Abstract
The left-handed Z-conformation of nucleic acids can be adopted by both DNA and RNA when bound by Zα domains found within a variety of innate immune response proteins. Zα domains stabilize this higher-energy conformation by making specific interactions with the unique geometry of Z-DNA/Z-RNA. However, the mechanism by which a right-handed helix contorts to become left-handed in the presence of proteins, including the intermediate steps involved, is poorly understood. Through a combination of Nuclear Magnetic Resonance (NMR) and other biophysical measurements, we have determined that in the absence of Zα, under low salt conditions at room temperature, d(CpG) and r(CpG) constructs show no observable evidence of transient Z-conformations greater than 0.5% on either the intermediate or slow NMR timescales. At higher temperatures, we observe a transient unfolded intermediate. The ease of melting a nucleic acid duplex correlates to Z-form adoption rates in the presence of Zα. The largest contributing factor to the activation energies of Z-form adoption as measured by Arrhenius plots is the ease of flipping the sugar pucker, as required for Z-DNA and Z-RNA. Together, these data validate the previously proposed ‘zipper model’ for Z-form adoption in the presence of Zα. Overall, Z-conformations are more likely to be adopted by double-stranded DNA and RNA regions flanked by less stable regions, and by RNAs experiencing torsional/mechanical stress.
Keywords: Z-DNA, Z-RNA, Zα, dynamics, NMR
Introduction
The most stable double-stranded helical conformations for DNA and RNA under physiological conditions are the B- and A-form, respectively. Both conformations are right-handed helices but otherwise differ in shape and geomertry1. Nucleic acid binding proteins often exploit this fact in order to selectively recognize DNA or RNA2,3. Interestingly, both DNA and RNA will adopt a higher-energy, left-handed double-stranded conformation known as the Z-form under certain conditions (Figure 1A), such as when recognized and stabilized by Z-DNA/Z-RNA-binding Zα domains4–8 or through chemical conditions/modifications (extensively reviewed here9). Other than the inverted helicity, the Z-conformation is more elongated compared to B-DNA/A-RNA and is composed of a repeating dinucleotide unit where the sugar puckers alternate between the C2′- and C3′-endo conformation along with the bases between the anti- and syn-conformations4,8–11. This arrangement leads to a lone pair–π contact only found within Z geometry (involving the O4′ of the C2′-endo sugar and the syn base12,13). The unique features of Z-form helices result in a jagged backbone conformation which “zig-zags” along the helical axis. This brings the phosphates closer together on average than in the B- or A-conformation, causing electrostatic repulsion and accounting for a significant contribution to the Z-form’s intrinsic instabillity14–18 (Figure 1A).
Figure 1.
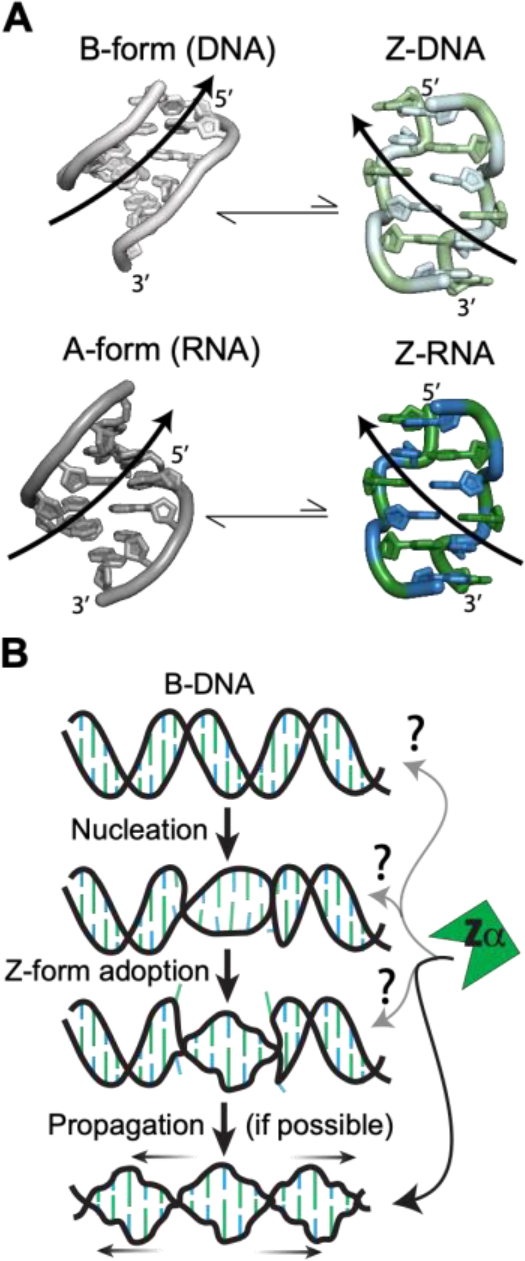
Characteristics of Z-DNA/RNA and Z-conformation adoption. (a) Both dsDNA (top) and dsRNA (bottom), normally in the B- and A-conformations, can adopt the higher-energy left-handed Z-form (right). Z-DNA and Z-RNA are structurally equivalent and will revert to the B/A-form in the absence of stabilizing factors. Pyrimidine (blue) – purine (green) repeats alternate between the C2′-endo and C3′-endo sugar pucker conformations along with the nucleobases between anti and the syn conformations. This leads to a zig-zagged backbone. B-DNA, A-RNA, Z-DNA, and Z-RNA models were made using PDBs 1N1K1, 1PBM2, 1QBJ3, and 2GXB4, respectively. (b) Zipper model for the conversion from B-DNA to Z-DNA5. First, a high-energy nucleation event allows for helical handedness conversion and a short Z-DNA stretch to be adopted. This can then propagate down the helix in a cooperative manner if the sequence allows it. At what points Zα plays a role in the zipper model is mostly unknown.
Despite these striking conformational changes, the Z-conformation retains Watson-Crick base-pairing8. Therefore, switching from the B/A-form to the Z-conformation requires both a 180° rotation of every nucleotide base in the helix about the glycosidic bond and a complete inversion of every other nucleotide (including the ribose), converting back to the anti-conformation19. This process is both topologically and thermodynamically challenging20. Over the years, different models have been proposed to theoretically address how this could occur8,19,21–31. The zipper model32,33, where an initial high-energy nucleation event allows a short Z-form segment enclosed between two B-Z junctions to be adopted, which then propagates through the helix in a cooperative manner, seems to fit the experimental evidence involving Z-DNA adoption well34,35 and is supported by molecular dynamics simulations36 (Figure 1B).
Zα domains are found within a variety of innate immune response proteins or viral proteins and have been demonstrated to play pivotal roles in these proteins’ ability to regulate the innate immune response or evade it, respectively37. These domains stabilize the Z-conformation of DNA and RNA by making key contacts with the unusual features present in Z-form helices6,38. Zα domains help to alleviate the steric repulsion of the closely placed, negatively charged phosphates by making charge-charge and water-mediated contacts with the Z-form backbone6,38. Previous Zα:nucleic acid structures have identified a number of key residues in the recognition and stabilization of the Z-conformation. Lys 1696,39 and Asn 17340 make contacts with the phosphate backbone. Pro 192 and Pro 193 position the beta-hairpin loop in a way which facilitates further interactions with kinked Z-RNA backbone40. Trp 195 forms part of the hydrophobic core in addition to making a water-mediated contact with the phosphate backbone6,41,42. Tyr 177 is one of the most important residues, which makes a CH–π interaction with a syn base in the Z-form helix and is necessary for Zα to stabilize the Z-conformation43,44 (Figure S1). The increasing number of discoveries of proteins containing functional Zα domains suggests that Z-conformations are being adopted in cells and play an important biological function. However, what sequences and under what conditions these conformations are being adopted are still mostly unknown37.
Understanding the steps by which Zα domains stabilize Z-conformations in DNA and RNA is crucial to be able to predict the ability of varying sequence contexts to adopt the Z-form. Previous Nuclear Magnetic Resonance (NMR) and Single-Molecule Förster Resonance Energy Transfer (smFRET) studies on Z-form adoption in the presence of Zα have shown the presence of multiple intermediate states between binding and Z-DNA/RNA adoption45–50. However, the identity of these states remains poorly understood (Figure 1B). One smFRET study showed that Zα bound to a pre-sampled Z-conformation in a DNA duplex containing 5-methyl dCs (which lowers the energy barrier of Z-form adoption51), with the authors proposing that Zα recognizes a pre-sampled Z-form state52. This is also supported by the fact that the Zα binding interface is pre-organized to recognize Z-DNA/Z-RNA53. However, we still do not have a clear picture of how Z-forms are adopted, and what role(s) Zα plays in the conversion. A better understanding of the Z-form adoption process via stabilization by Zα and the intermediate states involved would significantly improve our understanding of Z-form biology.
To help answer this question, we measured Off-Resonance R1ρ relaxation dispersion experiments54 on model Z-forming d(CpG) and r(CpG) constructs. This experiment aims to probe for low population, dynamic states (in the μs-ms timescale) under low salt conditions and in the absence of Zα. The Off-Resonance R1ρ relaxation dispersion experiment allows for the characterization of microsecond-to-millisecond chemical exchange processes in solution55,56, a timescale which usually coincides with adoption of excited states57. These motions can lead to formation of “excited states” that correspond to local minima in the free energy landscape58. Our experiments revealed evidence of transient duplex melting, suggesting that helix melting may play a role in Z-form adoption. To study this further, we then carried out circular dichroism time-courses and other biophysical measurements to investigate the energy barriers of Z-form adoption in the presence of the Zα domain from human ADAR159, one of the most well-characterized Zα domains6,38,39,60. We find that the ease of melting a duplex heavily correlates with its Z-form adoption rate and that the sugar pucker and nucleobase rearrangement is the rate-limiting step for Z-form adoption. Finally, we find that duplex RNA goes through a single-stranded intermediate before Z-form conversion and that Zα actively promotes the adoption of this single-stranded state, thereby having an active role in Z-form adoption. We speculate that while transient duplex melting is likely relatively frequent, transient Z-conformation likely only becomes populated when Zα is already bound to B/A-DNA/RNA and is therefore present to stabilize it. Consequently, we would predict that Z-conformations in the cell are quite stable, as they are either already in Z-form via stabilization by Zα domains or due to other mechanisms (such as helical torsion or chemical modifications).
Results
Evaluating the Z-Form Stability of Model Z-Adopting d(CpG) and r(CpG) Constructs.
To gain a better understanding of the Z-form adoption process and the intermediate states involved, we first looked for the existence of transient states in d(CpG) and r(CpG) sequences, which are well characterized Z-adopting sequences8,10,61–63, under low salt conditions (25 mM NaCl) where the Z-form would not be expected to form (Figure 2A). We chose these constructs because they each have a different energy barrier for adopting the Z-conformation, and, theoretically, should have different populations of transient Z-conformations. Because the riboses in Z-form helices alternate between the C2′-endo and C3′-endo conformations, the 2′ hydroxyl in RNA represents a significant energy barrier to Z-form adoption64–66. We quantified Z-form adoption using circular dichroism titrations which showed that in comparison to Z-DNA, Z-RNA needs more than double the salt concentration to be stabilized (4 and 5 M NaClO4 for r(CpG)3 and r(CpG)6 constructs, respectively, compared to 2 M NaClO4 for both the d(CpG)3 and d(CpG)6, Figure 2B, Figure S2, Table 1). The 5-methyl cytosine modifications in the d(5mCpG)3 construct destabilizes the B-conformation relative to the Z-form, thereby significantly decreasing the energy barrier for Z-DNA adoption35,67–71. Its Z-form NaClO4 midpoint is 800 mM which is 2.5x lower than the d(CpG)3 construct (Figure 2B, Table 1). The 8-methyl guanine modification destabilizes both the B- and A-conformations of DNA and RNA by sterically clashing with the ribose when the base is in anti, resulting in the methylated purine adopting the syn conformation and thereby significantly promoting Z-form adoption72–74. Achieving Z-form midpoints for the 8mG4 d(CpG)3 and 8mG4 r(CpG)3 (8-methyl guanine at position 4) constructs requires 500x and 250x less NaClO4 compared to the d(CpG)3 and r(CpG)3, with values of ~4 and ~16 mM, respectively (Figure 2B, Table 1). We also measured the melting temperatures (TM) of the duplexes to make sure they were in good agreement with predicted TM (Figure S3, Table 1). Overall, this data confirms that our selected constructs have different energy barriers for Z-form adoption and, therefore, are good candidates to probe for transient Z-conformations.
Figure 2.
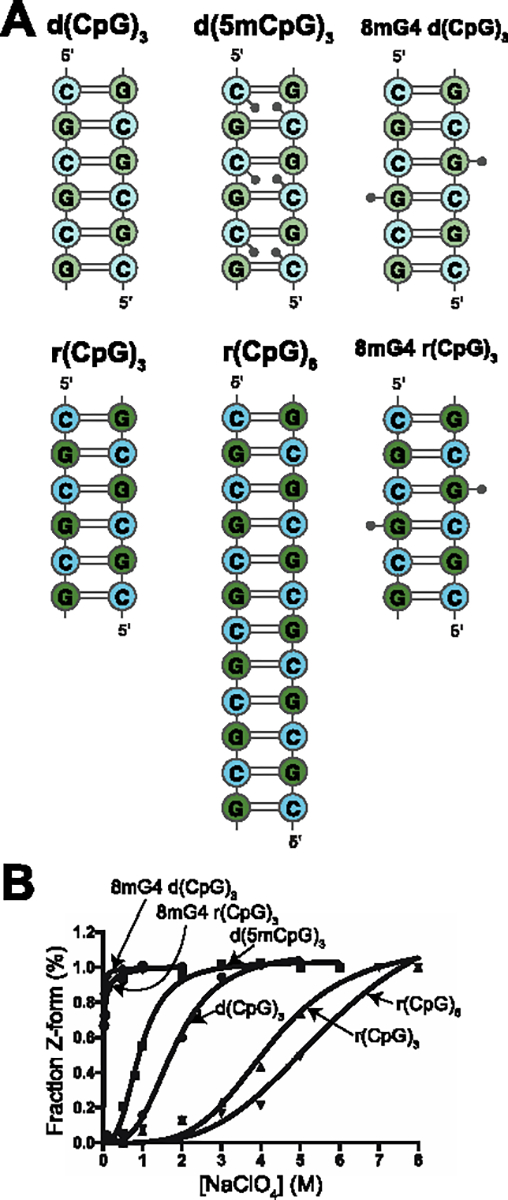
DNA and RNA constructs selected for NMR measurements. (a) 2D representations of the different d(CpG) and r(CpG) constructs used for NMR measurements in this study. DNA bases are more lightly shaded than RNA ones. Methyl groups for the modified constructs are depicted as small grey circles and indicate their relative position within the duplex. (b) Fits of circular dichroism titrations of NaClO4 into the DNA and RNA constructors shown in (a), with the fraction of the duplex in the Z-conformation on the y-axis and the concentration of NaClO4 (M) on the x-axis. The fraction of Z-DNA and Z-RNA was tracked by following the ellipticity at 266 and 285 nm, respectively, as described in the Methods sections.
Table 1.
