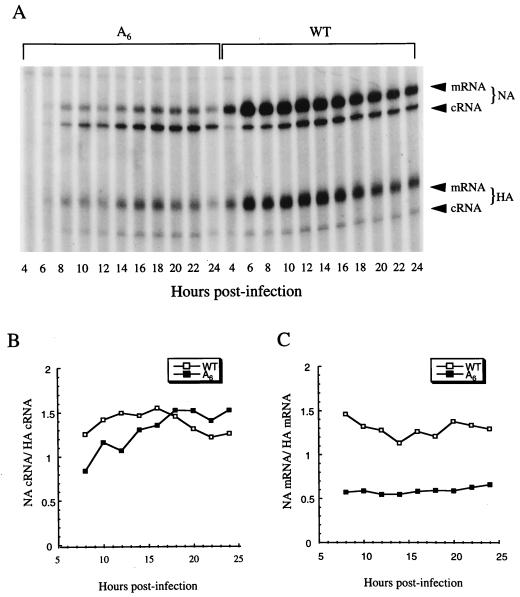
FIG. 6.
Primer extension assay for NA-specific cRNA and mRNA. (A) NA-specific cRNA and mRNA levels in cells infected with the A6 mutant (A6) and the wild-type virus (WT). Total RNA from infected cells was isolated at the indicated time points p.i. The expected products for NA cRNA (142 nt), NA mRNA (152 to 157 nt), HA cRNA (94 nt), and HA mRNA (104 to 109 nt) are indicated. Since influenza virus mRNAs are initiated by capped RNA primers, which are heterogeneous in size, primer extension products of the mRNA are expected to produce a broad band which is 10 to 15 nt longer than that produced by products of cRNA. (B) NA cRNA-to-HA cRNA ratios in cells infected with the A6 mutant or the wild-type virus. (C) NA mRNA-to-HA mRNA ratio in cells infected the A6 mutant or the wild-type virus.
To determine NA and nonstructural protein (NS) vRNA levels, an NA vRNA-specific primer (5′-GTGGCAATAACTAATCGGTCA-3′; complementary to nt 1151 to 1171 of NA vRNA) and a NS vRNA-specific primer (5′-GGGAACAATTAGGTCAGAAGT-3′; complementary to nt 695 to 715 of NS vRNA) were used in the primer extension assay. An NA cRNA- and mRNA-specific primer (5′-GTTGAGTCCTGCCCAGCAACAACT-3′; corresponding to nt 142 to 122 of the NA vRNA) and a hemagglutinin (HA) cRNA- and mRNA-specific primer (5′-CATATTGTGTCTGCATCTGTAGCT-3′; corresponding to nt 94 to 71 of the HA vRNA) were used to determine, respectively, levels of NA and HA mRNA and cRNA.
In situ hybridization.
MDBK cells grown on glass cover slides were washed with PBS and then infected with viruses at an MOI of 0.5. Infected cells were harvested at 8 h p.i., washed twice with PBS, and fixed in 4% paraformaldehyde in PBS. The fixed cells were then permeabilized by PBS containing 0.5% Triton X-100 for 15 min at room temperature. Cells were then washed twice in PBS, followed by two washes in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Samples were hybridized in 20 μl of hybridization buffer (50% formamide, 2× SSC, 10% dextran sulfate, 1 mg of Escherichia coli tRNA/ml) containing about 100 ng of riboprobe at 37°C overnight. After hybridization, cells were washed twice in hybridization buffer at 37°C, followed by successive washings for 15 min with 2×, 1×, 0.5×, and 0.2× SSC at room temperature. Samples were then incubated in 4× SSC containing 1 μg of anti-digoxigenin-fluorescein antibody (Boehringer Mannheim)/ml for 1 h at room temperature. After the incubation, samples were washed with 4× SSC containing 0.01% Triton X-100 three times and mounted in antibleach medium containing DAPI for DNA staining (Vector). Samples were examined by a Zeiss Axiophot microscope.
For NA cRNA and NA mRNA detection, a riboprobe (corresponding to nt 1 to 299 of the NA vRNA) was prepared by T3 RNA polymerase transcription of BamHI-linearized pT3NAM1. For poly(U) tail mRNA detection, a plasmid which contains a poly(A) sequence and part of 3′ end of the NA gene was constructed. The transcription product from this plasmid contained a sequence of 86 A residues [specific for the poly(U) tail] and the last 21 NA-specific residues of NA mRNA. Both riboprobes were labelled with digoxigenin-UTP (Boehringer Mannheim) during in vitro transcriptions.
RESULTS
Polymerase slippage is required for influenza virus gene expression.
