Abstract
Red clover mottle virus (RCMV) is a member of the comoviruses, a group of picornavirus-like plant viruses. The X-ray structure of RCMV strain S has been determined and refined to 2.4 Å. The overall structure of RCMV is similar to that of two other comoviruses, Cowpea mosaic virus (CPMV) and Bean pod mottle virus (BPMV). The sequence of the coat proteins of RCMV strain O were modeled into the capsid structure of strain S without causing any distortion, confirming the close resemblance between the two strains. By comparing the RCMV structure with that of other comoviruses, a structural fingerprint at the N terminus of the small subunit was identified which allowed subgrouping of comoviruses into CPMV-like and BPMV-like viruses.
Red clover mottle virus (RCMV) is a member of the comoviruses, a group of plant viruses in the picornavirus superfamily with nonenveloped, icosahedral capsids and bipartite, single-stranded, positive-sense RNA genomes. As with other comoviruses, the two RNA molecules (termed RNA 1 and RNA 2) are encapsidated separately in isometric particles made up of 60 copies each of a large (L) and a small (S) coat protein (31, 34, 47). A number of strains of RCMV have been characterized (reference 28 and references therein), and the complete nucleotide sequence of the genome of one of them, RCMV strain S (34), has been determined (41, 43, 44). Inspection of the RNA sequence together with in vitro translation data (42, 45) indicates that RCMV has a mode of gene expression similar to that of the type member of the group, Cowpea mosaic virus (CPMV). Comparison of the RNA 2 sequences of RCMV strain S with those of CPMV, Bean pod mottle virus (BPMV) and Cowpea severe mosaic virus (CPSMV) appears to divide comoviruses into two subgroups, with RCMV and CPMV forming one subgroup and CPSMV and BPMV forming the other (9).
One notable feature of RCMV is the ability of different strains of the virus to form viable pseudorecombinants (35). Pseudorecombinants between strains S and O have proved useful in mapping symptom and host range determinants (11, 36). The viability of the pseudorecombinants indicates that the coat proteins encoded by the RNA 2 of one strain can be processed correctly by the RNA 1-encoded 24K proteinase of another and are able to efficiently encapsidate the RNA 1 of the heterologous strain. This implies that any differences in the amino acid sequences of the coat proteins from the different strains do not dramatically alter their structure and assembly characteristics.
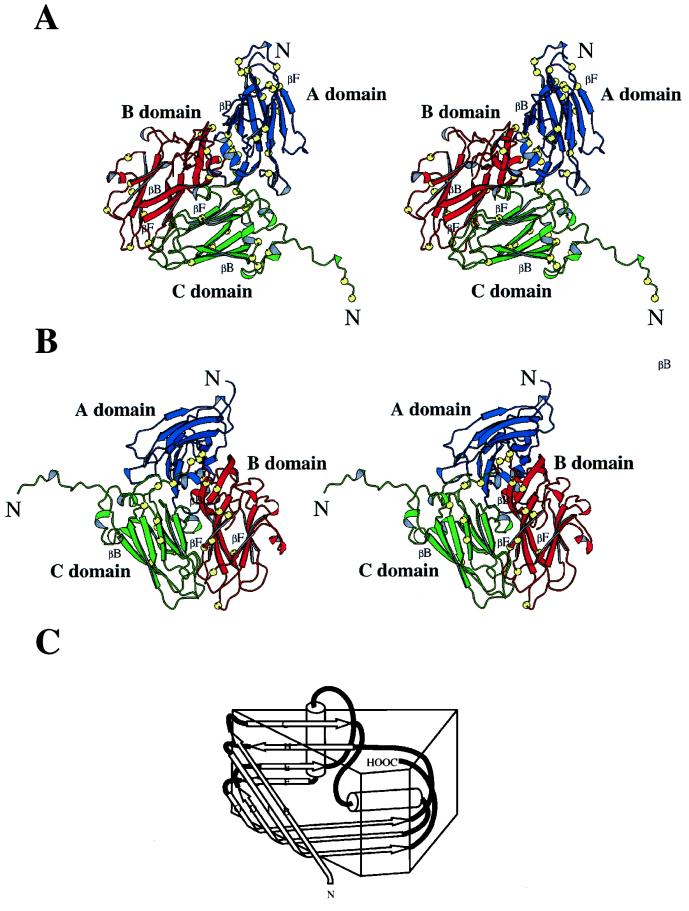
Our basic knowledge of comovirus structure has been derived from crystallographic studies of CPMV and BPMV (10, 31, 48; T. Lin, Z. Chen, R. Usha, C. V. Stauffaacher, J.-B. Dai, T. Schmidt, and J. E. Johnson, submitted for publication). In both cases the structures reveal architectures typical of P=3 plant and animal viruses, with the asymmetric unit consisting of three eight-stranded antiparallel β-sandwich domains (38) (Fig. 1). The A domain is located around the fivefold axes and comprises the S subunit of either 24 kDa (CPMV) or 22 kDa (BPMV). The B and C domains, located alternately around threefold axes, are formed by the L subunit of 41 kDa (in both cases). Despite the apparent overall similarity between the CPMV and BPMV structures, the viruses show different physical properties. For example, by ultracentrifugation on CsCl gradients, the RNA 1-containing component (bottom component) of CPMV could be separated into subcomponents, termed bottom-upper (BU) and bottom-lower (BL) components (4). This behavior was associated with the Cs ion permeability of the particle (53). By contrast, the bottom component of BPMV could not be separated into subcomponents by CsCl gradient centrifugation (4). In terms of its permeability to Cs ions, RCMV appears to be more similar to CPMV than to BPMV (1). This finding is consistent with the subgrouping based on the sequence alignments. Another difference between CPMV and BPMV is that while the crystal structure of the RNA 2-containing component (middle component) of BPMV revealed ordered ribonucleotides in an RNA binding pocket (10), no such ordered RNA was seen in the corresponding CPMV structure. The diverse properties of CPMV and BPMV provided an incentive for the further investigation of comovirus structures. To this end, crystals of RCMV strain S were prepared and used to solve the structure of the virus by molecular replacement.