Duplex constructs’ melting temperatures and NaClO4 Z-form midpoints measured from circular dichroism.
| Construct | TM (°C) | Z-form midpoint (M [NaClO4]) |
|---|---|---|
| d(CpG)3 | 50.24 ± 0.00 | 1.923 ± 0.121 |
| d(5mCpG)3 | 54.04 ± 0.24 | 0.824 ± 0.093 |
| 8mG4 d(CpG)3 | 26.01 ± 0.22 | 0.0044 ± 0.0005 |
| 8mG4 d(CpG)3 (4:1 Zα:RNA) | 59.80 ± 0.27 | |
| d(CpG)6 | 77.77 ± 0.74 | 1.867 ± 0.111 |
| d(CpG)3TG(CpG)2 | 80.78 ± 2.59 | 1.683 ± 0.052 |
| (dCprG)6 | 55.41 ± 0.14 | 1.064 ± 0.157 |
| d(CpG)12 | >100 | 1.770 ± 0.066 |
| r(CpG)3 | 49.22 ± 2.13 | 4.105 ± 0.162 |
| 8mG4 r(CpG)3 | A-form: 49.49 ± 5.38 Z-form: 22.76 ± 0.60 |
0.0164 ± 0.0066 |
| 8mG4 r(CpG)3 (4:1 Zα:RNA) | 64.32 ± 0.32 | |
| r(CpG)6 | 86.36 ± 0.86 | 5.218 ± 0.185 |
| r(CpG)3UG(CpG)2 | 54.02 ± 4.09 | 6.299 ± 0.207 |
| r(CpG)2CIUG(CpG)2 | 38.39 ± 0.46 | 5.013 ± 0.222 |
| r(CpG)12 | >100 | 5.513 ± 0.119 |
| (dCpLG)6 | N/A | N/A |
| r(CpG)3 cUUCGg tetraloop | N/A | >6 |
| r(CpG)3 6mer Ura loop | 77.72 ± 1.82 | >6 |
N/A means that the melting temperature could not be measured over the range measured.
Off-Resonance R1ρ Relaxation Dispersion Measurements Reveal an Excited State in Model Z-adopting d(CpG) and r(CpG) Constructs at 42°C.
We sought to probe for dynamic states during the A/B to Z transition. To this end, we recorded and assigned Off-Resonance 13C-R1ρ54 spectra of the d(CpG)3, d(5mCpG)3, 8mG4 d(CpG)3, r(CpG)3, and r(CpG)6 constructs. In addition, we also measured ZZ-exchange experiments (which probe dynamics in the second timescale75) on the 8mG4 d(CpG)3 and 8mG4 r(CpG)3 constructs which are in slow exchange between the A/B and the Z-conformation and therefore served as positive controls. The presence of an excited state can be identified in the R2 + Rex profile of the Off-Resonance R1ρ experiment by increased relaxation due to exchange at the excited state’s chemical shift position (relative to the ground state). As the power of the 13C spin-locking pulse is increased, the contribution of relaxation due to exchange is quenched, allowing for the exchange rate (kEX) and difference in chemical shift (Δω) between the ground and excited states, as well as their populations (pA and pB), to be extracted (a theoretical illustration is shown in Figure S4A, S4C). ZZ-exchange allows characterization of slow timescale exchange processes by observing the transfer of longitudinal relaxation between the ground and excited states during mixing time, allowing for a direct readout of the exchange rate between the two states76 (Figure S4B).
The 13C-1H HSQC and 1H-1H NOESY assignments and NOESY “walk” strategy for the assignment of B- and A-form helices are shown for the d(CpG)3 (Figures 3A, 3B, Table S1). Assignments for the other constructs are shown in Figures S5–S12, and chemical shift values can be found in Tables S2–S6. We note that due to the palindromic nature of all the constructs we tested, the two strands are chemically equivalent (Figures 3A, 3B). For example, the aromatic CH8 peak of G4 is actually two overlapped peaks with identical chemical shifts, one from G4 of one strand and the other from the second strand. Therefore, all NMR observables for these constructs represent an average of the residue in question from both strands.
Figure 3.
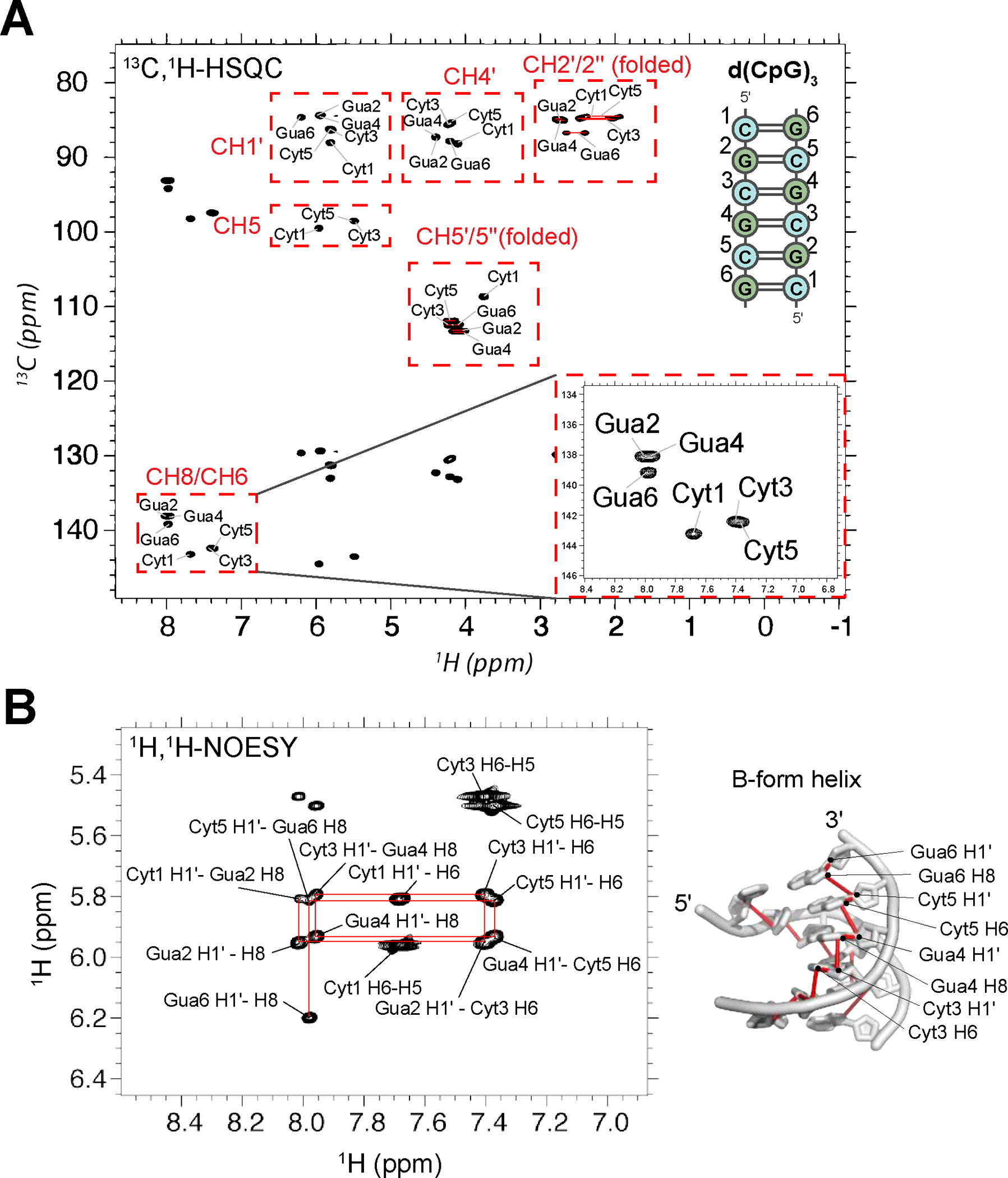
NMR assignment of the d(CpG)3 construct. (a) Full 13C-1H HSQC spectra assignments for the d(CpG)3 construct are shown (depicted on the right with assignment numbering). The CH2’/2” and CH5’/5” peak positions are folded in from their normal positions around 40 and 66 ppm, respectively. Their proper chemical shift values are indicated in Table S1. CH3’ resonances were not assignable due to water suppression. Inset shows a zoom in of the aromatic assignments. Note that the two strands of the duplex are chemically equivalent and, therefore, have identical chemical shifts. (b) 1H-1H NOESY experiment with a mixing time of 320 ms showing the aromatic H8/H6 to ribose H1′ connectivites. The NOESY “walk” through the B-form helix is indicated with red lines, an example of which is shown on the structure of a B-form helix (PDB: 1N1K1) to the right.
For the d(CpG)3 construct at 25°C in 25 mM NaCl, we obverse no μs-ms timescale exchange processes in our Off-Resonance R1ρ profiles that can be fit with any reasonable confidence (Figure 4, dispersion profiles for other residues are shown in Figure S13). This indicates that at 25°C, either the d(CpG)3 is not in exchange with a transient state on this timescale or that the excited state’s population and dynamics is beyond detection by our experimental procedure (< 0.5–1% population).
Figure 4.
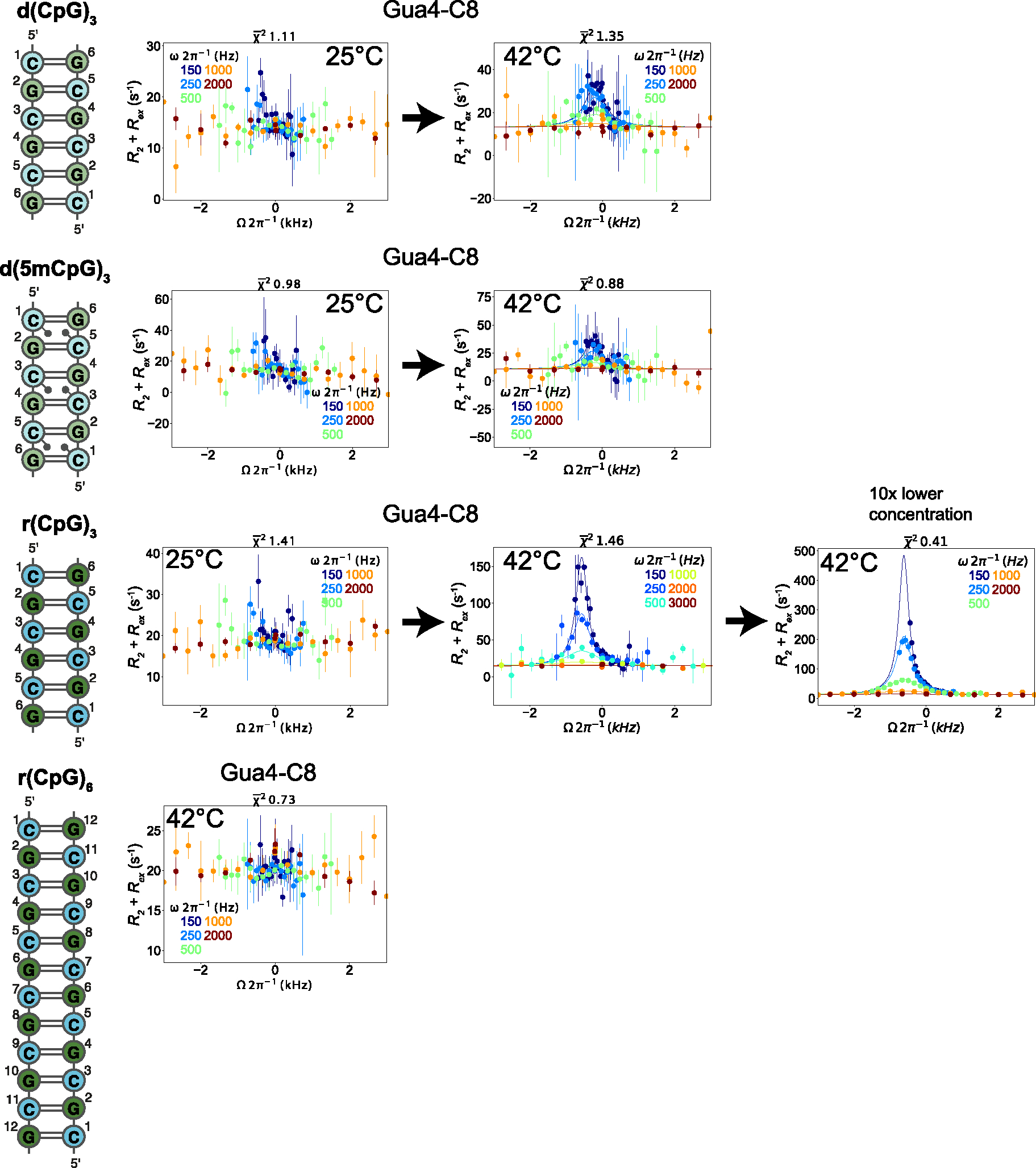
Off-Resonance R1ρ relaxation dispersion profiles of the different DNA and RNA constructs at 25°C and 42°C. 2D representations of the d(CpG) and r(CpG) constructs and corresponding Off-Resonance R1ρ relaxation dispersion profiles for the C8 atom of Gua4 carried out at five different spin-lock powers (150, 250, 500, 1000, and 2000 Hz, colored coded according to the legend within each plot) at 25°C and 42°C are shown to the right. The dispersion profile at 10x lower concentration of duplex for the r(CpG)3 construct is also shown. R2 + R2ex (= (R1ρ – R1cos2θ)/sin2θ, where θ = tan−1(lock power/offset)) values are given as a function of the resonance offset from the major state (Ωoff/2π). Error bars represent experimental uncertainty from a bootstrapping method, as described in the Methods section. The fits (solid lines) were carried out as described in the materials and methods, and fitted parameters are found in Tables 2 and 3.
Because Z-form adoption is an entropically driven process and known to be promoted as a function of temperature77, we measured the same experiment at 42°C (a commonly used incubation temperature with Zα77). The hypothesis we tested was that the increased temperature may promote the population of any transient Z-form state. At 42°C, a clear excited state is populated for all residues of the d(CpG)3 construct except for cytosine 1 and guanine 6 (Figure 4, Figure S13). Due to the similarity of the extracted exchange parameters for all residues, we fit them globally, which gave an exchange rate (kEX) of 1630 ± 140 s−1 and an excited state population (pB) of 4.0 ± 0.6 % (Table 2). In addition, the excited state chemical shift differences (Δω) for all residues were downfield (de-shielded) relative to their ground-state positions (Table 2), suggesting that the entire d(CpG)3 construct was experiencing the same exchange process and that this excited state resulted in a more open conformation of the duplex, as would be predicted for the aromatic purine C8 atoms of Z-conformation (Figure S14).
Table 2.
Conformational exchange parameters from Off-Resonance R1ρ relaxation dispersion experiments for tested constructs using a global fitting routine.
| Construct | Residue/Atom identity | Temperature (°C) | kex (s−1) | pE (%) | Δω (ppm) |
|---|---|---|---|---|---|
| d(CpG)3 | Gua2/C8 | 42 | 1630 ± 140 | 4.0 ± 0.6 | 1.35 ± 0.12 |
| Cyt3/C6 | 42 | 1630 ± 140 | 4.0 ± 0.6 | 0.87 ± 0.07 | |
| Gua4/C8 | 42 | 1630 ± 140 | 4.0 ± 0.6 | 1.11 ± 0.09 | |
| Cyt5/C6 | 42 | 1630 ± 140 | 4.0 ± 0.6 | 0.91 ± 0.08 | |
| d(5mCpG)3 | Cyt3/C6 | 42 | 1300 ± 280 | 2.0 ± 0.5 | 1.32 ± 0.25 |
| Gua4/C8 | 42 | 1300 ± 280 | 2.0 ± 0.5 | 1.69 ± 0.22 | |
| Cyt5/C6 | 42 | 1300 ± 280 | 1.93 ± 0.06 | 1.18 ± 0.21 | |
| r(CpG)3 | Gua2/C8 | 42 | 977 ± 53 | 1.93 ± 0.06 | 2.17 ± 0.08 |
| Cyt3/C6 | 42 | 977 ± 53 | 1.93 ± 0.06 | 2.20 ± 0.07 | |
| Gua4/C8 | 42 | 977 ± 53 | 1.93 ± 0.06 | 3.76 ± 0.07 | |
| Cyt5/C6 | 42 | 977 ± 53 | 1.93 ± 0.06 | 2.45 ± 0.06 | |
| Gua6/C8 | 42 | 977 ± 53 | 1.93 ± 0.06 | 2.47 ± 0.06 |
Only exchange parameters for profiles that could be fit reliably are shown.