The synthesis of a poly(A) tail occurs after the cleavage of the 3′ ends of pre-mRNAs by a common mechanism for all polyadenylated cellular mRNAs. Influenza virus, however, has evolved a different mechanism for polyadenylation of its mRNA. Recently, we demonstrated that a homopolymeric sequence (a U track) near the 5′ end of influenza virus vRNA is required for polymerase slippage and, consequently, for the synthesis of a homopolymeric tail (46). Replacing the U track (Fig. 1) with an A track in a model vRNA template resulted in synthesis of poly(U)-tailed mRNA both in vitro and in vivo. To test whether a poly(U)-tailed mRNA could be used for gene expression, we used a plasmid-based CAT reporter system. CAT vRNA expression plasmid pPOLI-CAT-RT (42) was transfected into human 293 kidney cells to synthesize a vRNA-like template containing a CAT gene in negative orientation, flanked by the 5′ and 3′ noncoding regions of segment 8 of influenza A/WSN/33 virus. Four protein expression plasmids which encode viral PB1, PB2, PA, and NP proteins were also cotransfected into the cell for the replication and transcription of the CAT vRNA template. To synthesize poly(U)-tailed mRNA, the polyadenylation site (U6 track) in the 5′ noncoding region of the CAT vRNA template was mutated into an A6 track. The presence of poly(U) sequences at the 3′ ends of mRNAs transcribed from this mutant template has been rigorously demonstrated elsewhere (46).
When cells were transfected with a wild-type CAT vRNA expression plasmid, strong CAT activity was detected (Fig. 2, lanes 1 to 4). However, when the construct with the U6 track mutated into an A6 track (U6→A6 construct; see above) was transfected into the cells, only low (3 ± 1.5% of that of the wild type) but significant CAT activity was detected (Fig. 2; compare lanes 5 and 6 with lanes 1 to 4). By contrast, cells transfected with a control plasmid which contained a U6→UCUACG mutation had no detectable CAT activity (Fig. 2, lanes 7 and 8). These results agreed with our previous findings that a homopolymeric track near the 5′ end of vRNA was required for the pausing of the polymerase, polymerase slippage, and synthesis of a homopolymeric tail (46). Since no gene expression was detected when the U6 track was mutated into a “random” sequence (Fig. 2, lanes 7 and 8), these results also demonstrated that a homopolymeric track (either U6 or A6) is essential for gene expression.
FIG. 2.
Effects of mutations of the U6 track on CAT expression. Human 293 kidney cells were transfected with four protein expression plasmids (pGT-h-PB1, pGT-h-PB2, pGT-h-PA, and pGT-h-NP) and pPOL-I-CAT-RT with a wild-type polyadenylation signal (lanes 1 to 4), a U6→A6 mutation (lanes 5 and 6), or a U6→UCUACG mutation (lanes 7 and 8). The CAT activities of the cell lysates were analyzed as described in Materials and Methods. Dilution factors of the cell lysates are indicated. Lane 9, mock transfection.
Rescue of an influenza virus encoding an NA-specific mRNA with a poly(U) tail.
Since the poly(U)-tailed mRNA transcribed from the CAT vRNA was translated, though at low efficiency, it was of interest to test whether we could rescue a transfectant influenza virus which could synthesize poly(U)-tailed mRNA. We decided to replace the polyadenylation signal (U6) with an A6 sequence in the vRNA of the NA segment, partly because of the availability of an efficient rescue system for this gene segment (8) and partly because influenza virus is known to tolerate a severe reduction in NA protein levels (10, 28). To construct a transfectant virus with an A track (hereafter called the A6 mutant) in the polyadenylation site of the NA segment, a synthetic NA vRNA containing a U6→A6 mutation in its 5′ noncoding region was first reconstituted into RNPs. The reconstituted RNPs were then transfected into cells infected with A/WSN/HK helper virus (7). In three independent transfections, both wild-type and A6 mutant viruses were rescued. However, when the U track was mutated into a random sequence (UCUACG), no transfectant virus could be rescued. The transfectant viruses were plaque purified three times, and a single plaque of each virus was used for further analysis.
To confirm the presence of the U6→A6 mutation in the NA vRNA of the A6 mutant, vRNA from purified virus was isolated, amplified by RT-PCR, cloned, and sequenced. Apart from the introduced U6→A6 mutation, no other mutation was detected in the 5′ and 3′ noncoding regions of this segment (data not shown). In addition, the U6→A6 mutation could be detected even after 10 passages of the A6 mutant on MDBK cells (data not shown), indicating that the U6→A6 mutation was stable at least over 10 passages.
Poly(U)-tailed NA mRNA is detected in cells infected with the A6 mutant.