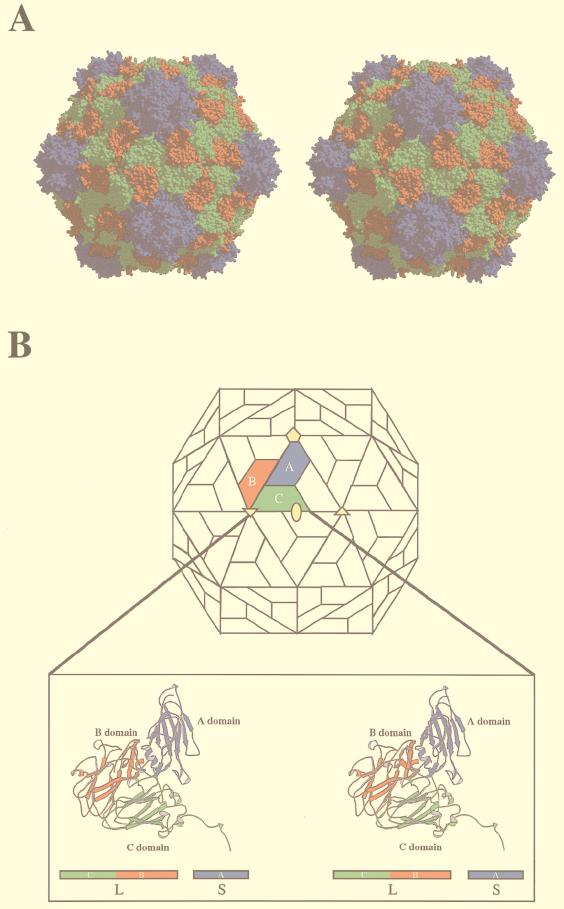
FIG. 1.
The structures of viral capsid and the icosahedral asymmetric unit of RCMV. Two proteins (S and L subunits) are in the RCMV capsid. The S subunit forms the A domain (in blue), while the L subunit forms the B (red) and C (green) domains. (A) Stereoview of a space-filling drawing of the RCMV capsid. All atoms are shown as spheres corresponding to a diameter of 1.8 Å. The pentameric S subunits form the protrusion. The CPK presentation was made with MidasPlus (14, 20). (B) At the top, the icosahedral asymmetric unit of the capsid is color coded in the schematic presentation of the CPMV capsid. The S subunit occupies the A position, forming the A domain around the fivefold axis; the two domains of the L subunit occupy the B and C positions. Positions A, B, and C are quasiequivalent positions of identical gene products in a T=3 surface lattice. At the bottom, a stereoview of a ribbon diagram of the icosahedral asymmetric unit is shown. All three domains are variations of the jelly-roll β-sandwich structure. The schematic presentation of composite proteins, L and S subunits, is also shown. The ribbon diagrams were drawn with the program MOLSCRIPT (26).
MATERIALS AND METHODS
Virus propagation and purification.
RCMV strains S and O (34) were propagated in Pisum sativum cv “Onward,” and virus particles were purified as described by Shanks et al. (41). RNA was extracted from virus particles by the method of Zimmern (55).
Determination of the structure of RCMV strain S.
Elongated RCMV crystals were produced by the sitting drop vapor diffusion method (33). The starting solution contained 10 mg of RCMV per ml in 10 mM sodium phosphate (pH 7). The reservoir solution contained 50 mM potassium phosphate (pH 7), 1.8% polyethylene glycol 8000, 0.3 M ammonium sulfate, 2 mM EDTA, and 1 mM sodium azide. Equal volumes of the virus and reservoir solution were mixed and equilibrated with the reservoir solution at room temperature. The crystals grew to 0.5 to 1 mm in all dimensions after 5 to 7 days.
The lifetime of RCMV crystals was about 40 h under CuKα radiation of a rotating anode operating at 35 kV and 40 mA. Screenless precession photographs with X-ray perpendicular to hk0 and h0l were taken with a μ angle of 1.5°. A complete data set, including 257 pairs of oscillation patterns, was recorded on photographic films in the A1 station of the Cornell High Energy Synchrotron Source with crystal-to-film distance of 90 or 100 mm, oscillation angle of 0.5°, and wavelength of 1.565 Å. The diffraction patterns were digitized at 50-μm intervals on a rotating-drum microdensitometer (Optronics model C-4100; Optronics International, Inc., Chelmsford, Mass.). The crystal orientations were determined by an autoindexing algorithm (25). The recorded reflection maxima were processed (37), scaled, and postrefined (39). A rotation function algorithm (50, 51) was used to resolve the ambiguity in the orientation. The initial structure factors were calculated from a poly(Ala) model of CPMV. The phase refinements were carried out as described previously (21, 40). The models were built by the programs FRODO and O (22, 23). The refinement scheme was similar to that adopted for the refinement of CPMV structures (Lin et al., submitted). Random shifts of 0.25 Å were applied to the coordinates before we calculated the structure factors from a refined model. These structure factors were used to calculate omit maps to differentiate ambiguities. Difference Fourier synthesis techniques with the resulting electron density map being averaged once were used to locate the water molecules by use of the programs PEAKMAX and WATPEAK of the CCP4 suite (12). Automatic procedures in program O were used to model the water molecules. The suite PROCHECK (29) was used to monitor the geometries of the models. The coordinates will be deposited in the Protein Data Bank.
Determination of the strain O coat protein sequences.
The virus-specific inserts in plasmids pRCOM-C27 and pRCOM-D1 (36) were excised by digestion with EcoRI and subcloned in EcoRI-digested M13mp18. Two subclones of pRCOM-C27, with inserts in the opposite orientation, were sequenced by the exonuclease III deletion method of Henikoff (16) by using the −40 universal primer. Sequencing was carried out either manually with Sequenase (U.S. Biochemicals) and [α-35S]dATP or automatically by using an Applied Biosystems 373 DNA sequencer. Gaps in the sequence were filled in by manual sequencing with specific primers within the virus-specific region. Additional sequence information in the region encoding the N terminus of the L protein was obtained from the subclones of pRCOM-D1 by priming with specific primers. Further RNA 2-specific cDNA clones were obtained by producing double-stranded cDNA to RNA 2 as described earlier (41), digesting the DNA with Sau3A, and ligating the products into BamHI-digested M13mp18. The resulting recombinants were sequenced manually as described above.