We observed a similar phenomenon for the d(5mCpG)3 and r(CpG)3 constructs, with no observable exchange processes at 25°C but a clear excited state at 42°C (Figures 4, S15, S16, Table 2). Again, we were able to fit the data from both constructs globally, which gave an exchange rate of 1300 ± 280 s−1 with a population of 2.0 ± 0.5 % for the d(5mCpG)3 construct and an exchange rate of 977 ± 53 s−1 with a population of 1.93 ± 0.06 % for the r(CpG)3 construct (Table 2). Similar to the d(CpG)3 case, the excited state chemical shift differences for the fit residues in the d(5mCpG)3 and r(CpG)3 constructs displayed downfield chemical shift values. In contrast, the r(CpG)6 duplex has no observable exchange process at 42°C (Figures 4, S17). This suggests that all three of the 6 bp constructs are sampling a similar state with different dynamics. However, the identity of this state is unknown without comparing its chemical shift difference (Δω) to what would be expected between the B/A- and Z-forms.
Excited State Chemical Shifts from Off-Resonance R1ρ Correlate with Melted Duplex better than with stabilized Z-DNA/Z-RNA.
To identify whether the excited states observed at 42°C in the d(CpG)3, d(5mCpG)3, and r(CpG)3 constructs were a transient Z-conformation or other exchanging states, we needed to determine the chemical shift difference between the aromatic residues in the B-form/A-form and in the Z-form for the DNA and RNA constructs. To this end, we assigned the 13C, 1H chemical shifts of the 8mG4 d(CpG)3 and 8mG4 r(CpG)3 constructs, the DNA version of which had been previously confirmed to be in slow exchange between the B- and Z-conformations72. To our knowledge, the Z-forming capability of the singly methylated 8mG4 r(CpG)3 has not been tested until now, although the construct is chemically similar to the double-methylated m8Gm (8-methyl and 2′-O-methyl guanosine) r(CpG)3 construct which has been studied previously73.
Assignment and peak analysis of the HSQC spectrum recorded on the 8mG4 d(CpG)3 construct (Figures S6, S7) confirmed that the construct is indeed in a slow exchange between the B- and Z-conformations (Figure 5A) being mostly Z-form with a B-form population of 8.3 ± 2.3 % at 25°C (as determined from peak volume integration, Table 3). The extracted excited state chemical shift differences from the Off-Resonance R1ρ experiments measured on the d(CpG)3 at 42°C exhibit a high degree of correlation with the chemical shift difference between the B- and Z-form peaks in 13C, 1H HSQC of the 8mG4 d(CpG)3 construct (Figure 5B, R2 = 0.89). However, in all cases except for Cyt5 C6, they agree much better with the chemical shift difference between the 13C, 1H HSQC peaks of the d(CpG)3 at 42°C (folded) and 70°C (unfolded, Figure 5B, R2 = 0.99).
Figure 5.
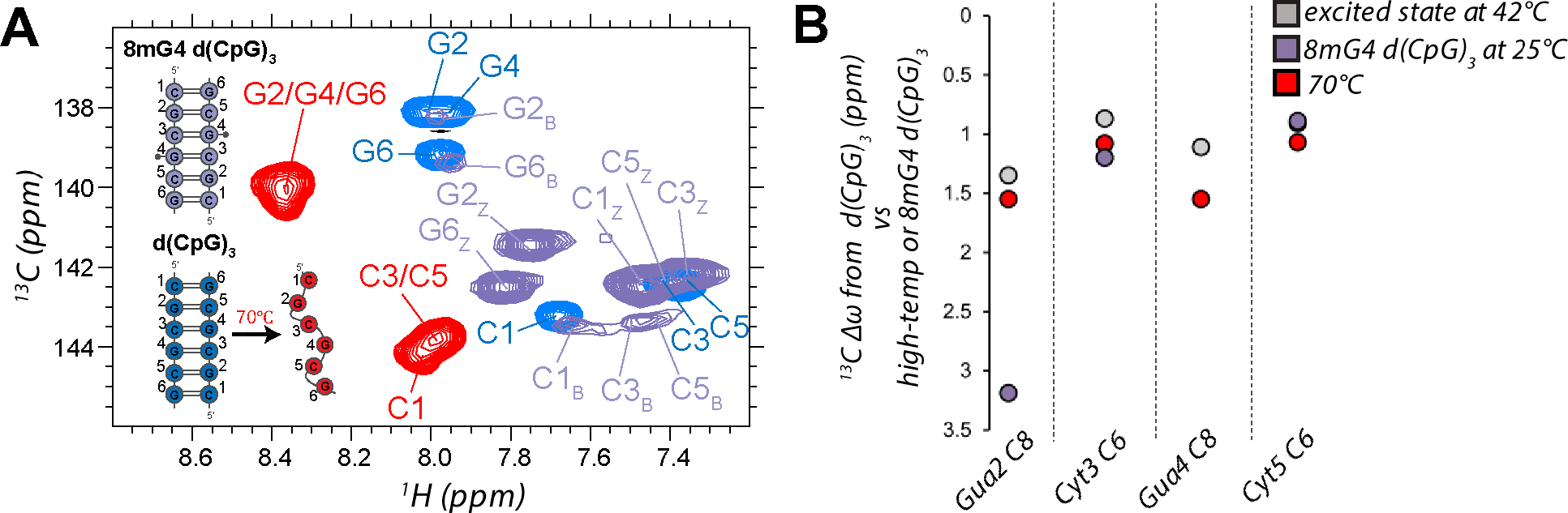
Identification of excited state chemical shift differences extracted from Off-Resonance R1ρ experiments measured on the d(CpG)3. (a) Aromatic 13C-1H HSQC (CH8 of purines and CH6 of pyrimidines) assignments are shown for 8mG4 d(CpG)3 (purple peaks, B-form and Z-form peaks are denoted by subscripts B or Z, respectively) compared to the d(CpG)3 construct at 42°C (folded, blue peaks) and 70°C (melted, red peaks). (b) Chemical shift differences (13C Δω) extracted from Off-Resonance R1ρ experiments measured on the d(CpG)3 construct at 42°C, the difference between the B-form and Z-form peaks in the 13C, 1H HSQC of the 8mG4 d(CpG)3 construct, and the difference between the folded and melted peaks in the 13C, 1H HSQC of the d(CpG)3 construct at 42°C and 70°C. The Z-form chemical shift position for Guanine 4 for the 8mG4 d(CpG)3 construct could not be compared due to the methyl modification.
Table 3.
B- and Z-form populations in the 8mG4 d(CpG)3 constructs at different temperatures determined from peak volume integration.
| Residue/Atom identity | B-form population (%) at 5°C | B-form population (%) at 15°C | B-form population (%) at 20°C | B-form population (%) at 25°C | B-form population (%) at 30°C | B-form population (%) at 35°C |
|---|---|---|---|---|---|---|
| Cyt1/C1’ | 12.5 | 14.0 | 13.3 | 9.3 | 3.9 | 2.9 |
| Gua2/C1’ | 17.3 | 14.8 | 16.5 | 10.3 | 9.4 | 2.7 |
| Gua2/C8 | 16.9 | 10.7 | 7.9 | 4.9 | 1.3 | 0.51 |
| Gua6/C8 | 11.1 | 9.6 | 8.8 | 8.8 | 0.82 | 0.36 |
| average | 14.4 ± 3.1 | 12.3 ± 2.5 | 11.6 ± 4.0 | 8.3 ± 2.3 | 3.9 ± 3.9 | 1.6 ± 1.4 |
The 8mG4 r(CpG)3 construct is also in slow exchange between the A- and Z-conformations (Figure 6A), albeit with an A-form population of 50.5 ± 5.3 % (Table 4). This significant decrease in the population of Z-form compared to the 8mG4 d(CpG)3 construct is likely a reflection of the differences in C2′-endo sugar pucker stability in DNA vs. RNA66. The addition of 100 mM NaClO4 stabilizes the Z-RNA state, decreasing the A-form population to 26.6 ± 4.0 % (Figure 6A, Table 4). Similar to the d(CpG)3 construct, the extracted excited state chemical shift differences from the Off-Resonance R1ρ experiments measured on the r(CpG)3 duplex at 42°C agree much better with a melted duplex (Figure 6B, R2 = 0.97) than with the stabilized Z-conformation (Figure 6B, R2 = 0.63). They also poorly correlate with the chemical shift differences between the r(CpG)3 construct in low- (25 mM NaCl, A-form) and high-salt (Z-form, 6 M NaClO4), showing that this poor correlation is not due to the chemical shift deviations due to the 8-methyl guanine modification (Figure S18, R2 = 0.87).
Figure 6.
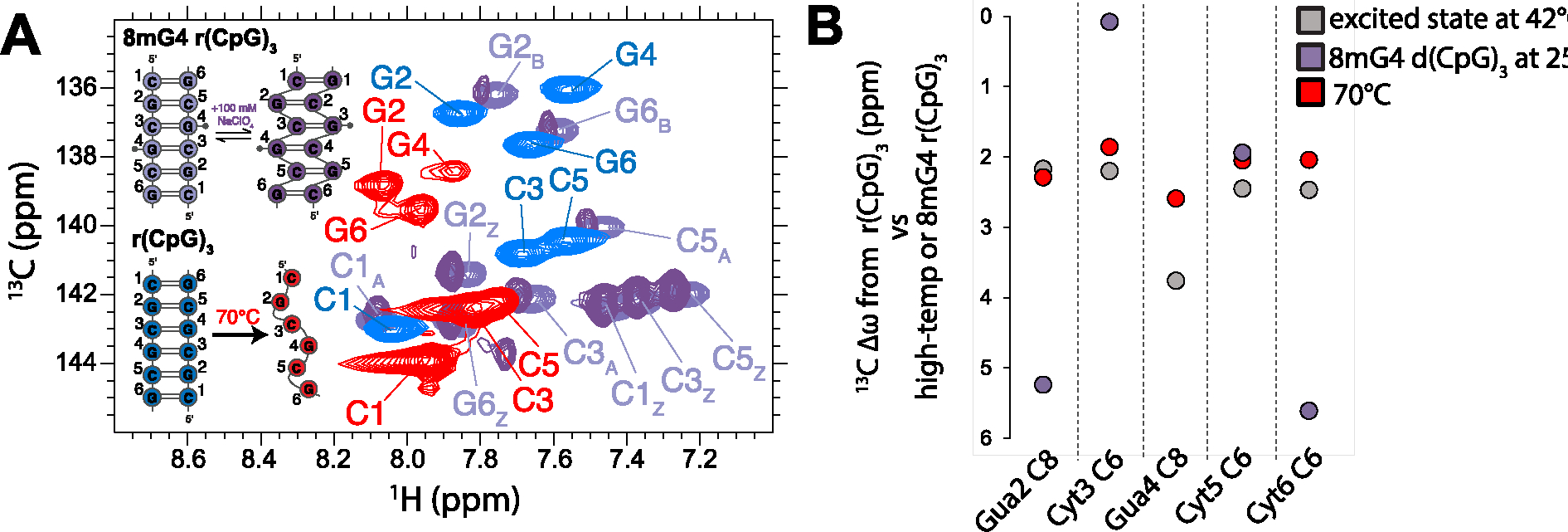
Identification of excited state chemical shift difference extracted from Off-Resonance R1ρ experiments measured on the r(CpG)3 construct. (a) Aromatic 13C-1H HSQC (CH8 of purines and CH6 of pyrimidines) assignments are shown for 8mG4 r(CpG)3 (purple peaks, A-form and Z-form peaks are denoted by subscript A and Z, respectively) compared to the r(CpG)3 construct at 42°C (folded, blue peaks) and 70°C (melted, red peaks). For the 8mG4 r(CpG)3 duplex, the addition of 100 mM NaClO4 promotes the population of Z-RNA while decreasing the population of A-RNA (dark purple peaks). (b) Chemical shift differences (13C Δω) extracted from Off-Resonance R1ρ experiments measured on the r(CpG)3 construct at 42°C, the difference between the A-form and Z-form peaks in the 13C, 1H HSQC of the 8mG4 r(CpG)3 construct, and the difference between the folded and melted peaks in the 13C, 1H HSQC of the r(CpG)3 construct at 42°C and 70°C. The Z-form chemical shift position for Guanine 4 for the 8mG4 r(CpG)3 construct could not be compared due to the methyl modification.
Table 4.
A- and Z-form populations in the 8mG4 r(CpG)3 constructs at different temperatures determined from peak volume integration.
| Residue/Atom identity | B-form population (%) at 5°C | B-form population (%) at 15°C | B-form population (%) at 25°C | B-form population (%) at 35°C | B-form population (%) at 45°C | B-form population (%) at 25°C (100 mM NaClO4) |
|---|---|---|---|---|---|---|
| Cyt1/C5 | 41.3 | 49.6 | 43.7 | 18.4 | 17.1 | 23.3 |
| Cyt1/C6 | 44.1 | 55.8 | 54.9 | 24.5 | 14.9 | 26.4 |
| Gua2/C8 | 40.6 | 40.3 | 50.8 | 39.0 | 26.3 | |
| Cyt3/C5 | 40.9 | 50.1 | 41.3 | 18.5 | 20.2 | |
| Cyt3/C6 | 48.1 | 55.8 | 47.7 | 42.4 | 28.3 | |
| Cyt5/C1’ | 67.7 | 56.72 | 49.7 | 45.9 | 34.2 | |
| Cyt5/C5’ | 48.2 | 51.4 | 50.0 | 15.9 | 27.4 | |
| Cyt5/C5” | 49.2 | 54.2 | 51.2 | 29.7 | 26.4 | |
| Cyt5/C5 | 51.6 | 51.2 | 56.2 | |||
| Cyt5/C6 | 44.5 | 54.7 | 60.0 | 49.5 | ||
| Gua6/C8 | 41.0 | 49.5 | 49.7 | 29.5 | ||
| average | 47.0 ± 7.84 | 51.8 ± 4.6 | 50.5 ± 5.3 | 31.3 ± 12.2 | 16.0 ± 1.5 | 26.6 ± 4.0 |
Overall, we conclude that the excited states observed in the Off-Resonance R1ρ experiments measured on the d(CpG)3 and r(CpG)3 duplexes at 42°C most likely represent transiently unfolded states and not Z-form adoption. This is supported by the observation that the excited state chemical shift values for the purine C8 and pyrimidine C6 atoms of the d(CpG)3, d(5mCpG)3, and r(CpG)3 constructs are all downfield by similar magnitudes (Table 2), which occurs for duplex melting (Figure 5A, 6A). This contrasts with the Z-conformation, where the aromatic C8 atoms of purines in the syn conformation are significantly more de-shielded compared to the C6 atoms of the cytosines (Figure 5A, 6B, Figure S14).
In addition, the 5-methyl cytosine modification has a well-known stabilizing effect on the temperature-dependent melting of DNA78–80. The excited state measured by Off-Resonance R1ρ for the d(5mCpG)3 duplex has a population of 2% compared to the 4% observed for its non-methylated counterpart (Table 2), again supporting that the identity of the minor state is indeed duplex melting. We also measured Off-Resonance R1ρ on a r(CpG)6 duplex at 42°C, which is double the length of the r(CpG)3 and, therefore has a significantly higher melting temperature (Figure 4, Figure S17). If the excited state observed for the r(CpG)3 was truly a Z-conformation, it would be plausible to anticipate an excited state for the r(CpG)6. This arises from the fact that the Z-form activation energy was previously determined to be independent of chain length81, whereas transient melting should no longer be observable. However, we observe no evidence of an excited state in the r(CpG)6 duplex at 42°C. Finally, the population and kEX of the excited state in the r(CpG)3 construct grows/decreases from 1. ± 0.1% and 977 ± 53 s−1 at 3.6 mM to 6.5 ± 0.6% and 513 ± 59 s−1 at 300 μM (Figure 4, Table 5), which is also in line with the known concentration dependence of the melting temperature in nucleic acid duplexes82.
Table 5.
Conformational exchange parameters from Off-Resonance R1ρ relaxation dispersion experiments for the r(CpG)3 construct at 3 mM and 300 μM.
| Construct | Residue/Atom identity | Temperature (°C) | kex (s−1) | pE (%) | Δω (ppm) |
|---|---|---|---|---|---|
| 3.6 mM r(CpG)3 | Gua4/C8 | 42 | 977 ± 53 | 1.9 ± 0.1 | 3.76 ± 0.07 |
| 300 μM 15N, 13C G4 r(CpG)3 | Gua4/C8 | 42 | 513 ± 59 | 6.5 ± 0.6 | 4.14 ± 0.07 |
Transient Melting Promotes Z-Conformation Adoption in the Presence of Zα.