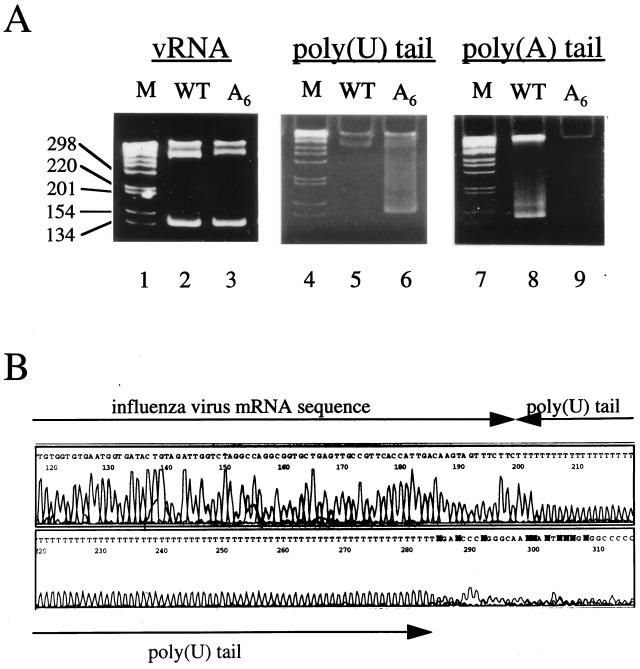
To confirm that the U6→A6 mutation in the NA vRNA resulted in the synthesis of poly(U)-tailed NA mRNA, we isolated total RNA from cells infected with the wild-type virus or the A6 mutant. Total RNA was then tested by an RT-PCR assay which is specific for the detection of either poly(A)-tailed or poly(U)-tailed mRNA (48). In reverse transcription reactions, 5′-GC-clamped T20 and 5′-GC-clamped A20 primers were used to detect poly(A)-tailed and poly(U)-tailed mRNA, respectively. The resultant positive signal appears as a characteristic smear as described before (48). As shown in Fig. 3A, poly(U)-tailed NA mRNA was detected in the total RNA isolated from cells infected with the A6 mutant (lane 6) but not in that isolated from cells infected with the wild-type virus (lane 5). By contrast, poly(A)-tailed NA mRNA could be detected only in RNA from cells infected with the wild-type virus (lane 8), not in the RNA from cells infected with the A6 mutant (lane 9). These results showed that poly(U)-tailed NA mRNA was specifically synthesized in cells infected with the A6 mutant and that it was not polyadenylated. Lanes 2 and 3 in Fig. 3A show RT-PCR products corresponding to the 5′ end of NA vRNA isolated from cells infected with the wild type and the A6 mutant, respectively. The strong signals comigrating with the 134-nt DNA marker indicate that cells used for RNA isolation were, as expected, infected with viruses. The RT-PCR products from poly(U)-tailed mRNAs (lane 6) were then cloned and sequenced (see Materials and Methods). Nine independent clones were found to contain poly(U) sequences in the range of 40 to 127 nt. Figure 3B shows one example with a poly(U) tail of 84 U residues.
FIG. 3.
Poly(U)-tailed NA mRNA is synthesized by the A6 transfectant virus. (A) RT-PCR assays. Total RNA from cells infected with the wild-type virus (lanes 2, 5, and 8) or the A6 mutant (lanes 3, 6, and 9) was tested by RT-PCR for vRNA (lanes 2 and 3), poly(U)-tailed NA mRNA (lanes 5 and 6), and poly(A)-tailed NA mRNA (lanes 8 and 9). RT-PCR products were analyzed on 8% native acrylamide gels. The high-molecular-weight bands near the wells are nonspecific PCR products. Lanes 1, 4, and 7 are DNA markers. (B) Poly(U) sequence n = 84) of a cDNA clone derived from poly(U)-tailed NA mRNA. The influenza virus mRNA sequence is indicated. N, either C, A, G, or T.
The A6 mutant is attenuated due to the reduction of NA protein expression.
The A6 mutant consistently produced small plaques when grown on MDBK cells (wild-type plaque diameter at day 2 p.i. = 1.1 ± 0.32 mm, n = 30; A6 mutant plaque diameter at day 2 p.i. = 0.5 ± 0.21 mm, n = 30), suggesting that this mutant was attenuated. To characterize the growth properties of the A6 mutant in more detail, confluent MDBK cells were infected at an MOI of 0.01 and the number of infectious viral particles released into the medium was assayed by plaque assay on MDBK cells. As predicted from the plaque size, the A6 mutant was attenuated. The maximum plaque titer of the A6 mutant was about 1 log unit lower than that of the wild type (Fig. 4A). The A6 mutant was also tested on Madin-Darby canine kidney (MDCK) cells, and there was, again, about a 1-log-unit reduction in maximum plaque titer compared to that for the wild-type virus (data not shown).
FIG. 4.
The A6 mutant is attenuated due to the reduction of NA expression. (A) Growth curves of transfectant viruses on MDBK cells. Cells were infected with the wild-type virus (WT) or the A6 mutant (A6) at an MOI of 0.01. Infectious particles released into the media at the indicated time points were titrated by plaque assay of MDBK cells. (B) NA activities of wild-type virus and the A6 mutant. NA activity was expressed as nanomoles of 4-methylumbelliferone released per minute per microgram of viral protein. (C) NA protein expression in infected cells. Cells infected with the A6 mutant (a and b) or the wild-type virus (c and d) and mock-infected cells (e and f) were tested for NA protein expression. NA protein expression was visualized by fluorescence microscopy with a fluorescein isothiocyanate (FITC)-labelled antibody. Cells were mounted in antibleach medium containing DAPI for DNA staining. The merged images of NA protein expression and cell nuclei (b, d, and f) are shown. Note that not all cells were infected with virus.