N-terminal sequence analysis of the strain O coat proteins.
A sample of purified strain O virions was denatured and electrophoresed on a polyacrylamide gel, and the proteins were transferred to Problott membranes as described by Achon et al. (2). The protein bands were visualized by staining with 1% (wt/vol) Amido Black, and individual bands were excised for N-terminal sequence analysis by using an Applied Biosystems model 492 gas-phase protein sequencer.
RESULTS AND DISCUSSION
Determination of the structure of RCMV strain S.
The space group of RCMV strain S crystals was determined by screenless precession photography. The precession photographs of the hk0 and h0l planes of RCMV crystals showed two perpendicular mirror planes. The same symmetry elements were also found in the higher layers of the diffraction patterns, suggesting 222 symmetry. All the reflections with indices of h+k+l=odd were systematically absent, indicating a centered lattice. The cell dimensions were not the same along three axes, indicating the space group I222. Packing considerations required that the three noncrystallographic twofold axes of the particle were coincident with the crystallographic twofold axes. Consequently, there was 15-fold noncrystallographic redundancy. There was no translation problem, but the particle orientation remained to be determined from two possibilities.
RCMV crystals diffracted X-rays to 2.2 Å with synchrotron radiation. A complete data set comprised of 257 A-B pairs of oscillation patterns recorded on photographic film was obtained. The lattice constants for the orthorhombic body-centered cell were a = 333.4, b = 305.0, and c = 315.0 Å. Fifty A films were initially processed, scaled, and postrefined. The cell dimensions were refined as a = 332.07, b = 303.87, and c = 314.31 Å. These parameters were used to reprocess the original 50 and the rest of the A films to 2.4 Å. Analysis of the intensity profile as a function of resolution (54) showed that reflections at the lower resolution were underestimated, probably due to the saturation of film by large numbers of high-intensity reflections. Subsequently, the B films were also processed, and the reflections were merged with the reflections of the A films. The final data set contained 451,303 reflections, with an Rmerg of 8.8%. The completeness of the data is listed in Table 1.
TABLE 1.
Data reduction and phase refinementa
| Resolution (Å) | Data completeness (%) | CC in phase refinement |
|---|---|---|
| 30.0 | ||
| 15.0 | 100 | 0.963 |
| 10.0 | 100 | 0.991 |
| 7.50 | 100 | 0.987 |
| 5.00 | 98 | 0.986 |
| 3.50 | 96 | 0.979 |
| 3.00 | 90 | 0.925 |
| 2.75 | 80 | 0.813 |
| 2.50 | 52 | 0.624 |
| 2.40 | 26 | 0.415 |
| Overall | 72 | 0.960 |
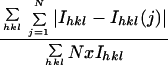 |
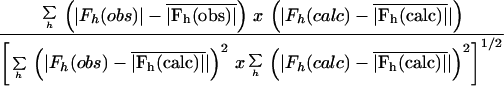 |
A self-rotation function (50, 51) was used to resolve the ambiguity in the particle orientation. CPMV was chosen as the initial phasing model. The direct sequence homology is 53% between RCMV and CPMV coat proteins (41), and the two virus particles share similar physical properties (reference 31 and references therein). The starting phases were calculated from a poly(Ala) CPMV model placed in the RCMV cell, and the phases were refined at a 3-Å resolution with 12 cycles of averaging. The final map was reasonably free of model bias as judged by side-chain density that matched the RCMV sequence (Fig. 2A). Manual building of an RCMV model was carried out based on the amino acid sequence of the coat proteins deduced from the nucleotide sequence of RNA 2 (41), taking into account the recently verified cleavage sites used to release the L and S capsid proteins (28). With the introduction of an atomic mask (40) and calculation of the structure factors from the new model, the final round of averaging had a correlation coefficient of 96% for all the data between 30 and 2.4 Å (Table 1). An equatorial section of the averaged electron density of the virus particle is shown in Fig. 2B. All residues were assigned in the final model except 32 C-terminal residues (residues 183 to 214) of the S subunit and 8 C-terminal residues (residues 369 to 376) of the L subunit, which were disordered. In light of the CPMV structure, the undefined density of the C terminus of the S subunit can also be attributed to a partial degradation (27, 31; Lin et al., submitted).
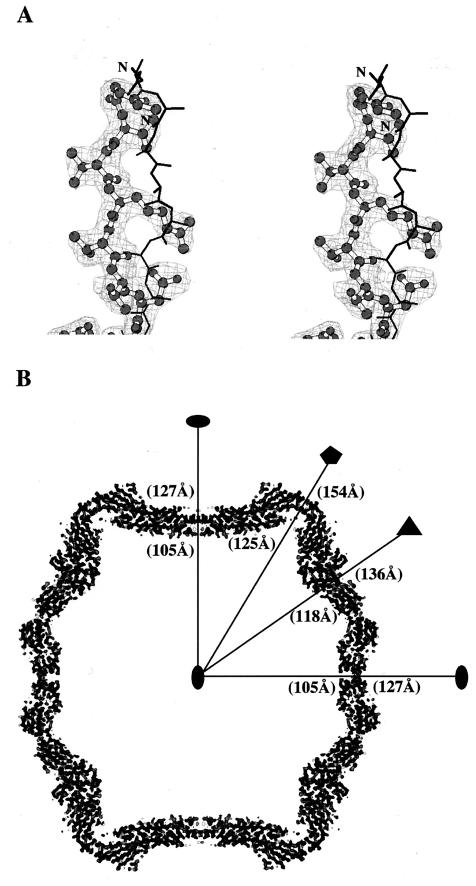
FIG. 2.
Averaged electron density of RCMV. (A) Stereoview of electron density around the N terminus of the S subunit. The initial phasing model (in “black tracing”) is overlapped with the averaged density (in “chickenwire”) and the final RCMV model (in “ball-and-stick”). The initial phasing model is derived from a poly(Ala) structure of CPMV. There was no indication of bias toward the initial phasing model in the averaged electron density. The RCMV sequence was modeled into the averaged electron density without ambiguity. The electron density shown was contoured at 1.5ς. This figure was drawn by using the program BOBSCRIPT (13). (B) An equatorial section of the averaged electron density of the virus particle. The outer and inner dimensions of the particle along each symmetry element are indicated. The density was contoured at 2ς. This figure was generated with the program O (22, 23).
