Abstract
Introduction:
Microbial contamination of drinking water, particularly by pathogens such as Escherichia coli O157: H7, is a significant public health concern worldwide, especially in regions with limited access to clean water like the Gaza Strip. However, few studies have quantified the disease burden associated with E. coli O157: H7 contamination in such challenging water management contexts.
Objective:
This study aimed to conduct a comprehensive Quantitative Microbial Risk Assessment to estimate the annual infection risk and disease burden attributed to E. coli O157: H7 in Gaza’s drinking water.
Methods:
Applying the typical four steps of the Quantitative Microbial Risk Assessment technique—hazard identification, exposure assessment, dose-response analysis, and risk characterization—the study assessed the microbial risk associated with E. coli O157: H7 contamination in Gaza’s drinking water supply. A total of 1317 water samples from various sources across Gaza were collected and analyzed for the presence of E. coli O157: H7. Using Microsoft ExcelTM and @RISKTM software, a Quantitative Microbial Risk Assessment model was constructed to quantify the risk of infection associated with E. coli O157: H7 contamination. Monte Carlo simulation techniques were employed to assess uncertainty surrounding input variables and generate probabilistic estimates of infection risk and disease burden.
Results:
Analysis of the water samples revealed the presence of E. coli O157: H7 in 6.9% of samples, with mean, standard deviation, and maximum values of 1.97, 9.74, and 112 MPN/100 ml, respectively. The risk model estimated a median infection risk of 3.21 × 10-01 per person per year and a median disease burden of 3.21 × 10-01 Disability-Adjusted Life Years per person per year, significantly exceeding acceptable thresholds set by the WHO.
Conclusion:
These findings emphasize the urgent need for proactive strategies to mitigate public health risks associated with waterborne pathogens in Gaza.
Keywords: Burden of disease, E. coli O157: H7, Gaza Strip, QMRA, waterborne diseases, water safety plan
Introduction
The persistent issue of microbiological contamination in drinking water supply systems (DWSSs) is a global concern, especially in developing countries where access to safe drinking water remains a challenge. 1 The Gaza Strip exemplifies this crisis with significant microbial contamination of its drinking water, as evidenced by numerous studies detecting coliform bacteria in Gaza’s water sources.2,3 Despite these findings, there is a lack of comprehensive assessments concerning the associated burden of diseases resulting from contaminated drinking water.
Several factors contribute to the compromised quality and quantity of groundwater in the region. The high population density, coupled with excessive groundwater extraction and improper wastewater disposal, exacerbates the issue.4,5 Despite efforts to address this challenge, such as the establishment of commercial small-scale brackish desalination plants, these solutions have vulnerabilities. Cross-contamination risks persist during water handling, transportation via tanker trucks, and storage, which contribute to ongoing health hazards. 6 Furthermore, outdated municipal water distribution systems and lapses in disinfection processes further jeopardize water quality. 7
The epidemiological landscape vividly reflects the consequences of this water quality crisis, particularly in terms of diarrheal diseases, which continue to be a leading cause of child morbidity in the Gaza Strip. Notably, there has been a concerning increase in the prevalence of diarrhea cases, highlighting the urgent need for effective interventions. 8
Recognizing the imperative for a comprehensive approach to water safety management, the World Health Organization (WHO) advocates for the implementation of Water Safety Plans (WSP) and quantitative microbial risk assessment (QMRA). These frameworks offer systematic methodologies for managing microbial risks throughout the water supply chain.9–11
QMRA, adapted from chemical risk assessment paradigms, encompasses hazard identification, exposure assessment, dose-response analysis, and risk characterization. This approach provides a robust tool for evaluating and mitigating health risks associated with waterborne pathogens. 12
The use of disability-adjusted life years (DALYs) in risk assessment provides a comprehensive measure of disease burden attributable to contaminated drinking water. DALYs offer insights into the potential impact of waterborne pathogens on public health, aiding in resource allocation and intervention prioritization.9,13
Escherichia coli (E. coli) serves as a critical indicator organism for assessing drinking water quality, with certain pathogenic strains posing significant health risks. Among these, E. coli O157: H7, a member of the Enterohemorrhagic E. coli (EHEC) group, has garnered particular attention due to its association with waterborne outbreaks worldwide.14,15 The fecal-oral transmission route underscores the importance of implementing multiple-barrier principles to safeguard drinking water from microbial contaminants.16,17
Infections with E. coli O157: H7 can lead to a spectrum of symptoms, ranging from mild diarrhea to severe, bloody diarrhea, often accompanied by abdominal cramping. Vulnerable populations, such as young children and the elderly, may experience systemic complications, including thrombotic thrombocytopenic purpura, hemolytic-uremic syndrome, and acute renal failure.18–20
Despite the significance of E. coli O157: H7 as a waterborne pathogen, limited studies have explored its quantitative microbial risk in drinking water systems globally. Moreover, specific investigations into the risk posed by this pathogen in Gaza’s drinking water are notably lacking. Therefore, this study aims to fill this critical knowledge gap by employing QMRA to estimate infection risks and disease burdens associated with E. coli O157: H7 contamination in Gaza’s drinking water. By providing decision-makers and stakeholders with actionable insights, this research endeavors to inform evidence-based interventions and optimize investment in DWSS infrastructure.
