Abstract
Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a distinct open reading frame called K15 at a position equivalent to the gene encoding LMP2A of Epstein-Barr virus (EBV). K15 isolates from body cavity-based lymphoma (BCBL) cells exhibited a dramatic sequence variation and a complex splicing pattern. However, all K15 alleles are organized similarly with the potential SH2 and SH3 binding motifs in their cytoplasmic regions. Northern blot analysis showed that K15 was weakly expressed in latently infected BCBL-1 cells, and the level of its expression was significantly induced by tetradecanoyl phorbol acetate stimulation. K15 encoded 40- to 55-kDa proteins, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and was localized at the cytoplasm and plasma membrane. To demonstrate the signal-transducing activity of the K15 protein, we constructed a chimeric protein in which the cytoplasmic tail of the human CD8α polypeptide was replaced with that of KSHV K15. While the CD8-K15 chimera was not capable of eliciting cellular signal transduction upon stimulation with an anti-CD8 antibody, it significantly inhibited B-cell receptor signaling, as evidenced by a suppression of tyrosine phosphorylation and intracellular calcium mobilization. This inhibition required the putative SH2 or SH3 binding motif in the cytoplasmic region of K15. Biochemical study of CD8-K15 chimeras showed that the cytoplasmic region of K15 was constitutively tyrosine phosphorylated and that the tyrosine residue within the putative SH2 binding motif of K15 was a primary site of phosphorylation. These results demonstrate that KSHV K15 resembles LMP2A in genomic location, splicing pattern, and protein structure and by the presence of functional signal-transducing motifs in the cytoplasmic region. Thus, KSHV K15 is likely a distant evolutionary relative of EBV LMP2A.
DNA sequences of a novel member of the herpesvirus group, called Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8, have been widely identified in Kaposi's sarcoma (KS) tumors from human immunodeficiency virus-positive and -negative patients (4, 5, 21, 32, 35). KSHV has also been identified in body cavity-based lymphoma (BCBL) and some forms of Castleman's disease (4, 5, 28). These are principally or exclusively of B-cell origin. Cell lines have been derived from some of the BCBL, and while some harbor both Epstein-Barr virus (EBV) and KSHV, others harbor KSHV only. The genomic sequence indicates that KSHV is a gammaherpesvirus that is closely related to herpesvirus saimiri (HVS) (25, 29) and the recently isolated rhesus monkey rhadinovirus (7, 33).
In primary lymphocytes, cross-linking the B-cell receptor (BCR) or T-cell receptor (TCR) leads to an intricate signal cascade including the recruitment and activation of the src family tyrosine kinases; the subsequent activation and recruitment of other kinases, phosphatases, or adapter proteins; the hydrolysis of phospholipids; the mobilization of intracellular calcium; the activation of protein kinase C; the activation of nuclear transcription factors; and the transcription of BCR or TCR signal-specific genes (3, 37). In contrast, these signal transduction cascades are blocked in EBV-transformed B cells and HVS-transformed T cells (13, 22, 24). Latent membrane protein 2A (LMP2A) of EBV and tyrosine kinase-interacting protein (tip) of HVS have been proposed to be responsible for this phenotype (12, 13, 20, 24).
LMP2A is one of nine viral proteins expressed in B cells latently infected with EBV in vitro (14, 18, 19). LMP2A contains 12 transmembrane domains and short stretches of amino and carboxyl termini and is expressed in aggregates at the plasma membranes of latently infected B cells. The amino terminus of LMP2A contains a functional immunoreceptor tyrosine-based activation motif (ITAM) (19). This motif is tyrosine phosphorylated and is necessary for association of LMP2A with the SH2 domain of the src family kinases and syk kinase. In addition, by using CD8 chimeras, this motif has been shown to be capable of triggering cellular signal transduction leading to intracellular calcium mobilization and cytokine production (1). Furthermore, a recent study with transgenic mice has demonstrated that LMP2A provides a constitutive survival signaling activity in primary B cells of transgenic mice (2). While LMP2A is not required for EBV-induced B-lymphocyte transformation, studies with EBV-transformed lymphoblastoid cell lines suggest that ITAM-mediated signaling of LMP2A is necessary for establishing or maintaining viral latency in vivo (19, 22–24).
In this report, we demonstrate that KSHV contains a distinct open reading frame called K15 at a position equivalent to the gene encoding LMP2A of EBV. Although K15 does not exhibit homology to LMP2A, both proteins contain a similar structural organization, including the amino-terminal multiple transmembrane domain and the carboxyl signal-transducing domain. Biochemical studies of CD8-K15 chimeras demonstrate that unlike LMP2A, K15 is not capable of eliciting cellular signal transduction. In the other hand, like LMP2A, K15 is capable of blocking BCR signal transduction. Thus, these results suggest that KSHV K15 modulates lymphocyte signaling in a manner similar to but distinct from EBV LMP2A.
MATERIALS AND METHODS
Cell culture and transfection.
BJAB, BC-1, and BCBL-1 cells were grown in RPMI medium supplemented with 10% fetal calf serum. COS-1 cells were grown in Dulbecco modified Eagle medium supplemented with 10% fetal calf serum. A fusin lipofection (Boehringer Mannheim) transfection procedure was used for transient expression in COS-1 cells. The pcDNA3-CD8-K15 chimeric constructs (20 μg) were introduced into BJAB cells by electroporation at 250 V and 960 μF in serum-free Dulbecco modified Eagle medium. After a 48-h incubation, the cells were cultured with selection medium containing 2 mg of neomycin per ml for 5 weeks.
Plasmid constructions.
KSHV K15 cDNA was cloned by reverse transcriptase PCR (RT-PCR). mRNA from BCBL-1 cells or BC-1 cells was isolated by using an mRNA isolation kit from Qiagen (Santa Clarita, Calif.). Approximately 0.3 μg of mRNA was reverse transcribed by murine leukemia virus RT in a 20-μl reaction mixture with poly(dT) primer for 20 min at 42°C. As a control, cDNA synthesis was performed without RT. One microliter of the same cDNA preparation was used for PCR amplification in a 50-μl-volume reaction mixture with 100 pmol of oligo(dT) per liter as the 3′ primer and 3652ATGAAGACACTCATATTCT3634 or 3623ATGGCTTTGGGCCCTACTGG3604 as the 5′ primer, which overlapped with the potential translational initiation site at the 5′ end of the gene (26). The primers used for PCR contain an EcoRI site at the 5′ end and a XhoI site at the 3′ end for subsequent cloning. Amplified DNA was directly cloned into the Topo-II vector (Invitrogen). Twenty independent clones were subsequently sequenced on both strands by using an ABI PRISM 377 automatic DNA sequencer. The cytoplasmic region of K15 was amplified by PCR and was fused in frame to the human CD8α containing a deletion of its carboxyl terminus (CD8Δ) in the pSP72 vector (Promega Biotech). For stable expression, the KpnI and BglII DNA fragment containing CD8-K15 chimera sequence was cloned into the KpnI and BamHI sites of pcDNA3-Neo (Invitrogen). All mutations in the K15 gene were generated by PCR using oligonucleotide-directed mutagenesis (8). The amplified DNA fragments containing mutations in K15 were purified and cloned into the pSP72 vector. Each K15 mutant was completely sequenced to verify the presence of the mutation and the absence of any other changes. After confirmation of the DNA sequence, DNA containing the desired K15 mutation was recloned into pcDNA3-Neo vector containing CD8Δ.
Recombinant K15 protein and antibodies.