In the light of the CAT reporter gene study (see above; Fig. 2), the most likely explanation for the attenuation of the A6 mutant was the reduction of NA protein expression. To test this possibility, a spectrofluorometric assay (see Materials and Methods) was used to analyze the NA activity associated with the purified virus (47). The NA activity of the A6 mutant was about 8% of that of the wild type (Fig. 4B). We then used a monoclonal anti-NA antibody to show that the reduction of NA protein in virions was due to the low level of expression of the NA protein in infected cells. As shown in Fig. 4C, d, NA was highly expressed in the cytoplasm of the cells infected with the wild-type virus. By contrast, cells infected with the A6 mutant expressed significantly less NA protein (Fig. 4C, b). Although the results of the immunofluorescence assay are not quantitative, they are in agreement with the results in Fig. 4B showing that reduced amounts of NA were incorporated into virions. In addition, these results suggest that the attenuation of the A6 mutant was due to the reduction of the NA protein expression in cells infected with the A6 mutant. However, these results cannot exclude the possibility that the A6 mutant virus contains secondary mutations in the genome which might contribute to the observed viral phenotype.
Levels of the NA vRNA and mRNA, but not cRNA, are specifically reduced.
The above results showed that the NA protein expression in cells infected with the A6 mutant was severely reduced. Such reduction could be due to low levels of poly(U)-tailed mRNA synthesis in infected cells. To test this hypothesis, total RNA was isolated from infected cells at different time points and the amounts of NA-specific vRNA, cRNA, and mRNA were quantified by primer extension assays. In the primer extension assay for the NA vRNA analysis, the NS vRNA was used as an internal control. As shown in Fig. 5A, at every time point tested, NA vRNA levels in cells infected with the A6 mutant were lower than those in cells infected with the wild-type virus. By contrast, the NS vRNA levels from cells infected with the A6 mutant were similar to those of the wild type. When the NA vRNA levels were normalized to the NS vRNA levels in infected cells, the steady-state NA vRNA levels of the A6 mutant were found to be between 40 and 53% of those of the wild type (Fig. 5C). We also quantified the amount of NA vRNA in virions (Fig. 5B). The levels of the NA vRNA in the transfectant virus (the A6 mutant) were about 60% of that of the wild type (Fig. 5B).
Since both cRNA and mRNA are positive sense, the same NA-specific primer was used to analyze both of these RNAs. In these primer extension reactions, an HA-specific primer was used as an internal control. As shown in Fig. 6A, cells infected with the wild-type virus showed stronger signals for NA mRNA than for NA cRNA. By contrast, the NA mRNA signals in cells infected with the A6 mutant were much weaker than NA cRNA signals. These results suggested that poly(U)-tailed mRNA was synthesized at a reduced level. When the levels of the NA cRNA were normalized to the internal control (i.e., HA cRNA), the steady-state cRNA levels of the A6 mutant were found to be not statistically different from those of the wild type on the basis of a Student t test (from three independent experiments) (Fig. 6B). By contrast, when we normalized the NA mRNA to the internal control (i.e., HA mRNA), the steady-state mRNA levels of the A6 mutant in infected cells were found to be only about 40 to 50% of those of the wild type (Fig. 6C). These results demonstrated that the NA mRNA, but not the NA cRNA, of the A6 mutant was synthesized at reduced levels.
Poly(U)-tailed NA mRNA is predominantly localized in the nucleus.
Although cells infected with the A6 mutant showed a reduction of the NA mRNA levels, such reduction could not fully explain the dramatic reduction of the NA activity of the A6 mutant (8% of the wild type; Fig. 4B). Since the influenza virus mRNAs are synthesized in the nuclei of infected cells (19, 22), it was possible that the poly(U)-tailed NA mRNA was defective in nuclear export. To address this question, we used fluorescent in situ hybridization to visualize the distribution of the poly(U)-tailed NA mRNA in infected cells (Fig. 7). We first used a riboprobe which is specific for the positive-sense NA RNAs (i.e., one that detects both mRNA and cRNA) (Fig. 7A to I). Since cRNAs are synthesized in small amounts compared to mRNAs during infection (17, 24) and since they remain in the nucleus throughout viral infection (56), one may assume that any positive signal in the cytoplasm represents mRNAs, while a signal in the nucleus represents predominantly mRNA, although cRNA might partly contribute to this signal. As shown in Fig. 7D to F, cells infected with the wild-type virus showed strong signals in the cytoplasm. These results showed that poly(A)-tailed mRNAs were transported from the nucleus to the cytoplasm, as expected. By contrast, in cells infected with the A6 mutant, positive-sense NA RNAs could only be detected in the nucleus (Fig. 7A to C), suggesting that poly(U)-tailed mRNAs have a defect in nuclear transport and remain in the nucleus. No signal was detected in mock infection (Fig. 7G to I).
FIG. 7.
Poly(U)-tailed NA mRNA is predominantly localized in the nucleus. Cells infected with the A6 mutant (A to C and J to L) or wild-type (D to F and M to O) virus and mock-infected cells (G to I and P to R) were hybridized with NA mRNA-specific (A to I) or poly(U)-tailed NA mRNA-specific (J to R) probes. Nuclei of cells were stained with DAPI (left column). The distribution of the corresponding probe was visualized by fluorescence microscopy with FITC (middle column). The right column consists of the merged images of the other two columns. Note that not all the cells were infected with virus.