Most Z-conformation adoption models assume a high-energy nucleation event before helical handedness reversal and Z-form stabilization35 (Figure 1B). In addition, we previously observed that Z-RNA adoption within the context of A-Z junctions occurred more readily when the Z-RNA stretch was flanked by internal loops or wobble base pairs43. Therefore, we wondered whether the transient melted state observed by NMR in the d(CpG)3, d(5mCpG)3, and r(CpG)3 constructs could play a significant role in Z-form adoption in the presence of Zα domains. Particularly, we hypothesized that increasing the probability of transient duplex melting would promote the rate of Z-form adoption by Zα. To investigate this question, we employed circular dichroism (CD) spectroscopy, a technique which has been used extensively to study both Z-DNA and Z-RNA conversion43,77,83,84. The B-form, A-form, and Z-form of nucleic acids have unique CD absorbance patterns85,86 in the 220–320 nm range with minimal interference from protein signal, making it ideal to track Z-form adoption in the presence of Zα (Figure S19).
We designed and tested a series of DNA and RNA duplexes which have different levels of duplex stability (depicted in Figure S20) and confirmed their ability or inability to adopt the Z-conformation in high-salt and in 1:2n (RNA:protein, where n is the number of binding sites) complex with Zα (Figure S21, salt mid-points can be found in Figure S2, Table 1). We next followed the conversion of these duplexes to the Z-form as a function of time and at different temperatures after adding saturating amounts of Zα (as is depicted in Figure S19, results in Figure 7, Figure S22 and Table 6).
Figure 7.
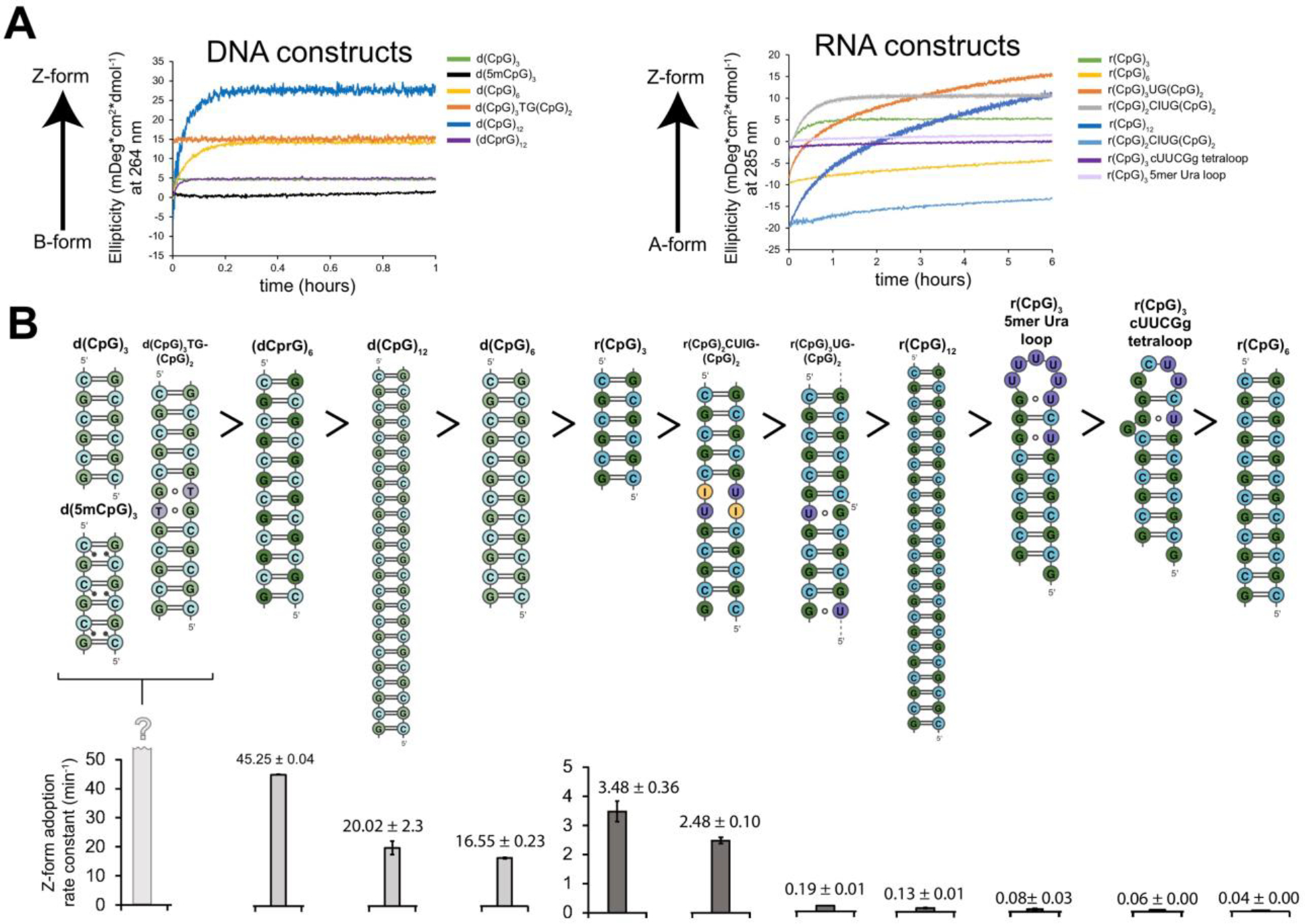
Z-form adoption rates in DNA and RNA constructs. (a) Circular dichroism time-course experiments showing the rate of Z-DNA (tracked at 266 nm) and Z-RNA (tracked at 285 nm) adoption after the addition of saturating concentrations of Zα at 25°C. The different constructs are color-coded according to the legend on the right-handed side. (b) 2D depictions of DNA and RNA constructs (above) are shown in descending order according to their Z-form adoption rates at 25°C (below), with their half-times indicated in minutes.
Table 6.
Rate constants and activation energies extracted from circular dichroism time course experiments.
| Construct | k (h−1) at 5°C | k (h−1) at 15°C | k (h−1) at 20°C | k (h−1) at 25°C | k (h−1) at 32°C | k (h−1) at 42°C | k (h−1) at 50°C | k (h−1) at 55°C | EA (kcal mol−1) |
|---|---|---|---|---|---|---|---|---|---|
| d(CpG)3 | * | * | * | ||||||
| d(5mCpG)3 | * | * | * | ||||||
| d(CpG)6 | 0.655 ± 0.009 | 2.768 ± 0.06 | 16.545 ± 0.23 | 26.562 ± 0.003 | |||||
| d(CpG)3TG(CpG)2 | * | * | * | ||||||
| (dCprG)6 | 0.732 ± 0.08 | 8.012 ± 0.11 | 45.27 ± 0.04 | 33.990 ± 0.909 | |||||
| d(CpG)12 | 0.716 ± 0.08 | 2.979 ± 0.39 | 20.015 ± 2.30 | 27.387 ± 0.045 | |||||
| r(CpG)3 | 0.464 ± 0.12 | 0.994 ± 0.002 | 3.480 ± 0.36 | 27.06 ± 0.845 | 150.6 ± 22.91 | 811.85 ± 179.96 | 40.584 ± 0.343 | ||
| r(CpG)6 | 0.035 ± 0.000 | 0.283 ± 0.002 | 1.814 ± 0.35 | 17.685 ± 2.072 | 25.145 ± 2.072 | 42.870 ± 0.719 | |||
| r(CpG)3UG(CpG)2 | 0.191 ± 0.007 | 40.625 ± 1.761 | 167.8 ± 57.70 | 44.787 ± 1.838 | |||||
| r(CpG)2CIUG(CpG)2 | 0.2038 ± 0.000 | 0.5982 ± 0.006 | 2.479 ± 0.103 | 17.865 ± 0.375 | 73.065 ± 22.154 | 40.735 ± 1.902 | |||
| r(CpG)12 | 0.129 ± 0.011 | 0.724 ± 0.003 | 7.03 ± 0.017 | 29.3 ± 0.024 | 73.75 ± 8.966 | 41.371 ± 0.340 | |||
| r(CpG)12 (2:1 Zα:RNA) | 0.117 ± 0.024 | 0.432 ± 0.045 | 3.351 ± 1.617 | 8.278 ± 0.002 | 26.465 ± 0.332 | 33.705 ± 1.021 | |||
| r(CpG)3 cUUCGg tetraloop | 0.058 ± 0.004 | 0.664 ± 0.021 | 3.099 ± 0.204 | 18.075 ± 1.181 | 42.95 ± 2.234 | 41.153 ± 0.507 | |||
| r(CpG)3 5mer Ura loop | 0.077 ± 0.026 | 0.654 ± 0.008 | 2.777 ± 0.026 | 17.74 ± 0.453 | 46.72 ± 0.084 | 40.050 ± 0.278 |
= construct already completely in the Z-conformation before wavelength monitoring began
Our NMR results showed that the r(CpG)3 construct displayed transient duplex unfolding while the r(CpG)6 construct did not. Therefore, we decided to test how duplex length (6, 12, and 24 bp CpG DNA and RNA duplexes, depicted in Figure S20) plays a role in Z-DNA and Z-RNA adoption rates in the presence of Zα. It has been previously shown that Z-form adoption is promoted as chain-length is increased in poly d(CpG)n constructs, with the rationale being that longer chains have more potential sites for high-energy nucleation events to occur thereby promoting Z-form adoption24. However, the duplex length was only able to be crudely estimated and only two lengths were tested (a 24 and 580 bp duplex), meaning that the potential effects of duplex melting due to temperature may have been missed.
We observe that the shorter 6 bp d(CpG)3 and r(CpG)3 constructs convert to the Z-form significantly faster compared to the 12 bp ones (Figure 7). The d(CpG)3 construct is already completely in the Z-conformation before we could begin the CD measurement (~5 s delay before measurement), while the d(CpG)6 converted to the Z-form with a rate constant (k) of 16.55 ± 0.18 h−1 at 25°C (Table 6). For the RNA case, a similar phenomenon was observed with the r(CpG)3 flipping to the Z-conformation 100x faster compared to the r(CpG)6 with rate constants of 3.480 ± 0.025 and 0.035 ± 0.000 h−1 at 25°C, respectively (Table 6). The slower kinetics observed for RNA is due to the higher energy barrier for flipping the sugar pucker into the C2′-endo conformation for RNA compared to DNA66,77. Interestingly, doubling the number of bps again from 12 to 24 appears to have the opposite effect, although more subtle, increasing the rates from 16.55 ± 0.18 and 0.035 ± 0.000 h−1 to 20.02 ± 0.38 and 0.129 ± 0.000 h−1, corresponding to an increase of 1.2x and 3.7x in the rate constants for DNA and RNA CpG repeats, respectively (Table 6). This increase is likely due to an increased likelihood of nucleation events occurring within the chain as it gets longer, as measured previously24. Indeed, using a sub-stoichiometric concentration of Zα for the r(CpG)12 construct (2:1 Zα:RNA where a total of 8 Zα can bind to the r(CpG)12) results in a similar rate constant to the fully saturated experiment at 25°C, but results in an overall lower final population of Z-form adoption, suggesting a high-level of cooperativity (Figure 7, Table 6). Interestingly, the rate constant becomes increasingly slower (relative to the fully saturated experiment) as the temperature increases, indicating that the lower stoichiometric amount of Zα cannot fully recapitulate the rates observed in the fully saturated experiment (Table 6). We also measured time-courses for the d(5mCpG)3 construct, but as with the d(CpG)3 duplex, it was already in the Z-conformation before measurement could begin. Therefore, Z-form adoption occurs quickly on shorter duplexes because they more easily melt compared to longer duplexes.
However, helix length is not the only factor which impacts duplex destabilization. Next, we wanted to test whether promoting base pair opening in the d(CpG)6 and r(CpG)6 constructs, which converted to the Z-form quite slowly, might increase their Z-adoption rate in the presence of Zα. It is well known that TG and UG wobble base pairs within the context of B- and A-form helices result in local helical distortions, which promote base pair dynamics87–90. Therefore, we replaced the fourth G in the d(CpG)6 and r(CpG)6 constructs with either a T or U nucleotide to create TG and UG wobble base pairs, which minimally perturb Z-form structure91. This resulted in a DNA construct with a tandem TG wobble (d(CpG)3TG(CpG)2) and an RNA one with two UG wobbles spaced apart by 4 bps (r(CpG)3UG(CpG)2) due to a register shift of the duplex (as depicted in Figure S20), as confirmed by NMR and melting temperature measurements (Figure S23, Table 1). We confirmed that Zα is still able to convert these non CpG sequences to the Z-conformation by circular dichroism (Figure S21), as anticipated from prior X-ray crystal structures of Z-DNA with non CpG sequences92.
The introduction of these TG and UG wobble base pairs into the d(CpG)6 and r(CpG)6 constructs increased their rate constants significantly (Figure 7. S22, Table 6). The Z-adoption rate of the d(CpG)3TG(CpG)2 duplex could no longer be measured as it was already in the Z-form before measurement could begin. The r(CpG)3UG(CpG)2 construct had a rate constant of 0.191 ± 0.000 h−1 at 25°C, corresponding to a 5.5x increase compared to the r(CpG)6 construct (Table 6).
Since A-to-I editing of AU base pairs by ADAR1 is well-known to destabilize dsRNA structures93–95, we also tested to see if a tandem inosine-uracil base pair insertion into the r(CpG)6 would also promote Z-form adoption rates by Zα. The r(CpG)2CUIG(CpG)2 construct (depicted in Figure S20) has a very low melting temperature of 38.39°C (Figure S3, Table 1), confirming the destabilizing effect of the tandem inosine insertion. The r(CpG)2CUIG(CpG)2 construct converted to the Z-conformation with a rate constant (k) of 2.48 ± 0.10 h−1 at 25°C (Figure 7, Figure S22, Table 6), which is 71x faster than the r(CpG)6 and only 0.7x slower than the r(CpG)3 constructs at the same temperature.
Finally, we wanted to investigate what the effects of capping a Z-forming sequence with loops of diverse stabilities has on its Z-form adoption rate. Therefore, we capped the r(CpG)3, which adopts the Z-form relatively quickly, to make two stem-loop constructs, one with a tetraloop having the cUUCGg sequence, and another with a 5mer loop containing uracils (Figure S20). Both constructs required a high concentration of salt to adopt the Z-conformation compared to the duplex constructs (adopting the Z-form around 8M NaClO4, Figure S2). Zα binding appears to promote only a partial growth in ellipticity at 285 nm, suggesting possible A-Z junction formation (Figure S21). Following the two stem-loop’s partial Z-form adoption by CD time courses showed that they have rates comparable to, albeit faster than, the r(CpG)6, with the 5mer uracil loop being slightly faster than the tetraloop construct (Figure 7, S22). Therefore, capping the r(CpG)3 construct is highly inhibitive to its Z-conformation adoption rate. We speculate that this is likely due to either sterically preventing reorganization of the stem into the correct Z-form geometry, or by making spontaneous melting events rarer.
Overall, these results suggest that the intrinsic ability of a duplex to melt has a significant effect on its Z-DNA/Z-RNA adoption rate. This is supported by several observations. First, doubling the base pairs from 6 to 12 (which depleted the population of transient duplex melting as seen by NMR) causes the Z-adoption rate to proceed significantly slower for both DNA and RNA. Second, converting Z-form duplexes to stem-loops has an inhibitory effect on their ability to adopt the Z-conformation. Third, promoting base pair opening dynamics by the introduction of TG, UG, and IU wobble base pairs into the B-form and A-form helices of the CpG constructs promoted an increase of the Z-form adoption rate, with the inosine insertion having a very pronounced effect. This posits the possibility that the A-to-I editing of an RNA by ADAR1 may promote Zα binding resulting in a positive feedback loop promoting further RNA editing, which we expand on in the discussion. These results are in line with one of our previous studies, which showed that Zα preferred to bind and convert dsRNA segments flanked by internal loops and wobble base pairs43.