Refinement and overall structure.
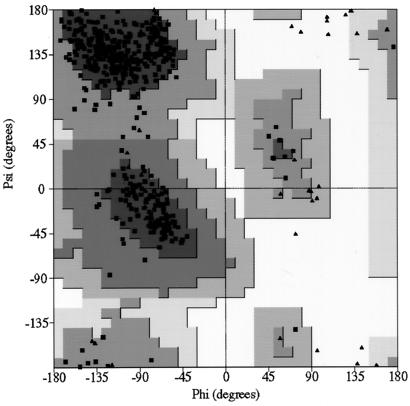
The RCMV model was initially refined by simulated annealing (5–7), which decreased the R factor from 26.4 to 16.3% for data between resolutions of 10.0 and 5.0 Å. Further simulated annealing followed by conjugate gradient minimizations and manual modeling with data between resolutions of 8.0 and 3.0 Å decreased the R factor from 28.2 to 22.4%. A difference map was calculated and averaged once for the identification of water molecules. The resulting model was further refined by conjugate gradient minimization with data between resolutions of 10.0 and 2.4 Å to an R factor of 22.2%. Refinement of temperature factors was performed. The final structure includes 550 amino acid residues (4,238 atoms) and 122 water molecules in the icosahedral asymmetric unit with a crystallographic R factor of 19.9% for 442,909 reflections and with I/ς(I) ≥ 2 in the resolution range of 10.0 to 2.4 Å. A Ramachandran plot of the main chain angles is shown in Fig. 3.
FIG. 3.
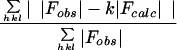 |
This figure was generated by the program PROCHECK (29).
The overall structure of RCMV is similar to CPMV (48; Lin et al., submitted) and BPMV (10). Each icosahedral asymmetric unit is occupied by one S and one L subunit. By fivefold, threefold, and twofold symmetry operations, 60 copies of the icosahedral asymmetric units are generated that constitute the virus capsid with protrusions around the fivefold axes (Fig. 1). The outer radius at the fivefold axes is 154 Å; the outer radius across the twofold axes is 127 Å, and the outer radius across the threefold axes is 136 Å. The average thickness of the capsid is about 40 Å. The S subunit is a simple β-barrel, while the L subunit contains two β-barrels in a single polypeptide chain. Comparison of the RCMV capsid with the T=3 surface lattice shows that the S subunit occupies the A (pentamer) position, while the N-terminal and C-terminal β-barrels of the L subunit occupy the C and B (quasihexamer) positions, respectively.
Comparison with the coat proteins of RCMV strain O.
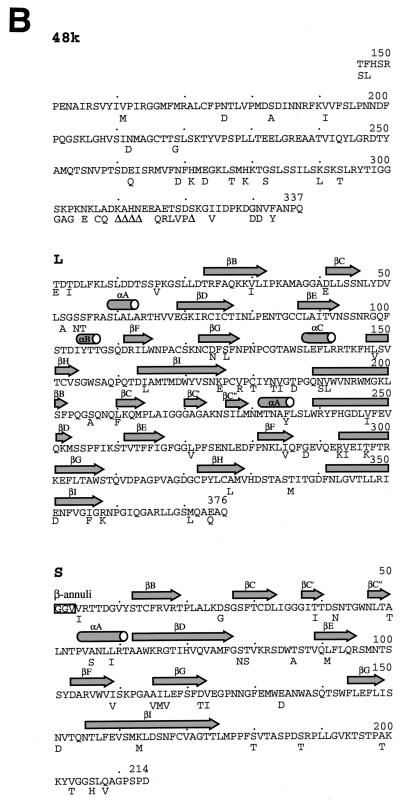
The structural implications of amino acid substitutions between the coat proteins of closely related strains of tobacco mosaic virus (TMV) have proved to be invaluable in assigning functions to various residues (3). To carry out a similar analysis on a comovirus, the amino acid sequences of the two coat proteins of RCMV strain O were determined by analysis of the nucleotide sequences of cDNA clones of the 3′-terminal region of strain O RNA 2 (36). In total, the sequence of the 3′-terminal 2,565 nucleotides was determined. This sequence contained a single long open reading frame and the entire 3′ noncoding region of RNA 2 (Fig. 4A). The nucleotide sequence identity between strains S and O in the coding region was 75.9%, considerably higher than the 44% previously found in the 3′ noncoding regions (36). The amino acid sequences of the L and S subunits of RCMV strain O (Fig. 4B) were deduced by comparison of the amino acid sequence of the translation product of strain O RNA 2 with its strain S equivalent. The proposed cleavage sites for the release of the mature coat proteins were confirmed by Edman degradation with proteins isolated from a sodium dodecyl sulfate-polyacrylamide gel. The sequence of the C-terminal 187 amino acids of the RCMV 48K protein, a protein involved in cell-to-cell movement of comoviruses, could also be deduced (Fig. 4B).
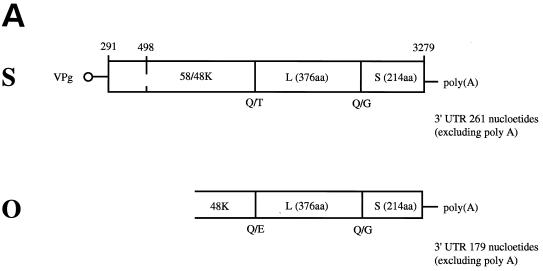
FIG. 4.
