Abstract
The UL25 gene of pseudorabies virus (PrV) can encode a protein of about 57 kDa which is well conserved among herpesviruses. The UL25 protein of herpes simplex virus type 1 is a capsid constituent involved in virus penetration and capsid maturation. To identify and characterize the UL25 gene product of PrV, polyclonal mouse anti-UL25 antibodies were raised to a bacterially expressed fusion protein. In immunoblotting and immunoprecipitation assays of PrV-infected cell lysates, these anti-UL25 antisera specifically recognized a protein of the expected size with late expression kinetics. This 57-kDa product was also present in purified virions and was found to be associated with all types of capsids. Synthesis of a protein migrating at the same size point was directed from the eukaryotic expression plasmid pCG-UL25. To determine the subcellular localization of UL25, immunofluorescence studies with anti-UL25 antisera were performed on Nonidet P-40-extracted COS-7 cells infected with PrV or transfected with pCG-UL25. In PrV-infected cells, newly synthesized UL25 is directed mainly to distinct nuclear compartments, whereas UL25 expressed in the absence of other viral proteins is distributed more uniformly in the nucleus and colocalizes also with microtubules. To study the fate of UL25 at very early stages of infection, immunofluorescence experiments were performed on invading PrV particles in the presence or absence of drugs that specifically depolymerize components of the cytoskeleton. We found that the incoming nucleocapsids colocalize with microtubules during their transport to the nucleus and that UL25 remains associated with nucleocapsids during this transport.
Pseudorabies virus (PrV), an alphaherpesvirus closely related to herpes simplex virus type 1 (HSV-1), is the etiologic agent of Aujeszky's disease, an illness involving prominent neurological and respiratory symptoms in pigs (32). Following an oronasal infection, PrV invades the peripheral endings of the primary sensory, sympathetic, and parasympathetic neurons and then proceeds to the corresponding ganglions and to the central nervous system, causing massive cell destruction (4, 18).
Herpesvirus penetration into cells is a complex process involving the interaction of many viral glycoproteins with components of the plasma membrane (reviewed in references 23 and 30). After fusion of the viral envelope with the cellular membrane, capsids are liberated into the cytosol, are dissociated from many of the tegument proteins, and migrate toward the nuclear pores, where the viral DNA is transferred into the nucleoplasm (5, 12, 20). Transcription, replication, and assembly of progeny capsids occur within the nucleus (27). In HSV-1-infected cells, the transport of capsids toward the nucleus proceeds efficiently along microtubules after binding of the capsids to dynein, a microtubule-dependent motor responsible for the retrograde transport of organelles (29). This active transport mechanism seems to be particularly important for neurotropic viruses because cell bodies of neurons are located far away from the viral entry sites. The viral protein(s) implicated in dynein binding has yet to be identified.
HSV-1 capsid shells assemble in presence of the major capsid protein VP5, the triplex-constituting proteins VP23 and VP19C, VP26 forming the capsomer tips, the scaffolding protein VP22a, and the protease VP24 and its cleavage product VP21. These proteins are encoded by the UL19, UL18, UL38, UL35, UL26.5, and UL26 genes, respectively (reviewed in reference 13). During nucleocapsid maturation, the viral DNA replaces the scaffold core of the intermediate capsid shells. In the nuclei of infected cells, therefore, three capsid types are found: the C or nucleocapsids containing the viral genome in place of the scaffold core, the B or intermediate capsid shells containing a core composed of the scaffolding protein, and the A or abortive shells carrying neither DNA nor the scaffolding protein. Studies performed with temperature-sensitive or deletion HSV-1 mutants have shown that the UL6, UL15, UL17, UL25, UL28, UL32, and UL33 gene products are essential for cleavage of concatemeric DNA into unit length viral genomes and/or its packaging into preformed B capsids (reviewed in references 13 and 28). The precise functions of these proteins are unknown. Furthermore, the efficiency of capsid maturation is greatly increased in the presence of the UL12 gene product, an alkaline nuclease involved in resolving complex DNA replication intermediates (21). In PrV, the UL21 gene product was shown to be also involved in capsid maturation (8). Since many of the identified genes encoding capsid assembly and maturation proteins share a high degree of homology among alphaherpesviruses, PrV capsid assembly is believed to be very similar to that of HSV-1 (9, 10, 16, 17, 24, 33).
The UL25 gene product of HSV-1 is expressed late in the replication cycle and is a minor yet essential constituent of viral capsids (2, 22). With the help of the temperature-sensitive HSV-1 mutant ts1204 harboring a defect in the region of the UL25 gene, it has been shown that, at the nonpermissive temperature, virions were capable of fixing to the plasma membrane but could not penetrate into the cell (1). In addition, the same mutant accumulated abortive capsid shells when the shift to the nonpermissive temperature occurred after penetration. Studies with a UL25 deletion mutant demonstrated that in the absence of UL25, replicated DNA can be cleaved normally yet is not packaged into capsids (22). These findings suggest that UL25 could play a role both at a very early step in the viral cycle and in capsid maturation.
The UL25 gene of PrV is located in the BamHI genome fragment 9 and can encode a protein of 534 amino acids (9). The predicted 57-kDa UL25 protein is particularly well conserved among alphaherpesviruses, suggesting that it might exert similar functions in these viruses. To identify and characterize the PrV UL25 gene product, polyclonal mouse anti-UL25 antibodies were raised to a bacterially expressed fusion protein. We report that these anti-UL25 antibodies specifically react with a 57-kDa capsid protein that is synthesized late in PrV infection. De novo-synthesized UL25 is directed to distinct nuclear compartments in PrV-infected cells, whereas plasmid-expressed UL25 is distributed throughout the cell and localizes specifically to the nucleus and microtubules. After penetration of PrV into the cell, UL25 remains associated with nucleocapsids during their microtubule-mediated transport to the nucleus.
MATERIALS AND METHODS
Cells and virus.
African green monkey kidney COS-7 cells were propagated in Dulbecco modified Eagle medium (DMEM; GIBCO BRL) containing 10% fetal calf serum (FCS), and BSR cells, derived from baby hamster kidney cells, were maintained in Glasgow's modified minimal essential medium supplemented with 9% calf serum at 37°C in a 5% CO2 incubator. The NG108-15 cell line, a hybrid of mouse neuroblastoma N18 and rat glioma C6 cell lines, was cultured in DMEM plus 10% FCS. The Kaplan strain of PrV was propagated in BSR cells by infecting them with a multiplicity of infection (MOI) of 0.001 PFU/cell. At 72 h postinfection, virions were concentrated by pelleting the infected cell culture supernatant through a cushion of 25% glycerol in 10 mM Tris-HCl (pH 7.5)–50 mM NaCl–1 mM EDTA for 1 h at 27,000 rpm in a Beckman SW28 rotor.