For purification of recombinant K15 protein from Escherichia coli, the K15 DNA fragment corresponding to the cytoplasmic region of K15 from BCBL-1 cells was amplified by PCR with primers containing BamHI and SalI recognition sequences at the ends and subcloned into BamHI and SalI cloning sites of the pQE-40 expression vector (Qiagen, San Diego, Calif.) with the potential of incorporating six histidines at the amino terminus. Clones were sequenced to ensure the presence of the exact desired sequence. When E. coli XL-1 Blue containing plasmid pQE40-K15 reached an optical density at 600 nm of approximately 0.6, 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added, and the cells were harvested 3 h after induction. Cells were solubilized with 6 M guanidine hydrochloride. Due to the presence of the affinity tail, six-histidine-tagged K15 protein was purified to virtual homogeneity in one step by Ni2+-chelate affinity chromatography. The purified recombinant His6-K15 proteins were used to generate polyclonal antibody in New Zealand White rabbits. A Ni2+-chelate affinity column containing K15 protein was used to purify the antigen-specific antibodies. Antibody specific for K15 was eluted with a high-pH solution (0.1 M triethylamine, pH 11.5).
Immunoprecipitation and immunoblotting.
Cells were harvested and lysed with lysis buffer (0.15 M NaCl, 1% Nonidet P-40, and 50 mM HEPES buffer [pH 8.0]) containing 1 mM Na2VO3, 1 mM NaF, and protease inhibitors (leupeptin, aprotinin, phenylmethylsulfonyl fluoride [PMSF], and bestatin). Immunoprecipitation was performed with 1:500-diluted antibody together with 30 μl of protein A/G-agarose beads. For protein immunoblots, polypeptides in cell lysates corresponding to 105 cells were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a nitrocellulose membrane filter. Protein detection was performed with a 1:1,000 or 1:3,000 dilution of primary antibody by using an enhanced chemiluminescence system (Amersham, Chicago, Ill.).
Northern blot analysis.
Northern blot analysis was performed under standard conditions with randomly labeled probes representing either full-length K15 or the 5′ half of K15. Total RNA was purified from BJAB, unstimulated BCBL-1, and tetradecanoyl phorbol myristate (TPA)-stimulated BCBL-1 cells according to the manufacturer's instructions (Qiagen), and 10 μg of total RNA was loaded in each lane. The filters were baked at 80°C for 2 h and then hybridized with radioactive probes.
Immunofluorescence.
Cells were fixed with 4% paraformaldehyde for 15 min, permeablized with 70% ethanol for 15 min, blocked with 10% goat serum in phosphate-buffered saline (PBS) for 30 min, and reacted with 1:100-diluted primary antibody in PBS for 30 min at room temperature. After incubation, cells were washed extensively with PBS, incubated with 1:100-diluted secondary antibody (Vector) in PBS for 30 min at room temperature, and washed three times with PBS. Protein staining was performed with 1:500-diluted Sypro (Molecular Probes) for 1 min. Immunofluorescence was detected with a Leica immunofluorescence confocal microscope.
Flow cytometry analysis.
A total of 5 × 105 cells were washed with RPMI medium containing 10% fetal calf serum and incubated with fluorescein isothiocyanate (FITC)-conjugated or phycoerythrin-conjugated monoclonal antibodies for 30 min at 4°C. After being washed, each sample was fixed with 1% formalin solution, and flow cytometry analysis was performed with a FACS Scan (Becton Dickinson Co., Mountainview, Calif.). For cell sorting, 2 × 107 cells were stained with FITC-conjugated CD8 (UCHT2; PharMingen) antibody for 30 min at 4°C. Stained cells were sorted based on CD8 surface expression, using a FACS Vantage (Becton Dickinson). After being sorted, cells were washed twice with PBS and cultured with RPMI–10% fetal calf serum medium. UCHT2 CD8 and G20-127 antibodies used for fluorescence-activated cell sorter (FACS) analysis were obtained from PharMingen, and OKT8 antibody used for stimulation was obtained from the American Type Culture Collection.
Antibody stimulation.
Next, 107 cells were incubated with 10 μg of anti-CD8 (OKT8) or anti-immunoglobulin M (IgM) antibody at 37°C for various times. After stimulation, cells were immediately frozen and lysed with cold lysis buffer containing 1 mM Na2VO3, 1 mM NaF, and protease inhibitors (leupeptin, aprotinin, PMSF, and bestatin). Precleared cell lysates were used for immunoblotting or for immunoprecipitation.
Calcium mobilization analysis.
A total of 2 × 106 cells were loaded with 1 μM Indo-1 in 2 ml of RPMI complete medium for 20 min at 37°C. A detailed protocol for this process has been described previously (24). Baseline calcium levels were established for 1 min prior to the addition of the antibody. Cells were stimulated with 10 μg of mouse anti-CD8 (OKT8) antibody followed by 10 μg of goat anti-mouse antibody, or 10 μg of mouse anti-IgM antibody alone, and data were then collected for 4 min. Baseline absolute intracellular calcium levels were determined by using an ionophore and EGTA. Data were collected and analyzed on a FACS Vantage (Becton Dickinson).
RESULTS
Cloning and sequence analysis of K15 from BCBL.
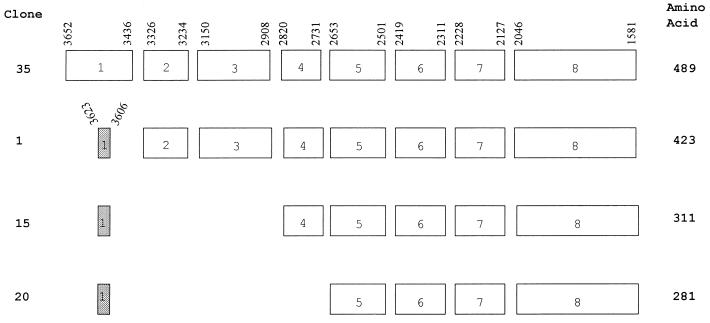
At a position and transcriptional direction equivalent to the LMP2A gene of EBV, KSHV contains a distinct reading frame called K15. Initial DNA sequence analysis of the KSHV genome did not reveal this open reading frame because of its multiple splicing (29). KSHV K15 cDNA was cloned from mRNA of unstimulated BCBL-1 cells by RT-PCR as described in Materials and Methods, using an oligo(dT) primer at the 3′ end and a gene-specific primer which overlapped with the potential translational initiation site at the 5′ end of the gene (26, 29). Sequence analysis of 20 independent cDNA clones identified at least four differently spliced forms of K15 in BCBL-1 cells (Fig. 1). Clones 35 and 1 contained eight exons, clone 15 contained six exons, and clone 20 contained five exons (Fig. 1). While all clones shared exons 5 to 8, clone 35 contained a first exon which was entirely different from that of clones 1, 15, and 20. The first exon of clone 35 encoded 72 amino acids, and the first exon of clones 1, 15, and 20 encoded only 6 amino acids. These spliced forms were predicted to encode products of from 281 to 489 amino acids (Fig. 1).
FIG. 1.
Overall splicing pattern of KSHV K15 gene. Sequence analysis with 20 independent cDNA clones from BCBL-1 cells identified four differently spliced forms of K15. The boxes indicate each coding exon, and the numbers above the boxes, which are based on the sequence data from Nicholas et al. (26), indicate the potential splicing junction sites.