(A) Organization of RCMV RNA 2. The top portion of the diagram represents the complete sequence of the RNA-2 of strain S (41), while the lower portion represents the region of strain O which has been sequenced to date. The genome-linked protein (VPg) and the 3′ polyadenylate [poly(A)] are indicated, as is the size of the 3′ untranslated region (3′ UTR) in each case. The open reading frame on each RNA 2 is shown as an open rectangle. For strain S the positions of the two initiation codons responsible for the synthesis of the 58K (position 291) and 48K (position 498) proteins are marked, as is the position of the termination codon at the end of the polyprotein (position 3279). The positions of the polyprotein processing sites are shown together with the dipeptide sequence which is cleaved (Q/T, Q/E, or Q/G). The identity of each final cleavage product (48K/58K; L and S) is also indicated. (B) Detailed comparisons of the amino acid sequences of the C-terminal portion of the 48K protein (48k), large coat protein (L), and small coat protein (S) of RCMV strains S and O. The upper sequence represents the sequence of strain S, with the differences found in strain O given underneath. Δ represents an amino acid which is deleted in the strain O protein. The secondary structure assignments are shown as arrows and cylinders.
The L and S subunits of strain O consist of 376 and 214 amino acids (Fig. 4B), respectively, exactly the same sizes as their strain S counterparts. Overall, the coat proteins of strains S and O are 90.2% (L) and 87.9% (S) identical. Visual inspection of the alignment between the strain S and O coat proteins indicates that the changes are not evenly spread throughout the polypeptide chains but tend to be clustered (Fig. 4B). One particularly notable cluster occurs between amino acids 175 and 191 of the L protein, where 8 of 17 amino acids have changed, and not necessarily in a conservative manner.
To investigate the structural implications of the sequence variations in the coat proteins of strains S and O, the residues which were changed in strain O were mapped onto the three-dimensional (3D) structure of strain S. The changes in the strain O sequence can be comfortably accommodated in the atomic model of S strain with no obvious disruption of the structure. This finding is consistent with the observation that the two strains are serologically indistinguishable (P. Oxelfelt, unpublished data) and can form viable pseudorecombinants. Many of the changes are found on the exterior and interior surfaces of the viral capsid (Fig. 5). Several of the changes found among the buried residues are compensatory. For example, residues Ile285 and Val294 of the L subunit are packed against each other in strain S. The coordinated sequence changes to Val285 and Ile294 in strain O result in the occupation of the same volume and thus do not alter the capsid structure. As shown in Fig. 5B, many changes cluster around a cleft on which RNA was located in the BPMV structure (10). The most variable sequence (eight changes between amino acids 175 and 191 of the L subunit) comprises the top half of this cluster and is in the region connecting the C and B domains. Changes in the sequence in this nucleotide-binding region may be related to the fact that strain O produces considerably less top component (empty, protein-only shells) than strain S (34).
FIG. 5.
Illustration of the difference between the strain S and O. The ribbon diagram is drawn through the Cα positions. The A domain is in blue, the B domain is in red, and the C domain is in green. (A) Stereoview of the icosahedral asymmetric unit of strain S, with all changes found in strain O labeled with yellow spheres. All of the differences can be accommodated in the context of the structure of strain S. (B) Stereoview of icosahedral asymmetric unit observed from the interior. Large numbers of changes (labeled as yellow spheres) are clustered around the cleft in which ordered ribonucleotides were observed in the crystal structure of BPMV. For clarity, only the changes around the cleft are shown. (C) Schematic presentation of the canonical β barrel for reference to the illustrations in panels A and B.
The identity between the C-terminal portions of the 48K movement proteins of strains S and O is 60.4%, considerably less than that found in the coat proteins. Most of the changes are concentrated near the C terminus of the 48K protein and include a number of deletions (Fig. 4B). These changes occur in precisely the region that has been implicated in the interaction between the tubules containing, and probably formed by, the 48K protein and virus particles in the case of CPMV (30). These changes in the 48K protein may be needed to compensate for differences in the surface properties of the virus particles of strains S and O.
A structural fingerprint for subgrouping comoviruses.
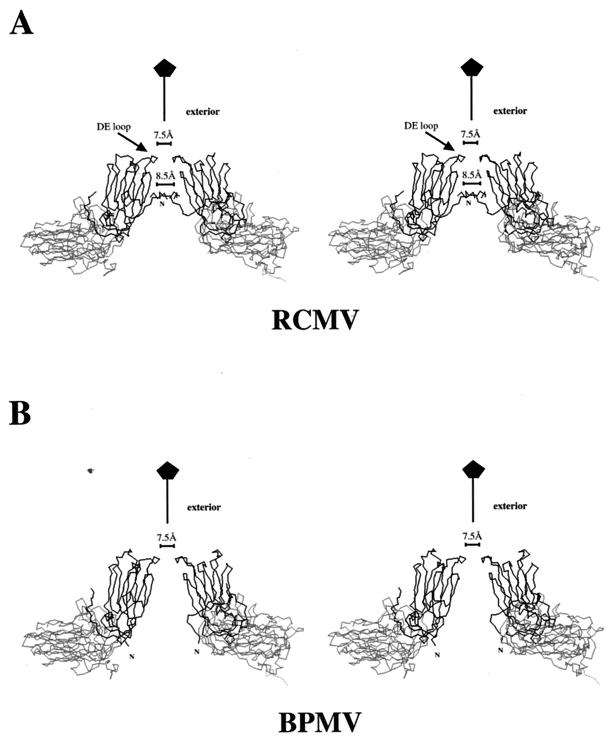
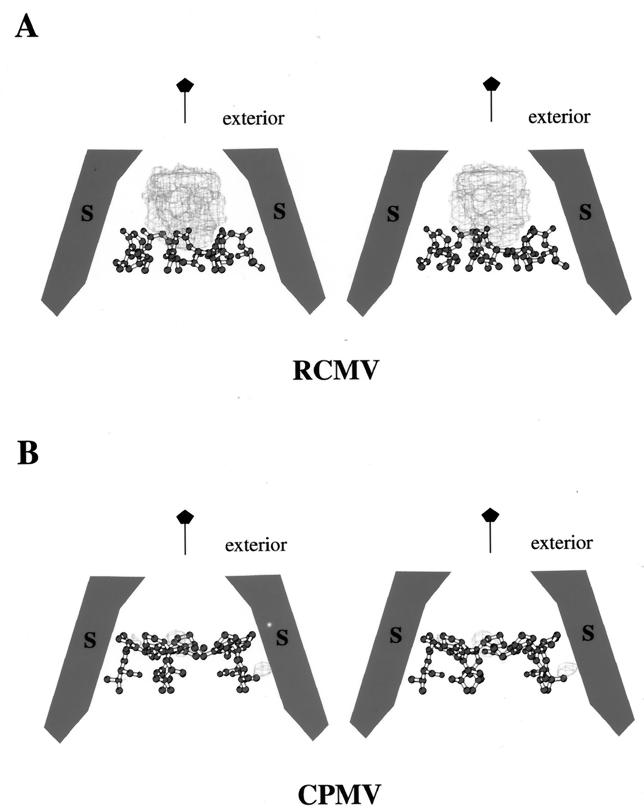
A noticeable surface feature of RCMV and other comoviruses is the protrusion centered around the viral fivefold axes (Fig. 1A). This protrusion is formed by the pentameric S subunits which are not tangential to the spherical capsid. Looking down the fivefold axis toward the viral interior, there is no part of the S subunit that approaches the particle symmetry axis. A channel is apparent along the fivefold axis running from the exterior to the interior (Fig. 6). RCMV bears more similarity to CPMV than to BPMV in this part of the structure.
