ABSTRACT
Europe has suffered unprecedented epizootics of high pathogenicity avian influenza (HPAI) clade 2.3.4.4b H5N1 since Autumn 2021. As well as impacting upon commercial and wild avian species, the virus has also infected mammalian species more than ever observed previously. Mammalian species involved in spill over events have primarily been scavenging terrestrial carnivores and farmed mammalian species although marine mammals have also been affected. Alongside reports of detections of mammalian species found dead through different surveillance schemes, several mass mortality events have been reported in farmed and wild animals. In November 2022, an unusual mortality event was reported in captive bush dogs (Speothos venaticus) with clade 2.3.4.4b H5N1 HPAIV of avian origin being the causative agent. The event involved an enclosure of 15 bush dogs, 10 of which succumbed during a nine-day period with some dogs exhibiting neurological disease. Ingestion of infected meat is proposed as the most likely infection route.
KEYWORDS: Avian influenza, systemic infection, bush dogs, conservation species, terrestrial carnivores, H5N1, high pathogenicity, zoonotic assessment
Introduction
High pathogenicity avian influenza virus (HPAIV) has caused significant mortalities across avian species since the emergence of H5Nx clade 2.3.4.4 viruses in Europe and the United Kingdom (UK) in 2014 [1]. In recent years, these viruses have caused repeated detections of H5Nx HPAIVs in poultry and wild birds. In Autumn 2021 the situation escalated significantly with clade 2.3.4.4b HPAIV H5N1 emerging to cause global outbreaks [1]. Between 2014 and 2020, spillover of avian influenza infection into mammals was uncommon although occasional reports have been described [1,2]. With the escalation of H5N1 outbreaks during 2020/21, through the summer of 2021 and into 2022/23, infection pressure in the environment, as a result of high mortality levels in wild birds led to an increase in the occurrence of spill over events globally [3,4]. To date, almost all detections in mammals have involved wild scavenging species that have most likely become infected following the ingestion of infected wild bird carcasses in the environment [5]. Importantly, with the vast majority of cases the infection was reported in a single mammal following an undefined exposure to infectious material. However, on occasion, mammalian infection with avian origin H5N1 HPAIV has been implicated as the cause of mass mortalities. This has occurred most notably in natural events whereby wild animals have been affected (e.g. seals in the United States and sea lions in Peru [3,4,6–8]) as well as in farmed species (farmed mink in Spain [9], farmed foxes in Finland [10]) and domesticated species (cats in Poland [11] and South Korea [12]). These events have triggered interest in the potential for mammal-to-mammal transmission and in any resultant genetic adaptation [13]. Whilst these outbreaks have shown concurrent infection of multiple individuals with the same virus, data supporting active transmission between mammals is lacking and a definitive conclusion on the ability of these viruses to spread directly from mammal to mammal has not been reached. On top of mortality events within the veterinary sector, this clade 2.3.4.4b H5N1 HPAIV has also been associated with human infection, with outcomes varying from asymptomatic to severe with hospitalization of infected individuals [14]. These features have raised the zoonotic profile of the currently circulating H5N1 clade 2.3.4.4b HPAIV, although again human-to-human transmission has not been demonstrated.
Here we report on the infection and severe mortality within a pack of bush dogs (Speothos venaticus) in captivity with avian origin H5N1 clade 2.3.4.4b HPAIV. Bush dogs are a near-threatened species of wild canids that are of conservation concern. Wild populations of these dogs range from northern regions of Panama (Central America) to northeastern Argentina and Paraguay; with populations also being present in Colombia, Venezuela, the Guianas, Brazil, and eastern Bolivia and Peru. This species is characterized by its gregarious and diurnal behaviour [15]. In this disease event which occurred in November 2022, two-thirds of the pack of bush dogs, held captive in a wildlife collection in the UK, became clinically unwell with a disease that had a short duration and led to death and/or the need for euthanasia on welfare grounds. Avian influenza was not suspected at first and several tests and analyses were undertaken to ascertain cause of death and to exclude the involvement of common canine pathogens. Inconclusive results led bush dog samples being assessed by shotgun metagenomics, which detected the presence of influenza type A virus sequences in internal organs. We describe the disease event, timeline, virological and pathological impact of the disease and sequence analysis of the causative agent.
Materials and methods
Clinical investigation, post-mortem examination, tissue sampling and histopathological analysis
Clinical records including blood chemistry data and mortality were recorded daily by the safari park personnel. All found dead of clinically euthanased bush dogs underwent full post-mortem examination (PME). Of the 10 animals, 6 were euthanised using intracardiac pentobarbitone (0.03 mg/kg, 1 mg/ml) under anaesthesia (medetomidine (0.03 mg/kg, 1 mg/ml) and ketamine (4.03–4.85 mg/kg, 100 mg/ml)) and 4 were found dead within the enclosure. PME included gross examination, and tissues selection. Tissue samples were submitted for bacteriological analysis to a private laboratory, frozen tissues were stored at −18°C or in 10% formalin for histology (Supplementary Table 1). Both haematoxylin and eosin (H&E) and immunohistochemistry (IHC) were undertaken as described previously [30] with antigen staining being measured semi-quantitatively as described previously [16]. The specificity of immunolabelling was assessed in positive control sections by replacing the primary antibody with a matching mouse IgG isotype; no non-specific cross-linking was observed. Frozen tissues for virological analysis (Supplementary Table 2) were prepared as a 10% (w/v) suspension in Leibovitz’s L-15 medium and incubated at room temperature for 60 min before using standard RNA extraction protocols [17].
Environmental and epidemiological investigation
Environmental samples were collected from 11 locations within or surrounding the bush dog enclosure (Supplementary Table 3a). Different matrices were investigated including silt, foam and faeces that were processed as described previously [18] and water samples that were tested without dilution.
Due to the retrospective nature of this case, samples from other animals which died following bush dog mortalities, were tested (Supplementary Table 3b). Swabs samples and brain samples were cut into 1 mL serum-free Leibovitz’s L-15 medium containing antibiotics (penicillin, streptomycin), incubated at room temperature for 10 min before standard viral RNA (vRNA) extraction. Brain tissues were prepared as a 10% (w/v) suspension in L-15 medium and incubated at room temperature for 60 min before using standard RNA extraction protocols [17].