Interestingly, we also note that the 8-methyl d(CpG)3 and r(CpG)3 constructs, which exist in slow exchange between the B-/A- and Z-conformations, had very low melting temperatures (Table 1). While the observation is only correlational, this supports that B-/A-form helices must be destabilized for the Z-conformation to become populated.
Z-DNA and Z-RNA formation is a complex process involving duplex melting and sugar pucker rearrangement.
Since the ease of melting an RNA or DNA duplex correlated with Z-form adoption rates in the presence of Zα, we wondered whether these constructs might have lower activation energies. The activation energy of Z-DNA and Z-RNA formation by Zα in a d(CpG)6 and r(CpG)6 construct was previously calculated using Arrhenius fits to be 24 and 38 kcal mol−1, respectively77. We carried out a similar analysis for all the constructs for which we could accurately measure the Z-adoption rate at different temperatures ranging from 5°C to 55°C (Figure S22, S24, Table 6; note that the trends observed at 25°C still hold for the other temperatures). We measured the activation energy of the d(CpG)6 and d(CpG)12 to be 26.562 ± 0.003 and 27.387 ± 0.045 kcalmol−1, respectively (Table 6). For the RNA constructs, activation energies were found to be 40.58 ± 0.34 (r(CpG)3; in agreement with the published value77), 42.87 ± 0.72 (r(CpG)6), 41.37 ± 0.34 (r(CpG)12), 44.79 ± 1.84 (r(CpG)3UG(CpG)2), 40.74 ± 1.90 (r(CpG)2CUIG(CpG)2), 41.15 ± 0.51 (r(CpG)3 cUUCGg tetraloop), and 40.05 ± 0.28 kcal mol−1 (r(CpG)3 5mer Ura loop) (Figure S24, Table 6). Therefore, despite the significant differences in the observed rate constants, the activation energies only differ significantly depending on whether the construct is DNA or RNA. These results are in agreement with an earlier study, which showed no difference in activation energy for different (CpG) chain lengths81 and suggest that there is a higher-energy process other than duplex melting that must occur before full Z-form adoption.
We reasoned this high-energy barrier is likely the rearrangement of the sugar puckers and bases as seen in the Z-conformation, which would explain the large activation energy difference between the DNA and RNA constructs, as has been previously hypothesized77. If this is true, we would expect that a DNA-RNA hybrid construct should have a lower activation energy barrier, with cytosines being deoxyribo (allowing them to more easily adopt the C2′-endo conformation), and guanines being ribo (which would favor the C3′-endo conformation). As predicted, we measured the activation energy of the (dCprG)6 construct to be ~10 kcal mol−1 lower than the r(CpG)6 construct at 33.99 ± 0.91 kcal mol−1 (Table 6). Interestingly, the (dCprG)6 construct also flips to the Z-conformation 2.7x faster than the d(CpG)6 at 25°C, which is likely due to its decreased stability as judged by its lowered TM of 55°C compared to the 78°C for the d(CpG)6, Table 1). In further support of the role of the sugar pucker conformation in the activation energy barrier of Z-form adoption, locking the guanosines into the C3′-endo by a methylene bridge between the 2′ oxygen and the 4′ carbon of the pentose ring (otherwise known as “locked” nucleic acid or LNA96) prevented the duplex (dCpLG)6 (depicted in Figure S20) from flipping to the Z-form (Figure S2, S21). This suggests that even though the guanines start and end in the C3′-endo conformation, some conformational flexibility is required during the switch from the A- to the Z-conformation, or that the locked bases prevent the duplex from melting in a way that allows for a Z-like state to be adopted.
Taken together, these data show that the major energetic barrier for Z-form adoption is the conformational rearrangement of the sugar puckers and bases and is length-independent over a 24 bp span. The large differences observed for the Z-form adoption rates can be rationalized by assuming that duplex melting represents a relatively low energy barrier process that is required prior to the high-energy conformational switch.
The conversion of r(CpG) RNA to the Z-form by Zα occurs on a slow timescale and proceeds through a single-stranded intermediate.
Our data so far suggests that Z-form adoption is a multi-step process involving duplex melting followed by sugar pucker rearrangement and stabilization. We wondered at what point Zα plays a role in this proposed model. Theoretically, Zα could only stabilize a Z-like state once it is adopted, or it could also promote duplex melting, thereby promoting the overall Z-adoption rate. We first attempted to gain insight into this question by measuring Off-Resonance R1ρ experiments on the r(CpG)3 where guanine 4 was isotopically labeled with 15N and 13C at increasing concentrations of Zα. We measured these experiments at 25°C to avoid convolution with transient duplex melting seen at 42°C. At all RNA:Zα tested ratios, we observed no evidence of dynamics on a μs-ms timescale (Figure 8A). Instead, Zα-dependent stabilization of Z-RNA occurs on a slow timescale, which was confirmed by the disappearance of guanine C8H8 from the A-form peak position and its reappearance at the Z-form position (without any observed chemical shift perturbations, Figure 8B). This finding is in agreement with earlier NMR titration studies which looked at the imino protons of d(CpG)3 and r(CpG)3 constructs upon titration of Zα, which also showed slow exchange49. For the 8mG4 d(CpG)3 construct, which is in slow exchange between the B- and Z-conformation, we were able to measure ZZ-exchange on the C8 atoms of guanine 2 and 6 (Figure S25, Table 7). From the fits of this measurement, we extracted an exchange rate from the B- to the Z-form of 24.7 ± 10.0 and 31.9 ± 11.6 s−1 and a backward rate (from the Z- to the B-form) of 5.4 ± 1.3 and 7.8 ± 1.5 s−1, for Gua2 and Gua6, respectively (Figure S25, Table 7). We carried out a similar analysis for the 8mG4 r(CpG)3 construct, which revealed an exchange rate from the A- to the Z-form of 5.4 ± 1.1, 4.6 ± 0.7, 4.8 ± 0.5 s−1 and a backward rate (from the Z- to the A-form) of 5.1 ± 0.7, 4.3 ± 1.0, 5.6 ± 0.4 s−1 for Gua2, Cyt3, and Gua6, respectively (Figure S26, Table 8). Therefore, conversion between the B-/A-form and the Z-conformation appears to occur on a slow timescale, independently of the stabilization due to chemical modification of the duplex or Zα binding.
Figure 8.
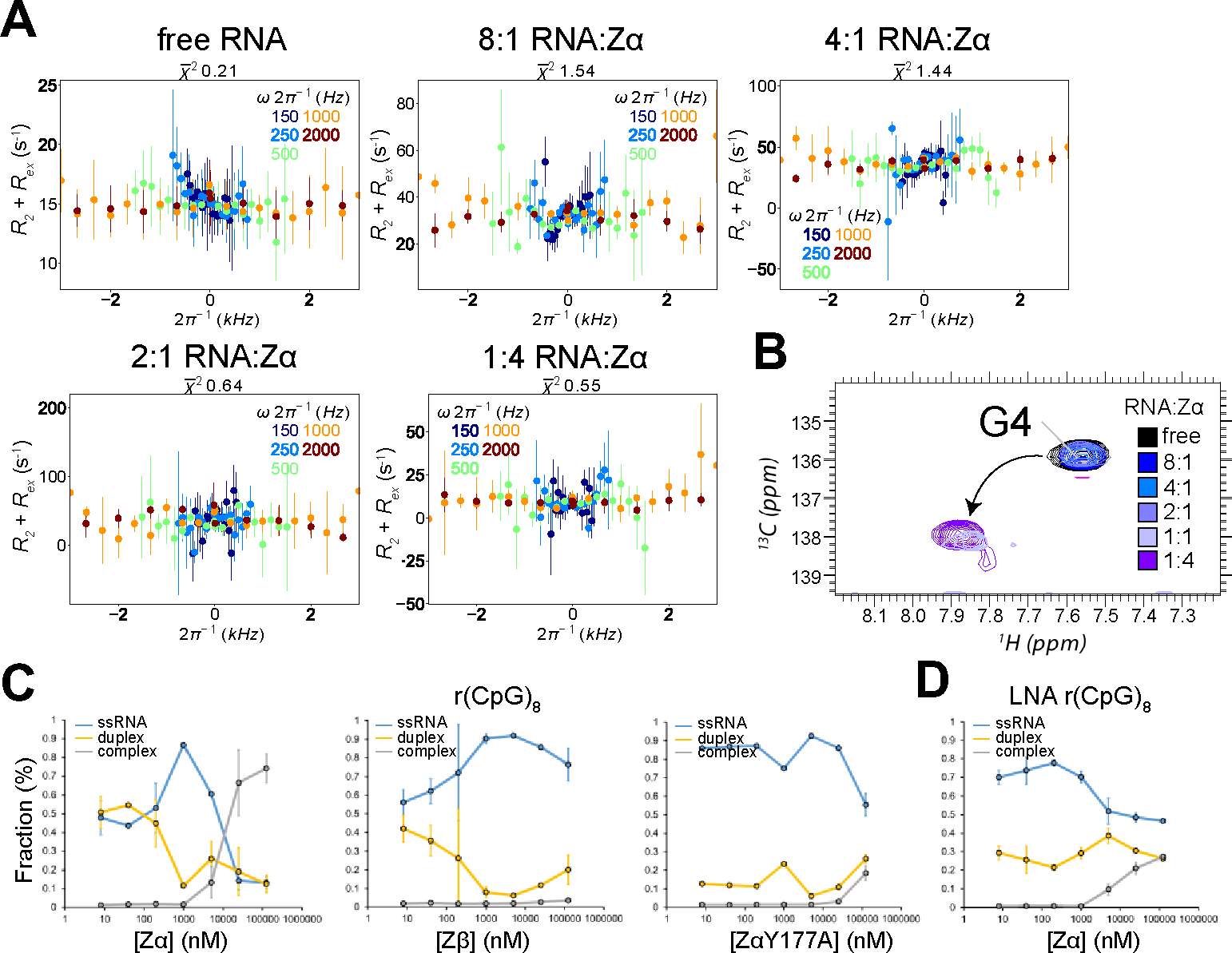
Characterization of exchange in the r(CpG)3 construct at increasing concentrations of Zα. (a) Off-Resonance R1ρ relaxation dispersion profiles for the C8 atom of isotopically labeled Gua4 in the r(CpG)3 construct measured with increasing concentrations of Zα. The molar ratio of RNA:Zα can be found at the top of the graphs. Off-resonance R1ρ experiments were carried out at five different spin-lock powers (150, 250, 500, 1000, and 2000 Hz, colored coded according to the legend on the right). R2 + Rex (= R1ρ) values are given as a function of the resonance offset from the major state (Ωoff/2π). Error bars represent experimental uncertainty. (b) 1H,13C HSQC spectra showing the C8 atom of isotopically labeled Gua4 in the r(CpG)3 construct is shown at increasing concentrations of Zα, color coded according to the legend on the right. (c) The fraction of ssRNA r(CpG)8, dsRNA r(CpG)8, and r(CpG)8 in complex with Zα, Zβ, and ZαY177A extracted from electrophoretic mobility shift assays (shown in Figure S25). The values shown are an average of two replicates plotted on a log scale. (d) Same as in (c), but with a (CpG)8 LNA construct which cannot adopt the Z-conformation.
Table 7.
Conformation exchange parameters for exchange between the B- and Z-form of the 8mG4 d(CpG)3 construct measured from ZZ-exchange.
| Residue | pB (%) at 25°C (from ratio of peak intensities) | pB (%) at 25°C (from ratio of kex) | kBZ (s−1) at 25°C | kZB (s−1) at 25°C |
|---|---|---|---|---|
| Gua2 C8 | 4.9 | 18.0 ± 11.3 | 24.7 ± 10.0 | 5.4 ± 1.3 |
| Gua6 C8 | 8.8 | 19.6 ± 11.1 | 31.9 ± 11.6 | 7.8 ± 1.5 |
Table 8.
Conformation exchange parameters for exchange between the A- and Z-form of the 8mG4 r(CpG)3 construct measured from ZZ-exchange.
| Residue | pA (%) at 25°C (from ratio of peak intensities) | pA (%) at 25°C (from ratio of Kex) | kAZ (s−1) at 25°C | kZA (s−1) at 25°C |
|---|---|---|---|---|
| Gua2 C8 | 42.0 | 48.6 | 5.4 ± 1.1 | 5.1 ± 0.7 |
| Cyt3 C6 | 40.2 | 48.4 | 4.6 ± 0.7 | 4.3 ± 1.0 |
| Gua6 C8 | 55.1 | 53.7 | 4.8 ± 0.5 | 5.6 ± 0.4 |
Serendipitously, we discovered while carrying out electrophoretic mobility shift assays (EMSA) that the binding of Zα to a r(CpG)8 construct causes the RNA to go through a single-stranded intermediate prior to Z-form adoption (Figure 8C, S27A). Under our experimental conditions, the r(CpG)8 in the free form exists in equilibrium as a single-stranded and double stranded species (Figure S27A). The two bands were assigned by titrating in excess amounts of unlabeled r(CpG)8, which increased the melting temperature TM and caused the upper band to increase in intensity (Figure S27E). In addition, running the same samples under denaturing conditions resulted in the upper and lower band merging, showing that they are not products of degradation (Figure S27F). This suggests that at the RNA concentration used for the EMSA (~24 pM), the r(CpG)8 is below the KD needed to form stable duplexes. As the concentration of Zα is increased up to 1 μM, the dsRNA species becomes further depopulated while the ssRNA species increases, without any measurable complex formation. After 1 μM, the ssRNA species disappears, and a much higher molecular weight band appears indicating complex formation (Figure 8C, S27A). We do not observe this phenomenon with an LNA version of (CpG)8 (which cannot adopt the Z-form as showed earlier, Figure S21), with little change in the relative populations of single- and double-stranded species (Figure 8D, S27D). These results further support that the DNA and RNA must go through an unfolded intermediate prior to Z-form adoption, but also that Zα is able to help promote this melting event at low concentrations. It is possible that we only observed this activity of Zα because the instability of the duplex species meant that melted duplexes could not immediately reform, allowing us to capture it on a gel.
Interestingly, duplex melting by the addition of Zα occurs relatively quickly as judged by an EMSA time-course, which showed no difference in the relative populations of dsRNA and ssRNA for the r(CpG)8 after the addition of 1 μM Zα from 5 to 35 minutes (Figure S27G). Quickly spiking in 5 and 25 μM Zα after the 30-minute mark does not lead to the productive formation of complex, as we observed for the full 30-minute incubation with Zα (compare Figure S27G to S27A). This supports our circular dichroism time-course results which suggested that a second, high-energy Z-like state must be adopted after duplex melting before productive Z-form adoption can occur.
Unexpectedly, we observe a similar behavior for Zβ (structurally conserved with Zα - isoelectric point is 8.1 compared to 9.8 - and incapable of promoting Z-form) and a Zα mutant (ZαY177A, which cannot stabilize Z-DNA/Z-RNA) but without noticeable complex formation at higher protein concentrations (Figures 8C, S27B). ZαY177A shows melting activity, but it is not observable until about 5 μM of protein (Figures 8C, S27C). Therefore, this suggests that the observed melting activity of Zα is not specific to its Z-form adoption activity, as it could be a general feature of positively charged helix-turn-helix domains.
Proposed Model for Z-Form Adoption by Zα.