Plasmids.
To construct pET-UL25/8, the PvuI-SacI segment (nucleotides 3593 to 2233) of BamHI-fragment 9 of PrV containing the UL25 gene was subcloned into the HindIII site of the bacterial expression vector pET22b(+) (Novagen) under the control of the bacteriophage T7 promoter. The expression product of pET-UL25/8 contains an amino-terminal periplasm localizing leader (pelB) sequence and six carboxy-terminal histidine residues fused to a truncated UL25 protein lacking the carboxy-terminal 107 amino acids of the putative 534-amino-acid gene product.
The coding regions of the UL25, UL26, UL26.5, UL6, UL18, UL19, and UL27-gB genes were subcloned under the control of the cytomegalovirus immediate-early (CMV-IE) promoter into the BamHI site of the eukaryotic expression vector pCG (7). Plasmids pCG-UL25, pCG-UL26, and pCG-UL26.5 carry the PvuI-NcoI (nucleotides 3593 to 1868), the PstI-BamHI (nucleotides 2021 to 1), or the PstI-BamHI (nucleotides 1199 to 1) segment of PrV BamHI fragment 9, respectively. UL6 was assembled from the 177-bp PvuI-BamHI and the 1,842-bp BamHI-DrdI segments of BamHI fragments 6 and 3, respectively. The UL18 and UL19 genes were liberated from BamHI fragment 4 by digestion with BsrGI and Bsu36I (nucleotides 7117 to 8249) and with NsiI and SphI (nucleotides 2922 to 7369), respectively. The UL27-gB gene was first reconstituted from the 2.8- and 1.8-kbp SphI-SphI segments of BamHI fragment 1. Following introduction of BamHI sites upstream and downstream of the UL27-gB coding region by site-directed mutagenesis, the UL27-gB gene was liberated by digestion with BamHI and transferred into pCG. The BamHI fragments 9, 6/3, 4, and 1 of Kaplan strain PrV were kindly provided by T. Ben-Porat and T. Mettenleiter, and the available EMBL databank accession numbers for their nucleotide sequences are X95710, X97257, and L00676, respectively (9, 10, 16).
Capsid purification.
Crude capsid pellets recovered after nonionic detergent treatment of concentrated intracellular PrV were generously provided by J. C. Audonnet (MERIAL, Lyon, France). The crude capsid material from about 2 × 1010 infected cells was resuspended in TNE (20 mM Tris-HCl, pH 7.6; 0.5 M NaCl; 1 mM EDTA), Dounce homogenized, and sonicated for 30 s in 6-s intervals. The cleared suspension was layered onto a 35% (wt/vol) sucrose cushion in TNE and centrifuged at 19,600 rpm for 1 h in a Beckman SW28 rotor (19). The pellet was resuspended in 12 ml of TNE, and one-tenth of this suspension was layered onto a continuous 60 to 20% (wt/vol) sucrose gradient in TNE; capsids were banded for 1 h at 24,000 rpm in a Beckman SW28 rotor. Fractions corresponding to the C, B, and A capsids were collected, gradient-purified for a second round, and then concentrated by pelleting in phosphate-buffered saline (PBS) for 1 h at 24,000 rpm in a Beckman SW41 rotor.
Antisera.
Induced expression of pET-UL25/8 in the Escherichia coli strain BL21(DE3) carrying the T7 RNA polymerase gene in its genome resulted in the accumulation of the expected 55-kDa pelB-UL25-His fusion protein in inclusion bodies despite its pelB amino-terminal signal sequence. The UL25 fusion protein was then solubilized and purified by metal chelation chromatography in buffers containing 6 M urea as instructed by the supplier (Novagen). For production of polyclonal anti-UL25 antibodies, 6-week-old female BALB/c mice were primed with pristane (2,6,10,14-tetramethyldecanoic acid) and immunized 6 days later by intraperitoneal injection of 50 μg of affinity-purified pelB-UL25-His protein in Freund's complete adjuvant (GIBCO BRL). The mice were challenged twice 2 and 4 weeks later with the same amount of fusion protein in Freund's incomplete adjuvant, and ascites were induced by injection of 106 SP2/0 myeloma cells. Nucleocapsid antisera were obtained by an initial intradermal injection of New Zealand White (saprophyte pathogen-free) rabbits with 1 mg of sucrose-gradient purified PrV nucleocapsids in Freund's complete adjuvant and by three subcutaneous booster immunizations at 2-week intervals with the same amount of antigen in Freund's incomplete adjuvant (AGRO-BIO). Polyclonal mouse anti-glycoprotein B (anti-gB) and anti-glycoprotein C (anti-gC) antibodies were raised to gB and gC expressed in Sf21 lepidopteran cells infected with the respective recombinant baculoviruses, and the polyclonal rabbit anti-PrV antibody was raised to purified and sodium dodecyl sulfate (SDS)-treated virions. Both the rabbit polyclonal anti-tubulin antibody and phalloidin were purchased from Sigma. Dichlorotriazinyl amino fluorescein (DTAF)-labeled donkey anti-rabbit immunoglobulin G (IgG) was obtained from Jackson ImmunoResearch Laboratories, and Texas red-coupled donkey anti-mouse IgG was from Interchim.
Virus infections, plasmid transfections, and drug treatments of cells.