To confirm that poly(U)-tailed NA mRNAs remained in the nucleus after being synthesized, we used a riboprobe specific for the poly(U)-tailed NA mRNA, but not for NA cRNA, in the in situ hybridization. As expected, no poly(U)-tailed NA mRNA was detected in cells infected with wild-type virus (Fig. 7M to O) or in mock infections (Fig. 7P to R). By contrast, poly(U)-tailed NA mRNAs were detected in cells infected with the A6 mutant (Fig. 7J to L). More importantly, the poly(U)-tailed NA mRNA was predominantly detected in nuclei of infected cells (Fig. 7J to L). In summary, these results demonstrate that the poly(U)-tailed mRNA remains predominantly in the nucleus after synthesis, which strongly suggests that the poly(U)-tailed mRNA is defective in nuclear export.
DISCUSSION
Here, we have engineered an influenza virus by mutation of the wild-type U6 polyadenylation signal (53) to an A6 mutant sequence at the 5′ end of the NA vRNA segment. This novel transfectant virus synthesized poly(U)-tailed NA mRNA instead of the usual poly(A)-tailed mRNA in infected cells. Since the poly(U)-tailed mRNA does not contain signals for the eukaryotic polyadenylation machinery (50, 59), it is unlikely that host cellular proteins would add a poly(A) sequence to this novel form of mRNA. Interestingly, the A6 mutant virus showed an attenuated phenotype and produced a reduced amount of NA. NA is known to promote the release of viral particles from infected cells (39). Therefore, the attenuation of the A6 mutant was most likely due to the reduced levels of NA expression in infected cells.
We detected reduced levels of poly(U)-tailed NA mRNA in infected cells, suggesting that the low level of NA expression could be due to the low levels of poly(U)-tailed NA mRNAs. Since model vRNA templates with either a U6 or A6 track could synthesize similar amounts of mRNAs in vitro (46), it was unlikely that the mutated NA vRNA of the A6 mutant was a less-efficient template for the synthesis of poly(U)-tailed mRNA. The observed reduction of poly(U)-tailed NA mRNA levels was more likely due to the reduction of the NA vRNA levels. When we analyzed the steady-state levels of vRNAs in infected cells, we found that there was about a 50% reduction of NA vRNA levels. It was shown that the nonconserved nucleotides at the 3′ and 5′ ends of vRNA play an important role in replication (61). It is possible that the U6-to-A6 mutation, which resulted in changing the nonconserved sequences of cRNA and vRNA, could somehow down-regulate the replication process. However, the NA cRNA level of the A6 mutant was not affected by the mutation. In wild-type virus infection, vRNA can be transported to the cytoplasm for packaging into progeny virions or used as a template for transcription and replication. It is possible that, for infection with the A6 mutant, NA vRNAs which are normally used for mRNA synthesis or transported to the cytoplasm are recruited to NA cRNA synthesis so as to maintain sufficient levels of NA cRNA templates for NA vRNA synthesis.
The severe reduction of the NA activity of the A6 mutant virus did not correlate with the reduced NA poly(U)-tailed mRNA level, suggesting that the reduced level of poly(U)-tailed mRNA might not be the only factor that affects NA expression. From the results of in situ hybridization (Fig. 7), poly(A)-tailed NA mRNAs were mainly distributed in cytoplasm. By contrast, poly(U)-tailed NA mRNAs were found predominantly in the nucleus. Since protein synthesis occurs in the cytoplasm, a defect in the transport of poly(U)-tailed NA mRNAs from nucleus to cytoplasm would lead to limited amounts of mRNAs for protein expression. Hence, the severe reduction of NA protein expression in infected cells was most likely caused by the retention of poly(U)-tailed mRNAs in the nucleus. We were not able to detect NA mRNA in the cytoplasm of A6 mutant-infected cells (Fig. 7), indicating that poly(U)-tailed NA mRNA was present in the cytoplasm at levels below our detection limit. On the other hand, the detection of NA protein in infected cells (Fig. 4B) and in the A6 mutant virus (Fig. 4C) clearly demonstrated that a certain amount of poly(U)-tailed NA mRNA was available for translation. Thus it had to be transported (or diffused) into the cytoplasm. Further characterization, such as the characterization of the stability and the quantitative nuclear and cytoplasmic distribution of the poly(U)-tailed mRNA, is in progress.
We do not know whether replacing the poly(A) tail of the NA mRNA with a poly(U) tail affects translation efficiency. However, since the majority of the poly(U)-tailed mRNA remains in the nucleus, the significant level of NA protein expression (8% of that of the wild type) in the A6 mutant-infected cells suggests that the poly(U)-tailed NA mRNA is a competent template for translation. Nonpolyadenylated influenza virus mRNAs are known to be translated under in vitro conditions (40). Results from the present study also show that polyadenylation is not required for translation. Furthermore, our results imply that other factors are involved in the control of viral translation. Thus, the 5′-untranslated region of the viral mRNA, the NS1 protein, and some cellular proteins are known to facilitate the translation of viral mRNAs (4, 14, 40). Hence, the poly(A) tail of the influenza virus mRNA is not essential for translation, and its most important function might be to direct influenza mRNA export from the nucleus to the cytoplasm. In addition, in support of this view, bunyaviruses, which are replicated solely in the cytoplasm, apparently have viral mRNAs which are not polyadenylated (54).