Synthesizing all the results from this study, we have constructed a model for Z-DNA and Z-RNA adoption in the presence of Zα Figure 9). First, Zα binds non-specifically to a B-form or A-form helix using a similar binding interface used for Z-form stabilization (Figure 9, step 1). From here, the B-/A-form helix must experience a transient duplex melting event, which is more likely to occur in proximity to helix ends and internal loops and which Zα binding may help to promote (Figure 9, step 2). Both of these steps occur on a relatively fast timescale, with duplex melting being in the intermediate time regime (as measured by Off-Resonance R1ρ). After this, the strands must rearrange into a Z-like state with alternating sugar pucker and nucleobase conformations, which occurs slowly and is rate-limiting (Figure 9, step 3). This state is high-energy and is thus a rare state which is easier to adopt in DNA compared to RNA due to the difficulty in adopting the C2′-endo conformation in RNA. At this point, Zα will bind to the Z-like state with an order of magnitude higher affinity, stabilizing it into the Z-conformation and promoting the binding of additional Zα domains in a cooperative manner (Figure 9, step 4). This model has implications for the role of Zα domains in biology, which we discuss below.
Figure 9.
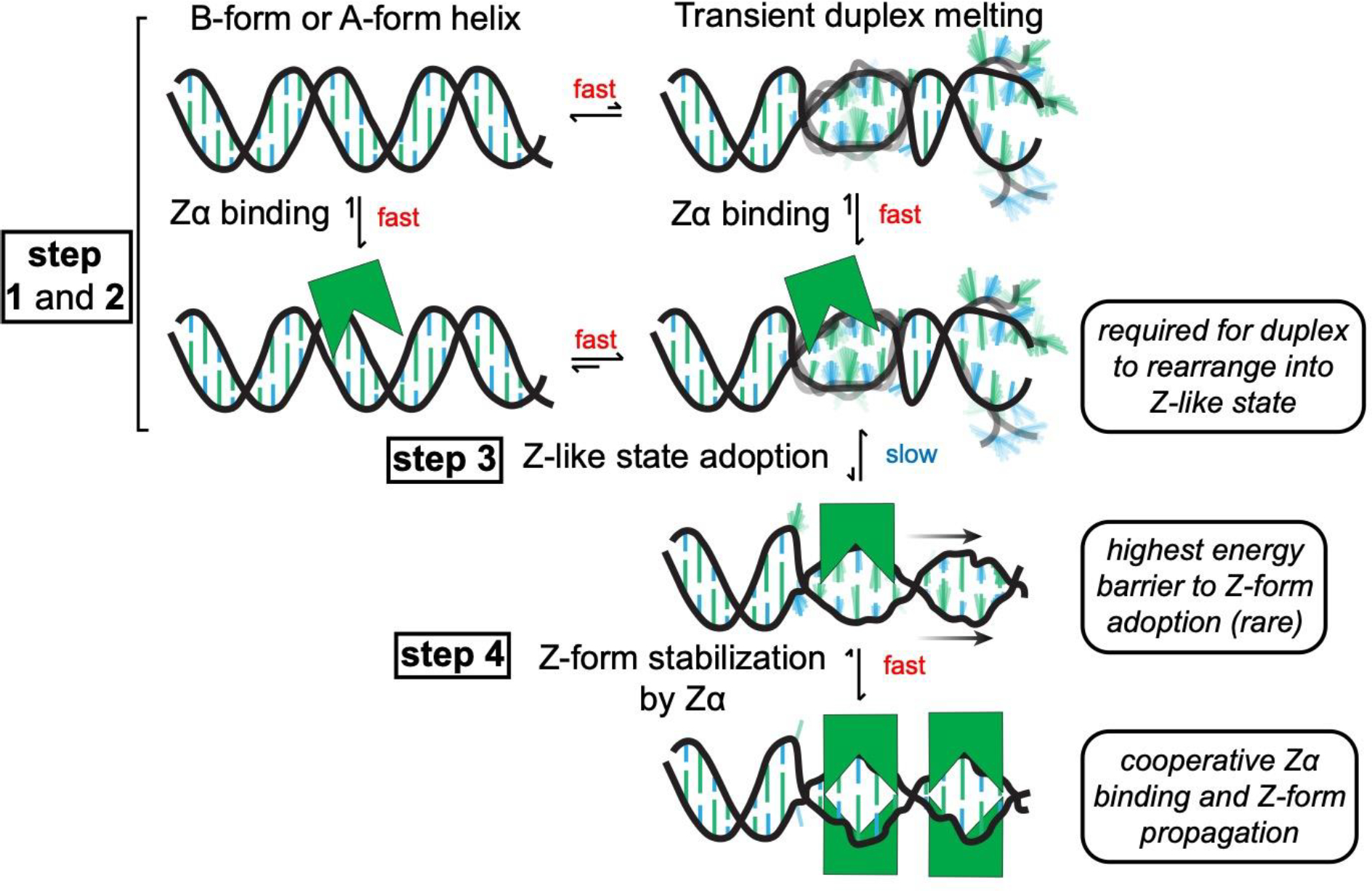
Model for Z-form adoption by Zα. First, Zα binds to a right-handed B- or A-form helix non-specifically (step 1). Next, the duplex transiently melts, which may be promoted through Zα binding (step 2). The dynamics of these steps occur on a relatively faster time scale (on the micro- to millisecond time regime). From here, the ribose sugar pucker and nucleosides have to rearrange into a high-energy, Z-like state (which occurs slowly taking seconds to hours, step 3). Then, Zα cooperatively binds to and stabilizes the Z-conformation (which occurs fast once a Z-like state is adopted, step 4)).
DISCUSSION
The conversion of right-handed helices to the left-handed Z-conformation is a thermodynamically challenging process20 involving complete helical reversal and alternation of the nucleobases and (deoxy)riboses between the syn/anti and C2′/C3′-endo conformations4,8–11. Zα domains, found within a variety of innate immune response proteins37, are able to promote this conversion in double-stranded DNA and RNA simply through binding to and stabilizing the unique Z-form geometry6,38. This process is known to involve intermediate steps45–50, but the identity of these steps has remained poorly characterized.
Our Off-Resonance R1ρ experiments, which probe low population dynamic states54–56,58, showed no evidence of transiently sampled Z-conformations but did reveal that short d/r(CpG)3 duplexes sampled a transiently unfolded state at 42°C whereas a r(CpG)6 construct did not (Figures 4–6). The transient melting observed in the shorter duplexes correlated with significantly faster Z-form adoption rates in the presence of Zα as measured by circular dichroism studies (Figure 7). We further investigated this phenomenon and showed that duplex stability generally played a major role in Z-form adoption rates, as was supported by our EMSA experiments (Figure 8). This was illustrated by the significantly faster Z-form adoption rates in the longer d/r(CpG)6 constructs with inserted non-canonical base pairs and inosine nucleobases (Figure 7).
Interestingly, despite the different nucleic acid constructs having significantly different rate constants, the only differences in measured activation energies were between DNA and RNA, with RNA having a significantly higher energy barrier (Figure S24). This suggests that there is another, rate-limiting, step after duplex melting that likely involves adopting the proper Z-form geometry (which requires more energy for RNA due to the difficulty in adopting the C2′-endo conformation77).
An Experimentally Validated Model for the Conversion of A/B to the Z-Conformation.
From these results, we propose a model whereby Zα binds to an A-/B-form helix non-specifically, the helix transiently melts and adopts a Z-like state, which is then followed by stabilization of the Z-form structure in a Zα-dependent manner (Figure 9). This model is very similar to the previously proposed zipper model32 but also takes into account the role of Zα domains and provides experimental evidence that the high-energy nucleation event described in the zipper model is likely duplex unfolding. We believe that this makes sense from a biochemical and structural perspective, as the intuitively most straight-forward path to convert from a right-handed to a left-handed helix would be to locally dissociate the two right-handed and base-paired strands from each other and reanneal them together in the left-handed conformation. This would also explain why B-Z junction adoption was previously shown to occur much more quickly than Z-DNA by itself97, as the favorable entropic energy gain from the creation of the junctions between B-DNA/A-RNA and Z-form sequences likely allows for the strands to dissociate and reform into the Z-conformation more easily.
Does Zα Stabilize Z-Conformations via an Active or Passive Mechanism?
One open question in the field is determining whether Zα domains play an “active” or “passive” role in Z-form adoption. That is, whether Zα domains recognize pre-sampled Z-conformations and subsequently stabilize them (the passive mechanism52) or whether binding of Zα to DNA/RNA pushes the helix into the Z-form (the active mechanism45–47,49). There is evidence for both models of Z-DNA/RNA adoption45–51,53. Our data suggest that two of the intermediate states in the pathway to Z-form adoption are duplex melting and conformational rearrangement into a Z-like state (Figure 9). In addition, we showed that Zα likely stabilizes a Z-like state which is adopted prior to full Z-conformation stabilization, which occurs on a slow timescale. Our data do not allow us to conclude definitively whether this state is adopted independently or if Zα is required to push the nucleic acid into this state once the duplex is melted. Through our Off Resonance R1ρ data, we only detected the presence of transient melting of our duplex constructs in the absence of Zα. This suggests that a possible transient Z-like state is either outside of the timescale measurable by Off Resonance R1ρ, or that the state is exceedingly rare and Zα is required for its adoption. However, by comparing the A-form peak intensities of the isotopically labeled r(CpG)3 construct to the noise level where the Z-form peak would be expected to be (under the slow exchange limit) at 25°C, the Z-conformation population cannot be more than 0.08%. If transient Z-form adoption occurs on a slow timescale, this would mean that it is a rare state under low salt conditions and without divalent metal ions.
Therefore, for nucleic acids that are not pre-stabilized into the Z-conformation by chemical modifications, buffer conditions, torsional/mechanical stress98,99, or other potential unknown factors, the presence of Zα is likely required for Z-DNA and Z-RNA to become populated at a significant level. In this way, Z-conformation adoption due to the presence of Zα can be thought of as an active mechanism, especially considering that Zα binds to A-form helices prior to Z-form adoption (as evidence by the binding of Zα to the (CpG)8 LNA, Figure S27D). However, that is not to say that Zα-dependent stabilization of Z-conformations always proceeds by an active mechanism. For pre-stabilized Z-form duplexes, Zα would likely immediately recognize and bind to the Z-form state, indicating a passive mechanism. For duplexes not already in the Z-form however, the active mechanism is more appropriate to describe how the helix transitions from the B/A-form to the Z-form.
Does A-to-I Editing by ADAR1 Induce Z-RNA?
Adenosine deaminase acting on RNA 1 (ADAR1) is an A-to-I editase which regulates the innate immune response by preventing activation of dsRNA sensors37,59. The longer isoform of ADAR1 contains an N-terminal Zα domain, which has been shown to play an important role in ADAR1’s function42,100,101. Our finding that inosylation of the r(CpG)6 construct increased its Z-form adoption rate to a comparable level as the r(CpG)3 was striking. Based on this result, we speculate that the A-to-I editing activity of ADAR1 could promote Z-RNA adoption in ADAR1’s substrates which could in turn facilitate Zα binding and further A-to-I editing, helping to alleviate the loss of A-to-I editing activity observed on shorter dsRNA segments102. This model could partially explain ADAR1p150’s higher overall A-to-I editing activity103–105 and its significantly enhanced editing of substrates containing (CpG) repeat sequences106.
Another mechanism which would lead to the same result is that the increased Z-RNA adoption due to A-to-I editing activity does not promote further editing, but instead helps further shield the RNA from dsRNA sensors107 by converting potential substrates into a conformation that is not readily recognizable by their typical A-form binding domains. Since the Zα domain would act in cis with the deaminase domain within ADAR1, it would likely also be able to outcompete ZBP1108–110 for binding to the Z-form targets it helps to create.
Under What Cellular Contexts Would Z-Conformations be Predicted to Form?
Extrapolating from our findings, we predict that Z-conformations would be more likely to form in dsRNAs containing many non-canonical base pairs, internal loops, or editing events that help to partially destabilize the double-stranded character of the RNA segment. This would allow for the Z-conformation to be adopted more easily and thus promote Zα binding. However, too many destabilizing base pairs would eventually cause the adopted Z-conformation to also become destabilized9. This would also be impacted by the number of pyrimidine-purine repeats, which determine the final stability of the adopted Z-conformation9. Therefore, there is likely a balance between the ease of helix melting and the stability of the adopted Z-conformation which determines the adoption rate and overall longevity of Z-conformations in the cell. In addition to the RNA sequence itself, many potential trans-acting factors would also be predicted to promote Z-form adoption, including helicase activity111 and torsional/mechanical stress98, which would facilitate duplex melting and therefore promote Z-form adoption as previously hypothesized99.
Should we revisit our outlook on Z-formation in cells?
A lot of focus has been put on identifying the RNA targets of Zα domains which have mostly revealed long dsRNA formed from repetitive elements109,110,112, such as Alu elements in humans, and viral RNAs113–115. We wonder whether the identity of the RNA targets is not as important as the result of converting those targets to the Z-conformation. Alu elements could be targeted because they represent one of the largest sources of dsRNA in the human transcriptome, not because they possess better Z-form adopting sequences. This could explain why Zα has been shown to target a variety of double-stranded fragments43,44. The question is if Alu elements are targeted for editing over proportionally compared to their relative abundance. The existence of evolutionarily conserved domains which facilitate Z-DNA and Z-RNA adoption suggests that there is a major advantage in converting double-stranded nucleic acids to the Z-conformation. For example, forcing long dsRNAs into the Z-form may be a potent mechanism to avoid triggering certain dsRNA sensors, which would have otherwise recognized the long tracts of A-form helices. Alternatively, perhaps converting dsRNA into the Z-form could act as a mechanism to evict A-form binding proteins, thereby promoting even further Zα binding and amplifying the signal of whichever Zα-containing protein is coating a particular RNA.
Supplementary Material
ACKNOWLEDGMENT
We would like to thank Atul Rangadurai for help implementing and analyzing the Off-Resonance R1ρ relaxation dispersion experiments, along with the NMR facility manager David Jones.
Funding Sources
We gratefully acknowledge for support Jeffrey Kieft (to P.N. and Q.V.), the National Science Foundation (Award #2153787 to Q.V. and B.V.), and the National Institutes of Health (Award #1F31AI167396 to P.N.).