Materials and methods
Description of the study area and water supply system
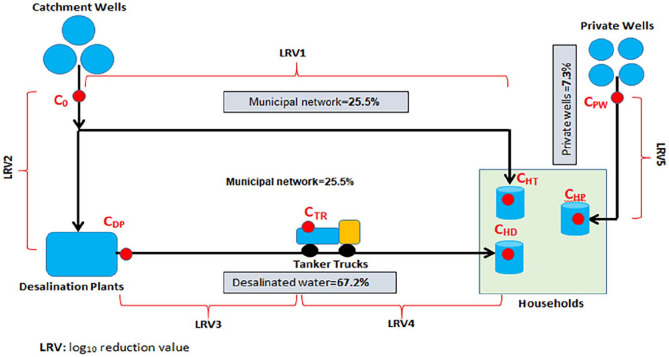
Gaza Strip is the southwestern gateway of Palestine, with around 1.9 million Palestinians on some 365 km2. Gaza ranks as the third most densely populated polity in the world. The territory borders Israel to the north and east, Egypt to the south, and the Mediterranean Sea to the West. Gaza Strip consists of five governorates namely: Northern, Gaza, Mid Zone, Khan Younis, and Rafah. 21 Gaza Strip has a unique DWSS. First, municipalities provide households and other facilities with groundwater via underground network pipes. Second, private commercial firms and Non-Governmental Organizations (NGOs) provide households and other facilities with trucked desalinated water from small-scale brackish water desalination plants. Third, households, particularly in rural areas, rely on their own private wells for drinking and domestic uses (Figure 1).
Figure 1.
Schematic illustration map of a typical Gaza Strip’s water supply system.
In the Gaza Strip, chlorination is the very popular method of water disinfection as insufficient amounts of chlorine are usually added to only two points of water supply system (after desalination and before pumping in the municipal network pipelines) mainly due to aesthetic issues such as odor and taste issues.22,23
Sampling and confirmation of E. coli O157: H7
In the current cross-sectional study, a total of 1317 water samples were collected from multiple points from catchment to consumer’s tap within the Gaza water supply system over the four seasons in the period between March 2018 and January 2019. Sampling frequency varied based on operational conditions and water quality monitoring protocols. Each sample was analyzed for E. coli O157:H7 concentrations using standard microbiological methods.
About 109, 109, 197, 384, 384, 67, and 67 water samples were taken from seven reference points of DWSS in the Gaza Strip, which were water wells, small-scale water desalination plants, tanker trucks, desalinated water at households, municipal water at households, private wells, private well water at households, respectively. Stratified random sampling was employed to warrant the representative distribution of the study sample into neighborhoods of the five Gaza Strip governorates in accordance with the number of households in each governorate. The standard methods for water microbiological examination were followed. 24
Water samples were sent to a private, well-equipped, licensed laboratory for microbiological analysis in a container containing ice cubes and analyzed immediately upon arrival at the laboratory. A community-based survey was conducted among 1857 Gaza households to collect information about drinking water sources and drinking water consumption per capita per day at households.
A Quanti-Tray/2000 with 49 large wells and 48 small wells was used without additional serotyping. The “Quanti-Tray/2000” system is manufactured by IDEXX Laboratories, Inc., a leading global provider of water testing solutions. The Quanti-Tray/2000 system is a widely used method for the enumeration of microbial contaminants, including E. coli O157: H7, in water samples. IDEXX Laboratories, Inc. is headquartered in Westbrook, Maine, USA, and has a global presence, providing innovative solutions for water quality testing and microbiological analysis.
A snap pack of metabolized Colisure’s nutrient-indicator MUG (4-Methylumbelliferyl β-D-glucopyranoside was added to 100 ml water sample and then poured into the Quanti-Tray. The Quanti-Tray thereafter was sealed using a Quanti-Tray sealer and incubated at 37°C for 24 h. To determine the number of E. coli, the Quanti-Tray was exposed to an ultra-violet lamp with 6-watt and 365-nanometer placed in a dark box, the red/magenta, and fluorescence wells mean the sample is positive for E. coli. Yello/Gold wells mean the sample is negative for E. coli. The most probable number (MPN) table was used to obtain the MPN of E. coli per 100 ml water sample.
For E. coli O157: H7 examination, a sample was taken from each confirmed Quanti-Tray well with positive E. coli individually and then struck on Petri dishes (60 × 15 mm) contains cefixime tellurite sorbitol-MacConkey (CT–SMAC) agar, thereafter incubated for 24 h at 37°C. The majority of E. coli isolates do ferment sorbitol and give characteristic pink colonies, therefore, they give colorless colonies on the medium. For the identification and confirmation of E. coli serogroup O157, an oxoid E. coli O157: H7 latex agglutination test kit was used. Also, the MPN table was used to estimate the E. coli O157: H7 densities per 100 ml water sample according to the number of positive large and small wells for E. coli O157: H7.25,26
Simulation methodology and implications of uncertainty and variability
In this study, we employed a two-dimensional simulation approach using @RISKTM software (Palisade, Ivybridge, UK) integrated with Microsoft ExcelTM to conduct a comprehensive Quantitative Microbial Risk Assessment (QMRA) for E. coli O157: H7 contamination in Gaza’s drinking water supply. The “two-dimensional simulation technique” involves considering uncertainty or variability across different model parameters and percentiles of the estimated infection risk distribution. This methodology allowed us to evaluate the complex interplay of multiple variables influencing microbial risk, including microbial concentrations, exposure pathways, and dose-response relationships.
Derivation of probability distributions
The probability distributions used in our simulation model were derived from a combination of empirical data collected during our sampling campaign and literature-based estimates. Specifically, we parameterized our model using measured microbial concentrations from water samples across various sources within the Gaza Strip, as well as dose-response relationships obtained from published epidemiological studies.
Addressing uncertainty and variability
Our study acknowledges and distinguishes between uncertainty (due to lack of precise knowledge about input parameters) and variability (inherent differences in microbial concentrations and exposure scenarios). The Monte Carlo simulation technique allowed us to quantify the uncertainty surrounding input variables and generate probabilistic estimates of infection risk and disease burden.
Central to our risk assessment methodology was the implementation of Monte Carlo simulation techniques. 27 Monte Carlo simulation provides a powerful framework for probabilistic modeling, generating numerous random samples from the specified probability distributions. Through the simulation of a large number of scenarios, Monte Carlo simulation allows us to quantitatively evaluate the likelihood of different outcomes and assess the associated risks with a high degree of confidence.