Following a positive diagnosis of HPAIV infection in dogs that developed clinical disease, samples from the five surviving dogs were collected in March 2023 (Table 3c), including oral and rectal swabs and blood samples for molecular and serological analysis. Sera were aspirated from clotted blood samples and heat-treated at 56°C for 30 min. All serum samples were assessed for H5-specific antibodies using established methods [19].
Virological investigation
RNA extraction and molecular analysis
RNA was extracted samples using either TRIzol (Invitrogen) or the MagMAX CORE Nucleic Acid Purification Kit (ThermoFisher Scientific) as previously described [17]. Extracted RNA was assessed for vRNA using an M gene-specific [20], an H5 HPAIV specific [21] and/or NA specific [22,23] detection RRT-PCR. RRT-PCR Cq values ≤ 36.00 were considered as AIV positive. Samples with Cq >36 were considered negative. A standard curve was generated using a 10-fold dilution series of titrated H5N1-21/22 HPAIV RNA as previously described [17].
Virus isolation and propagation
For each sample, 100 µl of material was added to 100 µl of phosphate-buffered saline (PBS) and inoculated into the allantoic cavity of specific pathogen-free (SPF) embryonated fowls’ eggs (EFE), as described previously [17]. At 2 days post inoculation (dpi), the allantoic fluid of one EFE was tested for the presence of a haemagglutinating agent as previously described [17]. If no haemagglutinating-activity was observed, allantoic fluid from the other egg was tested at 6 dpi. Haemagglutinating activity >1/4 at was considered positive for virus isolation. Conversely, HA activity <1/4 was considered negative for virus isolation.
Genomic analysis
The extracted RNA was converted to double-stranded cDNA and sequenced using either an Illumina MiSeq or NextSeq 550 [2]. Analysis was performed with SeqMan NGen Software 17.5 (DNASTAR, USA) and Reference-guided application using GenBank virus reference sequences with a genome assembly pipeline as described previously [2]. All influenza sequences generated (Supplementary table 2 and 3) and used in this study are available through the GISAID EpiFlu Database (https://www.gisaid.org). Comparison of the study-derived sequences and contemporary H5N1 sequences was undertaken against all avian H5N1 clade 2.3.4.4b sequences available on GISAID between 01-01-2020 and 17-07-23. All sequences were aligned on a per segment basis using MAFFT v7.520 and alignments were assessed using AliView version 1.26 with partial sequences being removed. Trimmed sequences were used to create a concatenated alignment using SeqKit and then used to a infer maximum-likelihood phylogenetic tree using IQ-Tree version 2.2.3. This resultant phylogenetic tree contained over 2,000 sequences and was therefore sub-sampled to cover 98% of the diversity within using PARNAS, which reduce the dataset down approximately 300 sequences whilst still containing representatives of the predominant UK genotypes. The sub-sampled dataset was then used to infer maximum-likelihood phylogenies for each gene segment using IQ-Tree along with ModelFinder and 1,000 ultrafast bootstraps. Once the genotype of the Bush dogs and associated sequences was determined, all sequences of the relevant genotype were retrieved from the original dataset (prior to sub-sampling) and used to generate time-resolved phylogenies using TreeTime. Phylogenetic trees were visualized as described previously [2]. Nucleotide identity between sequences was determined as described previously [24].
Results
Clinical setting
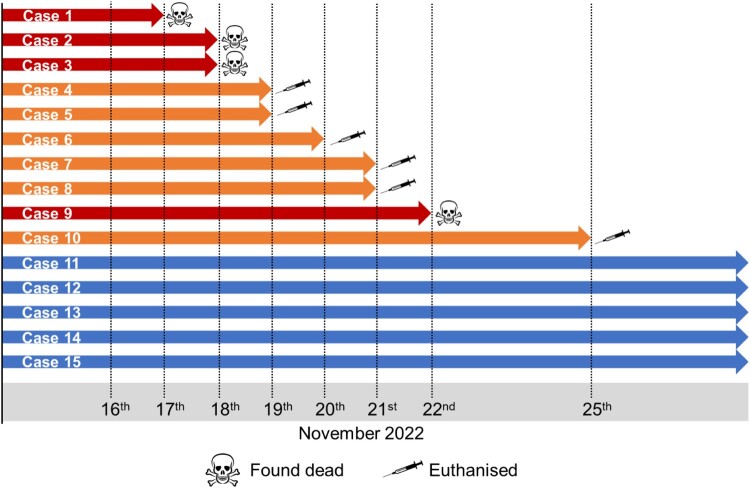
The bush dogs had been clinically healthy in the preceding 3 years and had been being fed a diet that included frozen shot wild birds and game. A timeline of disease status is presented in Figure 1. Case 1, Day 1, male, 9-year-old bush dog was found dead adjacent to the entrance of the nest box with a CCTV review showing hindlimb ataxia, forelimb hypermetria, depression and polyuria. Case 2, Day 1, male, 3-year-old was moribund and assessed under anaesthesia with uraemia, raised creatinine, hyperphosphataemia, total bilirubinaemia, and a raised alanine aminotransferase was noted, and was found dead the following morning, Day 2. Case 3, Day 2, 4-year-old female was found dead in the main pond within the exhibit. Case 4, Day 3, a 5-year-old female was euthanized due to severe ataxia combined with depressed responsiveness and polyuria. Case 5, Day 3, a 4-year-old male was euthanized due to severe ataxia combined with depressed responsiveness and polyuria. Case 6, Day 4, a 9-year-old female presented with severe hind-limb ataxia, weakness, reduced activity and polyuria, and was euthanized. Case 7, Day 5, a 4-year-old female appeared clinically healthy during the morning assessment but within 2 h was moribund and unresponsive and was euthanized on welfare grounds. Case 8, Day 5, a 3-year-old male was clinically normal at the morning assessment but within 2 h had developed severe ataxia and generalized weakness, vocalizing and was euthanized on welfare grounds. Case 9, Day 6, a 1-year-old female was found dead during the morning check. Case 10, Day 9, a 4-year-old male presented with severe hind limb weakness, knuckling, and ataxia, and was euthanized on welfare grounds. The remaining five animals, two males and three females, age range 18 months to 4 years were clinically normal during the same period and remained unaffected from the event.