REFERENCES
- (1).Wolfram S Principles of Nucleic Acid Structure, 1st ed.; Springer; New York: New York, 1984. [Google Scholar]
- (2).Stefl R; Skrisovska L; Allain FHT RNA Sequence- and Shape-Dependent Recognition by Proteins in the Ribonucleoprotein Particle. EMBO Reports. 2005. 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Battistini F; Hospital A; Buitrago D; Gallego D; Dans PD; Gelpí JL; Orozco M How B-DNA Dynamics Decipher Sequence-Selective Protein Recognition. J. Mol. Biol. 2019. 10.1016/j.jmb.2019.07.021. [DOI] [PubMed] [Google Scholar]
- (4).Hall K; Cruz P; Tinoco I; Jovin TM; Van De Sande JH “Z-RNA” - A Left-Handed RNA Double Helix. Nature 1984. 10.1038/311584a0. [DOI] [PubMed] [Google Scholar]
- (5).Schwartz T; Rould MA; Lowenhaupt K; Herbert A; Rich A Crystal Structure of the Zα Domain of the Human Editing Enzyme ADAR1 Bound to Left-Handed Z-DNA. Science (80-. ). 1999. 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- (6).Placido D; Brown BA 2nd.; Lowenhaupt K.; Rich A.; Athanasiadis, A. A Left-Handed RNA Double Helix Bound by the Z Alpha Domain of the RNA-Editing Enzyme ADAR1. Structure 2007, 15 (4), 395–404. 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Popenda M; Milecki J; Adamiak RW High Salt Solution Structure of a Left-Handed RNA Double Helix. Nucleic Acids Res 2004, 32 (13), 4044–4054. 10.1093/nar/gkh736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wang AHJ; Quigley GJ; Kolpak FJ; Crawford JL; Van Boom JH; Van Der Marel G; Rich A Molecular Structure of a Left-Handed Double Helical DNA Fragment at Atomic Resolution. Nature 1979. 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- (9).Krall JB; Nichols PJ; Henen MA; Vicens Q; Vögeli B Structure and Formation of Z-DNA and Z-RNA. Molecules 2023, 28 (2). 10.3390/molecules28020843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Ho PS; Mooers BH Z-DNA Crystallography. Biopolymers 1997, 14 (1), 65–90. [DOI] [PubMed] [Google Scholar]
- (11).Wang AJ; Quigley GJ; Kolpak FJ; van der Marel G; van Boom JH; Rich A Left-Handed Double Helical DNA: Variations in the Backbone Conformation. Science (80-. ). 1981, 211 (4478), 171–176. 10.1126/science.7444458. [DOI] [PubMed] [Google Scholar]
- (12).Zirbel CL; Auffinger P Lone Pair…π Contacts and Structure Signatures of r(UNCG) Tetraloops, Z-Turns, and Z-Steps: A WebFR3D Survey. Molecules 2022, 27 (14), 4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Kruse H; Mrazikova K; D’Ascenzo L; Sponer J; Auffinger P Short but Weak: The Z-DNA Lone-Pair⋅⋅⋅π Conundrum Challenges Standard Carbon Van Der Waals Radii. Angew. Chemie - Int. Ed. 2020. 10.1002/anie.202004201. [DOI] [PubMed] [Google Scholar]
- (14).Šponer J; Gabb HA; Leszczynski J; Hobza P Base-Base and Deoxyribose-Base Stacking Interactions in B-DNA and Z- DNA: A Quantum-Chemical Study. Biophys. J. 1997. 10.1016/S0006-3495(97)78049-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Kollman PA; Weiner PK; Dearing A THEORETICAL STUDIES OF THE STRUCTURE AND ENERGIES OF BASE-PAIRED NUCLEOTIDES AND THE DISSOCIATION KINETICS OF A PROFLAVINE-DINUCLEOTIDE COMPLEX. Ann. N. Y. Acad. Sci. 1981. 10.1111/j.1749-6632.1981.tb50572.x. [DOI] [PubMed] [Google Scholar]
- (16).Gueron M; Demaret JP A Simple Explanation of the Electrostatics of the B-to-Z Transition of DNA. Proc. Natl. Acad. Sci. U. S. A. 1992. 10.1073/pnas.89.13.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Misra VK; Honig B The Electrostatic Contribution to the B to Z Transition of DNA. Biochemistry 1996. 10.1021/bi951463y. [DOI] [PubMed] [Google Scholar]
- (18).Guéron M; Demaret JP; Filoche M A Unified Theory of the B-Z Transition of DNA in High and Low Concentrations of Multivalent Ions. Biophys. J. 2000. 10.1016/S0006-3495(00)76665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Harvey SC DNA Structural Dynamics: Longitudinal Breathing as a Possible Mechanism for the B in Equilibrium Z Transition. Nucleic Acids Res. 1983, 11 (14), 4867–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Jovin T The Transition Between B-DNA And Z-DNA. Annu. Rev. Phys. Chem. 1987. 10.1146/annurev.physchem.38.1.521. [DOI] [Google Scholar]
- (21).Ivanov V; Grzeskowiak K; Zocchi G Evidence for an Intermediate State in the B-to-Z Transition of DNA. J. Phys. Chem. B 2003. 10.1021/jp035593p. [DOI] [Google Scholar]
- (22).Fuertes MA; Cepeda V; Alonso C; Perez JM Molecular Mechanisms for the B-Z Transition in the Example of Poly[d(G-C) x d(G-C)] Polymers. A Critical Review. Chem Rev 2006, 106 (6), 2045–2064. 10.1021/cr050243f. [DOI] [PubMed] [Google Scholar]
- (23).Sung CH; Lowenhaupt K; Rich A; Kim YG; Kyeong KK Crystal Structure of a Junction between B-DNA and Z-DNA Reveals Two Extruded Bases. Nature 2005. 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- (24).Pohl FM; Jovin TM Salt-Induced Co-Operative Conformational Change of a Synthetic DNA: Equilibrium and Kinetic Studies with Poly (DG-DC). J Mol Biol 1972, 67 (3), 375–396. 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- (25).Pohl FM Salt-Induced Transition between Two Double-Helical Forms of Oligo (DC-DG). Cold Spring Harb Symp Quant Biol 1983, 47 Pt 1, 113–117. 10.1101/sqb.1983.047.01.014. [DOI] [PubMed] [Google Scholar]
- (26).Goto S Characterization of Intermediate Conformational States in the B in Equilibrium Z Transitions of Poly(DG-DC).Poly(DG-DC). Biopolymers 1984, 23 (11 Pt 1), 2211–2222. 10.1002/bip.360231108. [DOI] [PubMed] [Google Scholar]
- (27).Manzini G; Xodo LE; Quadrifoglio F; Van Boom JH; Van Der Marel GA Dc-Dg Alternating Oligonucleotides: Thermodynamic and Kinetic Aspects of the b-z Transformation. J. Biomol. Struct. Dyn. 1987. 10.1080/07391102.1987.10507666. [DOI] [PubMed] [Google Scholar]
- (28).Sarma RH; Sarma MH Understanding the Motiions of DNA. In Biomolecular Sterodynamics, Adenine; 1981; pp 327–343. [Google Scholar]
- (29).Sundaralingam M; Westhof E Structural Motifs of the Nucleotidyl Unit and the Handedness of Polynucleotide Helices. Int. J. Quantum Chem. 1981, 287–306. [Google Scholar]
- (30).Chaires JB; Sturtevant JM Thermodynamics of the B to Z Transition in Poly(DGdC). Biopolymers 1988. 10.1002/bip.360270905. [DOI] [PubMed] [Google Scholar]
- (31).Saenger W; Heinemann U Raison d’être and Structural Model for the B-Z Transition of Poly d(G-C)*poly d(G-C). FEBS Lett. 1989. 10.1016/0014-5793(89)81539-X. [DOI] [PubMed] [Google Scholar]
- (32).Ho PS The Non-B-DNA Structure of d(CA/TG)(n) Does Not Differ from That of Z-DNA. Proc. Natl. Acad. Sci. U. S. A. 1994. 10.1073/pnas.91.20.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lim W; Feng YP The Stretched Intermediate Model of B-Z DNA Transition. Biophys. J. 2005. 10.1529/biophysj.104.052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Czarny RS; Ho PS Thermogenomic Analysis of Left-Handed Z-DNA Propensities in Genomes. In Z-DNA: Methods and Protocols; Kim KK., Subramani VK., Eds.; Springer; US: New York, NY, 2023; pp 195–215. 10.1007/978-1-0716-3084-6_14. [DOI] [PubMed] [Google Scholar]
- (35).Fuertes MA; Cepeda V; Alonso C; Perez JM Molecular Mechanisms for the B-Z Transition in the Example of Poly[d(G—C)×d(G—C)] Polymers. A Critical Review. ChemInform 2006. 10.1002/chin.200637261. [DOI] [PubMed] [Google Scholar]
- (36).Kastenholz MA; Schwartz TU; Hünenberger PH The Transition between the B and Z Conformations of DNA Investigated by Targeted Molecular Dynamics Simulations with Explicit Solvation. Biophys. J. 2006. 10.1529/biophysj.106.083667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Nichols PJ; Krall JB; Henen MA; Vögeli B; Vicens Q Z-RNA Biology: A Central Role in the Innate Immune Response? RNA 2023. 10.1261/rna.079429.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Brown BA 2nd.; Lowenhaupt K.; Wilbert CM.; Hanlon EB.; Rich A. The Zalpha Domain of the Editing Enzyme DsRNA Adenosine Deaminase Binds Left-Handed Z-RNA as Well as Z-DNA. Proc Natl Acad Sci U S A 2000, 97 (25), 13532–13536. 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Schwartz T; Rould MA; Lowenhaupt K; Herbert A; Rich A Crystal Structure of the Zalpha Domain of the Human Editing Enzyme ADAR1 Bound to Left-Handed Z-DNA. Science (80-. ). 1999, 11 (284(5421)), 1841–1845. [DOI] [PubMed] [Google Scholar]
- (40).Langeberg CJ; Nichols PJ; Henen MA; Vicens Q; Vögeli B Differential Structural Features of Two Mutant ADAR1p150 Zα Domains Associated with Aicardi-Goutières Syndrome. J. Mol. Biol. 2023, 435 (8), 168040. 10.1016/j.jmb.2023.168040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Schade M; Turner CJ; Lowenhaupt K; Rich A; Herbert A Structure-Function Analysis of the Z-DNA-Binding Domain Zalpha of DsRNA Adenosine Deaminase Type I Reveals Similarity to the (Alpha + Beta) Family of Helix-Turn-Helix Proteins. EMBO J. 1999, 18 (2), 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Nakahama T; Kato Y; Shibuya T; Inoue M; Kim JI; Vongpipatana T; Todo H; Xing Y; Kawahara Y Mutations in the Adenosine Deaminase ADAR1 That Prevent Endogenous Z-RNA Binding Induce Aicardi-Goutieres-Syndrome-like Encephalopathy. Immunity 2021, 54 (9), 1976–1988 e7. 10.1016/j.immuni.2021.08.022. [DOI] [PubMed] [Google Scholar]
- (43).Nichols PJ; Bevers S; Henen M; Kieft JS; Vicens Q; Vögeli B Recognition of Non-CpG Repeats in Alu and Ribosomal RNAs by the Z-RNA Binding Domain of ADAR1 Induces A-Z Junctions. Nat. Commun. 2021. 10.1038/s41467-021-21039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Feng S; Li H; Zhao J; Pervushin K; Lowenhaupt K; Schwartz TU; Dröge P Alternate RRNA Secondary Structures as Regulators of Translation. Nat. Struct. Mol. Biol. 2011. 10.1038/nsmb.1962. [DOI] [PubMed] [Google Scholar]
- (45).Kang YM; Bang J; Lee EH; Ahn HC; Seo YJ; Kyeong KK; Kim YG; Choi BS; Lee JH NMR Spectroscopic Elucidation of the B-Z Transition of a DNA Double Helix Induced by the Zα Domain of Human ADAR1. J. Am. Chem. Soc. 2009. 10.1021/ja902654u. [DOI] [PubMed] [Google Scholar]
- (46).Lee AR; Park CJ; Cheong HK; Ryu KS; Park JW; Kwon MY; Lee J; Kim KK; Choi BS; Lee JH Solution Structure of the Z-DNA Binding Domain of PKR-like Protein Kinase from Carassius Auratus and Quantitative Analyses of the Intermediate Complex during B-Z Transition. Nucleic Acids Res. 2016. 10.1093/nar/gkw025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Lee EH; Seo YJ; Kim HE; Lee YM; Kim CM; Lee JH Population Analysis of the Intermediate Complex States During B-Z Transition of Non-CG-Repeat DNA Duplexes Induced by the Z Alpha Domain of Human ADAR1. Bull. Korean Chem. Soc. 2011, 32 (2), 719–721. 10.5012/bkcs.2011.32.2.719. [DOI] [Google Scholar]
- (48).Kim SH; Lim SH; Lee AR; Kwon DH; Song HK; Lee JH; Cho M; Johner A; Lee NK; Hong SC Unveiling the Pathway to Z-DNA in the Protein-Induced B-Z Transition. Nucleic Acids Res. 2018. 10.1093/nar/gky200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Lee AR; Hwang J; Hur JH; Ryu KS; Kim KK; Choi BS; Kim NK; Lee JH NMR Dynamics Study Reveals the Zα Domain of Human ADAR1 Associates with and Dissociates from Z-RNA More Slowly than Z-DNA. ACS Chem. Biol. 2019. 10.1021/acschembio.8b00914. [DOI] [PubMed] [Google Scholar]
- (50).Park C; Zheng X; Park CY; Kim J; Lee SK; Won H; Choi J; Kim YG; Choi HJ Dual Conformational Recognition by Z-DNA Binding Protein Is Important for the B-Z Transition Process. Nucleic Acids Res 2020, 48 (22), 12957–12971. 10.1093/nar/gkaa1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Behe M; Zimmerman S; Felsenfeld G Changes in the Helical Repeat of Poly(DG-M5 DC) · Poly(DG-M5dC) and Poly(DG-DC) · Poly(DG-DC) Associated with the B - Z Transition. Nature 1981. 10.1038/293233a0. [DOI] [PubMed] [Google Scholar]
- (52).Bae S; Kim D; Kim KK; Kim YG; Hohng S Intrinsic Z-DNA Is Stabilized by the Conformational Selection Mechanism of Z-DNA-Binding Proteins. J. Am. Chem. Soc. 2011. 10.1021/ja107498y. [DOI] [PubMed] [Google Scholar]
- (53).Schade M; Turner CJ; Kühne R; Schmieder P; Lowenhaupt K; Herbert A; Rich A; Oschkinat H The Solution Structure of the Za Domain of the Human RNA Editing Enzyme ADAR1 Reveals a Prepositioned Binding Surface for Z-DNA. Proc. Natl. Acad. Sci. U. S. A. 1999. 10.1073/pnas.96.22.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Rangadurai A; Szymaski ES; Kimsey IJ; Shi H; Al-Hashimi HM Characterizing Micro-to-Millisecond Chemical Exchange in Nucleic Acids Using off-Resonance R1ρ Relaxation Dispersion. Progress in Nuclear Magnetic Resonance Spectroscopy. 2019. 10.1016/j.pnmrs.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Palmer AG; Massi F Characterization of the Dynamics of Biomacromolecules Using Rotating-Frame Spin Relaxation NMR Spectroscopy. Chemical Reviews. 2006. 10.1021/cr0404287. [DOI] [PubMed] [Google Scholar]
- (56).Palmer AG Chemical Exchange Effects in Biological Macromolecules. eMagRes 2007. 10.1002/9780470034590.emrstm0068. [DOI] [Google Scholar]
- (57).Bothe JR; Nikolova EN; Eichhorn CD; Chugh J; Hansen AL; Al-Hashimi HM Characterizing RNA Dynamics at Atomic Resolution Using Solution-State NMR Spectroscopy. Nature Methods. 2011, pp 919–931. 10.1038/nmeth.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Mulder FAA; Mittermaier A; Hon B; Dahlquist FW; Kay LE Studying Excited States of Proteins by NMR Spectroscopy. Nat. Struct. Biol. 2001. 10.1038/nsb1101-932. [DOI] [PubMed] [Google Scholar]
- (59).Lamers MM; van den Hoogen BG; Haagmans BL ADAR1: “Editor-in-Chief” of Cytoplasmic Innate Immunity. Frontiers in immunology. 2019. 10.3389/fimmu.2019.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Herbert A; Schade M; Lowenhaupt K; Alfken J; Schwartz T; Shlyakhtenko LS; Lyubchenko YL; Rich A The Zalpha Domain from Human ADAR1 Binds to the Z-DNA Conformer of Many Different Sequences. Nucleic Acids Res 1998, 26 (15), 3486–3493. 10.1093/nar/26.15.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (61).Placido D; Brown BA; Lowenhaupt K; Rich A; Athanasiadis A A Left-Handed RNA Double Helix Bound by the Zα Domain of the RNA-Editing Enzyme ADAR1. Structure 2007. 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Davis PW; Hall K; Cruz P; Tinoco I; Neilson T The Tetraribonucleotide RCpGpCpG Forms a Left-Handed Z-RNA Double-Helix. Nucleic Acids Res. 1986. 10.1093/nar/14.3.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Rich A; Nordheim A; Wang AHJ The Chemistry and Biology of Left-Handed Z-DNA. Annu. Rev. Biochem. 1983. 10.1146/annurev.bi.53.070184.004043. [DOI] [PubMed] [Google Scholar]