In further strengthening the statistical rigor of our analysis, we drew upon recent advancements in the field of microbial risk assessment. Studies such as Custodio et al. 28 2023 have demonstrated the effectiveness of Monte Carlo simulation in evaluating microbial contamination levels in drinking water systems. By leveraging insights from these studies, we optimized our risk assessment approach, ensuring the reliability and validity of our results.
Calculation of log10 reduction values
To assess treatment effectiveness and attenuation of E. coli O157:H7 concentrations throughout the water distribution system, Log10 Reduction Values (LRVs) were calculated. LRVs were determined by comparing the log reduction in E. coli O157:H7 concentrations after treatment to the initial concentrations before treatment for each specific sampling event.
LRVs were computed using paired sample analysis, where the difference in log-transformed concentrations (post-treatment minus pre-treatment) was calculated for each sample point and sampling date. Individual LRVs were then used to construct a distribution, capturing the variability and uncertainty in treatment efficacy across different sampling events and locations.
Stochastic assessment
The stochastic nature of treatment effectiveness and microbial attenuation was characterized by analyzing the distribution of LRVs. Each measured LRV represented a data point in the distribution, allowing for statistical analysis to quantify variability and uncertainty associated with treatment processes within the water supply system.
Statistical methods, including descriptive statistics and probability distributions, were employed to assess the effectiveness of treatment interventions and the overall microbial risk reduction achieved at different stages of the water distribution network.
Gamma distributions derivation
The Gamma distributions were derived to represent the observed concentrations of E. coli and E. coli O157: H7 in the water samples, facilitating the analysis and interpretation of microbial contamination levels in the study; the following steps were followed:
- 1. Data Collection and Pre-processing:
- ○ Data on the concentrations of E. coli and E. coli O157: H7 in water samples were collected from the laboratory experiments.
- ○ These concentrations typically represent counts or levels of microbial contamination per unit volume (e.g., CFU/ml or MPN/100 ml).
- 2. Fitting Gamma Distributions:
- ○ The collected concentration data were fitted to Gamma distributions using statistical software.
- ○ The Gamma distribution is commonly used to model positive-valued continuous data, such as microbial counts, where the shape (α) and rate (β) parameters of the distribution can be estimated based on the observed data.
- ○ The shape parameter (α) of the Gamma distribution is related to the variance and skewness of the data, while the rate parameter (β) influences the scale and location of the distribution.
- 3. Parameter Estimation:
- ○ The parameters (α and β) of the Gamma distribution were estimated based on the concentration data.
- ○ Maximum likelihood estimation (MLE) approach was employed to estimate these parameters.
- 4. Model Validation:
- ○ After fitting the Gamma distribution to the concentration data, model validation techniques were employed to assess the goodness-of-fit.
- ○ This validation step ensures that the Gamma distribution adequately describes the observed data and provides reliable estimates of the underlying distribution of microbial concentrations.
- 5. Interpretation and Application:
- ○ The derived Gamma distributions were then used to characterize the variability and uncertainty associated with the concentrations of E. coli and E. coli O157: H7 in the water samples.
- ○ These distributions provide valuable insights into the statistical properties of the microbial data, enabling researchers to make informed decisions and conduct further analyses based on the fitted models.
QMRA model for E. coli O157: H7
In this study, we employed the standard four-step QMRA approach, encompassing hazard identification, exposure assessment, dose-response analysis, and risk characterization, to evaluate the health risks associated with E. coli O157: H7 contamination in Gaza’s drinking water. Our methodology aligns with similar investigations conducted by researchers worldwide, including studies by Smith et al. 29 and Jones et al., 30 who utilized the QMRA framework to assess microbial risks in various waterborne disease outbreaks. The input parameters for our model were derived from both the data collected in our study and relevant scientific literature, ensuring a comprehensive and evidence-based analysis (Table 1). This approach is consistent with methodologies employed in studies by Brown et al. 31 and García et al., 32 underscoring the importance of incorporating both primary data and existing research findings to enhance the accuracy and reliability of QMRA models. By adopting established QMRA methodologies and parameterization techniques, our study provides valuable insights into the microbial risks associated with drinking water sources in Gaza, contributing to the broader body of literature on waterborne disease risk assessment and management.
Table 1.
Input model parameters.
| Variable | Symbol | Unit | Distributional assumption | Reference |
|---|---|---|---|---|
| E.coli O157:H7 concentration | C0 | MPN/100 ml | Gamma.inv (0.0634, 6.920) × Sum(C0) | This study |
| CDP | MPN/100 ml | Gamma.inv (0.0323, 3.526) × Sum(CDP) | This study | |
| CTR | MPN/100 ml | Gamma.inv (0.0365, 7.209) × Sum(CTR) | This study | |
| CHD | MPN/100 ml | Gamma.inv (0.0212, 8.168) × Sum(CHD) | This study | |
| CHT | MPN/100 ml | Gamma.inv (0.0472, 18.149) × Sum(CHT) | This study | |
| CPW | MPN/100 ml | Gamma.inv (0.113, 7.574) × Sum(CPW) | This study | |
| CHP | MPN/100 ml | Gamma.inv (0.181, 12.170) × Sum(CHP) | This study | |
| Calculation of log reduction values (LRV) | LRV1 | Log | −log10 (CHT/C0) | This study |
| LRV2 | Log | −log10 (CDP/C0) | This study | |
| LRV3 | Log | −log10 (CTR/CDP) | This study | |
| LRV4 | Log | −log10 (CHD/CTR) | This study | |
| LRV5 | Log | −log10 (CHP/CPW) | This study | |
| Water intake rate (IR) | IRHT | L/capita.d | Norm.inv (8.56, 1.57) × 0.24 | 33 |
| IRHD | L/capita.d | Norm.inv (8.46, 1.60) × 0.24 | 33 | |
| IRHP | L/capita.d | Norm.inv (8.54, 1.71) × 0.24 | 33 | |
| Parameters for the β-Poisson model | K | — | Lognorm.inv (−8.431, 0.515) | 34 |
| Disease burden (B) | B | DALY per case | PERT (49.23, 54.7, 60.17)/1000 | 35 |
| Illness: infection (Iill/inf) | I | Proportion | PERT (0.69, 0.845, 1) | 36–38 |
| Susceptible fraction of the population (S) | S | Proportion | Constant (1) | This study |
Most probable number (MPN) was assumed as a surrogate for the colony-forming unit (CFU).