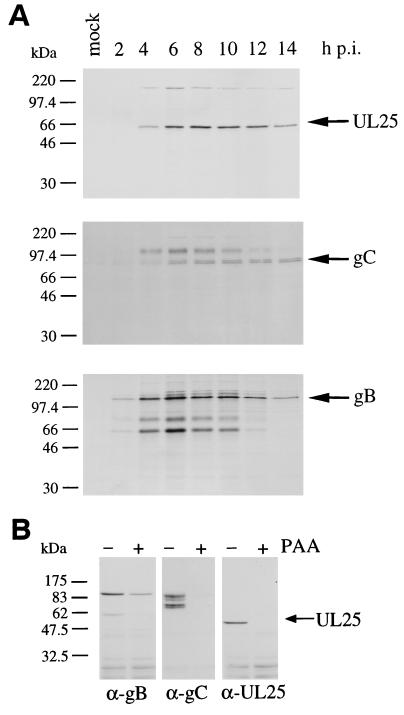
To analyze de novo synthesis of viral proteins, cells were washed with OptiMEM (GIBCO BRL) and then infected with PrV at an MOI of 5 PFU/cell. After 1 h of adsorption at 37°C, the inoculum was replaced by culture medium, and incubation was continued at 37°C. In experiments analyzing incoming virus particles, washed cells were inoculated with PrV at an MOI of 50 at 4°C. Infection was initiated by shifting the cells to 37°C. For transfection of COS-7 or BSR cells in 35-mm culture dishes, we used either the Lipofectin (GIBCO BRL) or FuGENE 6 (Boehringer Mannheim) transfection reagents and 2 μg of plasmid DNA according to the instructions of the suppliers. Incubations at 37°C were continued for 24 h for the immunofluorescence studies and for 48 h for the immunoblot analyses. For drug treatment of transfected cells, the culture medium was replaced 24 h after transfection with growth medium containing 2 μM nocodazole (Sigma) or 0.5 μM cytochalasin D (Sigma), and incubation was continued for 2 h at 37°C. In infection experiments, cells were kept in OptiMEM containing these drugs for 1 h before virus inoculation and during infection. To determine the kinetics class of UL25, cells were kept in 300 μg of phosphonoacetic acid (PAA; ICN) per ml throughout infection.
Immunoblot analyses.
Cells in 35-mm culture dishes were washed twice with PBS (137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4; pH 7.3) and lysed for 20 min on ice in 50 mM Tris-HCl (pH 8)–62.5 mM EDTA–1% Nonidet P-40 (NP40)–0.4% sodium deoxycholate supplemented with 15 μg of antipain-dihydrochloride, 2.5 μg of aprotinin, 2.5 μg of pepstatin, 5 μg of chymostatin, 2.5 μg of leupeptin, and 100 μg of pefabloc (protease inhibitors; Boehringer Mannheim) per ml. About one-tenth of the lysates was run on denaturing SDS-polyacrylamide gels and blotted onto nitrocellulose membranes. Before incubation with the primary antibody at a dilution of 1:1,000 in TBST (10 mM Tris-HCl, pH 8; 150 mM NaCl; 0.05% Tween 20) plus 1% bovine serum albumin (and 3% FCS for the anti-PrV and anti-capsid antibodies), the membranes were saturated with 5% nonfat milk in TBST. The secondary antibodies, goat anti-mouse IgG or goat anti-rabbit IgG coupled to horseradish peroxidase (Sigma), were diluted according to the supplier's instructions. The immunoblots were revealed by enhanced chemiluminescence (Amersham Life Sciences).
Immunoprecipitation.
BSR cells in 60-mm dishes were infected with PrV at an MOI of 5. Cells were starved for 1 h in methionine- and cysteine-free minimum essential medium (ICN), radiolabeled for 1 h with 290 μCi of Pro-Mix L-[35S] cell-labeling mix (Amersham Life Sciences), and then chased with regular culture medium for 30 min at 37°C. Lysis was performed for 20 min on ice in 50 mM Tris-HCl (pH 8)–150 mM NaCl–1% NP-40 in the presence of the protease inhibitors as detailed above. The lysate of each 60-mm dish was divided into three parts for use in the immunoprecipitation with the anti-UL25, anti-gC, and anti-gB antibodies at a dilution of 1/500 according to a method described earlier (14). Half of the immunoprecipitated proteins were then analyzed by SDS–10% polyacrylamide gel electrophoresis (PAGE).
Indirect immunofluorescence.
Infected or transfected cells on glass coverslips were fixed with 4% (wt/vol) paraformaldehyde in PBS for 20 min at room temperature and then permeabilized for 1 min with 0.1% Triton X-100. For detergent extraction, cells were washed once with PBS and twice with CSK buffer (10 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8; 100 mM KCl, 300 mM sucrose, 2.5 mM MgCl2) before extraction with 1% NP-40 in CSK buffer for 3 min on ice (31). The detergent-insoluble cell components remaining on the coverslips were then washed with CSK buffer and fixed with methanol for 4 min at −20°C. Before they were immunolabeled for 1 h at 37°C, cells were washed three times with PBS and then blocked in 10% donkey serum in PBS for 30 min at room temperature. The anti-UL25 and anti-capsid antisera were diluted 1:100, and the rabbit anti-tubulin antiserum was diluted 1:50 in PBS containing 10% donkey serum. Coverslips were extensively washed with PBS before labeling with the secondary antibodies in 10% donkey serum for 45 min. After three rinses with PBS and one rinse with water, the coverslips were mounted onto glass slides by using Immu-Mount (Shandon) and then analyzed with an Olympus BX-FLA fluorescence microscope.
RESULTS
UL25 is a structural protein of PrV.
To identify and characterize the PrV UL25 gene product, polyclonal mouse anti-UL25 antibodies were raised to a bacterially expressed and affinity-purified 55-kDa pelB-UL25-His fusion polypeptide as described in Materials and Methods. All five mice immunized with the fusion protein produced antibodies recognizing the UL25 antigen on immunoblots (result with anti-UL25 a1 shown in Fig. 1, lane 1). These anti-UL25 antibodies specifically recognized a protein correlating with the predicted size of UL25 (57 kDa) in PrV-infected BSR cell extracts (Fig. 1, lane 3). Synthesis of a protein of the same size was also directed from the eukaryotic expression plasmid pCG-UL25 carrying the UL25 gene under the control of the CMV-IE promoter in COS-7 cells (Fig. 1, lane 2), whereas no protein bands were detected in mock-infected or transfected cell lysates (Fig. 1, lane 5). The 57-kDa product was also present in virions purified from supernatants of PrV-infected cells (Fig. 1, lane 4), indicating that UL25 is a structural component of PrV.
FIG. 1.
Immunoblot analysis of UL25 expression. Affinity-purified pelB-UL25-His fusion polypeptide (lane 1), protein extracts of COS-7 cells transfected with pCG-UL25 (lane 2) or BSR cells infected with PrV (lane 3), purified PrV Kaplan strain virions (7 × 105 PFU; lane 4), and mock-infected BSR cell extracts (lane 5) were subjected to denaturing SDS–12% PAGE and then immunoblotted with polyclonal mouse anti-UL25 antibodies.