Figure 1.
Timeline of mortality event within the bush dog enclosure
Pathological investigation and preliminary collateral analysis were performed at private laboratories. On gross examination, the primary findings were diffuse mild to moderate hepatomegaly (8/10), with multifocal to diffuse pale tan discolouration within liver lobes (7/10) and mild to moderate ascites (7/10). Intracardiac euthanasia made gross assessment of the lungs challenging (6/10), but mild to moderate multifocal pulmonary haemorrhage was observed in three of the remaining four cases. Less common gross findings included bilateral adrenomegaly (6/10), diffuse splenomegaly (3/10), and segmental small intestinal congestion and haemorrhage (4/10).
Vascular changes were observed microscopically, ranging from occasional subacute multifocal moderate to marked fibrinoid necrosis of small arterioles without associated inflammation, to acute, multifocal, mild to severe necrotizing and sometimes leukocytoclastic phlebitis of vessel walls, with fibrin thrombi, and necrosis of adjacent structures. In the liver, random mild to moderate acute multifocal necrosis (Figure 2(a)) and inflammation affecting both the parenchyma, and blood vessels and bile ducts within portal triads, was seen in all 10 cases. Foci of hepatocellular necrosis were rarely surrounded by minimal neutrophilic and histiocytic inflammation.
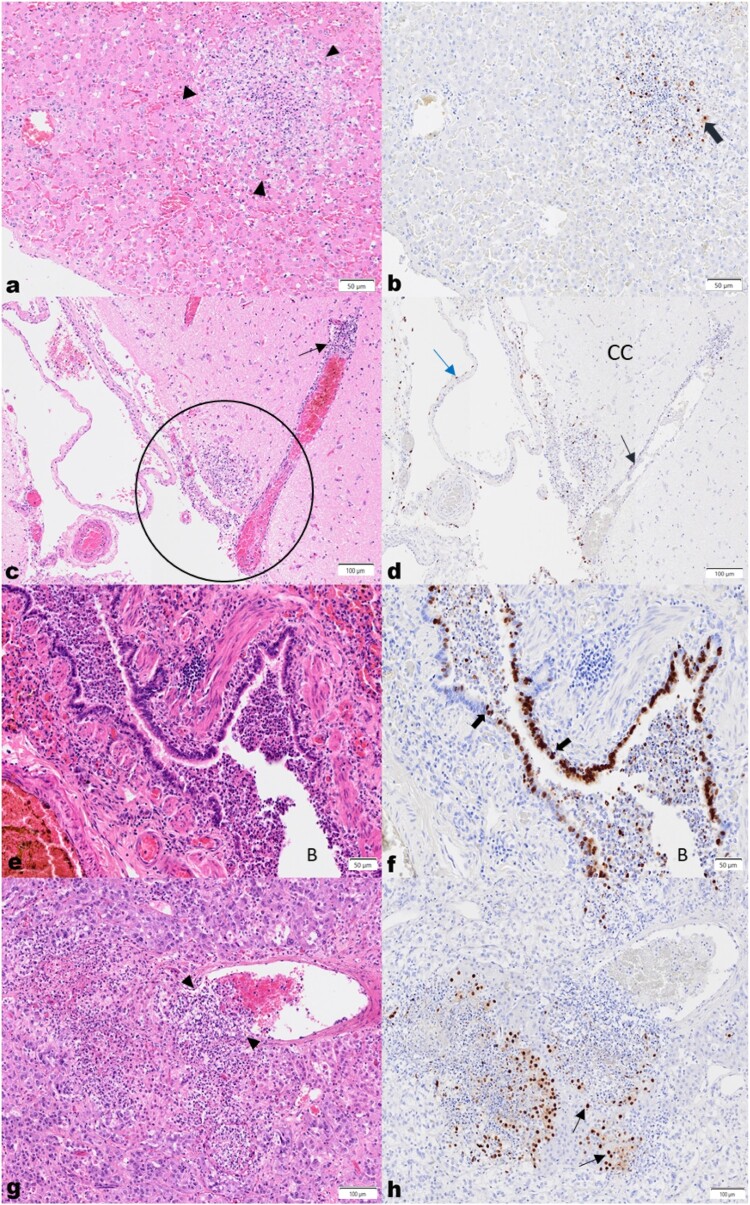
Figure 2.
High pathogenicity avian influenza virus H5N1 in bush dogs. (a) Liver, H&E. Acute, random focal hepatocellular necrosis, comprised of karyorrhectic debris and fibrin exudation with rare neutrophils and histiocytes. (b) IHC shows viral antigen in hepatocytes (thick brown arrow) on the periphery of the lesion, and in cellular debris. (c) Brain, cerebral cortex (CC), H&E. Vasculitis with segmental multifocal infiltration of a venule wall with neutrophils and histiocytes, is shown (thin black arrows). Multifocal to locally extensive necrosis and inflammation extends from the meninges into the adjacent neuropil (black circle). (d) IHC demonstrates antigen in vascular endothelial (black arrow) and arachnoid epithelial cells (blue arrow). (e) Lung, H&E. The bronchiolar lumen (B) is filled with degenerate and viable neutrophils, macrophages, eosinophilic material and shed epithelial cells, and the lamina propria and submucosa of the bronchiolar wall is diffusely mildly infiltrated by lymphocytes, histiocytes and fewer neutrophils. Adjacent alveolar spaces are collapsed and alveolar septae are congested and mildly expanded by neutrophils and macrophages. (f) IHC labelling is present in bronchiolar epithelial cells (black arrows) and within shed epithelial cells and macrophages in the lumen. (g) Adrenal gland, H&E. A subacute focal severe necrotizing vasculitis is seen with expansion of the vascular wall with neutrophils, pyknotic cellular debris and fewer mononuclear cells (black arrowheads), with subacute multifocal severe necrosis and inflammation of the adjacent epithelial cells in the acini of the adrenal medulla and cortex. (h) IHC highlights viral antigen in the acinar epithelial cells (black arrows).