Results
Microbial contamination analysis
Analysis of 1317 water samples from various sources across the Gaza Strip revealed the presence of E. coli O157: H7 in 6.9% of samples. The mean, standard deviation, and maximum values of E. coli O157: H7 concentrations were 1.97, 9.74, and 112 MPN/100 ml, respectively.
Hazard identification
E. coli O157: H7 was selected as the reference waterborne organism for the QMRA model due to its documented role in waterborne disease outbreaks globally. This decision was informed by extensive literature and recommendations from international health organizations such as the WHO and the Centers for Disease Control and Prevention (CDC).
Exposure assessment
Our study quantitatively determined the magnitude of E. coli O157: H7 exposure through drinking water consumption from various sources. Log10 reduction values (LRVs) were calculated for different points within Gaza’s water supply system to assess the attenuation of E. coli O157: H7 concentrations (Table 2).
Table 2.
Concentration of E. coli O157: H7 in drinking water and log10 reduction values.
| Sample source | E. coli O157: H7 (MPN/100 ml) | ||||||
|---|---|---|---|---|---|---|---|
| Mean | SE | SD | Left | Right | Max | Min | |
| Water wells (C0) | 4.355 | 3.297 | 17.364 | 1.058 | 7.652 | 112 | 0 |
| Desalination plants (CDP) | 0.541 | 0.574 | 3.023 | −0.033 | 1.115 | 20.9 | 0 |
| Tanker trucks (CTR) | 0.93 | 0.685 | 4.876 | 0.245 | 1.615 | 39.5 | 0 |
| Desalinated water at households (CHD) | 0.598 | 0.412 | 4.107 | 0.186 | 1.01 | 39.2 | 0 |
| Municipal water at households (CHT) | 1.725 | 0.797 | 7.944 | 0.928 | 2.522 | 72.9 | 0 |
| Private wells (CPW) | 5.696 | 4.163 | 17.067 | 1.533 | 9.859 | 112 | 0 |
| Private well water at households (CHP) | 9.097 | 5.246 | 21.506 | 3.851 | 14.343 | 101.2 | 0 |
| DWSS reference points | log10 reduction value (Log) | ||||||
| Mean | SE | SD | Left | Right | Max | Min | |
| LRV1 | 2.715 | 0.221 | 11.25 | 2.494 | 2.936 | 82.202 | −46.233 |
| LRV2 | 7.09 | 0.29 | 14.817 | 6.8 | 7.38 | 111.907 | −52.24 |
| LRV3 | −1.465* | 0.348 | 17.75 | −1.813 | −1.117 | 92.552 | −107.684 |
| LRV4 | 8.5 | 0.473 | 24.137 | 8.027 | 8.973 | 208.069 | −95.721 |
| LRV5 | −1.465* | 0.088 | 4.513 | −1.553 | −1.377 | 22.915 | −36.819 |
Dose-response assessment
The Beta-Poisson dose-response model was employed to estimate the risk of E. coli O157: H7 infection, incorporating two-dimensional Monte Carlo simulation techniques to account for uncertainties in model parameters (Tables 3 and 4).
Table 3.
Summary statistics for estimated infection risk across percentiles.
| Statistic | Mean | SD | Min | 2.50% | 25% | 50% | 75% | 97.50% | Max |
|---|---|---|---|---|---|---|---|---|---|
| HT | |||||||||
| Median | 3.16E-01 | 4.55E-01 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 8.17E-07 | 1.00E+00 | 1.00E+00 | 1.00E+00 |
| Mean | 3.11E-01 | 4.51E-01 | 0.00E+00 | 0.00E+00 | 3.11E-13 | 2.23E-05 | 8.98E-01 | 1.00E+00 | 1.00E+00 |
| 2.50% | 2.15E-01 | 4.02E-01 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 7.38E-12 | 2.28E-02 | 1.00E+00 | 1.00E+00 |
| 97.50% | 3.85E-01 | 4.76E-01 | 0.00E+00 | 0.00E+00 | 2.31E-12 | 1.80E-04 | 1.00E+00 | 1.00E+00 | 1.00E+00 |
| HD | |||||||||
| Median | 3.00E-01 | 4.55E-01 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 1.00E+00 | 1.00E+00 | 1.00E+00 |
| Mean | 2.98E-01 | 4.53E-01 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 4.05E-16 | 9.56E-01 | 1.00E+00 | 1.00E+00 |
| 2.50% | 2.45E-01 | 4.26E-01 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 1.29E-01 | 1.00E+00 | 1.00E+00 |
| 97.50% | 3.39E-01 | 4.70E-01 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 1.00E+00 | 1.00E+00 | 1.00E+00 |
| HP | |||||||||
| Median | 5.38E-01 | 4.74E-01 | 0.00E+00 | 4.70E-12 | 4.48E-04 | 8.49E-01 | 1.00E+00 | 1.00E+00 | 1.00E+00 |
| Mean | 5.32E-01 | 4.72E-01 | 3.44E-15 | 9.46E-10 | 2.26E-03 | 6.75E-01 | 1.00E+00 | 1.00E+00 | 1.00E+00 |
| 2.50% | 3.82E-01 | 4.55E-01 | 0.00E+00 | 0.00E+00 | 4.86E-08 | 6.71E-03 | 1.00E+00 | 1.00E+00 | 1.00E+00 |
| 97.50% | 6.40E-01 | 4.81E-01 | 0.00E+00 | 7.94E-09 | 1.67E-02 | 1.00E+00 | 1.00E+00 | 1.00E+00 | 1.00E+00 |
| Avg | |||||||||
| Median | 3.21E-01 | 3.32E-01 | 0.00E+00 | 3.32E-09 | 7.18E-02 | 2.55E-01 | 6.72E-01 | 1.00E+00 | 1.00E+00 |
| Mean | 3.18E-01 | 3.31E-01 | 3.93E-13 | 5.64E-08 | 5.62E-02 | 1.92E-01 | 6.58E-01 | 9.98E-01 | 1.00E+00 |
| 2.50% | 2.60E-01 | 3.12E-01 | 0.00E+00 | 1.34E-13 | 2.40E-03 | 7.30E-02 | 3.28E-01 | 9.76E-01 | 1.00E+00 |
| 97.50% | 3.59E-01 | 3.44E-01 | 1.23E-12 | 4.71E-07 | 7.30E-02 | 2.56E-01 | 6.99E-01 | 1.00E+00 | 1.00E+00 |
Table 4.