In general, little is known about the mechanism of viral mRNA nuclear export. Several influenza virus proteins have been shown to be involved in processes related to RNA transport. For example, nucleoprotein and matrix protein 1 are known to bind to vRNA and facilitate its nuclear export (31, 36, 60). Recently, NEP (formerly known as NS2) was identified as having a function similar to that of the Rev protein of human immunodeficiency virus type 1 in mediating viral RNP export (37). However, these proteins seem to be restricted to mediating nuclear export of vRNA only. The NS1 protein appears to interact with the cellular 3′-end-processing machinery (2, 35) and inhibits the nuclear export of cellular poly(A)-containing mRNAs (1, 11, 30, 51, 52). However, new evidence suggests that these functions of NS1 are dispensable for viral replication under certain conditions (13). Since influenza virus mRNAs have a structure similar to that of cellular mRNAs [e.g., a 5′ cap and a poly(A) tail], it would not be surprising if they utilized the cellular mRNA transport system for nuclear export.
For eukaryotic cells, there is a growing body of evidence supporting the idea that maturation of cellular pre-mRNAs is a prerequisite for nuclear export (6, 16, 20, 26). In agreement with this idea, the addition of a poly(A) tail was found to stimulate mRNA export (20). For nonpolyadenylated histone mRNAs, the generation of mature 3′ ends is also known to be required for nuclear export (6). Here, in the absence of a characteristic signal of mature mRNA [e.g., a poly(A) tail], cellular nuclear export factors might regard the poly(U)-tailed NA mRNA of the A6 mutant as a pre-mRNA and retain the poly(U)-tailed mRNA in the nucleus. Since the synthesis of the poly(A) tail by the influenza virus RNA polymerase does not require the cellular cleavage/polyadenylation machinery (43, 45, 46, 48, 49), our results suggest that a poly(A) sequence itself is required for efficient mRNA export.
On the other hand, it is possible that the poly(U) tail itself is a signal for nuclear retention. In eukaryotic cells, several cellular proteins that are associated with the mRNA in the nucleus are selectively removed prior to mRNA nuclear export (26), indicating that some of these cellular proteins might be involved in nuclear retention. Interestingly, hnRNP C protein, a protein which is involved in mRNA nuclear retention, has been shown to have avid binding properties for a poly(U) sequence (33, 57). We speculate that hnRNP C protein or perhaps other nuclear retention factors (cellular and viral) might specifically bind to the poly(U) sequence of the poly(U)-tailed mRNA and impede the export of poly(U)-tailed mRNA from the nucleus to the cytoplasm.
Finally, this novel form of influenza virus could have several potential applications. The mutant virus is apparently stable, and mutants of this type could be an example of a new way of producing live attenuated influenza virus vaccines. In addition, this novel virus might be a useful model in further elucidating basic mechanisms of mRNA nuclear export and the function of the poly(A) tail in other mRNA processes, such as translation.
ACKNOWLEDGMENTS
L.L.M.P. was supported by the Croucher Foundation. G.G.B. and E.F. were supported by the MRC (program grant G9523972).
We thank Peter Palese for plasmids, Walter Gerhard for the anti-NA antibody, and David Pritlove for many helpful discussions. We also thank Shona Murphy for critical reading of the manuscript.
REFERENCES
- 1.Alonso-Caplen F V, Nemeroff M E, Qiu Y, Krug R M. Nucleocytoplasmic transport: the influenza virus NS1 protein regulates the transport of spliced NS2 mRNA and its precursor NS1 mRNA. Genes Dev. 1992;6:255–267. doi: 10.1101/gad.6.2.255. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Li Y, Krug R M. Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J. 1999;18:2273–2283. doi: 10.1093/emboj/18.8.2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colgan D F, Manley J L. Mechanism and regulation of mRNA polyadenylation. Genes Dev. 1997;11:2755–2766. doi: 10.1101/gad.11.21.2755. [DOI] [PubMed] [Google Scholar]
- 4.de la Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 6.Eckner R, Ellmeier W, Birnstiel M L. Mature messenger RNA 3′ end formation stimulates RNA export from the nucleus. EMBO J. 1991;10:3513–3522. doi: 10.1002/j.1460-2075.1991.tb04915.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Enami M, Palese P. High-efficiency formation of influenza virus transfectant. J Virol. 1991;65:2711–2713. doi: 10.1128/jvi.65.5.2711-2713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Enami M, Luytjes W, Krystal M, Palese P. Introduction of site specific mutations into the genome of influenza virus. Proc Natl Acad Sci USA. 1990;87:3802–3805. doi: 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fodor E, Pritlove D C, Brownlee G G. The influenza virus panhandle is involved in the initiation of transcription. J Virol. 1994;68:4092–4096. doi: 10.1128/jvi.68.6.4092-4096.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fodor E, Palese P, Brownlee G G, García-Sastre A. Attenuation of influenza A virus mRNA levels by promoter mutations. J Virol. 