The UL25 protein is a minor capsid constituent.
To determine the localization of UL25 within the virion, capsids were purified by banding twice on continuous 60 to 20% sucrose gradients. Bands corresponding to the C, B, and A capsid types were isolated, concentrated, subjected to SDS-PAGE, and analyzed by immunoblotting with the anti-PrV antibodies raised to virions or the anti-UL25 antibodies (Fig. 2). As a control, lysates of COS-7 cells transiently transfected with plasmids pCG-UL19 and pCG-UL18 were analyzed by immunoblotting with the rabbit anti-capsid antibody which had been raised to gradient-purified PrV nucleocapsids. The capsid antiserum detected the 142-kDa UL19 and the 32-kDa UL18 major capsid proteins in transfected cells, which corresponded to the respective protein band in the C capsids (Fig. 2, α-capsid panel). There are several other proteins present in the C capsids that most likely represent other capsid constituents. The rabbit PrV antiserum recognized the UL19 and the UL18 proteins on immunoblots of C, B, and A capsids (Fig. 2, α-PrV panel). After the stripping and rehybridization of the same membrane with anti-UL25 antibodies, UL25 can be revealed in all three capsid types (Fig. 2, α-UL25 panel). There is no protein of the expected size of UL25 (57 kDa) distinctly discernible among Coomassie blue-stained capsid proteins (Fig. 2, stained panel), indicating that the UL25 protein of PrV constitutes a minor component of capsids. Furthermore, the anti-PrV and anti-capsid sera recognized UL25 only after long exposure times.
FIG. 2.
Analysis of capsid proteins. (Left panel) C capsids isolated from 60 to 20% sucrose gradients were subjected to SDS–12% PAGE and analyzed by Coomassie blue staining. (Right panels) Lysates of COS-7 cells transfected with pCG-UL19 (UL19) or pCG-UL18 (UL18) and gradient-purified C, B, and A capsids were analyzed by SDS–10% PAGE, followed by immunoblotting with anti-capsid, anti-PrV, or anti-UL25 antibodies as indicated below each blot. Positions of the UL19, UL18, and UL25 proteins are indicated on the right with respective arrows.
Expression kinetics of the UL25 protein.
To monitor the synthesis of UL25 in the course of a PrV replication cycle, BSR cells were infected at an MOI of 5 and, starting at the times indicated, proteins were radiolabeled for 1 h, chased for 30 min, and harvested for immunoprecipitation. The cell lysate of each time point served for precipitation with the polyclonal mouse anti-UL25, anti-gC, and anti-gB antibodies (Fig. 3A). The de novo synthesis of UL25 was thus monitored in parallel with that of gC and gB, which have late and early-late expression kinetics, respectively (6, 25, 26). Synthesis of both UL25 and gC was detectable after 4 h of infection and showed an expression peak at 6 to 9 h postinfection (Fig. 3A, upper and middle panels), whereas gB and its maturation products appeared as early as 2 h following PrV infection and showed a high level of synthesis from 4 to 11 h after infection (Fig. 3A, lower panel). The UL25 protein, therefore, appears to be expressed late in the PrV replication cycle. Note that a high-molecular-weight protein was coprecipitated with UL25 in PrV-infected cells. This appears to be due to a nonspecific interaction, since the same protein was also precipitated with anti-gC and anti-gB, as well as with other nonrelated antibodies (not shown). To confirm the kinetics class of UL25 expression, PrV-infected BSR cells were maintained in medium containing PAA, an inhibitor of DNA synthesis. At 6 h postinfection, nontreated and PAA-treated cells were harvested for immunoblot analysis with anti-gB, anti-gC, and anti-UL25 antibodies (Fig. 3B). The expression of gB was slightly affected by the presence of PAA (Fig. 3B, left panel), whereas the synthesis of both gC and UL25 was drastically reduced in the absence of viral DNA replication (Fig. 3B, middle and right panels). PrV UL25 is thus expressed with truly late kinetics.
FIG. 3.
Expression kinetics of the UL25 protein. (A) BSR cells were infected with PrV at an MOI of 5, radiolabeled for 1 h starting at the times indicated in hours postinfection (h p.i.), and then chased for 30 min and harvested for immunoprecipitation with the polyclonal mouse antibodies anti-UL25 (upper panel), anti-gC (middle panel), and anti-gB (lower panel). The immunoprecipitated proteins were separated by SDS–10% PAGE. (B) BSR cells were infected at an MOI of 5 in the absence or presence of 300 μg of PAA per ml, and at 6 h postinfection cells were harvested for immunoblot analysis with the anti-gB, anti-gC, and anti-UL25 antibodies as indicated below each panel.
Subcellular localization of de novo-synthesized UL25.
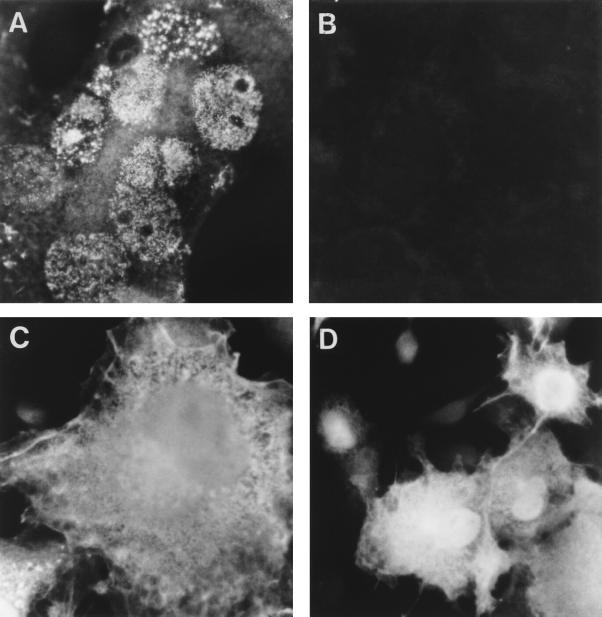
To determine the subcellular localization of UL25, indirect immunofluorescence studies with anti-UL25 antibodies were performed on cells infected with PrV or transfected with pCG-UL25 (Fig. 4). As early as 4 h after infection, the newly synthesized UL25 was detected in the nucleus with a spot- or patch-like distribution (Fig. 4A). In addition, a lighter cytoplasmic fluorescence was observed. Note that PrV infection induces syncytium formation in COS-7 cells. This predominantly nuclear localization was the same in BSR and NG108-15 cells (not shown). In mock-infected and mock-transfected cells, no specific staining was observed with the anti-UL25 antibodies (Fig. 4B). UL25 expressed from a plasmid vector in COS-7 and BSR cells in the absence of other PrV proteins was found to be distributed throughout the cell, localizing both to the nucleus, excluding the nucleoles, and to the cytoplasm (Fig. 4C and D). Its distribution within the nucleus, however, is more uniform than was observed in infected cells. The staining pattern of UL25 in transfected cells was found unaltered after its coexpression with the capsid proteins UL6, UL18, UL19, UL26, or UL26.5 (not shown).