Summary statistics for estimated burden of disease across percentiles.
| Statistic | Mean | SD | Min | 2.50% | 25% | 50% | 75% | 97.50% | Max |
|---|---|---|---|---|---|---|---|---|---|
| HT | |||||||||
| Median | 1.50E-02 | 2.16E-02 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 3.84E-08 | 4.68E-02 | 4.77E-02 | 4.77E-02 |
| Mean | 1.48E-02 | 2.15E-02 | 0.00E+00 | 0.00E+00 | 1.51E-14 | 1.08E-06 | 4.28E-02 | 4.78E-02 | 4.78E-02 |
| 2.50% | 9.86E-03 | 1.76E-02 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 3.10E-13 | 8.28E-04 | 4.09E-02 | 4.09E-02 |
| 97.50% | 1.93E-02 | 2.51E-02 | 0.00E+00 | 0.00E+00 | 1.14E-13 | 8.65E-06 | 5.45E-02 | 5.47E-02 | 5.47E-02 |
| HD | |||||||||
| Median | 1.43E-02 | 2.17E-02 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 4.74E-02 | 4.77E-02 | 4.77E-02 |
| Mean | 1.42E-02 | 2.17E-02 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 2.03E-17 | 4.57E-02 | 4.78E-02 | 4.78E-02 |
| 2.50% | 1.09E-02 | 1.83E-02 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 6.45E-03 | 4.09E-02 | 4.09E-02 |
| 97.50% | 1.74E-02 | 2.50E-02 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 0.00E+00 | 5.47E-02 | 5.47E-02 | 5.47E-02 |
| HP | |||||||||
| Median | 2.54E-02 | 2.25E-02 | 0.00E+00 | 2.23E-13 | 2.05E-05 | 3.89E-02 | 4.77E-02 | 4.77E-02 | 4.77E-02 |
| Mean | 2.54E-02 | 2.26E-02 | 1.79E-16 | 4.56E-11 | 1.08E-04 | 3.22E-02 | 4.78E-02 | 4.78E-02 | 4.78E-02 |
| 2.50% | 1.79E-02 | 1.93E-02 | 0.00E+00 | 0.00E+00 | 2.44E-09 | 3.44E-04 | 4.09E-02 | 4.09E-02 | 4.09E-02 |
| 97.50% | 3.22E-02 | 2.59E-02 | 0.00E+00 | 3.64E-10 | 8.24E-04 | 5.32E-02 | 5.47E-02 | 5.47E-02 | 5.47E-02 |
| Avg | |||||||||
| Median | 1.52E-02 | 1.58E-02 | 0.00E+00 | 1.54E-10 | 3.18E-03 | 1.11E-02 | 3.20E-02 | 4.76E-02 | 4.77E-02 |
| Mean | 1.52E-02 | 1.58E-02 | 1.51E-14 | 2.69E-09 | 2.68E-03 | 9.18E-03 | 3.14E-02 | 4.77E-02 | 4.78E-02 |
| 2.50% | 1.17E-02 | 1.34E-02 | 0.00E+00 | 6.65E-15 | 1.14E-04 | 3.16E-03 | 1.60E-02 | 4.08E-02 | 4.09E-02 |
| 97.50% | 1.84E-02 | 1.82E-02 | 5.56E-14 | 2.17E-08 | 3.90E-03 | 1.38E-02 | 3.71E-02 | 5.47E-02 | 5.47E-02 |
Risk characterization
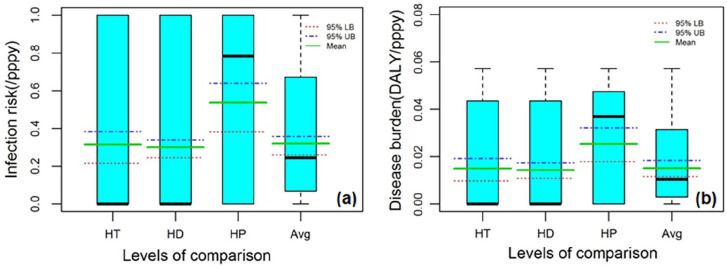
The study evaluated the probability of adverse health effects among Gaza’s population resulting from E. coli O157: H7 contamination, comparing findings with U.S. EPA and WHO benchmarks for infection risk and DALYs. The boxplot represents different percentiles of the estimated infection risk distribution across different drinking water supply sources (Figure 2).
Figure 2.
The box plots of estimated (a) infection risk (Inf; case pppy) and (b) burden of disease (DB; DALY pppy) for municipal network (HT), desalinated water (HD), private wells (HP) at households and drinking water supplies on average (Avg).