Variable changes in the lungs were present in six bush dogs. The bronchioles and adjacent alveoli were predominantly involved in four cases where the bronchiolar lumina were filled with degenerate neutrophils and epithelial cells with attenuation and necrosis of bronchiolar epithelium. (Figure 2(e)). Mild neutrophilic, lymphocytic and histiocytic infiltration of the lamina propria and submucosa was also seen. Adjacent to the affected airways, alveoli were variably filled with fibrin, oedema, alveolar macrophages, neutrophils and erythrocytes. Occasionally mild to moderate expansion of the alveolar septae with macrophages, lymphocytes and neutrophils was observed alongside vascular pathology.
The brain was collected in four cases, and in all, the leptomeninges were expanded by mild to marked multifocal subacute inflammation. In three cases (7, 9 and 10), the infiltrate was composed mainly of viable and degenerate neutrophils, with rarefication and inflammation of the adjacent neuropil (Figure 2(c)). In the fourth case (6), the infiltrate consisted predominantly of lymphocytes, macrophages, and plasma cells with fewer neutrophils, and locally extensive involvement of the choroid plexus on the section examined. In all cases, randomly within the cerebral cortex and brainstem, and rarely in the cerebellum, few to multiple foci of mild to moderate necrosis and inflammation consisting mainly of aggregates of macrophages and glial cells, necrotic debris, and rare neuronal necrosis, were seen. Grey matter and white matter were both affected. In the neuropil, vascular changes were mild and characterized by an acute multifocal vasculitis with infrequent mild histiocytic perivascular cuffing. However, in the leptomeninges, vascular lesions ranged from a mild to severe, and acute to subacute multifocal necrotizing vasculitis (Figure 2(c)).
In three cases, multifocal marked acute cortical and medullary epithelial necrosis with neutrophilic infiltration, congestion, and focal subacute severe leukocytoclastic vasculitis (Figure 2(g,h)) was observed in the adrenal glands.
In lymph nodes examined from three bush dogs, findings varied from moderate multifocal lymphadenitis with predominantly subcapsular sinus neutrophilia and histiocytosis, and multifocal necrosis, to severe, diffuse acute lymphadenitis. Severe changes were characterized by expansion of the sinuses with viable and degenerate neutrophils, histiocytes and fewer erythrocytes, and multifocal necrosis of the cortex and paracortex with infiltration of large numbers of degenerate neutrophils. Within adjacent small blood vessels, the walls were multifocally infiltrated by degenerate neutrophils and karyorrhectic debris, and in the lumina, multifocal thrombi comprised of fibrin, macrophages and neutrophils were present.
The spleen was minimally affected, but in all four reported cases of mild acute splenitis, multifocal clusters of viable and degenerate neutrophils and occasionally karyorrhectic debris and histiocytes, were seen in the white pulp. Haematopoietic precursors (extramedullary haematopoiesis) and haemosiderin-laden macrophages were scattered within the red pulp. No vascular changes were apparent.
In the intestinal sections examined, inflammation was mostly mild, multifocal, diffuse and chronic. However, in the small intestine, evidence of mild to moderate active inflammation and necrosis was noted sporadically, with crypt abscessation, and foci of necrosis and monocytic inflammation within and between the inner circular and outer longitudinal smooth muscle layers, often associated with capillaries and in the submucosal and myenteric plexuses. In one case, acute, moderate multifocal necrotizing typhlitis and colitis were identified, with lymphoid depletion and mild acute multifocal vasculitis rarely present in the small vessels of the submucosa.
Where the eye was sampled, significant widespread and occasionally severe inflammation was found centred around vessels, with resultant anterior uveitis, optic neuritis and infrequently, retinitis. Multifocal necrotizing neutrophilic inflammation of the third eyelid gland (nictitans gland) was captured in one section.
Interestingly, the heart (three cases) and pancreas (two cases) appeared mildly affected in the sections examined, displaying mild multifocal acute degeneration and necrosis with minimal associated inflammation. Incidental mild, chronic, multifocal interstitial inflammation was reported in the kidneys but a mild to moderate multifocal acute vasculitis was present in some sections. On the serosal surface of many abdominal organs, particularly the liver, a mild to moderate acute multifocal to diffuse fibrinous peritonitis was seen. A detailed summary of the gross and histopathology reports provided by the park owner and the local specialist laboratory is provided in the supplemental material (Supplementary Table 1).
The changes described were suggestive of a severe acute systemic infection, which was initially thought to be bacterial, but a viral or parasitic aetiology could not be ruled out. Collateral analysis was carried out to exclude common causes of death. Secondary or contaminant bacteria were isolated from affected organs, such as Escherichia coli, Staphyloccus aureus, Clostridium perfringes and Clostridium sordelli, Haemophilus parainfluenzae and Proteus spp. Molecular tests (PCRs) performed at a private laboratory excluded the presence of SARS-CoV-2, Leptospirosis, RHDV type 1 and Canine Adenovirus. Toxicology analysis for Ethylene glycol were negative. Despite this further ancillary testing, no aetiologic agent was identified and preliminary clinical investigation was inconclusive.
Immunohistochemistry
Immunohistochemistry was performed using an anti-influenza A antibody targeting viral nucleoprotein, on formalin-fixed paraffin embedded tissues previously submitted for routine histopathologic analysis (Supplementary Table 1). Generally, infrequent viral antigen was detected in macrophages and in cellular debris within and on the periphery of necrosis and inflammation in the many affected organs. Angiocentric necrosis and inflammation were sometimes accompanied by sporadic labelling of the vessel walls, particularly the endothelium and tunica media.
More specifically, within organs, occasional viral antigen was present in hepatocytes (Figure 2(b)), and in bile duct epithelium in the liver. In the brain, viral antigen was abundant in ependymal cells lining the lateral and third ventricles, and less frequently, multifocally in neurons, axons, and glial cells in the neuropil. Labelling was also present in the meningeal epithelial cells in the subarachnoid space (Figure 2(d)). Moderate sporadic labelling was seen in bronchiolar epithelium (Figure 2(f)) but was minimally visible in the alveolar septae.