FIG. 4.
Subcellular localization of UL25. Indirect immunofluorescence studies with anti-UL25 and Texas red-labeled secondary antibodies were performed on COS-7 cells (i) infected with PrV at an MOI of 5 after 6 h of infection (A), (ii) mock-infected (B), or (iii) transfected with pCG-UL25 (C and D). Note that PrV infection induces syncytium formation in COS-7 cells. Immunofluorescence was visualized with an Olympus BX-FLA fluorescence microscope at a magnification of ×750 (A, B, and C) or ×300 (D).
To further define the distribution of UL25, immunofluorescence experiments with anti-UL25 antibodies were carried out on PrV-infected or plasmid-transfected COS-7 cells that have been extracted with 1% NP-40 before fixation. This detergent extraction strips the cell of its membrane and cytosolic proteins, partially solubilizes proteins of the Golgi apparatus, and yet leaves the nucleus and cytoskeleton intact. The predominantly nuclear localization of UL25 in infected COS-7 cells was not modified after detergent extraction (not shown). Following detergent extraction of transfected cells, UL25 still localized to the nucleus (Fig. 5A). In addition, UL25 appeared to associate with the cytoskeleton, with an accumulation of the protein near the nucleus. These experiments were carried out in parallel with either a polyclonal anti-tubulin antibody recognizing microtubules or with phalloidin which specifically interacts with actin filaments. UL25 in transfected cells appeared to colocalize with microtubules with an accumulation at the microtubule-organizing center (not shown). Both microtubules and actin fibers were left intact in cells synthesizing UL25 in the absence of other viral proteins, whereas actin fibers were completely disrupted in PrV-infected cells (not shown).
FIG. 5.
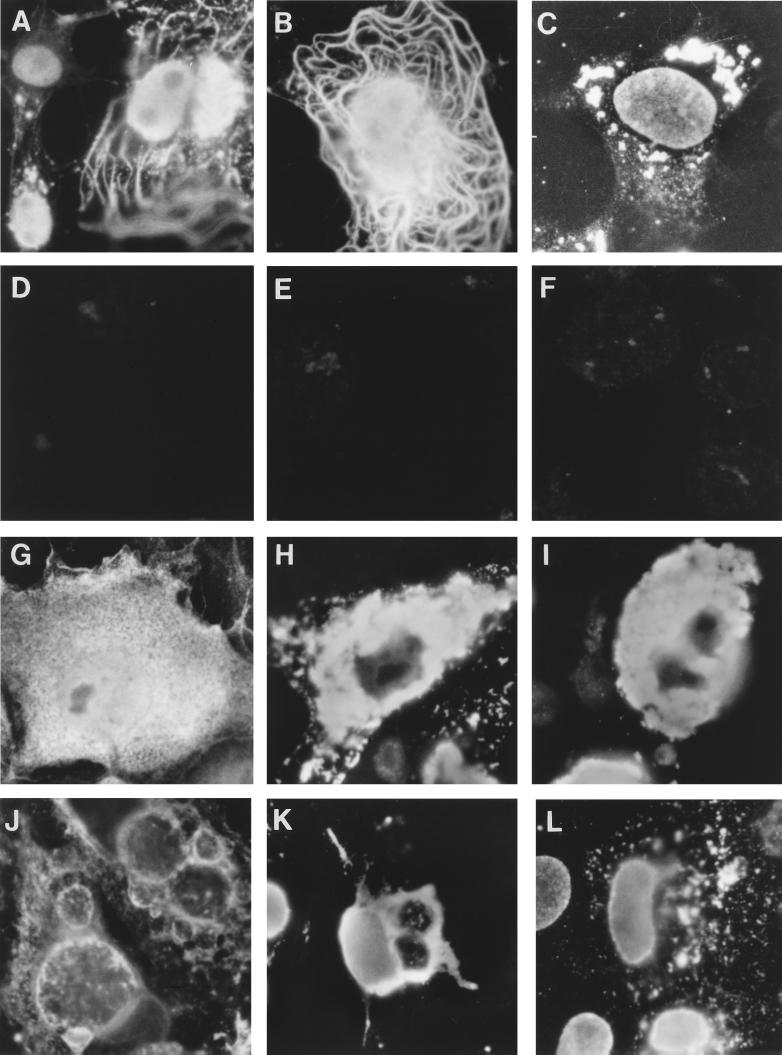
Effect of cytoskeletal drugs on the localization of UL25, UL19, and gB in transiently transfected COS-7 cells. At 24 h after mock transfection (D, E, and F) or after transfection with pCG-UL25 (A, B, and C), pCG-UL19 (G, H, and I), and pCG-gB (J, K, and L), cells were incubated for 2 h with OptiMEM alone (A, D, G, H, J, and K) or with OptiMEM containing either cytochalasin D (B and E) or nocodazole (C, F, I, and L). With the exception of cells in panels G and J, which were directly fixed, cells were extracted with 1% NP-40 before fixation and analyzed with anti-UL25 (A to F), anti-capsid (G, H, and I), and anti-gB (J, K, and L) antibodies. Secondary antibodies were donkey anti-rabbit antibody coupled to DTAF and anti-mouse antibody coupled to Texas red. Magnification, ×750.
To roughly estimate the fraction of overexpressed UL25 protein remaining associated with the nucleus and the cytoskeleton after NP-40 extraction of the cell, immunoblot analysis was performed on lysates of the detergent-soluble and insoluble fractions of pCG-UL25-transfected COS-7 cells that were subjected to the same extraction procedures as in the above-described immunofluorescence studies. The results indicated that a higher proportion of the overexpressed UL25 localizes to the insoluble fraction of the cell (not shown).