Figure 3 displays cumulative distribution plots illustrating the variability in estimated E. coli O157: H7 infection risk for each drinking water supply source at households in Gaza, as well as for the average of all three drinking water sources. The dark gray bands represent the 50% uncertainty range, while the light gray bands indicate the 95% uncertainty range on each quantile of variability. The Y-axis (proportion of population) corresponds to the X-axis (Yearly probability of infection), providing estimates of infection risk. For instance, for the average of all three drinking water sources at households (Av), the 20th percentile of variability for infection risk is approximately 0.03 pppy, with a credible interval between 0.00 and 0.07 pppy.
Figure 3.
Tornado chart in the variability and uncertainty dimension for median estimates of the spearman’s rank correlation between the input variables and burden of disease for (a) municipal network, (b) desalinated water, (c) private wells at households and (d) drinking water supplies on average.
Tornado diagram (Figure 4) illustrates the sensitivity of a mathematical model’s results to fluctuations in selected variables, showcasing how different sources of uncertainty in inputs impact the outputs of the system. The main factors affecting the variability of risks for the population and the estimated burden of disease include municipal water at households (HT), desalinated water at households (HD), private well water at households (HP), the average of all three drinking water sources at households (Av), and the 25% concentration of E. coli O157: H7 in water wells. Additionally, significant negative associations with LRV1-5 were observed.
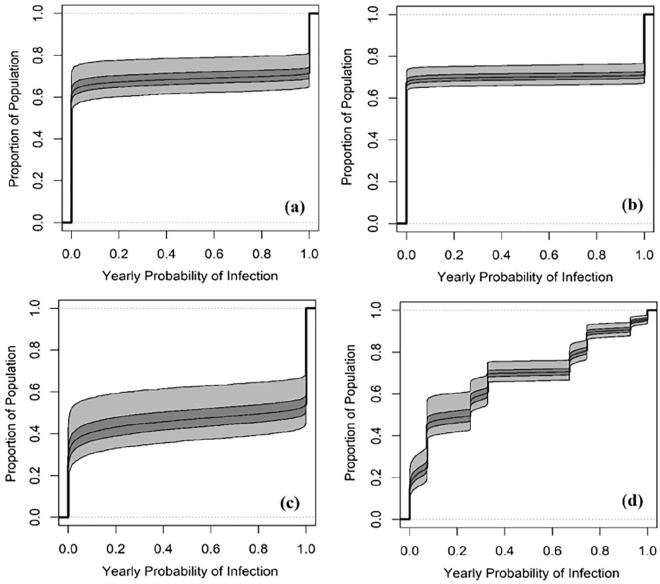
Figure 4.
Variability cumulative distribution plots of infection risk for (a) HT, (b) HD, (c) HP, and (d) average.
Discussion
Our analysis of 1317 water samples from various sources across the Gaza Strip revealed a notable presence of E. coli O157: H7, with 6.9% of samples testing positive. The mean, standard deviation, and maximum concentrations of E. coli O157: H7 were calculated at 1.97, 9.74, and 112 MPN/100 ml, respectively, indicating variability in contamination levels within Gaza’s water supply.
The selection of E. coli O157: H7 as the reference organism for our QMRA model was informed by its documented role in global waterborne disease outbreaks.38–40 Recommendations from international health organizations such as the WHO and CDC underscore the importance of monitoring and controlling this pathogen in drinking water systems.32,41 By leveraging extensive literature on E. coli O157: H7 epidemiology, our study contributes to understanding microbial risks and emphasizes the need for targeted interventions to improve drinking water quality.
Building upon previous research highlighting microbiological contamination in Gaza’s drinking water,42,43 our study quantitatively assessed E. coli O157: H7 exposure through water consumption. This approach aligns with methodologies used in similar investigations.44,45 and included calculating LRVs to evaluate attenuation of E. coli O157: H7 concentrations in Gaza’s water supply system.46,47
Employing the Beta-Poisson dose-response model integrated with Monte Carlo simulation techniques aligns with established methodologies for assessing infection risks from microbial contamination.29,48 The comparison of risk estimates with U.S. EPA and WHO benchmarks provides valuable insights into potential health impacts.49,50
The cumulative distribution plots for estimated E. coli O157: H7 infection risks demonstrate variability across different drinking water sources. Our analysis aligns with findings from comparable studies,29,48 highlighting the robustness and consistency of our results. The tornado diagram underscores key factors influencing population risks and disease burden estimates, supported by sensitivity analyses conducted in similar contexts.29,30
Despite providing critical insights, our study has limitations that warrant consideration. The cross-sectional design limits causal inference, emphasizing the need for longitudinal studies. Sampling strategies may introduce biases, and the absence of a formal power analysis affects result interpretation. Future research should address broader microbial contaminants and validate findings in diverse settings to enhance generalizability.
Conclusion
Our study enhances understanding of E. coli O157: H7 contamination risks in Gaza’s drinking water. By integrating quantitative assessments with real-world data and leveraging established methodologies, our findings contribute to evidence-based interventions aimed at improving water safety and safeguarding public health in similar settings globally.
Acknowledgments
We would like to acknowledge the abstract (pre-print) of our manuscript, available at https://doi.org/10.1016/S0140-6736(22)01139-4. 51 Additionally, we acknowledge the presentation of the content of this manuscript as a poster at The Lancet Palestinian Health Alliance (LPHA) 12th Annual Conference, as detailed in the referenced publication. This abstract provides an early glimpse into the findings and scope of our research. Furthermore, we extend our gratitude to the Palestinian Ministry of Health and Palestinian Water Authority for granting us permission to conduct this study in the Gaza Strip. We are also thankful to the desalination plants, tanker truck operators, and household owners for their cooperation and assistance in facilitating the implementation of this work.
Footnotes
Author contributions statement: SA and MH designed the study. SN supervised the work. SA collected the data and carried out the implementation. MH performed the data analysis and carried out the simulations. AHM and SHM wrote the manuscript with input from all authors. KZ and RN contributed to the design and implementation of the research, to the analysis of the results and to the writing of the manuscript. All authors reviewed the manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethics approval: The study protocol was approved by the Ethics Committee of Tehran University of Medical Sciences (Code: IR. TUMS.REC.1396.3917) and by the Helsinki Ethical Committee in the Gaza Strip (Code: PHRC/HC/288/17).