- (64).Rozners E; Moulder J Hydration of Short DNA, RNA and 2’-OMe Oligonucleotides Determined by Osmotic Stressing. Nucleic Acids Res 2004, 32 (1), 248–254. 10.1093/nar/gkh175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Teng M. kun; Liaw YC.; van der Marel GA.; van Boom JH.; Wang AHJ. Effects of the O2’ Hydroxyl Group on Z-DNA Conformation: Structure of Z-RNA and (AraC)-[Z-DNA]. Biochemistry 1989. 10.1021/bi00438a001. [DOI] [PubMed] [Google Scholar]
- (66).Foloppe N; MacKerell AD Conformational Properties of the Deoxyribose and Ribose Moieties of Nucleic Acids: A Quantum Mechanical Study. J. Phys. Chem. B 1998. 10.1021/jp9818683. [DOI] [Google Scholar]
- (67).Egli M; Gessner RV Stereoelectronic Effects of Deoxyribose O4’ on DNA Conformation. Proc Natl Acad Sci U S A 1995, 92 (1), 180–184. 10.1073/pnas.92.1.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Mráziková K; Šponer J; Mlýnský V; Auffinger P; Kruse H Short-Range Imbalances in the AMBER Lennard-Jones Potential for (Deoxy)Ribose···Nucleobase Lone-Pair···pContacts in Nucleic Acids. J. Chem. Inf. Model. 2021. 10.1021/acs.jcim.1c01047. [DOI] [PubMed] [Google Scholar]
- (69).Berger I; Egli M; Rich A Inter-Strand C-H...O Hydrogen Bonds Stabilizing Four-Stranded Intercalated Molecules: Stereoelectronic Effects of O4’ in Cytosine-Rich DNA. Proc Natl Acad Sci U S A 1996, 93 (22), 12116–12121. 10.1073/pnas.93.22.12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Desiraju GR The C-h...o Hydrogen Bond: Structural Implications and Supramolecular Design. Acc Chem Res 1996, 29 (9), 441–449. 10.1021/ar950135n. [DOI] [PubMed] [Google Scholar]
- (71).Behe M; Felsenfeld G Effects of Methylation on a Synthetic Polynucleotide: The B--Z Transition in Poly(DG-M5dC).Poly(DG-M5dC). Proc Natl Acad Sci U S A 1981, 78 (3), 1619–1623. 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Sugiyama H; Kawai K; Matsunaga A; Fujimoto K; Saito I; Robinson H; Wang AHJ Synthesis, Structure and Thermodynamic Properties of 8-Methylguanine-Containing Oligonucleotides: Z-DNA under Physiological Salt Conditions. Nucleic Acids Res. 1996. 10.1093/nar/24.7.1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Balasubramaniyam T; Ishizuka T; Xiao C. Da; Bao HL.; Xu Y. 2-O-Methyl-8-Methylguanosine as a Z-Form RNA Stabilizer for Structural and Functional Study of Z-RNA. Molecules 2018. 10.3390/molecules23102572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Xu Y; Ikeda R; Sugiyama H 8-Methylguanosine: A Powerful Z-DNA Stabilizer. J Am Chem Soc 2003, 125 (44), 13519–13524. 10.1021/ja036233i. [DOI] [PubMed] [Google Scholar]
- (75).Mittermaier A ZZ-Exchange. In Encyclopedia of Biophysics; Roberts GCK., Ed.; Springer; Berlin Heidelberg: Berlin, Heidelberg, 2013; pp 2796–2797. 10.1007/978-3-642-16712-6_354. [DOI] [Google Scholar]
- (76).Palmer AG Chemical Exchange in Biomacromolecules: Past, Present, and Future. J. Magn. Reson. 2014. 10.1016/j.jmr.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Brown BA; Lowenhaupt K; Wilbert CM; Hanlon EB; Rich A The Zα Domain of the Editing Enzyme DsRNA Adenosine Deaminase Binds Left-Handed Z-RNA as Well as Z-DNA. Proc. Natl. Acad. Sci. U. S. A. 2000. 10.1073/pnas.240464097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Nardo L; Lamperti M; Salerno D; Cassina V; Missana N; Bondani M; Tempestini A; Mantegazza F Effects of Non-CpG Site Methylation on DNA Thermal Stability: A Fluorescence Study. Nucleic Acids Res. 2015. 10.1093/nar/gkv884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Rausch C; Zhang P; Casas-Delucchi CS; Daiß JL; Engel C; Coster G; Hastert FD; Weber P; Cristina Cardoso M Cytosine Base Modifications Regulate DNA Duplex Stability and Metabolism. Nucleic Acids Res. 2021. 10.1093/nar/gkab509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Rodríguez López CM; Lloyd AJ; Leonard K; Wilkinson MJ Differential Effect of Three Base Modifications on DNA Thermostability Revealed by High Resolution Melting. Anal. Chem. 2012. 10.1021/ac301459x. [DOI] [PubMed] [Google Scholar]
- (81).Pohl FM; Jovin TM Salt-Induced Co-Operative Conformational Change of a Synthetic DNA: Equilibrium and Kinetic Studies with Poly(DG-DC). J. Mol. Biol. 1972. 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- (82).Freier SM; Kierzek R; Jaeger JA; Sugimoto N; Caruthers MH; Neilson T; Turner DH Improved Free-Energy Parameters for Predictions of RNA Duplex Stability. Proc. Natl. Acad. Sci. U. S. A. 1986. 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Kypr J; Kejnovská I; Renčiuk D; Vorlíčková M Circular Dichroism and Conformational Polymorphism of DNA. Nucleic Acids Research. 2009. 10.1093/nar/gkp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Subramani VK; Kim KK Characterization of Z-DNA Using Circular Dichroism. In Methods in Molecular Biology; 2023. 10.1007/978-1-0716-3084-6_2. [DOI] [PubMed] [Google Scholar]
- (85).Miyahara T; Nakatsuji H; Sugiyama H Helical Structure and Circular Dichroism Spectra of DNA: A Theoretical Study. J Phys Chem A 2013, 117 (1), 42–55. 10.1021/jp3085556. [DOI] [PubMed] [Google Scholar]
- (86).Nichols PJ; Bevers S; Henen MA; Kieft JS; Vicens Q; Vögeli B Adoption of A–Z Junctions in RNAs by Binding of Zα Domains. In Methods in Molecular Biology; 2023. 10.1007/978-1-0716-3084-6_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (87).Chong YE; Guo M; Yang XL; Kuhle B; Naganuma M; Sekine S. ichi; Yokoyama S.; Schimmel P. Distinct Ways of G:U Recognition by Conserved TRNA Binding Motifs. Proc. Natl. Acad. Sci. U. S. A. 2018. 10.1073/pnas.1807109115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Masquida B; Westhof E On the Wobble GoU and Related Pairs. RNA. 2000. 10.1017/S1355838200992082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Imhof P; Zahran M The Effect of a G:T Mispair on the Dynamics of DNA. PLoS One 2013. 10.1371/journal.pone.0053305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Patel DJ; Kozlowski SA; Marky LA; Rice JA; Broka C; Dallas J; Itakura K; Breslauer KJ Structure, Dynamics, and Energetics of Deoxyguanosine-Thymidine Wobble Base Pair Formation in the Self-Complementary d(CGTGAATTCGCG) Duplex in Solution. Biochemistry 1982, 21 (3), 437–444. 10.1021/bi00532a003. [DOI] [PubMed] [Google Scholar]
- (91).Ho PS; Frederick CA; Quigley GJ; van der Marel GA; van Boom JH; Wang AH; Rich AG T Wobble Base-Pairing in Z-DNA at 1.0 A Atomic Resolution: The Crystal Structure of d(CGCGTG). EMBO J 1985, 4 (13A), 3617–3623. 10.1002/j.1460-2075.1985.tb04125.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Ha SC; Choi J; Hwang HY; Rich A; Kim YG; Kim KK The Structures of Non-CG-Repeat Z-DNAs Co-Crystallized with the Z-DNA-Binding Domain, HZα(ADAR1). Nucleic Acids Res. 2009. 10.1093/nar/gkn976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (93).Serra MJ; Smolter PE; Westhof E Pronounced Instability of Tandem IU Base Pairs in RNA. Nucleic Acids Res. 2004, 32 (5), 1824–1828. 10.1093/nar/gkh501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Yu Z; Chen T; Cao X RNA Editing by ADAR1 Marks DsRNA as “Self.” Cell Research. 2015. 10.1038/cr.2015.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Solomon O; Di Segni A; Cesarkas K; Porath HT; Marcu-Malina V; Mizrahi O; Stern-Ginossar N; Kol N; Farage-Barhom S; Glick-Saar E; Lerenthal Y; Levanon EY; Amariglio N; Unger R; Goldstein I; Eyal E; Rechavi G RNA Editing by ADAR1 Leads to Context-Dependent Transcriptome-Wide Changes in RNA Secondary Structure. Nat. Commun. 2017. 10.1038/s41467-017-01458-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Braasch DA; Corey DR Locked Nucleic Acid (LNA): Fine-Tuning the Recognition of DNA and RNA. Chemistry and Biology. 2001. 10.1016/S1074-5521(00)00058-2. [DOI] [PubMed] [Google Scholar]
- (97).Dumat B; Larsen AF; Wilhelmsson LM Studying Z-DNA and B-to Z-DNA Transitions Using a Cytosine Analogue FRET-Pair. Nucleic Acids Res. 2016. 10.1093/nar/gkw114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Yi J; Yeou S; Lee NK DNA Bending Force Facilitates Z-DNA Formation under Physiological Salt Conditions. J. Am. Chem. Soc. 2022, 144 (29), 13137–13145. 10.1021/jacs.2c02466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Herbert A To “Z” or Not to “Z”: Z-RNA, Self-Recognition, and the MDA5 Helicase. PLoS Genetics. 2021. 10.1371/journal.pgen.1009513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Guo X; Liu S; Sheng Y; Zenati M; Billiar T; Herbert A; Wang Q ADAR1 Zα Domain P195A Mutation Activates the MDA5-Dependent RNA-Sensing Signaling Pathway in Brain without Decreasing Overall RNA Editing. Cell Rep. 2023, 42 (7), 112733. 10.1016/j.celrep.2023.112733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Mannion NM; Greenwood SM; Young R; Cox S; Brindle J; Read D; Nellåker C; Vesely C; Ponting CP; McLaughlin PJ; Jantsch MF; Dorin J; Adams IR; Scadden ADJ; Öhman M; Keegan LP; O’Connell MA The RNA-Editing Enzyme ADAR1 Controls Innate Immune Responses to RNA. Cell Rep. 2014. 10.1016/j.celrep.2014.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Lehmann KA; Bass BL The Importance of Internal Loops within RNA Substrates of ADAR1. J. Mol. Biol. 1999. 10.1006/jmbi.1999.2914. [DOI] [PubMed] [Google Scholar]
- (103).Chung H; Calis JJA; Wu X; Sun T; Yu Y; Sarbanes SL; Dao Thi VL; Shilvock AR; Hoffmann HH; Rosenberg BR; Rice CM Human ADAR1 Prevents Endogenous RNA from Triggering Translational Shutdown. Cell 2018. 10.1016/j.cell.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).Wong SK; Sato S; Lazinski DW Elevated Activity of the Large Form of ADAR1 in Vivo: Very Efficient RNA Editing Occurs in the Cytoplasm. RNA 2003. 10.1261/rna.5160403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (105).Pfaller CK; Donohue RC; Nersisyan S; Brodsky L; Cattaneo R Extensive Editing of Cellular and Viral Double-Stranded RNA Structures Accounts for Innate Immunity Suppression and the Proviral Activity of ADAR1p150. PLoS Biol 2018, 16 (11), e2006577. 10.1371/journal.pbio.2006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (106).Koeris M; Funke L; Shrestha J; Rich A; Maas S Modulation of ADAR1 Editing Activity by Z-RNA in Vitro. Nucleic Acids Res. 2005. 10.1093/nar/gki849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Chen YG; Hur S Cellular Origins of DsRNA, Their Recognition and Consequences. Nat. Rev. Mol. Cell Biol. 2022, 23 (4), 286–301. 10.1038/s41580-021-00430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (108).Karki R; Sundaram B; Sharma BR; Lee SJ; Malireddi RKS; Nguyen LN; Christgen S; Zheng M; Wang Y; Samir P; Neale G; Vogel P; Kanneganti TD ADAR1 Restricts ZBP1-Mediated Immune Response and PANoptosis to Promote Tumorigenesis. Cell Rep. 2021. 10.1016/j.celrep.2021.109858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (109).de Reuver R; Verdonck S; Dierick E; Nemegeer J; Hessmann E; Ahmad S; Jans M; Blancke G; Van Nieuwerburgh F; Botzki A; Vereecke L; van Loo G; Declercq W; Hur S; Vandenabeele P; Maelfait J ADAR1 Prevents Autoinflammation by Suppressing Spontaneous ZBP1 Activation. Nature 2022, 631 (32). [DOI] [PubMed] [Google Scholar]
- (110).Zhang T; Yin C; Fedorov A; Qiao L; Bao H; Beknazarov N; Wang S; Gautam A; Williams RM; Crawford JC; Peri S; Vasily S; Beg AA; Thomas PG; Walkley C; Xu Y; Poptsova M; Herbert A; Balachandran S ADAR1 Masks the Cancer Immunotherapeutic Promise of ZBP1-Driven Necroptosis. Nature 2022, 606 (7914), 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (111).Wittig B; Dorbic T; Rich A Transcription Is Associated with Z-DNA Formation in Metabolically Active Permeabilized Mammalian Cell Nuclei. Proc Natl Acad Sci U S A 1991, 88 (6), 2259–2263. 10.1073/pnas.88.6.2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (112).Jiao H; Wachsmuth L; Kumari S; Schwarzer R; Lin J; Eren RO; Fisher A; Lane R; Young GR; Kassiotis G; Kaiser WJ; Pasparakis M Z-Nucleic-Acid Sensing Triggers ZBP1-Dependent Necroptosis and Inflammation. Nature 2020. 10.1038/s41586-020-2129-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (113).Balachandran S; Mocarski ES Viral Z-RNA Triggers ZBP1-Dependent Cell Death. Current Opinion in Virology. 2021. 10.1016/j.coviro.2021.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (114).Thapa RJ; Ingram JP; Ragan KB; Nogusa S; Boyd DF; Benitez AA; Sridharan H; Kosoff R; Shubina M; Landsteiner VJ; Andrake M; Vogel P; Sigal LJ; tenOever BR; Thomas PG; Upton JW; Balachandran S DAI Senses Influenza A Virus Genomic RNA and Activates RIPK3-Dependent Cell Death. Cell Host Microbe 2016, 20 (5), 674–681. 10.1016/j.chom.2016.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (115).Zhang T; Yin C; Boyd DF; Quarato G; Ingram JP; Shubina M; Ragan KB; Ishizuka T; Crawford JC; Tummers B; Rodriguez DA; Xue J; Peri S; Kaiser WJ; López CB; Xu Y; Upton JW; Thomas PG; Green DR; Balachandran S Influenza Virus Z-RNAs Induce ZBP1-Mediated Necroptosis. Cell 2020. 10.1016/j.cell.2020.02.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
Figure references
- (1).Monleon D; Esteve V; Celda B. NMR Study of Hexanucleotide d(CCGCGG)2 Containing Two Triplet Repeats of Fragile X Syndrome. Biochem Biophys Res Commun 2003, 303 (1), 81–90. 10.1016/s0006-291x(03)00304-8. [DOI] [PubMed] [Google Scholar]
- (2).Popenda M; Biala E; Milecki J; Adamiak RW Solution Structure of RNA Duplexes Containing Alternating CG Base Pairs: NMR Study of r(CGCGCG)2 and 2’-O-Me(CGCGCG)2 under Low Salt Conditions. Nucleic Acids Res 1997, 25 (22), 4589–4598. 10.1093/nar/25.22.4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Schwartz T; Rould MA; Lowenhaupt K; Herbert A; Rich A Crystal Structure of the Zalpha Domain of the Human Editing Enzyme ADAR1 Bound to Left-Handed Z-DNA. Science (80-. ). 1999, 11 (284(5421)), 1841–1845. [DOI] [PubMed] [Google Scholar]
- (4).Placido D; Brown BA 2nd; Lowenhaupt K; Rich A; Athanasiadis A. A Left-Handed RNA Double Helix Bound by the Z Alpha Domain of the RNA-Editing Enzyme ADAR1. Structure 2007, 15 (4), 395–404. 10.1016/j.str.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Ho PS The Non-B-DNA Structure of d(CA/TG)(n) Does Not Differ from That of Z-DNA. Proc. Natl. Acad. Sci. U. S. A. 1994. 10.1073/pnas.91.20.9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