FIG. 6.
Stereoviews of comovirus subunits in the formation of a channel along the fivefold symmetry axis. The S subunit is in dark gray tracing; the L subunit is in light gray tracing. (A) RCMV. Tracings of three N-terminal residues from all five S subunits, which form the pentameric annulus, are shown. For clarity, however, only tracings of two subunits related by 144° around the viral fivefold axis are drawn. The narrowest opening located on the surface has a diameter of ca. 7.5 Å and is comprised of the DE loops of the S subunits. The second constriction (∼8.5 Å in diameter) is roughly in the middle of the channel and is formed by the pentameric annulus. A similar annulus was also identified in the CPMV structure. (B) BPMV. Two subunits related by 144° around the viral fivefold axis are shown. The N termini fold in the direction opposite that of RCMV and CPMV, and there is no annular structure.
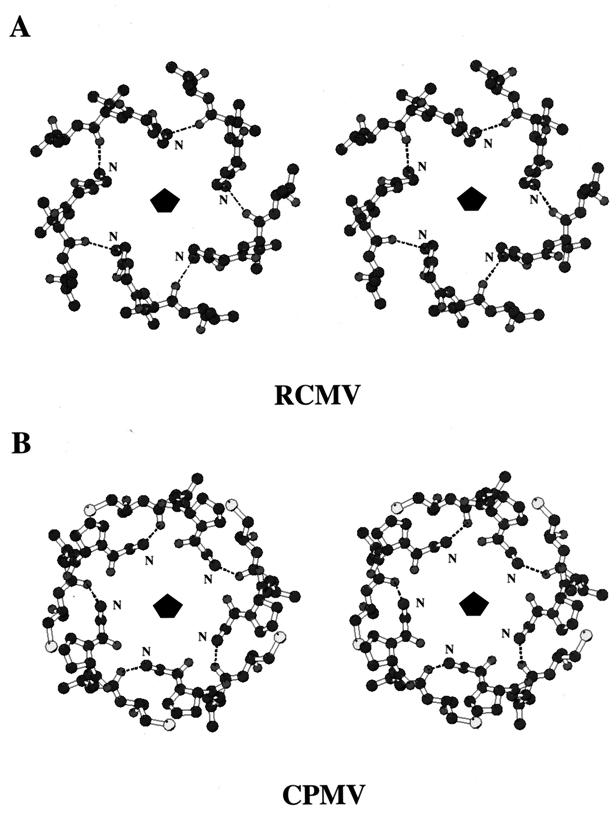
The channel has an overall funnel shape, with the narrow end at the outer surface and the wider end in the interior. The narrow end is comprised of the DE loops of the S subunits reaching over the innermost FG loop of the jelly-roll β-sandwich structure and clustering around the fivefold axis (Fig. 6A). The opening at this end is about 7.5 Å. Farther down the fivefold axis, a second constriction can be found in RCMV and CPMV, but not in BPMV (Fig. 6). This occurs as a result of the three N-terminal residues of the S subunits forming a pentameric annulus structure (Fig. 7). Similar annular structures were found in a number of icosahedral viruses (reference 17 and references therein). In CPMV and RCMV, the amino group of the N terminus forms a hydrogen bond with the main chain carbonyl oxygen of the neighboring third residue. The opening is ca. 8.5 Å. No constriction is found in BPMV since it lacks the pentameric annuli (Fig. 6B).
FIG. 7.
Stereoviews of the pentameric annuli of comoviruses looking down the fivefold axis from the viral exterior. All of the atoms are drawn as black spheres, except for the sulfur atoms, which are gray spheres. Hydrogen bonds are shown as dashed lines. (A) Pentameric annulus of RCMV. It is formed by hydrogen bonding with the first three N-terminal residues from each of the pentameric S subunits. The amino group of each N terminus forms a hydrogen bond with the main chain carbonyl oxygen of the neighboring third residue. The annulus is star-shaped, with each peptide extending upward and then downward. The opening is about 8.5 Å. (B) Pentameric annulus of CPMV. It adopts a hydrogen bonding pattern similar to that found in RCMV. Each N terminus extends upward toward the exterior, and the annulus is crown-shaped.
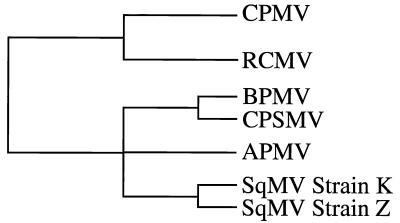
The original subgrouping of RCMV with CPMV and of BPMV with CPSMV was based on the degree of similarity between the proteins (48K, L, and S) encoded by their RNA 2 molecules (9). However, the extensions of this approach to other comoviruses were not particularly revealing, and no obvious discriminating criteria could be derived for subgrouping Andean potato mottle virus (APMV) (46) and several strains of squash mosaic viruses (SqMV) (15, 19). The results were especially ambiguous in the case of APMV, which shares a high degree of similarity with BPMV but a comparatively low degree of similarity with CPSMV (15). This discrepancy is inconsistent with the previous placement of BPMV and CPSMV in the same subgroup (9).