Significant immunolabelling was observed in acinar cells of the adrenal cortex (zona fasciculata and reticularis), and less commonly in cells of the adrenal medulla (Figure 2(h)). In the eye, viral antigen was detected in macrophages around vessels and within necrotic debris, particularly in the iris, uvea and meninges of the optic nerve. Moderate viral antigen was found in the acinar cells of the nictitans gland of the eye. Strong labelling was seen in the nuclei of smooth muscle cells of one or both the smooth muscle layers of the small and large intestines, but infrequently within macrophages, in the vascular endothelium of the capillaries, and in ganglion cells of the plexuses between them. Rarely, viral antigen was visible in mucosal epithelial cells of the stomach, and enterocytes in the small and large intestines. Occasional viral antigen was present in the serosal epithelium on the surfaces of the intestines, abdominal organs, and urinary bladder. Minimal antigen was rarely seen in cardiomyocytes.
Virological assessment
vRNA was detected across a broad range of tissues sampled (Supplementary Table 2) in all bush dog cases. Due to the retrospective nature of the detection, and the working practices of the team on location during the disease event, a significant number of tissues were collected at PME and these were assessed following retrospective confirmation of influenza A virus of avian origin as the causative agent. All samples tested positive across the suite of molecular tests used to detect the influenza A: M-gene, the high pathogenicity H5 haemagglutinin (H5 HP) and the N1 neuraminidase gene. Exceptions to this were oral and rectal swabs from Case 6 and the intestine from Case 10, all of which tested negative across all three molecular assays. Of significant interest were the positive results obtained from a urine sample that was aspirated directly from the bladder during the PME of Case 5. Virus isolation and propagation were successful, resulting in the harvesting of one viral isolate per dog.
Environmental and epidemiological investigation samples
During the mortality events, staff had also collected a suite of environmental samples (Supplementary Table 3) from their enclosure which could be assessed for evidence of environmental contamination. An additional sample set included samples from animals at the park that were considered at risk from the agent, and which died or were euthanised during or shortly after the canid mortality event. Interestingly, from all these samples, only one sample of water taken from a drinking trough within the bush dog enclosure tested positive for vRNA although an isolate was not successfully recovered from the sample (Supplementary Table 3). Samples from the five surviving dogs, collected approximately four months after the disease event, were also negative at RRT-PCR. However, mild seroconversion was detected in 2 out of 5 dogs following the HI test, demonstrating exposure to virus or viral antigen.
Genomic analyses
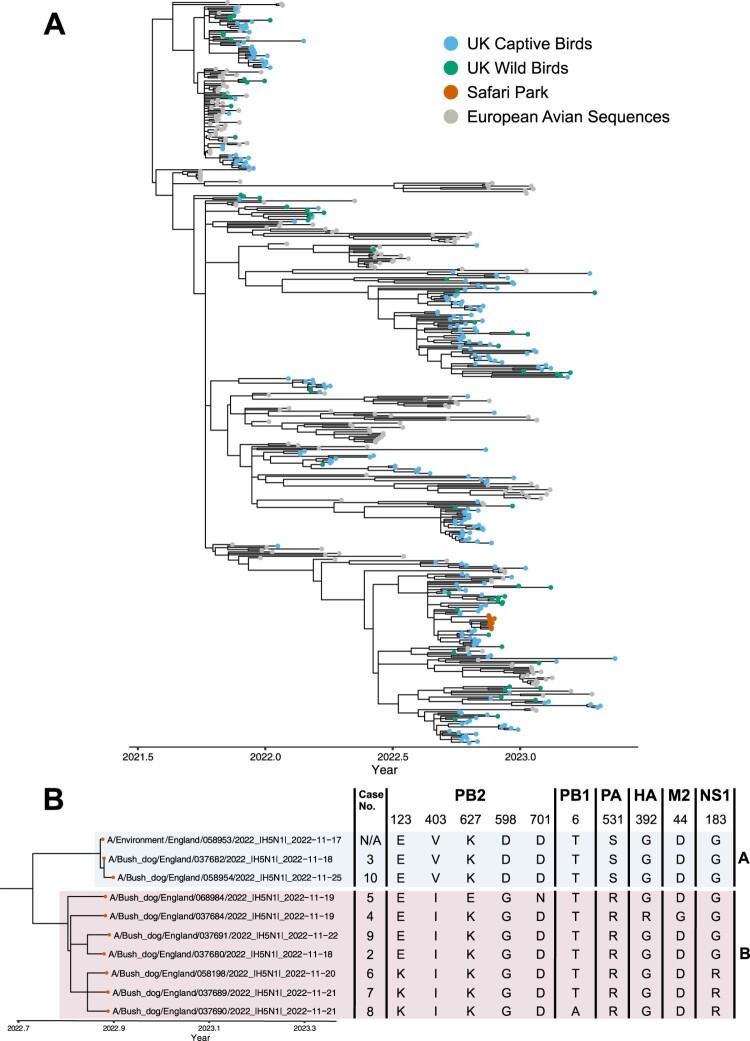
Initially, samples across Cases 6, 8 and 9 were subjected to metagenomic analyses; this included liver, kidney and brain samples from Case 6 and three tissue pools of liver, kidney and spleen from Cases 8 and 9. The analysis revealed sequence reads matching those of influenza A virus. This initial metagenomic findings in combination with the RRT-PCR results prompted further investigation of the samples obtained from the Bush dogs and WGS was attempted on tissue samples or viral isolates from the deceased animals and the positive environmental sample, resulting in 11 full H5N1 HPAIV genomes being sequenced. The 11 genomes demonstrated 99.4-100% nucleotide identity to each other across all eight influenza virus gene segments. Initial phylogenetic analysis revealed that the genomes obtained from the bush dog samples clustered with those of the UK AIV09 genotype [2] (also known as the AB genotype according to the EU Reference Laboratory schema), which was the predominant genotype in the UK from October until May 2023. To further investigate potential incursion routes for H5N1 into the captive Bush dog population, 10 of the 11 genomes from the safari park were concentrated and combined with a more fulsome dataset containing only genomes of the AIV09 (AB) genotype which were then used to infer time-resolved phylogenies. The omitted sequence (A/Bush dog/England/037675/2022) contained a number of gaps and was removed from these subsequent analyses. From these analyses, it was demonstrated that the bush dog genomes showed high similarity with AIV09 (AB) H5N1 HPAIV sequences from the UK, but formed a distinct group (Figure 3(a)), with a time to the most common recent ancestor (tMRCA) estimate of 24th September 2022 (range: 9th September 2022 to 13th October 2022). Within this group, the 10 bush dog sequences were divided into two separate sub-groups. Sub-group A contained the environmental sequence generated from the water trough in the Bush dog enclosure (A/Environment/England/058953/2022), along with two sequences from Bush dogs (A/Bush dog/England/037682/2022 (Case 3) and A/Bush dog/058954/2022 (Case 10)) and were generated from samples collected from the 17th of November through to the 25th of November, covering the entire sampling period. Sub-group B contained the remaining seven Bush dog sequences, generated from samples collected between the 18th of November and 22nd November. When looking at the amino acid changes across the bush dog sequences, 10 variable sites were observed across: polymerase basic protein-1 (PB1) and -2 (PB2), polymerase acidic protein (PA), HA, matrix protein 2 (M2) and non-structural protein-1 (NS1) (Figure 3(b)). The PB2 protein contained the most variant sites (five), which included changes at positions 627 and 701 which have both been reported to be involved in the adaptation of AIVs to mammalian hosts [13]. Interestingly, whilst nine of the 10 sequences, including the environmental sample, possessed a lysine (K) at position 627, one sequence (A/Bush dog/England/068984/2022 (Case 5)) did not. However, this sequence did contain an asparagine (N) at position 701, whereas the other sequences possessed an aspartic acid (D). When comparing the phylogenetic and amino acid variability, it was found that sub-groups A and B were distinguished by three variable sites in the PB2 (position 403 and 598) and PA (position 531) proteins. Within sub-group B, there were also three sequences (A/Bush dog/England/058198/2022, A/Bush dog/England/037689/2022 and A/Bush dog/England/037690/2022) that showed different amino acid substitution from the other sequences in sub-group B (PB2 position 123 and NS1 position 183), however, there were also other changes seen within this sub-group at different positions.