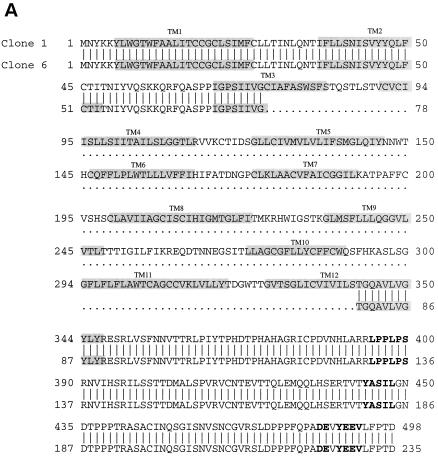
A comparison of the primary amino acid sequences of the four K15 isolates revealed extensive size variation in the amino-terminal hydrophobic region because of complex splicing (Fig. 2). All cDNA clone products are predicted to be large integral membrane proteins consisting of 4 to 12 hydrophobic membrane-spanning domains (Fig. 2). Regardless of the number of transmembrane domains, all cDNA clone products have the same carboxyl-terminal region, which is predicted to contain 142 amino acids. The presence of potential motifs in the cytoplasmic region was assessed based on the primary amino acid sequence. All spliced forms of K15 contained a conserved YEEV sequence at amino acids 480 to 483 (based on the primary amino acid sequence of clone 35), which is preceded by two negatively charged glutamic acid residues (Fig. 2). This motif matches very well with the consensus sequence for SH2 binding (EExxYEEV/I) to src family kinases (34). This is designated as an SH2-binding (SH2-B) motif. In addition, a proline-rich region from amino acid residues 385 to 390 of the clone 35 product shows homology with the consensus sequences for binding to the SH3 domains of signal-transducing proteins (6, 27, 38). This is designated as an SH3-binding (SH3-B) motif. In addition, while the YASIL sequence at amino acids 431 to 434 is consistent with an SH2-binding motif, it is not preceded by negatively charged amino acids, suggesting that it may be involved in activities other than SH2 binding. Thus, besides a considerable resemblance to LMP2A of EBV in genomic location, overall protein structure, and complex splicing pattern, K15 also contains potential SH2-B and SH3-B motifs in the cytoplasmic region as has been shown in LMP2A (19).
FIG. 2.
Amino acid sequence and structural organization of KSHV K15 from BCBL-1 cells. The amino acid sequences of four K15 clones from BCBL-1 cells were aligned to demonstrate similarities in structural organization. The gray boxes indicate the transmembrane (TM) region, and the boldface letters indicate the potential signal-transducing modules.
Genetic variation of K15 alleles.
Primary amino acid sequences of K15 from BC-1 cells were also determined by RT-PCR and DNA sequence analysis (Fig. 3). Sequence analysis of 20 independent cDNA clones detected two major spliced forms of K15 in BC-1 cells (Fig. 3A). K15 clones 1 and 6 from BC-1 cells are predicted to encode 498 and 235 amino acids, respectively (Fig. 3A). To our surprise, a remarkable sequence variation was detected between K15 genes derived from BCBL-1 and BC-1 cells (Fig. 3B). K15 genes from BC-1 and BCBL-1 cell lines exhibited only 30% identity and 40% homology at the amino acid level. Despite this dramatic sequence variation, both K15 proteins are predicted to have a similar structure: an amino-terminal multiple transmembrane region and a carboxyl-terminal cytoplasmic region. In addition, the three potential motifs, SH2-B, SH3-B, and YASIL, were completely conserved in the cytoplasmic region of both K15 proteins (Fig. 3B). This suggests that the conserved SH2-B, SH3-B, and YASIL motifs of the cytoplasmic region are likely important for K15 function.
FIG. 3.
Sequence comparison between BCBL-1 K15 and BC-1 K15. (A) The predicted amino acid sequence of K15 clones 1 and 6 of BC-1 cells. The amino acid sequences of two K15 clones from BC-1 cells were aligned to demonstrate similarities in structural organization. The gray boxes indicate the transmembrane (TM) region, and the boldface letters indicate the potential signal-transducing modules. (B) Sequence comparison between BCBL-1 K15 and BC-1 K15. The predicted protein sequences of K15 clone 35 from BCBL-1 cells and K15 clone 1 from BC-1 cells were aligned for comparison. The vertical and dotted lines indicate identity and similarity, respectively. Boldface sequences indicate motifs conserved between the two K15 clones.
Latent and lytic expression of K15.
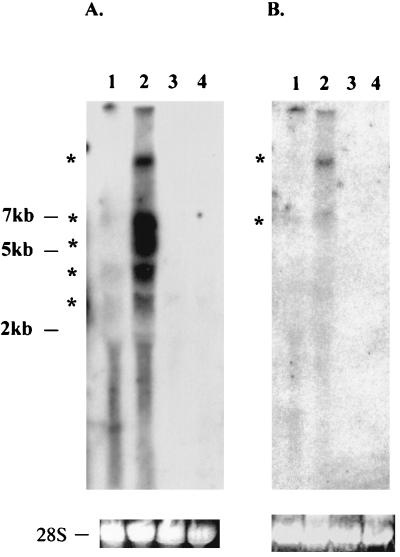
To determine K15 expression, Northern blot analysis was performed with total RNA from BCBL-1 cells with or without TPA stimulation for 48 h. When sequences representing the full-length K15 gene were used as a probe, multiple transcripts of 3, 4, 5.5, 7, and 10 kb were detected in BCBL-1 cells. These transcripts were weakly detected in unstimulated BCBL-1 cells, and their level of expression was significantly increased after TPA stimulation (Fig. 4A, lanes 1 and 2). No specific transcripts were detected in control BC-1 and BJAB cells (Fig. 4A, lanes 3 and 4). To further determine specificity, sequences representing the 5′ half of the K15 gene were used as a probe in Northern blot analysis. This probe specifically detected only the 7- and 10-kb transcripts among the five transcripts which were identified by the full-length K15 probe (Fig. 4B, lanes 1 and 2). These results suggest that the 7- and 10-kb transcripts likely encode the K15 gene and the 3-, 4-, and 5.5-kb transcripts may encode other KSHV genes or unidentified alternatively spliced forms of K15. These results indicate that K15 is weakly expressed during latency but that its expression is significantly induced during lytic viral replication.
FIG. 4.
Northern blot analysis of KSHV K15. A 10-μg portion of total RNA purified from BCBL-1 cells with (lane 2) or without (lane 1) TPA stimulation for 48 h, BC-1 cells (lane 3), or BJAB cells (lane 4) was separated through an agarose gel, transferred to a nylon membrane, and reacted with 32P-labeled probes representing either full-length K15 (A) or the 5′ half of K15 (B). Autoradiography was performed with a Fuji phosphorimager. Stars indicate the potential K15 specific transcripts.
Identification of K15 protein.
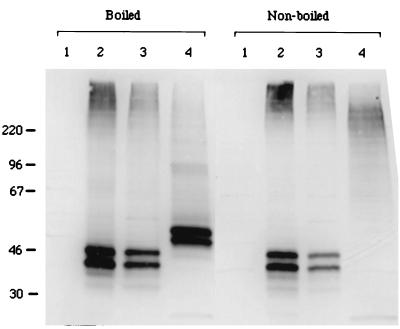
To analyze the K15 gene product of BCBL-1 cells, we generated a rabbit polyclonal antibody against a purified bacterial six-histidine-tagged K15 fusion protein which contained the putative cytoplasmic portion of BCBL-1 K15 as described in Materials and Methods. Expression of K15 clones 35 and 20 derived from BCBL-1 cells was then demonstrated. At 48 h posttransfection, heat-treated COS-1 cell lysates were used for an immunoblot assay with antibody specific for BCBL-1 K15. This antibody reacted specifically with the 50- and 55-kDa protein species of BCBL-1 K15 clone 35 and with 40- and 45-kDa protein species of BCBL-1 K15 clone 20 (Fig. 5). In contrast, these proteins were not detected in control COS-1 cells not expressing the K15 gene (Fig. 5).
FIG. 5.
Identification of the KSHV K15 protein. COS-1 cells were transfected with transient expression vector pFJ containing BCBL-1 K15 clone 35 or clone 20. Boiled or nonboiled cell lysates were fractionated by SDS-PAGE, transferred to nitrocellulose, and reacted with an anti-K15 antibody. Lane 1, COS-1 cells transfected with vector; lane 2, COS-1 cells transfected with 10 μg of pFJ-K15 clone 20; lane 3, COS-1 cells transfected with 5 μg of pFJ-K15 clone 20; lane 4, COS-1 cells transfected with 5 μg of pFJ-K15 clone 35.