Structural alignment (i.e., protein sequence alignment based on superposition of 3D structures) of RCMV, CPMV, and BPMV indicates that one of the discriminating sequence differences between the two subgroups is at the N termini of the S subunits. The N-terminal residue of the RCMV and CPMV S subunits is glycine in both cases, instead of serine as found in BPMV. Moreover, the N-terminal sequences of both the CPMV and RCMV S proteins are three residues longer than that of the corresponding BPMV protein. These three residues form the pentameric annuli observed in the X-ray crystal structures. Glycine residues are preferred as the first residue to avoid space constraints in the formation of the annular structure. The shorter N termini of the S subunits of BPMV prevent the formation of the annulus, and they are folded in the opposite direction (Fig. 6B). Thus, the longer N termini, with a glycine as the terminal residue, can serve as a fingerprint for the subgrouping of comoviruses between CPMV-like and BPMV-like viruses.
The above fingerprint is used to verify the subgrouping of CPSMV with BPMV based on sequence homology criteria (9). Although protein sequence alignments suggest that the CPSMV S subunit would adopt an overall architecture similar to those of CPMV, RCMV, and BPMV (Fig. 8), the N terminus of the CPSMV S subunits resembles that of BPMV in both length and terminal residue. It is therefore reasonable to suggest that, as in the case of BPMV, a pentameric annular structure is not formed, supporting the previous subgrouping of CPSMV with BPMV. When the structural alignments are extended to include the sequences of the S proteins of APMV (46) and SqMV (15, 19), subgrouping with BPMV is predicted in each case (Fig. 8). The N terminus of the APMV S subunit is two residues shorter than the length required for the formation of the annular structure. Moreover, the N-terminal residue is phenylalanine, which would not be conducive to the formation of the annular structure. Similarly, the N terminus of the SqMV S subunit is one residue too short, and, like BPMV, the S subunit of SqMV has a serine residue at its N terminus. It can therefore be concluded that, based on structural criteria, there are two major subgroups of comoviruses. One group includes CPMV and RCMV; the other includes BPMV, CPSMV, APMV, and SqMV. This conclusion is consistent with the proposed subgrouping of SqMV based on the calculation of a parsimonious phylogenetic tree made by using RNA 2 nucleotide sequences (15). Moreover, structural alignment clearly places AMPV in the subgroup of BPMV-like viruses.
FIG. 8.
Sequence alignment of comovirus S subunits based on the 3D structures of RCMV, CPMV, and BPMV. The secondary structure assignments are shown at the top of the sequences. The boxed residues at the beginning of CPMV and RCMV sequence are those involved in the formation of the pentameric annuli. The boxed residues in the C termini of CPMV, RCMV, and BPMV are not visible in the X-ray structure. The uppercase letters represent identities. The overall similarity in the alignment suggests that all six viruses adopt similar capsid structures. The most notable differences between the CPMV and RCMV S subunits and those of the other comoviruses reside in the N and C termini (see text). The sequences used in the alignment are CPMV (52), BPMV (32), CPSMV (9), APMV (46), strain S of RCMV (41), and strain Z of SqMV (15).
Another noticeable feature in the structural alignment of the S proteins of different comoviruses is that the length of sequence on the C-terminal side of the βI strand for BPMV, CPSMV, APMV, and SqMV (22, 22, 20, and 8 amino acids, respectively) is considerably less than it is for CPMV and RCMV (38 and 40 residues) (Fig. 8). Particularly striking is the fact that the C-terminal region of the SqMV S subunit is 32 residues shorter than its RCMV equivalent, having a total length equivalent to that of the ordered C-terminal polypeptide seen in the crystal structures of RCMV and BPMV. Though the full implications of these findings await further investigation, the C-terminal region of the S subunit of CPMV has been implicated in the packaging of the viral RNA (49). Taking into account all of the data from the structural and nucleotide sequence alignments, a scheme for subgrouping comoviruses is suggested as shown in Fig. 9.
FIG. 9.
Subgrouping of comoviruses. Comoviruses are placed in two major subgroups, CPMV-like viruses and BPMV-like viruses, based on a discriminating structural element, the pentameric annulus. A further classification of BPMV-like viruses is by the structural element, the pentameric annulus. Further classification of BPMV-like viruses is based on nucleotide sequence homology calculated by Haudenshield and Palukaitis (15). Similarity at the C termini, however, places APMV closer to BPMV and CPSMV in the BPMV-like subgroup.
Comparison of the pentameric annuli of RCMV and CPMV.
Despite sharing a similar pattern of hydrogen bonding, there is a noticeable difference between the annular structures of RCMV and CPMV (Fig. 7). The annulus of RCMV is star-shaped, with each N-terminal sequence extending first upward and then downward. By contrast, that of CPMV is crown-shaped, with each N-terminal sequence extending upward toward the exterior of the virion. Of greater significance, however, is the difference in the electron densities associated with the annuli in the two viruses, differences which seem to critically influence the annular structure. While spherical density, suggesting the presence of metal ions, is found in CPMV, a much bulkier density is found in RCMV (Fig. 10). This difference is most probably due to the fact that additional purification by CsCl gradient was employed in the preparation of CPMV for the structural studies (Lin et al., submitted). The conformation of the CPMV annular structure also allows the carbonyl oxygen of the N-terminal glycine to interact with the putative metal ion (Fig. 7B). The bulky density seen in RCMV probably represents some native material which can be replaced with metal ions, as seen in CPMV. The location of the putative metal ion in CPMV suggests that it can diffuse further into the interior of the capsid; it is situated inside the opening of the annulus, and there is no further barrier toward the interior (Fig. 10B). This is consistent with the findings that significant amounts of Cs ions diffuse into the CPMV BL component (53) and that the annular structure was perturbed by the flux of large amounts of Cs ions into the CPMV capsid (Lin et al., submitted). By contrast, the bulky density found in RCMV is situated above the annulus and seems to be prevented from further diffusion into the capsid (Fig. 10A). It seems quite possible that the channel is a conduit, with the pentameric annulus playing a discriminatory role for the exchange of material between the viral interior and the environmental exterior. No density is found in the equivalent region of the BPMV capsid, which does not possess annuli (10) and is not Cs permeable.