Figure 3.
(a) Time-resolved phylogenetic analysis of H5N1 HPAIV genotype AIV09 (AB) concatenated from genomes. Sequences from UK captive and wild birds, as well as the sequences obtained from the bush dogs are coloured accordingly. (b) A subset of the phylogenetic tree shown in A focussing on the bush dog sequences, case number and the amino acid changes therein. Sub-groups A and B described in the text are highlighted.
Discussion
The detection of influenza A virus of avian origin H5N1 as the causative agent of mortality in bush dogs was an unusual and unexpected event. Sequence reads for influenza A virus led to the described diagnostic pathway that conclusively defined clade 2.3.4.4b H5N1 influenza A virus of avian origin as the cause of disease. The internal distribution of the virus demonstrated systemic infection affecting multiple organs and clearly demonstrated that HPAI was the primary cause of death. Viral excretion through the respiratory and intestinal tract could be speculated although swabs (oral and rectal) collected from Case 6 did not detect the virus. The detection of vRNA from the urine sample for Case 5 may indicate possible horizontal transmission routes although there is no further evidence to substantiate this hypothesis.
Histopathologic findings demonstrated a severe acute systemic disease characterized by vasculitis, and widespread necrosis and inflammation in many organs, specifically the liver, brain, lung, and adrenal glands. Immunohistochemistry confirmed the presence of viral antigen most often seen in the brain, lung, adrenal glands, lymph nodes and liver, but also within vascular walls. The brain and lung are common targets in spill over mammalian HPAI H5N1 infections [4,25–27], with the virus potentially gaining entry following inhalation into the respiratory tract. Other routes, including vascular infection following ingestion of infectious material and intestinal entry have been described in H5N1 infection of cats, with subsequent viraemia [28]. In the current study, both lung and intestinal lesions were seen supporting a multifactorial pathogenesis. Dissemination of the virus to the brain with penetration of the blood–brain-barrier into the cerebrospinal fluid (CSF) has been proposed [29] and would account for virus detected in the meninges and spread to the adjacent neuropil. Vasculitis, and in particular, phlebitis was a significant feature of infection in this species. Recent reports in red foxes [3,25] and in historical and recent cases of HPAI H5N1 in naturally infected cats [30] report endotheliotropic behaviour of the virus, with endothelial damage and vasculitis in seen in multiple organs, which resembles the endotheliotropism seen in the pathogenesis of HPAI H5N1 infections in terrestrial poultry [28]. However, leukocytoclastic vasculitis, defined as a small-vessel neutrophilic vasculitis with fragmented nuclei present [31] has only recently been described in naturally infected cats [30].
The exposure route to influenza A virus of avian origin in this case is hard to conclusively define. The bush dogs had been fed a diet that included frozen shot wild birds and game. In the absence of local disease events that may have been transferred to the bush dogs in the enclosure, infection through ingestion of infected meat / offal would appear to be the most likely route of infection. Another potential infection route is through scavenging of any wild bird carcases/any sick wild birds landing in the un-netted pen. Other routes of infection including indirect contact (e.g. wild bird faeces) are possible but less likely and would not fit with the rapid onset of infection across a number of dogs within a short time frame. Wild bird activity was observed on the site during epidemiological investigations and black-headed gulls, greylag geese, pink-footed geese as well as corvids, pigeons and pheasants had been observed in the vicinity. The shot wild game supplied as food had originated from the neighbouring shooting estate but no reports of clinical signs or die-offs in avian species had been reported. Further, the stream/pond in the bush dog enclosure was supplied with water from a reservoir upstream that was frequented by wild bird species, although a sluice was in place to prevent carcasses being washed down from the reservoir and the water was relatively fast-flowing, again making this an unlikely infection route. Critically, the rapid development of disease and acute clinical outcome suggests that the dogs received a high dose of infectious material and so ingestion of material must be considered the most likely route of infection. Genetic assessment from tissues positive for each animal was undertaken to try and evaluate the possibility of dog-to-dog transfer. Local atmospheric conditions are known to influence environmental virus survival with lower temperatures promoting persistence [18,32]. In early October 2022, the day-night high-low temperature range near this site was 17–11°C. For samples collected on 17–22 November 2022 and 06 March 2023, day-night readings were consistent at 8–11°C and 4–8°C respectively, hence favourable to virus survival. Other factors can impact the detection of environmental vRNA from water, dilution (rain), volume (lake) and flowrate (river) [32]. This might account for a single M-gene signal detection from the bush dog water trough (17/11/2022) (Supplementary Table 3). All bush dogs had access to the water trough where we detected vRNA. Since five bush dogs survived this lowers the likelihood of water being the route or main source of virus introduction and trough contamination by infected bush dogs being a plausible explanation for this observation.