To test if UL25 specifically colocalizes with microtubules, COS-7 cells transiently transfected with pCG-UL25 were treated with either cytochalasin D, which specifically depolymerizes actin fibers, or nocodazole, which depolymerizes microtubules and fragments the Golgi apparatus, causing the dispersal of its derived vesicles throughout the cytoplasm (3, 15). Following detergent extraction, the localization of UL25 was analyzed with anti-UL25 antibodies. In cells treated with cytochalasin D (Fig. 5B), UL25 still localized to the nucleus and to microtubules with an accumulation of the protein at the microtubule-organizing center. In cells treated with nocodazole, however, UL25 was found in the nucleus and in aggregates dispersed throughout the remainder of the cell (Fig. 5C). The effect of nocodazole could be partly reversed when cells were washed and left to recuperate in regular growth medium overnight. UL25 then accumulated again in proximity to the nucleus (not shown). Nonspecific interactions of the UL25 antibodies were not observed after drug treatments and detergent extraction of mock-transfected cells (Fig. 5D to F).
To control whether the staining pattern observed is specific for UL25, COS-7 cells were transiently transfected with pCG-UL19 and pCG-gB, subjected to nocodazole treatment and NP-40 extraction as described above, and analyzed with anti-capsid and anti-gB antibodies, respectively (Fig. 5G to L). In fixed and permeabilized cells, UL19 was distributed throughout the cell (Fig. 5G). In nontreated or nocodazole-treated and NP-40-extracted cells, the UL19 protein aggregated in clumps near the nucleus (Fig. 5H and I, respectively). Overexpression of gB induced the formation of large, probably Golgi-derived, vesicles. It localized to the nuclear and plasma membranes and to the Golgi apparatus and was found distributed in patches throughout the cytoplasm (Fig. 5J). After detergent extraction, gB localized to the nuclear membrane and to residual Golgi vesicles (Fig. 5K). In nocodazole-treated and detergent-extracted cells, gB localized to the nuclear membrane and to dispersed Golgi vesicles. Similar distribution patterns were found for gB of HSV-1 (3, 11).
The staining patterns observed after nocodazole treatment and detergent extraction are unique for each of the tested proteins, and UL25 shows the specific property of colocalizing to the nucleus and to microtubules.
UL25 remains associated with incoming PrV nucleocapsids during their transport to the nucleus.
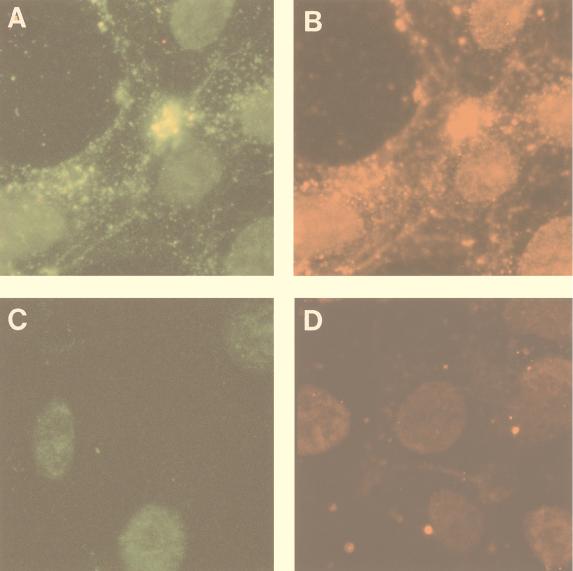
The fate of UL25 after penetration of the virus into the cell and during nucleocapsid transport to the nucleus was studied by indirect immunofluorescence (Fig. 6 and 7). Following infection of COS-7 cells with PrV at an MOI of 50, cells were extracted with 1% NP-40 at 10, 30, and 60 min after infection and analyzed by double immunofluorescence with the mouse anti-UL25 and either the rabbit anti-capsid (Fig. 6) or the rabbit anti-tubulin (Fig. 7) antibodies. The capsid antiserum revealed invading nucleocapsids as small bright spots that matched the fluorescent spots seen with the anti-UL25 antibodies on the same cells (Fig. 6A and B, respectively). These spots were found in proximity to the nucleus by as early as 30 min after infection. UL25, therefore, remains associated with the invading nucleocapsid. In double-labeling experiments with UL25 and tubulin antisera, capsids appeared to colocalize with microtubules (Fig. 7A and B) and were found to migrate toward the nucleus, where they accumulate at the microtubule-organizing center by as early as 10 min after infection (Fig. 7B). At 30 and 60 min postinfection, the fraction of nucleocapsids found on or near the nucleus increased (Fig. 7C).
FIG. 6.
Immunofluorescence microscopy of incoming PrV nucleocapsids. COS-7 cells were either mock infected (C and D) or were infected with PrV at an MOI of 50 (A and B). Cells were extracted with 1% NP-40 at 30 min postinfection, fixed, and analyzed by double immunofluorescence with the rabbit anti-capsid antibody (A) and the mouse anti-UL25 antibody (B). To prevent occlusion of the UL25 sites on the capsid by the capsid sera, cells were incubated first with the UL25 antiserum alone and then with both antibodies. Mock-infected cells were analyzed with either the anti-capsid antibody (C) or anti-UL25 antibody alone (D). Secondary antibodies were donkey anti-rabbit antibody coupled to DTAF and anti-mouse antibody coupled to Texas red. Texas red and fluorescein signals were visualized by using narrow-band Texas red or fluorescein isothiocyanate filters. The same fields were photographed in panels A and B. Magnification, ×750.
FIG. 7.
Effect of cytochalasin D and nocodazole on the localization of UL25 in PrV-infected cells. COS-7 cells in absence of drugs (A to C) or treated with cytochalasin D (D to F) or nocodazole (G to I) were infected with PrV at an MOI of 50, and at 10 min (A, B, D, E, G, and H) or 30 min (C, F, and I) postinfection, the cells were extracted with 1% NP-40, fixed, and analyzed by double immunofluorescence with the rabbit anti-tubulin antiserum (A, D, and G) and the mouse anti-UL25 antibody (B, C, E, F, H, and I). Arrows in panels A and D indicate the microtubule-organizing center. Secondary antibodies and visualization were as described in the legend to Fig. 6. The same fields were photographed in panels A and B, D and E, and G and H. Magnification, ×750.