LMP2A, which contains 12 transmembrane domains, is present as aggregates in plasma membranes (19). DNA sequence analysis predicts that K15 clones 20 and 35 have 4 and 12 transmembrane domains, respectively. To determine the potential aggregation of K15, cell lysates containing K15 protein were subjected to SDS-PAGE without heat treatment and reacted with an anti-K15 antibody in an immunoblot analysis. This analysis showed that the migration of 50- and 55-kDa protein species of K15 clone 35 was greatly retarded to ca. 200 kDa in SDS-PAGE (Fig. 5). In striking contrast, the migration of 40- and 45-kDa species of the K15 clone 20 was not altered in SDS-PAGE under the same conditions (Fig. 5). These data suggest that K15 clone 35, which contains 12 transmembrane domains, is primarily present as aggregates, whereas K15 clone 20, which contains 3 transmembrane domains, is far less abundant as aggregates.
Localization of KSHV K15.
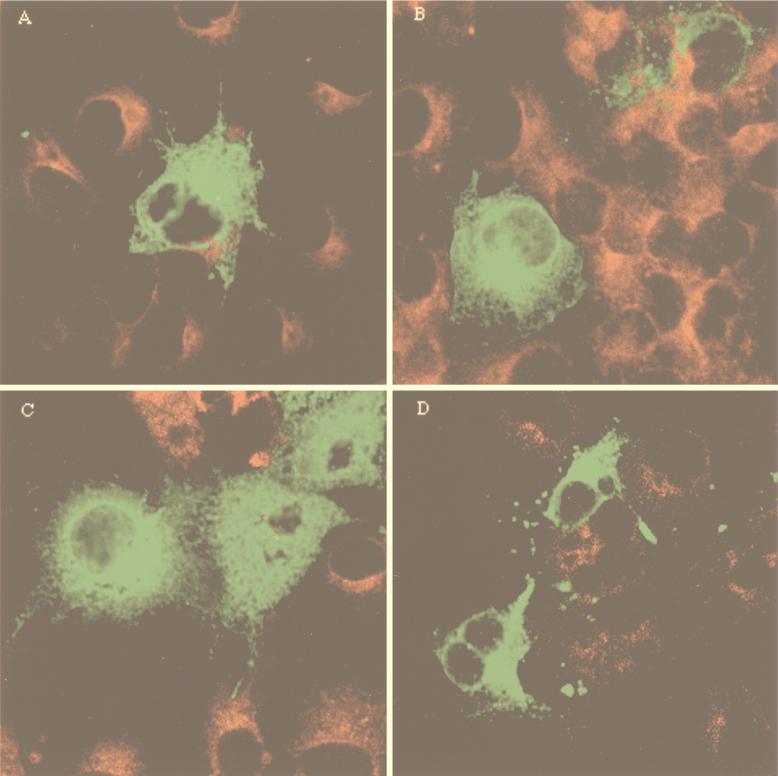
To determine the subcellular localization of K15 by indirect immunofluorescence tests, COS-1 cells transfected with K15 clones 1, 15, 20, and 35 from BCBL-1 cells were reacted with an anti-K15 antibody. The staining pattern in COS-1 cells suggested that all K15 clones localized primarily in the cytoplasm and at the plasma membrane (Fig. 6). In addition, a strong fluorescence was detected in the perinuclear region. This staining pattern is similar to that obtained when the Golgi is visualized. Thus, immunofluorescence tests demonstrated that, independent of the number of transmembrane domains, K15 alleles were located principally in the cytoplasm, Golgi, and plasma membrane.
FIG. 6.
Localization of K15 alleles. COS-1 cells were transfected with pFJ-K15 clone 1 (A), clone 15 (B), clone 20 (C), and clone 35 (D). These cells were permeabilized with 70% ethanol and reacted with 1:100-diluted rabbit anti-K15 antibody, followed by incubation with 1 μg of fluoresceinated goat anti-rabbit antibody. Immunofluorescence was detected using a Leica confocal immunofluorescence microscope.
Construction of a CD8α chimera with the cytoplasmic region of K15.
To determine if the cytoplasmic region of K15 is capable of inducing signals or altering cellular signal transduction, we analyzed the signaling capacity of the cytoplasmic tail independent of the transmembrane domains of K15. Antibody cross-linking of chimeric molecules composed of the extracellular and the transmembrane domains of the CD8α molecule and the cytoplasmic region of KSHV K1 or EBV LMP2A has been shown to be sufficient to elicit early and late signal-transducing events (1, 16). We constructed a chimeric protein in which 27 amino acids of the cytoplasmic tail of human CD8α protein were replaced with 142 amino acids of the cytoplasmic tail of BCBL-1 K15 (CD8-K15). CD8Δ, from which the cytoplasmic region has been deleted, was used as a control. Since mutations in the signal-transducing modules of cellular proteins abrogate their capacity to induce signaling (3, 9), we also introduced mutations at the SH2-B and SH3-B motifs as follows: CD8-K15 P→G contained mutations at positions 386, 387, 388, 390, and 341 of prolines to glycines; CD8-K15 Y481F contained a mutation at position 481 of tyrosine to phenylalanine; and CD8-K15 P→G/Y481F contained both mutations in the SH2-B and SH3-B motifs.
Construction of BJAB cell lines expressing CD8-K15 chimeras.
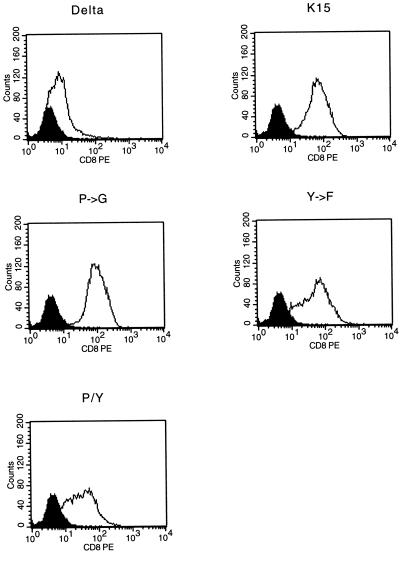
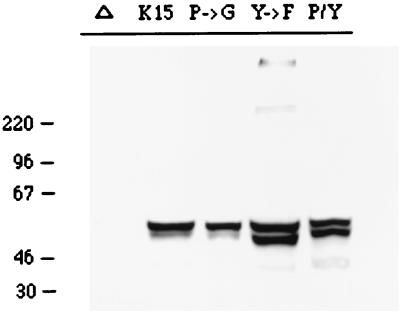
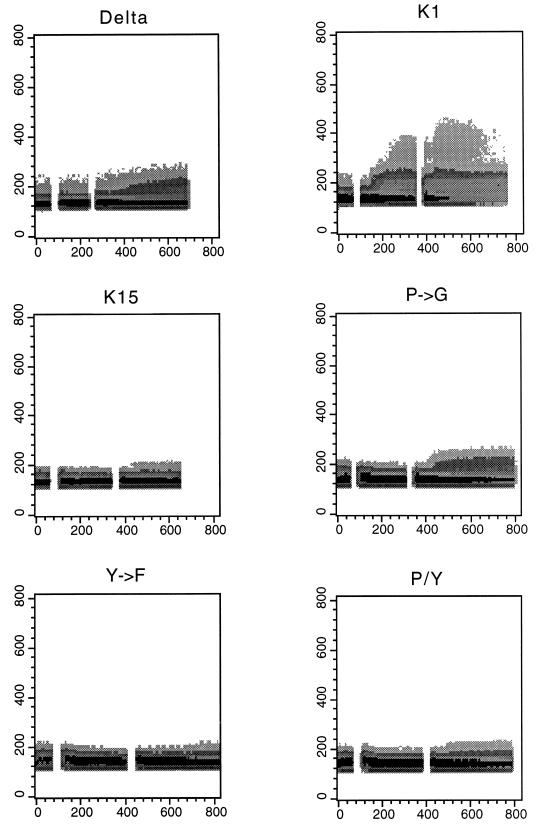
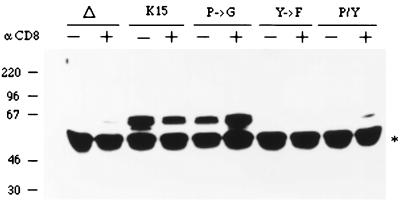
To assess the signal-transducing activity of CD8-K15 chimeras, BJAB cells (KSHV and EBV negative) were used to establish stable lines expressing the CD8-K15 chimeric genes. The CD8Δ, CD8-K15, CD8-K15 P→G, CD8-Y481F, and CD8-K15 P→G/Y481F chimeric genes were cloned into the expression vector pcDNA3-Neo. After electroporation of the expression vector into BJAB cells, cell lines were selected by growth in medium containing 2 mg of neomycin per ml for 5 weeks. Since CD8 is not expressed on the surface of BJAB cells, neomycin-resistant cells were sorted by FACS analysis based on the surface expression of the CD8. Comparable levels of CD8 surface expression of FACS-sorted cells were detected in most of the cells expressing the CD8-K15 chimeras, with the exception of CD8Δ cells (Fig. 7). The reduced level of CD8 surface expression in CD8Δ cells was likely caused by the absence of its cytoplasmic region. To demonstrate the expression of these chimeras, BJAB cells expressing the CD8-K15 chimeras were used for immunoblot analysis with an anti-K15 antibody. This assay revealed that CD8-K15 chimeras were expressed at somewhat variable but still comparable levels in BJAB cells (Fig. 8).