The genetics of the virus demonstrated a high level of viral genetic homogeneity across all eight viral segments of the sequences from the bush dogs. Phylogenetic analysis demonstrated that these sequences were consistent with the AIV09 (EURL: AB) H5N1 HPAIV genotype which predominated in the UK during the 2022–2023 autumn/winter period [2]. Time-resolved phylogenetic and amino acid analysis found that all the sequences from the bush dogs were the result of a single introduction, however, while there were amino acid substitutions, these do not appear to have been consistently maintained. Taken together, this suggests that transfer between dogs is unlikely and that a common source of infection is responsible, although it is impossible to definitively conclude whether dog-to-dog transmission occurred. Critically, five animals survived remaining clinically normal throughout. This may indicate that these dogs had not received a dose of virus sufficient to drive a productive infection. This is further supported by low-level serological responses being detected in two of the animals (Cases 11 and 12) that may indicate exposure to antigen or a low-level infection that was cleared by the host immune response. From the perspective of zoonotic risk, the well-established marker of mammalian adaptation (E627K) was detected in all but one of the bush dog sequences generated. This mutation alone is insufficient to drive an increase in zoonotic risk and so the risk to human populations must be considered very low.
Infection of unusual species with influenza A of avian origin raises important questions about the implications for infection of conservation species. The detection of mammalian infection with these viruses is of interest, through the potential for mammalian adaptation and establishment of mammal-to-mammal transmission. This factor is being monitored globally wherever mammalian infection is detected, and potential adaptive mutations scored for relevance to potential viral adaptation. Clearly, the feeding of wild shot birds to captive carnivores whilst infection pressure is high in wild birds should be discouraged in line with similar recommendations given to keepers of birds of prey or in alternative a rigorous risk assessment should be carried out before any carcass is fed to any of these animals.
Supplementary Material
Acknowledgements
We thank the scientific and support staff of the Pathology and Virology departments of APHA Weybridge and of the Surveillance and Laboratory Services department of the APHA Veterinary Investigation Centres. We acknowledge the authors, originating and submitting laboratories of the sequences from GISAID’s EpiFlu Database on which this research is based, Veterinary Pathology Group and PALS for their histopathology expertise in initial reporting, and analyses described in text. All submitters of the data may be contacted directly via the GISAID website (www.gisaid.org).
Funding Statement
MF, SMR, AMPB, SMR, NM, CJW, SST, JJ and ACB were partly funded by the UK Department for the Environment, Food and Rural Affairs (Defra) and the devolved Scottish and Welsh governments under grants SE2213, SV3400 and SV3006. ACB and JJ were also partly funded by the Biotechnology and Biological Sciences Research Council (BBSRC) and Department for Environment, Food and Rural Affairs (Defra, UK) research initiatives ‘FluMAP’ [grant number BB/X006204/1] and ‘FluTrailMap’ [grant number BB/Y007271/1], and the Medical Research Council (MRC) and Defra research initiative ‘FluTrailMap-One Health’ [grant number MR/Y03368X/1] as well as by Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services (USA), under contract no. 75N93021C00015. This investigation was also partly funded by the Defra GAP-DC project (SE0565).
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Adlhoch C, Fusaro A, Gonzales JL, et al. . Avian influenza overview February - May 2021. EFSA J Eur Food Saf Authority. 2021 Dec;19(12):e06951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Byrne AMP, James J, Mollett BC, et al. . Investigating the genetic diversity of H5 avian influenza viruses in the United Kingdom from 2020-2022. Microbiol Spectr. 2023 Aug 17;11(4):e0477622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rijks JM, Hesselink H, Lollinga P, et al. . Highly pathogenic avian influenza A(H5N1) virus in wild red foxes, The Netherlands, 2021. Emerg Infect Dis. 2021 Nov;27(11):2960–2962. doi: 10.3201/eid2711.211281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Floyd T, Banyard AC, Lean FZX, et al. . Encephalitis and death in wild mammals at a rehabilitation center after infection with highly pathogenic avian influenza A(H5N8) virus, United Kingdom. Emerg Infect Dis. 2021 Nov;27(11):2856–2863. doi: 10.3201/eid2711.211225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention . Technical report: highly pathogenic avian influenza A(H5N1) viruses; 2023.
- 6.Adlhoch C, Fusaro A, Gonzales JL, et al. . Avian influenza overview March - June 2022. EFSA J Eur Food Saf Authority. 2022 Apr;20(64):e07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Postel A, King J, Kaiser FK, et al. . Infections with highly pathogenic avian influenza A virus (HPAIV) H5N8 in harbor seals at the German North Sea coast, 2021. Emerg Microbes Infect. 2022;11(1):725–729. doi: 10.1080/22221751.2022.2043726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puryear W, Sawatzki K, Hill N, et al. Outbreak of highly pathogenic avian influenza H5N1 in New England Seals. bioRxiv. 2022:2022.07.29.501155. [DOI] [PMC free article] [PubMed]
- 9.Agüero M, Monne I, Sánchez A, et al. . Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022. Euro Surveill. 2023;28(3):2300001. doi: 10.2807/1560-7917.ES.2023.28.3.2300001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tammiranta N, Isomursu M, Fusaro A, et al. . Highly pathogenic avian influenza A (H5N1) virus infections in wild carnivores connected to mass mortalities of pheasants in Finland. Infect Genet Evol. 2023;111:105423. doi: 10.1016/j.meegid.2023.105423 [DOI] [PubMed] [Google Scholar]
- 11.Domańska-Blicharz K, Świętoń E, Świątalska A, et al. . Outbreak of highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus in cats, Poland, June to July 2023. Euro Surveill. 2023;28(31):pii=2300366. 10.2807/1560-7917.ES.2023.28.31.2300366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reuters . South Korea detects H5N1 bird flu in two cats at shelter 2023 [cited 2023 Aug 24]. Available from: https://www.reuters.com/business/healthcare-pharmaceuticals/south-korea-detects-h5n1-bird-flu-two-cats-shelter-2023-07-26/.