To test if capsids colocalize specifically with microtubules, the above experiments were performed on COS-7 cells treated with either cytochalasin D (Fig. 7D, E, and F) or nocodazole (Fig. 7G, H, and I). In the presence of cytochalasin D, nucleocapsids still associated with microtubules and concentrated at the microtubule-organizing center already by 10 min after infection (Fig. 7D and E). At 30 min postinfection, most capsids localized on or near the nucleus (Fig. 7F). In cells treated with nocodazole, however, microtubules and their organizing center were found to be disrupted (Fig. 7G), and nucleocapsids were dispersed in the cell after 10 min (Fig. 7H) and accumulated in aggregates after 30 min of infection (Fig. 7I). These observations suggest that incoming nucleocapsids of PrV, such as those of HSV-1, localize to microtubules and that this network may provide a means for their transport to the nucleus. During this transport, UL25 remains associated with nucleocapsids.
DISCUSSION
Polyclonal antibodies directed against a UL25 fusion protein specifically react with a pseudorabies virion constituent that is synthesized late in infection, as well as with UL25 expressed from plasmid vectors. UL25 was found in association with all three capsid types and appears to constitute a minor capsid protein.
The de novo-synthesized UL25 protein is directed to the nucleus in the absence of other viral proteins. In plasmid-transfected cells, UL25 is uniformly distributed in the nucleus, whereas in PrV-infected cells UL25 is directed to distinct nuclear compartments. In HSV-1-infected cells, the assembly of B capsids and subsequent maturation of nucleocapsids are thought to occur in different sites in the nucleus. It has been shown that the HSV-1 UL32 gene product is implicated in the transfer of preformed B capsids to replication compartments, probable sites of capsid maturation (19). In analogy to its HSV-1 counterpart, UL25 of PrV might exert its role in capsid maturation and might thus be targeted to such sites of capsid maturation after association with B capsids.
In addition to the nuclear localization, the UL25 protein appears to colocalize with microtubules. This localization is different from the capsid proteins UL18 (unpublished results) and UL19 and poses some intriguing questions regarding its biological significance. The HSV-1 UL25 protein appears to be implicated both in invasion of the virus into the cell and in packaging of cleaved viral DNA during maturation of capsids (1, 22). It has also been proposed that UL25 might be part of a portal vertex (13). To reconcile the different phenotypes observed for UL25 HSV-1 mutants, it could be hypothesized that as a constituent of a portal structure of the capsid, UL25 interacts, on one hand, directly or indirectly with microtubules at the moment of virus penetration and during nucleocapsid transport from the cell periphery to the nucleus and, on the other hand, fulfills its function in capsid maturation at a late stage of infection. In a first attempt to test this hypothesis, we studied the fate of UL25 after virus penetration and found that UL25 remains associated with nucleocapsids during their transport to the nucleus. Incoming PrV capsids, such as those of HSV-1 (29), colocalize with microtubules and seem to follow this network for their transport to the nucleus. Sodeik et al. have further concluded from their studies that this retrograde transport occurs after attachment of capsids to dynein, a microtubule-dependent motor (29). The viral protein(s) implicated in dynein binding has not yet been identified, and it would be interesting to test whether UL25 is the candidate sought for this role. To determine if the affinity of capsids for microtubules or dynein is indeed mediated by UL25, experiments need to be performed with virus mutants in which the UL25 gene has been altered or deleted.
ACKNOWLEDGMENTS
We would like to thank Laurent Bertrand and Sébastien Chardon for excellent technical assistance in plasmid constructions, Patrice Coulon for advice in anti-UL25 antibody production, and Brigitte Simonet, Vincent Quintin, and Nathalie Babic for providing the anti-gB, anti-gC, and anti-PrV antibodies, respectively. We also greatly appreciate receiving crude capsid material from Jean-Christophe Audonnet, and we thank Christine Tuffereau for helpful discussions.
This work was financed by the Centre National de la Recherche Scientifique (UPR A 9053) and the EC grant BMH4CT972573. K. Kaelin was supported by the Training and Mobility of Researchers grant 83EU-046333 of the Swiss National Science Foundation.
REFERENCES
- 1.Addison C, Rixon F J, Palfreyman J W, O'Hara M, Preston V G. Characterisation of a herpes simplex virus type 1 mutant which has a temperature-sensitive defect in penetration of cells and assembly of capsids. Virology. 1984;138:246–259. doi: 10.1016/0042-6822(84)90349-0. [DOI] [PubMed] [Google Scholar]
- 2.Ali M A, Forghani B, Cantin E M. Characterization of an essential HSV-1 protein encoded by the UL25 gene reported to be involved in virus penetration and capsid assembly. Virology. 1996;216:278–283. doi: 10.1006/viro.1996.0061. [DOI] [PubMed] [Google Scholar]
- 3.Avitabile E, Di Gaeta S, Torrisi M R, Ward P L, Roizman B, Campadelli-Fiume G. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J Virol. 1995;69:7472–7482. doi: 10.1128/jvi.69.12.7472-7482.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Babic N, Mettenleiter T C, Ugolini G, Flamand A, Coulon P. Propagation of pseudorabies virus in the nervous system of the mouse after intranasal inoculation. Virology. 1994;204:616–625. doi: 10.1006/viro.1994.1576. [DOI] [PubMed] [Google Scholar]
- 5.Batterson W, Furlong D, Roizman B. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J Virol. 1983;45:397–407. doi: 10.1128/jvi.45.1.397-407.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben-Porat T, Kaplan A S. Molecular biology of pseudorabies virus. In: Roizman B, editor. The herpesviruses. Vol. 3. New York, N.Y: Plenum Press; 1985. pp. 105–173. [Google Scholar]
- 7.Cathomen T, Buchholz C J, Spielhofer P, Cattaneo R. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology. 1995;214:628–632. doi: 10.1006/viro.1995.0075. [DOI] [PubMed] [Google Scholar]