FIG. 7.
Flow cytometric analysis of surface CD8-K15 chimera expression on BJAB cell lines. Live cells were stained for the surface expression of CD8 as described in Materials and Methods. Two hundred thousand events were collected by FACS Scan flow cytometry. As a control, a histogram of each cell line (solid line) is overlaid with a dark-shaded histogram of the control BJAB cells in the solid line. Delta, CD8Δ; K15, CD8-K15; P→G, CD8-K15 P→G; Y→F, CD8-K15 Y481F; P/Y, CD8-K15 P→G/Y481F.
FIG. 8.
Expression of CD8-K15 chimeras. Expression of CD8-K15 chimeras in BJAB cells was detected by immunoblot analysis with an anti-K15 antibody. Δ, CD8Δ; K15, CD8-K15; P→G, CD8-K15 P→G; Y→F, CD8-K15 Y481F; P/Y, CD8-K15 P→G/Y481F.
Lack of cellular signal-transducing ability of CD8-K15 chimeras upon antibody stimulation.
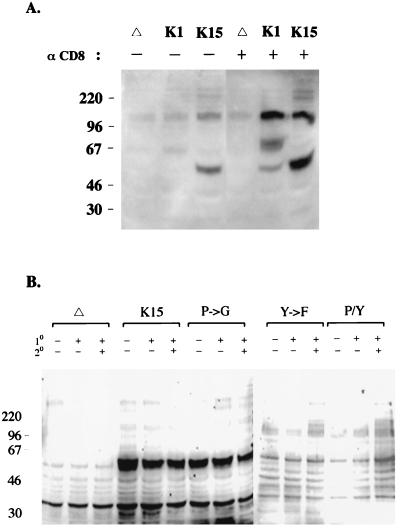
The early biochemical event subsequent to TCR or BCR stimulation is the induction of tyrosine phosphorylation of a number of cellular proteins (3, 36). We examined the effects of CD8-K15 chimera expression on cellular tyrosine phosphorylation upon stimulation with an anti-CD8 antibody. BJAB cells expressing CD8Δ or CD8-K15 chimera were stimulated with an anti-CD8 antibody and the level of tyrosine phosphorylation induction was observed by immunoblot assay with an antiphosphotyrosine antibody (Fig. 9). CD8-K1, which has been shown to induce cellular tyrosine phosphorylation upon stimulation with an anti-CD8 antibody (16), was included as a positive control. The stimulation of BJAB cells expressing the CD8-K15 chimera with a mouse anti-CD8 antibody did not induce cellular tyrosine phosphorylation (Fig. 9A). In contrast, tyrosine phosphorylation of a 60-kDa protein was strongly detected in BJAB cells expressing CD8-K15 independent of antibody stimulation (Fig. 9A). To further potentiate stimulating activity, an additional ligation with a goat anti-mouse antibody was included in the stimulation conditions. This experiment also revealed that expression of the CD8-K15 chimera did not induce tyrosine phosphorylation of cellular proteins upon stimulation except for a 60-kDa protein (Fig. 9B). Under the same conditions, an enhanced level of tyrosine phosphorylation induction was detected in CD8-K1-expressing cells (Fig. 9A). Mutant forms of the CD8-K15 chimera were also examined for their ability to induce cellular tyrosine phosphorylation under the same conditions. Like CD8-K15, none of the CD8-K15 mutants induced cellular tyrosine phosphorylation upon stimulation (Fig. 9B). These results showed that the cytoplasmic region of K15 was not capable of inducing tyrosine phosphorylation upon antibody stimulation. However, tyrosine phosphorylation of a 60-kDa protein was strongly detected in BJAB cells expressing CD8-K15 or CD8-K15 P→G independent of antibody stimulation.
FIG. 9.
Induction of cellular tyrosine phosphorylation after stimulation with an anti-CD8 antibody. (A) A comparison of tyrosine phosphorylation induction of CD8-K1- and CD8-K15-expressing cells after antibody stimulation. A total of 5 × 106 cells were incubated with (+) or without (−) an anti-CD8 (OKT8) antibody at 37°C for 2 min and then lysed with lysis buffer. Precleared cell lysates were used for immunoblot analysis with antiphosphotyrosine antibody. Δ, CD8Δ; K1, CD8-K1; K15, CD8-K15. (B) Induction of tyrosine phosphorylation of cells expressing CD8-K15 mutants. A total of 5 × 106 BJAB cells expressing CD8Δ, CD8-K15, or mutant forms of CD8-K15 were incubated without (−) or with (+) an anti-CD8 antibody (1°) alone or also with an additional anti-mouse antibody (2°) at 37°C for 2 min each as indicated at the top of the figure. Precleared cell lysates were used for immunoblot analysis with an antiphosphotyrosine antibody. Δ, CD8Δ; K15, CD8-K15; P→G, CD8-K15 P→G; Y→F, CD8-K15 Y481F; P/Y, CD8-K15 P→G/Y481F.
To further determine the potential ability of the cytoplasmic region of K15 to induce late signaling events, the cytoplasmic free calcium concentration was examined for increases after antibody stimulation. BJAB cells expressing CD8-K15 chimeras were treated with mouse anti-CD8 antibody, followed by a goat anti-mouse antibody, and intracellular free calcium levels were monitored by flow cytometry in three independent experiments. CD8-K1, which has been shown to induce intracellular calcium mobilization upon stimulation with an anti-CD8 antibody (16), was used as a positive control. Control CD8-K1 cells exhibited a rapid increase in intracellular calcium concentration immediately after anti-CD8 treatment, whereas none of CD8-K15 chimeras induced an increase of intracellular free calcium concentration upon stimulation with anti-CD8 alone or together with a goat anti-mouse antibody (Fig. 10). Thus, unlike those of KSHV K1 (16) and EBV LMP2A (1), the cytoplasmic region of K15 is not capable of transducing a signal to induce tyrosine phosphorylation or intracellular calcium mobilization.