Informed consent: Not applicable.
Trial registration: Not applicable.
ORCID iD: Samer Abuzerr  https://orcid.org/0000-0001-8950-3293
https://orcid.org/0000-0001-8950-3293
References
- 1. WHO/UNICEF. Drinking-water. Geneva, Switzerland: World Health Organization, 2019. [Google Scholar]
- 2. Amr SSA, Yassin MM. Microbial contamination of the drinking water distribution system and its impact on human health in Khan Yunis Governorate, Gaza Strip: seven years of monitoring (2000–2006). Public Health 2008; 122(11): 1275–1283. [DOI] [PubMed] [Google Scholar]
- 3. Yassin MM, Amr SSA, Al-Najar HM. Assessment of microbiological water quality and its relation to human health in Gaza Governorate, Gaza Strip. Public Health 2006; 120(12): 1177–1187. [DOI] [PubMed] [Google Scholar]
- 4. Abu Mayla YS, Amr AS. Chemical and microbiological quality of drinking water in Gaza Strip, Palestine. Sci Vision 2010; 10: 80–88. [Google Scholar]
- 5. Assaf K. Water as a human right: the understanding of water in Palestine: the understanding of water in the Arab countries of the Middle East. Berlin, Germany: Heinrich Boll Stiftung Foundation, 2004, pp. 136–165. [Google Scholar]
- 6. Aish AM. Water quality evaluation of small scale desalination plants in the Gaza Strip, Palestine. Desalin Water Treat 2011; 29(1–3): 164–173. [Google Scholar]
- 7. Yassin MM, Amr SSA, Al-Najar HM. Assessment of microbiological water quality and its relation to human health in Gaza Governorate, Gaza Strip. Public Health 2006; 120(12): 1177–1187. [DOI] [PubMed] [Google Scholar]
- 8. MOH. Epidemiological bulletin. Ministry of Health, 2013; 3(1). http://www.moh.gov.ps [Google Scholar]
- 9. WHO. Guidelines for drinking-water quality. Geneva: World Health Organization, 2004. [Google Scholar]
- 10. Howard G, Pedley S, Tibatemwa S. Quantitative microbial risk assessment to estimate health risks attributable to water supply: can the technique be applied in developing countries with limited data? J Water Health 2006; 4(1): 49–65. [PubMed] [Google Scholar]
- 11. Haas CN, Rose JB, Gerba CP. Quantitative microbial risk assessment. Hoboken, NJ: John Wiley & Sons, 1999. [Google Scholar]
- 12. WHO. Quantitative microbial risk assessment: application for water safety management. Geneva, Switzerland: World Health Organization, 2016. [Google Scholar]
- 13. Havelaar A, Melse J. Quantifying public health risk in the WHO Guidelines for drinking-water quality: a burden of disease approach. RIVM report: 734301022. Bilthoven: RIVM, 2003. [Google Scholar]
- 14. CDC. Isolation of E. coli O157: H7 from sporadic cases of hemorrhagic colitis—United States. MMWR Morb Mortal Wkly Rep 1982; 46(30): 700. [PubMed] [Google Scholar]
- 15. Pedritis H, Kidder G, Ogram A. E. coli O157: H7, a potential healt concern. Gainesville SL: IFAS Extension University of Florida, 2002, pp. 1461–1464. [Google Scholar]
- 16. Matthews L, Low J, Gally D, et al. Heterogeneous shedding of Escherichia coli O157 in cattle and its implications for control. Proc Natl Acad Sci U S A 2006; 103(3): 547–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Betancourt WQ, Rose JB. Drinking water treatment processes for removal of Cryptosporidium and Giardia. Vet Parasitol 2004; 126(1–2): 219–234. [DOI] [PubMed] [Google Scholar]
- 18. Erickson MC, Liao JY, Payton AS, et al. Survival and internalization of Salmonella and Escherichia coli O157: H7 sprayed onto different cabbage cultivars during cultivation in growth chambers. J Sci Food Agri 2019; 99(7): 3530–3537. [DOI] [PubMed] [Google Scholar]
- 19. Pollari F, Christidis T, Pintar KD, et al. Evidence for the benefits of food chain interventions on E. coli O157: H7/NM prevalence in retail ground beef and human disease incidence: a success story. Can J Public Health 2017; 108(1): e71–e78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Atnafie B, Paulos D, Abera M, et al. Occurrence of Escherichia coli O157: H7 in cattle feces and contamination of carcass and various contact surfaces in abattoir and butcher shops of Hawassa, Ethiopia. BMC Microbiol 2017; 17(1): 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. PCBS. Statistic brief in the international population day. Ramallah, Palestine: Palestinian Central Bureau of Statistics, 2017, p. 2. [Google Scholar]
- 22. Al-Safady M, Al-Najar H. Disinfection process of water supply system in the Gaza strip between real practice and WHO limitations. Res J Environ Sci 2011; 5(5): 414. [Google Scholar]
- 23. Abuzerr S, Hadi M, Zinszer K, et al. Comprehensive risk assessment of health-related hazardous events in the drinking water supply system from source to tap in Gaza Strip, Palestine. J Environ Public Health 2020; 2020: 7194780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Eaton AD, Clesceri LS, Franson MAH, et al. Standard methods for the examination of water and wastewater. Washington, DC: American Public Health Association, 2005. [Google Scholar]
- 25. March SB, Ratnam S. Latex agglutination test for detection of Escherichia coli serotype O157. J Clin Microbiol 1989; 27(7): 1675–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kinzelman J, Singh A, Ng C, et al. Use of IDEXX Colilert-18 registered and Quanti-Tray/2000 as a rapid and simple enumeration method for the implementation of recreational water monitoring and notification programs. Lake Reserv Manag 2005; 21(1): 73–77. [Google Scholar]