1998;72:6283–6290. doi: 10.1128/jvi.72.8.6283-6290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein inhibits mRNA splicing and blocks mRNA nucleocytoplasmic transport. EMBO J. 1994;13:704–712. doi: 10.1002/j.1460-2075.1994.tb06310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Sastre A, Muster T, Barclay W S, Percy N, Palese P. Use of mammalian internal ribosomal entry site element for expression of a foreign protein by a transfectant influenza virus. J Virol. 1994;68:6252–6261. doi: 10.1128/jvi.68.10.6254-6261.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.García-Sastre A, Egorov A, Matassov D, Bramdt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 14.Garfinkel M S, Katze M G. Translational control by influenza virus. Selective translation is mediated by sequences within the viral mRNA 5′-untranslated region. J Biol Chem. 1993;268:22223–22226. [PubMed] [Google Scholar]
- 15.Gorman M, Moffat L F, Howard B H. Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol. 1982;2:1044–1051. doi: 10.1128/mcb.2.9.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamm J, Mattaj I W. Monomethylated cap structures facilitate RNA export from the nucleus. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 17.Hay A J, Lomniczi B, Bellamy A R, Skehel J J. Transcription of the influenza virus genome. Virology. 1977;83:337–355. doi: 10.1016/0042-6822(77)90179-9. [DOI] [PubMed] [Google Scholar]
- 18.Hay A J, Skehel J J, McCauley J. Characterization of influenza virus RNA complete transcripts. Virology. 1982;116:517–522. doi: 10.1016/0042-6822(82)90144-1. [DOI] [PubMed] [Google Scholar]
- 19.Herz C, Stavnezer E, Krug R M, Gurney T., Jr Influenza virus, an RNA virus, synthesizes its messenger RNA in the nucleus of infected cells. Cell. 1981;26:391–400. doi: 10.1016/0092-8674(81)90208-7. [DOI] [PubMed] [Google Scholar]
- 20.Huang Y, Carmichael G G. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol Cell Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Izaurralde E, Adam S. Transport of macromolecules between the nucleus and the cytoplasm. RNA. 1998;4:351–364. [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson D A, Caton A J, McCready S J, Cook P R. Influenza virus RNA is synthesized at a fixed site in the nucleus. Nature. 1982;296:366–368. doi: 10.1038/296366a0. [DOI] [PubMed] [Google Scholar]
- 23.Jarmolowski A, Boelens W C, Izaurralde E, Mattaj I W. Nuclear export of different classes of RNA is mediated by specific factors. J Cell Biol. 1994;124:627–635. doi: 10.1083/jcb.124.5.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lamb R A, Choppin P W. The gene structure and replication of influenza virus. Annu Rev Biochem. 1983;52:467–506. doi: 10.1146/annurev.bi.52.070183.002343. [DOI] [PubMed] [Google Scholar]
- 25.Lamond A I. Nuclear RNA processing. Curr Opin Cell Biol. 1991;3:493–501. doi: 10.1016/0955-0674(91)90078-d. [DOI] [PubMed] [Google Scholar]
- 26.Legrain P, Rosbash M. Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell. 1989;57:573–583. doi: 10.1016/0092-8674(89)90127-x. [DOI] [PubMed] [Google Scholar]
- 27.Luo G, Luytjes W, Enami M, Palese P. The polyadenylation signal of influenza virus RNA involves a stretch of uridines followed by the RNA duplex of the panhandle structure. J Virol. 1991;65:2861–2867. doi: 10.1128/jvi.65.6.2861-2867.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo G, Bergmann M, García-Sastre A, Palese P. Mechanism of attenuation of a chimeric influenza A/B transfectant virus. J Virol. 1992;66:4679–4685. doi: 10.1128/jvi.66.8.4679-4685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malim M H, Hauber J, Le S, Maizel J V, Cullen B R. The HIV-1 Rev trans-activator acts through a structural target sequence to activate nuclear export of unspliced viral mRNA. Nature. 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 30.Marión R M, Aragón T, Beloso A, Nieto A, Ortin J. The N-terminal half of the influenza virus NS1 protein is sufficient for nuclear retention of mRNA and enhancement of viral mRNA translation. Nucleic Acids Res. 1997;25:4271–4377. doi: 10.1093/nar/25.21.4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 32.Mattaj I W, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 33.Nakielny S, Dreyfuss G. The hnRNP C proteins contain a nuclear retention sequence that can override nuclear export signals. J Cell Biol. 1996;134:1365–1373. doi: 10.1083/jcb.134.6.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nakielny S, Dreyfuss G. Nuclear export of proteins and RNAs. Curr Opin Cell Biol. 1997;9:420–429. doi: 10.1016/s0955-0674(97)80016-6. [DOI] [PubMed] [Google Scholar]
- 35.Nemeroff M E, Barabino S L M, Li Y, Keller W, Krug R M. Influenza virus NS1 protein inhibits 3′ end formation of cellular premRNAs. Mol Cell. 1999;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 36.Neumann G, Castrucci M R, Kawaoka Y. Nuclear import and export of influenza virus nucleoprotein. J Virol. 1997;71:9690–9700. doi: 10.1128/jvi.71.12.9690-9700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Neill R E, Talon J, Palese P. The influenza virus NEP (NS2 protein) mediates the nuclear export of viral ribonucleoproteins. EMBO J. 1998;17:288–296. doi: 10.1093/emboj/17.1.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Palese P. The genes of influenza virus. Cell. 1977;10:1–10. doi: 10.1016/0092-8674(77)90133-7. [DOI] [PubMed] [Google Scholar]