FIG. 10.
Intracellular free calcium mobilization after stimulation with anti-CD8 antibody. Calcium mobilization was monitored over time by changes in the ratio of violet to blue (405 to 485 nm) fluorescence of cells loaded with the calcium-sensitive dye Indo-1 and analyzed by flow cytometry. Data are presented as a histogram of the number of cells with a particular ratio to blue fluorescence (y axis) over time after anti-CD8 cross-linking, followed by an additional ligation with anti-mouse antibody cross-linking (x axis, second). Data were reproduced in three independent experiments. The breaks in the graph indicate the interval during the addition of antibodies. Delta, CD8Δ; K1, CD8-K1; K15, CD8-K15; P→G, CD8-K15 P→G; Y→F, CD8-K15 Y481F; P/Y, CD8-K15 P→G/Y481F.
Phosphorylation of tyrosine residues in the cytoplasmic region of K15.
Although CD8-K15 chimeras did not induce cellular signal transduction upon stimulation, tyrosine phosphorylation of a 60-kDa protein was readily detected in BJAB cells expressing CD8-K15 or CD8-K15 P→G independent of antibody stimulation (Fig. 9). Since this protein has the same molecular weight as CD8-K15 and CD8-K15 P→G, we investigated whether it was encoded by the CD8-K15 chimeras. Lysates of cells expressing the CD8-K15 chimeras were used for immunoprecipitation with an anti-CD8 antibody, which was followed by immunoblotting with an antiphosphotyrosine antibody. This showed that the CD8-K15 and CD8-K15 P→G chimeras were strongly phosphorylated and that the level of tyrosine phosphorylation of these proteins remained unchanged after stimulation (Fig. 11). In striking contrast, tyrosine phosphorylation of mutant CD8-K15 Y481F and CD8-K15 P→G/Y481F chimeras was not detected under the same conditions (Fig. 11). These results indicate that the cytoplasmic region of K15 is constitutively tyrosine phosphorylated and that the tyrosine residue within the putative SH2-B motif of K15 is a primary site of phosphorylation.
FIG. 11.
Tyrosine phosphorylation of CD8-K15 chimeras independent of stimulation. A total of 5 × 106 BJAB cells expressing CD8Δ, CD8-K15, or mutant forms of CD8-K15 were incubated without (−) or with (+) an anti-CD8 at 37°C for 2 min. Precleared cell lysates were used for immunoprecipitation with an anti-CD8 antibody. Immunoprecipitates were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with an antiphosphotyrosine antibody. Δ, CD8Δ; K15, CD8-K15; P→G, CD8-K15 P→G; Y→F, CD8-K15 Y481F; P/Y, CD8-K15 P→G/Y481F. The star indicates the immunoglobulin heavy chain.
Downregulation of BCR signaling by CD8-K15 chimeras.
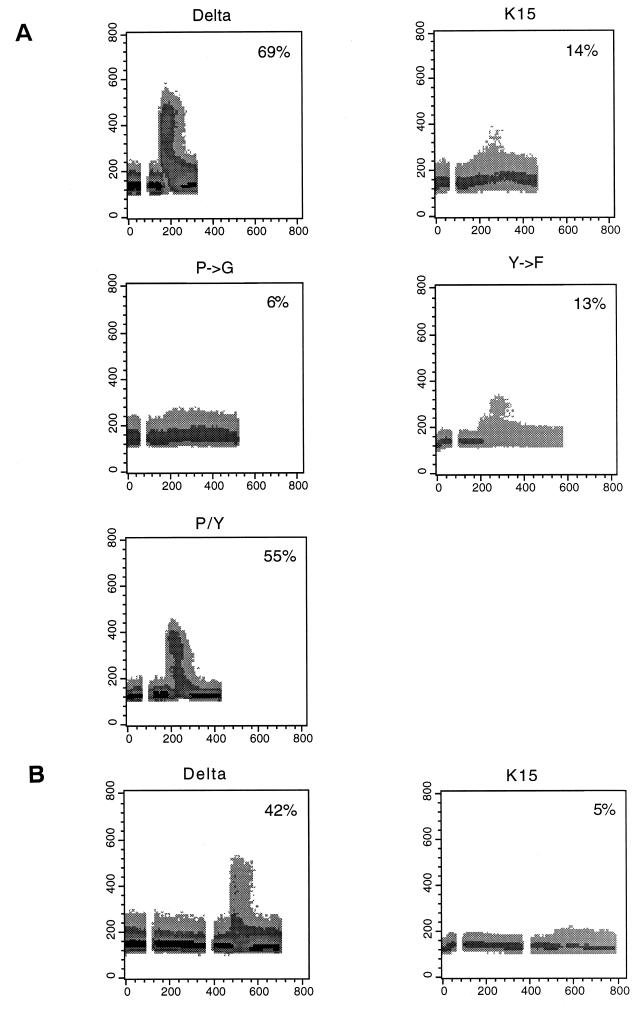
LMP2A expression has been shown to block normal BCR signal transduction in EBV-negative B cells, as evidenced by a decrease in both tyrosine phosphorylation induction and intracellular calcium mobilization after BCR cross-linking with bivalent anti-IgM antibody (19, 22–24). To assess the effect of the cytoplasmic region of K15 on BCR signal transduction, we first examined the level of IgM surface expression on BJAB cells expressing CD8-K15 chimeras by flow cytometry. It showed equivalent levels of IgM surface expression on BJAB cells expressing CD8Δ or CD8-K15 chimeras (data not shown). After stimulation with 10 μg of anti-IgM antibody, intracellular free calcium levels of these cells were monitored by flow cytometry. While a rapid increase of intracellular free calcium concentration was detected in BJAB CD8Δ cells in response to anti-IgM antibody, this increase was significantly diminished in BJAB CD8-K15 cells (Fig. 12A). BJAB CD8-K15 P→G cells and BJAB CD8-K15 Y282F cells also showed a dramatic reduction of intracellular free calcium mobilization upon anti-IgM stimulation (Fig. 12A). In contrast, BJAB CD8-K15 P→G/Y282F cells exhibited a rapid rise in the level of intracellular free calcium upon anti-IgM stimulation, as seen with BJAB CD8Δ cells (Fig. 12A). Furthermore, pretreatment of BJAB CD8-K15 cells with anti-CD8 antibody, which potentially oligomerized the CD8-K15 chimera on the cell surface, blocked anti-IgM-mediated intracellular free calcium mobilization to a greater degree (Fig. 12B).
FIG. 12.
Effect of CD8-K15 chimeras on intracellular free calcium mobilization induced by anti-IgM antibody. (A) Downregulation of IgM-mediated intracellular free calcium mobilization by CD8-K15 chimeras. The detailed procedure was described in the legend to Fig. 10 except that 10 μg of anti-IgM antibody was used. Delta, CD8Δ; K15, CD8-K15; P→G, CD8-K15 P→G; Y→F, CD8-K15 Y481F; P/Y, CD8-K15 P→G/Y481F. (B) Augmented inhibition of IgM-mediated intracellular free calcium mobilization by pretreatment with anti-CD8 antibody. CD8Δ cells (Delta) or CD8-K15 cells (K15) were stimulated with 10 μg of anti-CD8 antibody, followed by 10 μg of anti-IgM antibody. The breaks in the graph on the left side indicate the interval during the addition of each antibody. The percentage of cells responding to antibody treatment with a change in intracellular calcium level at the point of maximal change is presented inside each panel.