- 8.de Wind N, Wagenaar F, Pol J, Kimman T, Berns A. The pseudorabies virus homolog of the herpes simplex virus UL21 gene product is a capsid protein which is involved in capsid maturation. J Virol. 1992;66:7096–7103. doi: 10.1128/jvi.66.12.7096-7103.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dezélée S, Bras F, Vende P, Simonet B, Nguyen X, Flamand A, Masse M J. The BamHI fragment 9 of pseudorabies virus contains genes homologous to the UL24, UL25, UL26, and UL26.5 genes of herpes simplex virus type 1. Virus Res. 1996;42:27–39. doi: 10.1016/0168-1702(96)01293-2. [DOI] [PubMed] [Google Scholar]
- 10.Dijkstra J M, Fuchs W, Mettenleiter T C, Klupp B G. Identification and transcriptional analysis of pseudorabies virus UL6 to UL12 genes. Arch Virol. 1997;142:17–35. doi: 10.1007/s007050050056. [DOI] [PubMed] [Google Scholar]
- 11.Gilbert R, Ghosh K, Rasile L, Ghosh H P. Membrane anchoring domain of herpes simplex virus glycoprotein gB is sufficient for nuclear envelope localization. J Virol. 1994;68:2272–2285. doi: 10.1128/jvi.68.4.2272-2285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Granzow H, Weiland F, Jons A, Klupp B G, Karger A, Mettenleiter T C. Ultrastructural analysis of the replication cycle of pseudorabies virus in cell culture: a reassessment. J Virol. 1997;71:2072–2082. doi: 10.1128/jvi.71.3.2072-2082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 14.Huber M, Cattaneo R, Spielhofer P, Oervell C, Norrby E, Messerli M, Perriard J C, Billeter M A. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology. 1991;185:299–308. doi: 10.1016/0042-6822(91)90777-9. [DOI] [PubMed] [Google Scholar]
- 15.Joshi H C, Chu D, Buxbaum R E, Heidemann S R. Tension and compression in the cytoskeleton of PC 12 neurites. J Cell Biol. 1985;101:697–705. doi: 10.1083/jcb.101.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klupp B G, Kern H, Mettenleiter T C. The virulence-determining genomic BamHI fragment 4 of pseudorabies virus contains genes corresponding to the UL15 (partial), UL18, UL19, UL20, and UL21 genes of herpes simplex virus and a putative origin of replication. Virology. 1992;191:900–908. doi: 10.1016/0042-6822(92)90265-q. [DOI] [PubMed] [Google Scholar]
- 17.Koslowski K M, Shaver P R, Wang X Y, Tenney D J, Pederson N E. The pseudorabies virus UL28 protein enters the nucleus after coexpression with the herpes simplex virus UL15 protein. J Virol. 1997;71:9118–9123. doi: 10.1128/jvi.71.12.9118-9123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kritas S K, Pensaert M B, Mettenleiter T C. Role of envelope glycoproteins gI, gp63 and gIII in the invasion and spread of Aujeszky's disease virus in the olfactory nervous pathway of the pig. J Gen Virol. 1994;75:2319–2327. doi: 10.1099/0022-1317-75-9-2319. [DOI] [PubMed] [Google Scholar]
- 19.Lamberti C, Weller S K. The herpes simplex virus type 1 cleavage/packaging protein, UL32, is involved in efficient localization of capsids to replication compartments. J Virol. 1998;72:2463–2473. doi: 10.1128/jvi.72.3.2463-2473.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lycke E, Hamark B, Johansson M, Krotochwil A, Lycke J, Svennerholm B. Herpes simplex virus infection of the human sensory neuron. An electron microscopy study. Arch Virol. 1988;101:87–104. doi: 10.1007/BF01314654. [DOI] [PubMed] [Google Scholar]
- 21.Martinez R, Sarisky R T, Weber P C, Weller S K. Herpes simplex virus type 1 alkaline nuclease is required for efficient processing of viral DNA replication intermediates. J Virol. 1996;70:2075–2085. doi: 10.1128/jvi.70.4.2075-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNab A R, Desai P, Person S, Roof L L, Thomson D R, Newcomb W W, Brown J C, Homa F L. The product of the herpes simplex virus type 1 UL25 gene is required for encapsidation but not for cleavage of replicated viral DNA. J Virol. 1998;72:1060–1070. doi: 10.1128/jvi.72.2.1060-1070.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mettenleiter T C. Pseudorabies (Aujeszky's disease) virus: state of the art. August 1993. Acta Vet Hung. 1994;42:153–177. [PubMed] [Google Scholar]
- 24.Mettenleiter T C, Saalmuller A, Weiland F. Pseudorabies virus protein homologous to herpes simplex virus type 1 ICP18.5 is necessary for capsid maturation. J Virol. 1993;67:1236–1245. doi: 10.1128/jvi.67.3.1236-1245.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robbins A K, Dorney D J, Wathen M W, Whealy M E, Gold C, Watson R J, Holland L E, Weed S D, Levine M, Glorioso J C, Enquist L W. The pseudorabies virus gII gene is closely related to the gB glycoprotein gene of herpes simplex virus. J Virol. 1987;61:2691–2701. doi: 10.1128/jvi.61.9.2691-2701.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robbins A K, Watson R J, Whealy M E, Hays W W, Enquist L W. Characterization of a pseudorabies virus glycoprotein gene with homology to herpes simplex virus type 1 and type 2 glycoprotein C. J Virol. 1986;58:339–347. doi: 10.1128/jvi.58.2.339-347.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roizman B, Sears A E. Herpes simplex viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2231–2295. [Google Scholar]
- 28.Salmon B, Cunningham C, Davison A J, Harris W J, Baines J D. The herpes simplex virus type 1 UL17 gene encodes virion tegument proteins that are required for cleavage and packaging of viral DNA. J Virol. 1998;72:3779–3788. doi: 10.1128/jvi.72.5.3779-3788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spear P G. Membrane fusion induced by herpes simplex virus. In: Bentz J, editor. Viral fusion mechanisms. Ann Arbor, Mich: CRC Press Inc.; 1993. pp. 201–232. [Google Scholar]
- 31.Tuffereau C, Fischer S, Flamand A. Phosphorylation of the N and M1 proteins of rabies virus. J Gen Virol. 1985;66:2285–2289. doi: 10.1099/0022-1317-66-10-2285. [DOI] [PubMed] [Google Scholar]
- 32.Wittmann G, Rziha H J. Aujeszky's disease (pseudorabies) in pigs. In: Wittman G, editor. Herpesvirus diseases of cattle, horses and pigs. Boston, Mass: Kluwer Academic Publishers; 1989. pp. 230–325. [Google Scholar]
- 33.Yamada S, Imada T, Watanabe W, Honda Y, Nakajima-Iijima S, Shimizu Y, Sekikawa K. Nucleotide sequence and transcriptional mapping of the major capsid protein gene of pseudorabies virus. Virology. 1991;185:56–66. doi: 10.1016/0042-6822(91)90753-x. [DOI] [PubMed] [Google Scholar]