- 13.Suttie A, Deng YM, Greenhill AR, et al. . Inventory of molecular markers affecting biological characteristics of avian influenza A viruses. Virus Genes. 2019 Dec;55(6):739–768. doi: 10.1007/s11262-019-01700-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.UK Government . Investigation into the risk to human health of avian influenza (influenza A H5N1) in England 2023 [cited 2023 Aug 25]. Available from: https://www.gov.uk/government/publications/avian-influenza-influenza-a-h5n1-technical-briefings/investigation-into-the-risk-to-human-health-of-avian-influenza-influenza-a-h5n1-in-england-technical-briefing-5.
- 15.Tiepolo LM, Quadros J, Pitman MR.. A review of bush dog Speothos venaticus (Lund, 1842) (Carnivora, Canidae) occurrences in Paraná state, subtropical Brazil. Braz J Biol. 2016 Jun;76(2):444–449. doi: 10.1590/1519-6984.20914 [DOI] [PubMed] [Google Scholar]
- 16.Löndt BZ, Nunez A, Banks J, et al. . Pathogenesis of highly pathogenic avian influenza A/turkey/Turkey/1/2005 H5N1 in Pekin ducks (Anas platyrhynchos) infected experimentally. Avian Pathol. 2008 Dec;37(6):619–627. doi: 10.1080/03079450802499126 [DOI] [PubMed] [Google Scholar]
- 17.James J, Billington E, Warren CJ, et al. . Clade 2.3.4.4b H5N1 high pathogenicity avian influenza virus (HPAIV) from the 2021/22 epizootic is highly duck adapted and poorly adapted to chickens. J Gen Virol. 2023;104(5). doi: 10.1099/jgv.0.001852 [DOI] [PubMed] [Google Scholar]
- 18.James J, Warren CJ, De Silva D, et al. . The role of airborne particles in the epidemiology of clade 2.3.4.4b H5N1 high pathogenicity avian influenza virus in commercial poultry production units. Viruses. 2023;15(4):1002. doi: 10.3390/v15041002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.World Organisation for Animal Health (WOAH) . Terrestrial manual: chapter 3.3.4. Avian Influenza (Including infection with High Pathogenicity Avian Influenza Viruses); 2021.
- 20.Nagy A, Černíková L, Kunteová K, et al. . A universal RT-qPCR assay for “One Health” detection of influenza A viruses. PLoS One. 2021;16(1):e0244669. doi: 10.1371/journal.pone.0244669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.James J, Seekings AH, Skinner P, et al. . Rapid and sensitive detection of high pathogenicity Eurasian clade 2.3.4.4b avian influenza viruses in wild birds and poultry. J Virol Methods. 2022 Mar;301:114454. doi: 10.1016/j.jviromet.2022.114454 [DOI] [PubMed] [Google Scholar]
- 22.James J, Slomka MJ, Reid SM, et al. . Development and application of real-time PCR assays for specific detection of contemporary avian influenza virus subtypes N5, N6, N7, N8, and N9. Avian Dis. 2018;63(sp1):209–218, 10. doi: 10.1637/11900-051518-Reg.1 [DOI] [PubMed] [Google Scholar]
- 23.Slomka MJ, To TL, Tong HH, et al. . Challenges for accurate and prompt molecular diagnosis of clades of highly pathogenic avian influenza H5N1 viruses emerging in Vietnam. Avian Pathol. 2012;41(2):177–193. doi: 10.1080/03079457.2012.656578 [DOI] [PubMed] [Google Scholar]
- 24.Lean FZX, Leblond AL, Byrne AMP, et al. . Subclinical hepatitis E virus infection in laboratory ferrets in the UK. J Gen Virol. 2022 Nov;103(11). https://www.microbiologyresearch.org/content/journal/jgv/ 10.1099/jgv.0.001803 [DOI] [PubMed] [Google Scholar]
- 25.Bordes L, Vreman S, Heutink R, et al. . Highly pathogenic avian influenza H5N1 virus infections in wild Red foxes (Vulpes vulpes) show neurotropism and adaptive virus mutations. Microbiol Spectr. 2023 Feb 14;11(1):e0286722. doi: 10.1128/spectrum.02867-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lagan P, McKenna R, Baleed S, et al. . Highly pathogenic avian influenza A(H5N1) virus infection in foxes with PB2-M535I identified as a novel mammalian adaptation, Northern Ireland, July 2023. Eurosurveillance. 2023;28(42):2300526. doi: 10.2807/1560-7917.ES.2023.28.42.2300526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cronk BD, Caserta LC, Laverack M, et al. . Infection and tissue distribution of highly pathogenic avian influenza A type H5N1 (clade 2.3.4.4b) in red fox kits (Vulpes vulpes). Emerg Microbes Infect. 2023 Dec;12(2):2249554. doi: 10.1080/22221751.2023.2249554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reperant LA, van de Bildt MW, van Amerongen G, et al. . Marked endotheliotropism of highly pathogenic avian influenza virus H5N1 following intestinal inoculation in cats. J Virol. 2012 Jan;86(2):1158–1165. doi: 10.1128/JVI.06375-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Songserm T, Amonsin A, Jam-on R, et al. . Avian influenza H5N1 in naturally infected domestic cat. Emerg Infect Dis. 2006 Apr;12(4):681–683. doi: 10.3201/eid1204.051396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sillman SJ, Drozd M, Loy D, et al. . Naturally occurring highly pathogenic avian influenza virus H5N1 clade 2.3.4.4b infection in three domestic cats in North America during 2023. J Comp Pathol. 2023 Aug;205:17–23. doi: 10.1016/j.jcpa.2023.07.001 [DOI] [PubMed] [Google Scholar]
- 31.Maxie MG. Jubb, Kennedy & Palmer's pathology of domestic animals. Oxford: W.B. Saunders; 2015. [Google Scholar]
- 32.Furness RW, Gear SC, Camphuysen KCJ, et al. . Environmental samples test negative for avian influenza virus H5N1 four months after mass mortality at a seabird colony. Pathogens. 2023;12(4):584. doi: 10.3390/pathogens12040584 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.