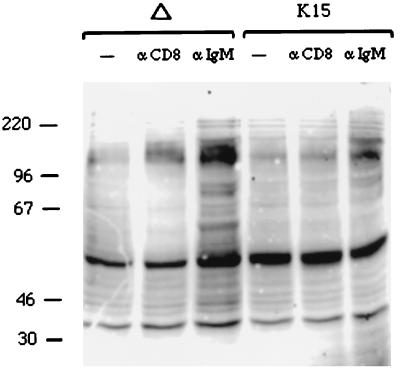
To further investigate the downregulation of BCR signaling induced by CD8-K15 expression, we evaluated the effect of anti-IgM stimulation on cellular tyrosine phosphorylation. Proteins of 50, 60, 70, 85, 120, and 160 kDa displayed an increase in tyrosine phosphorylation after anti-IgM stimulation in BJAB CD8Δ cells (Fig. 13). In marked contrast, in unstimulated BJAB CD8-K15 cells, the 60-kDa CD8-K15 protein was constitutively tyrosine phosphorylated, and this pattern remained almost unchanged after anti-IgM stimulation (Fig. 13). These results demonstrate that the cytoplasmic region of K15 is capable of blocking BCR signal transduction in BJAB cells. In addition, our experiments indicate that this activity requires the presence of either the SH2-B or the SH3-B motif.
FIG. 13.
K15 expression downregulates cellular tyrosine phosphorylation induced by anti-IgM antibody. BJAB cells expressing CD8Δ or CD8-K15 chimera were stimulated with 10 μg of anti-CD8 (αCD8) or anti-IgM antibody (αIgM) for 2 min. Unstimulated cell lysates were included as controls (−). Precleared cell lysates were subjected to an immunoblot analysis with an antiphosphotyrosine antibody.
DISCUSSION
In this report, we demonstrate that KSHV contains a novel open reading frame called K15 at the rightmost end of the viral genome. While K15 does not exhibit homology to LMP2A, it resembles LMP2A in genomic location, splicing pattern, and protein structure and by the presence of signal-transducing modules in its cytoplasmic region. Furthermore, biochemical studies with CD8 chimeras suggest that, like LMP2A, K15 downregulates B-cell antigen receptor signal transduction.
While the viral genome in herpesvirus particles is linear double-stranded DNA, it circularizes through its terminal repeats after infection and is maintained as an episomal molecule in latently infected cells. LMP2A has been shown to be transcribed across the fused terminal repeats of the EBV episome (15, 30). Thus, the intact LMP2A gene is created only after the circularization of the linear viral DNA at the terminal repeats. In addition, LMP2A is transcribed from the opposite strand from which LMP1 is transcribed; the LMP2A transcript is antisense to the LMP1 transcript (15). Since K15 is located at a genomic location and in a transcriptional direction equivalent to LMP2A, it is possible that K15 transcripts initiate from the opposite side of the viral genome, where the K1 gene resides. In fact, sequence analysis predicts that the approximately 300 bp between the initiation codon of the K15 gene and the terminal repeats does not contain any known promoter property. In addition, while the terminal repeat sequence in the EBV genome is approximately 2 to 3 kb, it extends to approximately 30 to 40 kb in the KSHV genome. Furthermore, while LMP2A is expressed latently, the K15 is weakly expressed during latency and its expression is significantly induced during lytic viral replication. Thus, the regulation of gene expression of KSHV K15 is likely distinct from that of EBV LMP2A.
Based on the sequence variation of the K1 gene, KSHV has been divided into four groups: A, B, C, and D (39). Surprisingly, we found more dramatic sequence variation in the K15 gene than in the K1 gene. K15 genes from the BC-1 and BCBL-1 cell lines exhibited only 30% amino acid identity, whereas K1 genes from both cells exhibit 94% amino acid identity (17, 39). No discernible similarity was detected in the K15 proteins with the exception of three potential signal-transducing motifs in the carboxyl cytoplasmic region: the SH2-B and SH3-B motifs and the YASIL sequence. Biochemical studies with CD8-K15 chimeras demonstrated that the tyrosine residue within the SH2-B motif is a major site of phosphorylation by cellular tyrosine kinases and that this tyrosine phosphorylation is independent of antibody stimulation. The proline-rich motif shows homology with consensus sequences for binding to the SH3 domains of signal-transducing proteins (6, 27, 38). The conserved YASIL sequence mimics the SH2-binding motif, but it is not preceded by negatively charged amino acids. In fact, the tyrosine residue in this motif is not significantly phosphorylated by cellular tyrosine kinases, suggesting that this motif is likely involved in activities other than SH2 binding. Nevertheless, despite the dramatic sequence variation, the conservation of the SH2-B and SH3-B motifs and YASIL sequence in the cytoplasmic region indicates their importance in K15 functions.
Studies with recombinant EBV demonstrate that LMP2A is not required for EBV transformation (14, 19). The amino-terminal ITAM of LMP2A has been shown to target different protein kinases in different cell types: src and syk family kinases in B lymphocytes (19) and csk in epithelial cells (31). LMP2A has been shown to function as a dominant-negative modulator of normal BCR signal transduction through an interaction of the amino-terminal ITAM with lyn and syk (22–24). In addition, studies with EBV-transformed lymphoblastoid cell lines demonstrate that the LMP2A-mediated signaling block prevents the activation of lytic replication (19, 23). In this report, we demonstrate that while the cytoplasmic region of K15 is not capable of eliciting cellular signal transduction, it has an inhibitory effect on BCR signal transduction. However, unlike LMP2A, in which the SH2-binding motif is a single important module for the inhibition of BCR signal transduction (10, 11, 19), K15 employs either the putative SH2- or SH3-binding motif for this activity. This suggests that K15 modulates lymphocyte signaling in a manner analogous to but distinct from LMP2A. In addition to KS, which is of endothelial cell origin, KSHV is associated with specific lymphoproliferative diseases, including BCBL and multicentric Castleman's disease (32, 35). These principally or exclusively originate in B cells. This suggests that K15 may target different cellular signaling molecules in KS endothelial cells versus BCBL cells. Thus, the identification of cellular targets and the expression pattern of K15 from different sources are likely to be important clues for elucidating the role of K15 in the KSHV life cycle and pathogenicity in different disease states.
ACKNOWLEDGMENTS
We especially thank B. Damania, L. Alexander, and R. Means for discussion and critical reading of the manuscript. We also thank Kristen Toohey for photography support and Maryann DeMaria for flow cytometry analysis.
This work was supported by U.S. Public Health Service grants CA31363, CA82057, and RR00168.
ADDENDUM
After the manuscript was submitted, Poole et al. (26a) and Glenn et al. (11a) published similar results.