- 39.Palese P, Compans R W. Inhibition of influenza virus replication in tissue culture by 2-deoxy-2,3-dehydro-N-trifluoro-acetyl-neuraminic acid (FANA): mechanism of action. J Gen Virol. 1976;33:159–164. doi: 10.1099/0022-1317-33-1-159. [DOI] [PubMed] [Google Scholar]
- 40.Park Y W, Wilusz J, Katze M. Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc Natl Acad Sci USA. 1999;96:6694–6699. doi: 10.1073/pnas.96.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between the nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 42.Pleschka S, Jaskunas S R, Engelhardt O G, Zürcher T, Palese P, García-Sastre A. A plasmid-based reverse genetics system for influenza A virus. J Virol. 1996;70:4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plotch S J, Krug R M. Influenza virion transcriptase: synthesis in vitro of large, polyadenylic acid-containing complementary RNA. J Virol. 1977;21:24–34. doi: 10.1128/jvi.21.1.24-34.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Plotch S J, Bouloy M, Ulmanen I, Krug R M. A unique cap (m7GpppXm) dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 45.Poon L L M, Pritlove D C, Sharps J, Brownlee G G. The RNA polymerase of influenza virus, bound to the 5′ end of virion RNA, acts in cis to polyadenylate mRNA. J Virol. 1998;72:8214–8219. doi: 10.1128/jvi.72.10.8214-8219.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Poon L L M, Pritlove D C, Fodor E, Brownlee G G. Direct evidence that the poly(A) tail of influenza A virus mRNA is synthesized by reiterative copying of a U track in the virion RNA template. J Virol. 1999;73:3473–3476. doi: 10.1128/jvi.73.4.3473-3476.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Potier M, Mameli L, Bélisle M, Dallaire L, Melancon S B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-α-D-N-acetylneuraminate) substrate. Anal Biochem. 1979;94:287–296. doi: 10.1016/0003-2697(79)90362-2. [DOI] [PubMed] [Google Scholar]
- 48.Pritlove D C, Poon L L M, Fodor E, Sharps J, Brownlee G G. Polyadenylation of influenza virus mRNA transcribed in vitro from model virion RNA templates: requirement for 5′ conserved sequences. J Virol. 1998;72:1280–1286. doi: 10.1128/jvi.72.2.1280-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pritlove D C, Poon L L M, Devenish L J, Leahy M B, Brownlee G G. A hairpin loop at the 5′ end of influenza A virus virion RNA is required for synthesis of poly(A)+ mRNA in vitro. J Virol. 1999;73:2109–2114. doi: 10.1128/jvi.73.3.2109-2114.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Proudfoot N J, Brownlee G G. 3′ non-coding region sequences in eukaryotic messenger RNA. Nature. 1976;263:211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- 51.Qian X, Alonso-Caplen F, Krug R M. Two functional domains of the influenza virus NS1 protein are required for regulation of nuclear export of mRNA. J Virol. 1994;68:2433–2441. doi: 10.1128/jvi.68.4.2433-2441.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Qiu Y, Krug R M. The influenza virus NS1 protein is a poly(A)-binding protein that inhibits nuclear export of mRNAs containing poly(A) J Virol. 1994;68:2423–2432. doi: 10.1128/jvi.68.4.2425-2432.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson J S, Schubert S M, Lazzarini R A. Polyadenylation sites for influenza virus mRNA. J Virol. 1981;32:550–559. doi: 10.1128/jvi.38.1.157-163.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmaljohn C S. Bunyaviridae: the virus and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1447–1471. [Google Scholar]
- 55.Seong B L, Brownlee G G. A new method for reconstituting influenza polymerase and RNA in vitro: a study of the promoter elements for cRNA and vRNA synthesis in vitro and viral rescue in vivo. Virology. 1992;186:247–260. doi: 10.1016/0042-6822(92)90079-5. [DOI] [PubMed] [Google Scholar]
- 56.Shapiro G I, Gurney T, Jr, Krug R M. Influenza virus gene expression: control mechanisms at early and late times of infection and nuclear-cytoplasmic transport of virus-specific RNAs. J Virol. 1987;61:764–773. doi: 10.1128/jvi.61.3.764-773.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swanson M S, Dreyfuss G. Classification and purification of proteins of heterogeneous nuclear ribonucleoprotein particles by RNA-binding specificities. Mol Cell Biol. 1988;8:2237–2241. doi: 10.1128/mcb.8.5.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tiley L S, Hagen M, Matthews J T, Krystal M. Sequence-specific binding of the influenza virus RNA polymerase to sequences located at the 5′ ends of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Verrotti A C, Thompson S R, Wreden C, Strickland S. Evolutionary coservation of sequence elements controlling cytoplasmic polyadenylation. Proc Natl Acad Sci USA. 1996;93:9027–9032. doi: 10.1073/pnas.93.17.9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Whittaker G, Bui M, Helenius A. Nuclear trafficking of influenza virus ribonucleoproteins in heterokaryons. J Virol. 1996;70:2743–2756. doi: 10.1128/jvi.70.5.2743-2756.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zheng H, Palese P, García-Sastre A. Nonconserved nucleotides at the 3′ and 5′ ends of an influenza A virus RNA play an important role in viral RNA replication. Virology. 1996;217:242–251. doi: 10.1006/viro.1996.0111. [DOI] [PubMed] [Google Scholar]