- 27. Cassin MH, Paoli GM, Lammerding AM. Simulation modeling for microbial risk assessment. J Food Protect 1998; 61(11): 1560–1566. [DOI] [PubMed] [Google Scholar]
- 28. Custodio M, Peñaloza R, Ochoa S, et al. Microbial and potentially toxic elements risk assessment in high Andean river water based on Monte Carlo simulation, Peru. Scient Rep 2023; 13(1): 21473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Smith JL, Fratamico PM, Gunther NW. Shiga toxin-producing Escherichia coli. Adv Appl Microbiol 2014; 86: 145–197. [DOI] [PubMed] [Google Scholar]
- 30. Jones CH, Wylie V, Ford H, et al. A robust scenario analysis approach to water recycling quantitative microbial risk assessment. J Appl Microbiol 2023; 134(3): lxad029. [DOI] [PubMed] [Google Scholar]
- 31. Brown CA, Harmon BG, Zhao T, et al. Experimental Escherichia coli O157: H7 carriage in calves. Appl Environ Microbiol 1997; 63(1): 27–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. García A, Fox JG, Besser TE. Zoonotic enterohemorrhagic Escherichia coli: a one health perspective. ILAR J 2010; 51(3): 221–232. [DOI] [PubMed] [Google Scholar]
- 33. Abuzerr S, Nasseri S, Yunesian M, et al. Household drinking water safety among the population of Gaza Strip, Palestine: knowledge, attitudes, practices, and satisfaction. J Water Sanit Hygiene Develop 2019; 9(3): 500–512. [Google Scholar]
- 34. Cornick NA, Helgerson A. Transmission and infectious dose of Escherichia coli O157: H7 in swine. Appl Environ Microbiol 2004; 70(9): 5331–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Seidu R, Abubakari A, Dennis IA, et al. A probabilistic assessment of the contribution of wastewater-irrigated lettuce to Escherichia coli O157: H7 infection risk and disease burden in Kumasi, Ghana. J Water Health 2015; 13(1): 217–229. [DOI] [PubMed] [Google Scholar]
- 36. Strachan NJ, Doyle MP, Kasuga F, et al. Dose response modelling of Escherichia coli O157 incorporating data from foodborne and environmental outbreaks. Int J Food Microbiol 2005; 103(1): 35–47. [DOI] [PubMed] [Google Scholar]
- 37. Teunis P, Ogden I, Strachan N. Hierarchical dose response of E. coli O157: H7 from human outbreaks incorporating heterogeneity in exposure. Epidemiol Infect 2008; 136(6): 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Saxena T, Kaushik P, Mohan MK. Prevalence of E. coli O157: H7 in water sources: an overview on associated diseases, outbreaks and detection methods. Diagn Microbiol Infect Dis 2015; 82(3): 249–264. [DOI] [PubMed] [Google Scholar]
- 39. Müller EE, Ehlers MM, Grabow WO. The occurrence of E. coli O157: H7 in South African water sources intended for direct and indirect human consumption. Water Res 2001; 35(13): 3085–3088. [DOI] [PubMed] [Google Scholar]
- 40. Olsen SJ, Miller G, Breuer T, et al. A waterborne outbreak of Escherichia coli O157: H7 infections and hemolytic uremic syndrome: implications for rural water systems. Emerg Infect Dis 2002; 8(4): 370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patel J, Sharma M, Ravishakar S. Effect of curli expression and hydrophobicity of Escherichia coli O157: H7 on attachment to fresh produce surfaces. J Appl Microbiol 2011; 110(3): 737–745. [DOI] [PubMed] [Google Scholar]
- 42. Sharif FA, Arafa HZ. Occurrence of Escherichia coli O157 in Gaza Strip: a preliminary study. IUG J Nat Stud 2015; 12(1): 81–90. [Google Scholar]
- 43. Abuzerr S, Nasseri S, Yunesian M, et al. Microbiological quality of drinking water and prevalence of waterborne diseases in the Gaza strip, Palestine: a narrative review. J Geosci Environ Protect 2019; 7(04): 122. [Google Scholar]
- 44. Ahmed W, Neller R, Katouli M. Evidence of septic system failure determined by a bacterial biochemical fingerprinting method. J Appl Microbiol 2005; 98(4): 910–920. [DOI] [PubMed] [Google Scholar]
- 45. Mahmoud NE, Altayb HN, Gurashi RM. Detection of carbapenem-resistant genes in Escherichia coli isolated from drinking water in Khartoum, Sudan. J Environ Public Health 2020; 2020: 2571293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hassan FM, Mahmood AR. Evaluate the efficiency of drinking water treatment plants in Baghdad City–Iraq. J Appl Environ Microbiol 2018; 6(1): 1–9. [Google Scholar]
- 47. Saleh TA. Protocols for synthesis of nanomaterials, polymers, and green materials as adsorbents for water treatment technologies. Environ Technol Innov 2021; 24: 101821. [Google Scholar]
- 48. Johnson PT, Paull SH. The ecology and emergence of diseases in fresh waters. Freshwater Biol 2011; 56(4): 638–657. [Google Scholar]
- 49. EPA US. Occurrence and exposure assessment for the final Long Term 2 Enhanced Surface Water Treatment Rule. United States Environmental Protection Agency. 2005; EPA 815-R-06-002. Washington, DC: Office of Water. [Google Scholar]
- 50. WHO. Guidelines for drinking-water quality: incorporating the first and second addenda. Geneva: World Health Organization, 2008. [PubMed] [Google Scholar]
- 51. Abuzerr S, Hadi M, Zinszer K, et al. Quantitative microbial risk assessment to estimate annual infection risk and disease burden attributable to Escherichia coli O157: H7 in drinking water in the Gaza Strip: a prospective study. Lancet 2022; 399: S4. [Google Scholar]