REFERENCES
- 1.Beaufils P, Choquet D, Mamoun R Z, Malissen B. The (YXXL/I)2 signalling motif found in the cytoplasmic segments of the bovine leukaemia virus envelope protein and Epstein-Barr virus latent membrane protein 2A can elicit early and late lymphocyte activation events. EMBO J. 1993;12:5105–5112. doi: 10.1002/j.1460-2075.1993.tb06205.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caldwell R G, Wilson J B, Anderson S J, Longnecker R. Epstein-Barr virus LMP2A drives B cell development and survival in the absence of normal B cell receptor signals. Immunity. 1998;9:405–411. doi: 10.1016/s1074-7613(00)80623-8. [DOI] [PubMed] [Google Scholar]
- 3.Cambier J C. Antigen and Fc receptor signaling. The awesome power of the immunoreceptor tyrosine-based activation motif (ITAM) J Immunol. 1995;155:3281–3285. [PubMed] [Google Scholar]
- 4.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du Z, Regier D A, Desrosiers R C. Improved recombinant PCR mutagenesis procedure that uses alkaline-denatured plasmid template. BioTechniques. 1995;18:376–378. [PubMed] [Google Scholar]
- 9.Flaswinkel H, Reth M. Dual role of the tyrosine activation motif of the Ig-α protein during signal transduction via the B cell antigen receptor. EMBO J. 1994;13:83–89. doi: 10.1002/j.1460-2075.1994.tb06237.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fruehling S, Lee S K, Herrold R, Frech B, Laux G, Kremmer E, Grasser F A, Longnecker R. Identification of latent membrane protein 2A (LMP2A) domains essential for the LMP2A dominant-negative effect on B-lymphocyte surface immunoglobulin signal transduction. J Virol. 1996;70:6216–6226. doi: 10.1128/jvi.70.9.6216-6226.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fruehling S, Longnecker R. The immunoreceptor tyrosine-based activation motif of Epstein-Barr virus LMP2A is essential for blocking BCR-mediated signal transduction. Virology. 1997;235:241–251. doi: 10.1006/viro.1997.8690. [DOI] [PubMed] [Google Scholar]
- 11a.Glenn M, Rainbow L, Aurad F, Davison A, Schulz T F. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J Virol. 1999;73:6953–6963. doi: 10.1128/jvi.73.8.6953-6963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jung J U, Lang S M, Friedrich U, Jun T, Roberts T M, Desrosiers R C, Biesinger B. Identification of lck-binding elements in Tip of herpesvirus saimiri. J Biol Chem. 1995;270:20660–20667. doi: 10.1074/jbc.270.35.20660. [DOI] [PubMed] [Google Scholar]
- 13.Jung J U, Lang S M, Jun T, Roberts T M, Veillette A, Desrosiers R C. Downregulation of Lck-mediated signal transduction by tip of herpesvirus saimiri. J Virol. 1995;69:7814–7822. doi: 10.1128/jvi.69.12.7814-7822.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Vol. 74. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 15.Laux G, Perricaudet M, Farrell P. A spliced Epstein-Barr virus gene expressed in immortalized lymphocytes is created by circularization of the linear viral genome. EMBO J. 1988;7:769–774. doi: 10.1002/j.1460-2075.1988.tb02874.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee H, Guo J, Li M, Choi J-K, DeMaria M, Rosenzweig M, Jung J U. Identification of an immunoreceptor tyrosine-based activation motif of K1 transforming protein of Kaposi's sarcoma-associated herpesvirus. Mol Cell Biol. 1998;18:5219–5228. doi: 10.1128/mcb.18.9.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A A, Desrosiers R C, Jung J U. Deregulation of cell growth by the Kaposi's sarcoma-associated herpesvirus K1 gene. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 18.Longnecker R, Kieff E. A second Epstein-Barr virus membrane protein (LMP2) is expressed in latent infection and colocalizes with LMP1. J Virol. 1990;64:2319–2326. doi: 10.1128/jvi.64.5.2319-2326.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Longnecker R, Miller C L. Regulation of Epstein-Barr virus latency by latent membrane protein 2. Trends Microbiol. 1996;4:38–42. doi: 10.1016/0966-842x(96)81504-6. [DOI] [PubMed] [Google Scholar]
- 20.Longnecker R, Miller C L, Miao X-Q, Tomkinson B, Kieff E. The last seven transmembrane and carboxy-terminal cytoplasmic domains of Epstein-Barr virus latent membrane protein 2 (LMP2) are dispensable for lymphocyte infection and growth transformation in vitro. J Virol. 1993;67:2006–2013. doi: 10.1128/jvi.67.4.2006-2013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mesri E A, Cesarman E, Arvanitakis L, Rafii S, Moore M A S, Posnett D N, Knowles D M, Asch A S. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183:2385–2389. doi: 10.1084/jem.183.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller C L, Burkhard A L, Lee J H, Stealey B, Longnecker R, Bolen J B, Kieff E. Integral membrane protein 2 of Epstein-Barr virus regulates reactivation from latency through dominant negative effects on protein-tyrosine kinases. Immunity. 1995;2:155–166. doi: 10.1016/s1074-7613(95)80040-9. [DOI] [PubMed] [Google Scholar]
- 23.Miller C L, Lee J H, Kieff E, Longnecker R. An integral membrane protein (LMP2) blocks reactivation of Epstein-Barr virus from latency following surface immunoglobulin crosslinking. Proc Natl Acad Sci USA. 1994;91:772–776. doi: 10.1073/pnas.91.2.772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller C L, Longnecker R, Kieff E. Epstein-Barr virus latent membrane protein 2A blocks calcium mobilization in B lymphocytes. J Virol. 1993;67:3087–3094. doi: 10.1128/jvi.67.6.3087-3094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neipel F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholas J, Zong J-C, Alcendor D J, Ciufo D M, Poole L J, Sarisky R T, Chiou C-J, Zhang X, Wan X, Guo H-G, Reitz M S, Hayward G S. Novel organizational features, captured cellular genes, and strain variability within the genome of KSHV/HHV8. J Natl Cancer Inst Monogr. 1998;23:79–88. doi: 10.1093/oxfordjournals.jncimonographs.a024179. [DOI] [PubMed] [Google Scholar]
- 26a.Poole L J, Zong J C, Ciufo D, Alcendor D, Cannon J S, Ambinder R, Orenstein J M, Reitz M S, Hayward G S. Comparison of genetic variability at multiple loci across the genomes of the major subtypes of Kaposi's sarcoma-associated herpesvirus reveals evidence for recombination and for two distinct types of open reading frame K15 alleles at the right-hand end. J Virol. 1999;73:6646–6660. doi: 10.1128/jvi.73.8.6646-6660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ren R, Mayer B J, Cicchetti P, Baltimore D. Identification of a ten-amino acid proline-rich SH3 binding site. Science. 1993;259:1159–1161. doi: 10.1126/science.8438166. [DOI] [PubMed] [Google Scholar]
- 28.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Ganem D. Lytic growth of Kaposi's sarcoma-associate herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 29.Russo J J, Bohenzxy R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sample J, Liebowitz D, Kieff E. Two related Epstein-Barr virus membrane proteins are encoded by separate genes. J Virol. 1989;63:933–937. doi: 10.1128/jvi.63.2.933-937.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scholle F, Longnecker R, Raab-Traub N. Epithelial cell adhesion to extracellular matrix proteins induces tyrosine phosphorylation of the Epstein-Barr virus latent membrane protein 2: a role for C-terminal Src kinase. J Virol. 1999;73:4767–4775. doi: 10.1128/jvi.73.6.4767-4775.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulz T F, Chang Y, Moore P S. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8). Washington, D.C.: American Society for Microbiology; 1998. [Google Scholar]
- 33.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Songyang Z, Shoelson S E, Chaudhuri M, Gish G, Pawson T, Haser W G, King F, Roberts T, Ratnofsky S, Lechleider R J, Neel B G, Birge R B, Fajardo J E, Chou M M, Hanafusa H, Schaffhausen B, Cantley L C. SH2 domains recognize specific phosphopeptide sequences. Cell. 1993;72:767–778. doi: 10.1016/0092-8674(93)90404-e. [DOI] [PubMed] [Google Scholar]
- 35.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, Sigaux F. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1997;86:1276–1280. [PubMed] [Google Scholar]
- 36.Wange R L, Samelson L E. Complex complexes: signaling at the TCR. Immunity. 1996;5:197–205. doi: 10.1016/s1074-7613(00)80315-5. [DOI] [PubMed] [Google Scholar]
- 37.Weiss A, Littman D R. Signal transduction by lymphocyte antigen receptors. Cell. 1994;76:263–274. doi: 10.1016/0092-8674(94)90334-4. [DOI] [PubMed] [Google Scholar]
- 38.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 39.Zong J C, Ciufo D M, Alcendor D J, Wan X, Nicholas J, Browning P J, Rady P L, Tyring S K, Orenstein J M, Rabkin C S, Su I J, Powell K F, Croxson M, Foreman K E, Nickoloff B J, Alkan S, Hayward G S. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–4170. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]