FIG. 10.
Stereoviews of the nonprotein density and the pentameric annuli observed in the channel along the viral fivefold axis. The annuli are shown in “ball-and-stick,” and the electron density is shown in “chickenwire.” The difference electron density maps are calculated with Fourier coefficients of Foeiα(ave) − Fmodeleiα(model). Any density shown in these maps would be that which has not been included in the atomic model. (A) RCMV. A bulky density is observed atop the annulus. It appears that this bulky density cannot penetrate further into the capsid due to the blockage by the annular structure. The virus was prepared from infected plant leaves without ultracentrifugation in a CsCl gradient. (B) CPMV. The density is spherical, suggesting metal ion, and it is situated inside the opening of the annulus. There is no obvious barrier between the spherical density and the viral interior. The preparation of the virus used in the structure determination involved ultracentrifugation with CsCl gradients. Both electron density maps are contoured at 3.5ς. This figure was drawn by using the program BOBSCRIPT (13).
Structural evolution.
Structural studies have suggested that picorna-like viruses (nepoviruses, comoviruses, and picornaviruses) have evolved from T=3 viruses by gene triplication, independent evolution of each β-barrel sandwich domain, and the development of cleavage sites in the polyprotein (8, 10, 18). Consideration of the complexity of the capsid structures places nepoviruses in the lower, comoviruses in the middle, and picornaviruses in the higher part of the evolution tree. Structural alignments suggest that the N terminus of the BPMV S subunit is not much different in structure from the equivalent polypeptide in a nepovirus, tobacco ringspot virus (18). It points away from the fivefold axis and is structurally unsophisticated (Fig. 6B). By contrast, the N termini of the CPMV and RCMV S subunits point in the opposite direction, with the extra residues allowing the formation of the pentameric annular structure that seems to correlate with Cs permeability. It therefore seems reasonable to suggest that the BPMV-like comoviruses are closer to nepoviruses in evolution, with the separation of L and S subunits. On the other hand, CPMV-like viruses are closer to picornaviruses in evolution, with more sophisticated features developed at the N termini of the S subunits.
ACKNOWLEDGMENTS
We thank Pat Barker (Babraham Institute, Cambridge, United Kingdom) for N-terminal sequence analysis of the RCMV strain O coat proteins. T.L. thanks Anette Schneemann for reviewing the manuscript before publication and Lars Liljas for encouragement in the preparation of the manuscript.
A.J.C. was supported by a BBSRC (United Kingdom) research studentship, and G.P.L. acknowledges the support of an EMBO short-term fellowship for part of this work. The crystallographic studies were supported by grant GM54076 to J.E.J.
Footnotes
Dedicated to the memory of J.-B. Dai.
REFERENCES
- 1.Abdelmoeti M A H. Red clover mottle virus (RCMV)—purification, stability, separation of components and genetic studies. Ph.D. thesis. Uppsala, Sweden: Swedish University of Agricultural Sciences; 1979. [Google Scholar]
- 2.Achon M A, Medina V, Shanks M, Markham P G, Lomonossoff G P. Characterisation of a maize-infecting potyvirus from Spain. Eur J Plant Pathol. 1994;100:157–165. [Google Scholar]
- 3.Altschuh D, Lesk A M, Bloomer A C, Klug A. Correlation of coordinated amino acid substitutions with function in virus related to tobacco mosaic virus. J Mol Biol. 1987;193:693–707. doi: 10.1016/0022-2836(87)90352-4. [DOI] [PubMed] [Google Scholar]
- 4.Bancroft J B. Purification and properties of beanpod mottle virus and associated centrifugal and electrophoretic components. Virology. 1962;16:419. doi: 10.1016/0042-6822(62)90222-2. [DOI] [PubMed] [Google Scholar]
- 5.Brunger A T. X-PLOR version 3.1. A system for X-ray crystallography and NMR. New Haven, Conn: Yale University Press; 1992. [Google Scholar]
- 6.Brunger A T, Rice L M. Crystallographic refinement by simulated annealing: methods and applications. Methods Enzymol. 1997;277:243–269. doi: 10.1016/s0076-6879(97)77015-0. [DOI] [PubMed] [Google Scholar]
- 7.Brunger A T, Krukowski A, Erickson J W. Slow-cooling protocols for crystallographic refinement by simulating annealing. Acta Crystallogr. 1990;A46:585–593. doi: 10.1107/s0108767390002355. [DOI] [PubMed] [Google Scholar]
- 8.Chandrasekar V, Johnson J E. The structure of tobacco ringspot virus: a link in the evolution of icosahedral capsids in the picornavirus superfamily. Structure. 1997;6:157–171. doi: 10.1016/s0969-2126(98)00018-5. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Bruening G. Nucleotide sequence and genetic map of cowpea severe mosaic virus RNA 2 and comparison with RNA 2 of other comoviruses. Virology. 1992;187:682–692. doi: 10.1016/0042-6822(92)90471-z. [DOI] [PubMed] [Google Scholar]
- 10.Chen Z, Stauffacher C, Li Y, Schmidt T, Bomu W, Kamer G, Shanks M, Lomonossoff G, Johnson J E. Protein-RNA interactions in an icosahedral virus at 3.0 Å resolution. Science. 1989;245:154–159. doi: 10.1126/science.2749253. [DOI] [PubMed] [Google Scholar]
- 11.Clark A J. Production and characterization of hybrid comoviruses. Ph.D. thesis. East Anglia, United Kingdom: University of East Anglia; 1997. [Google Scholar]
- 12.Collaborative Computational Project, Number 4. The CCP4 suites: programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 13.Esnouf R M. An extensively modified version of Moscript that includes greatly enhanced coloring capacities. J Mol Graphics. 1997;15:132–134. doi: 10.1016/S1093-3263(97)00021-1. [DOI] [PubMed] [Google Scholar]
- 14.Ferrin T E, Huang C C, Jarvis L E, Langridge R. The MIDAS display system. J Mol Graphics. 1988;9:13–27. [Google Scholar]
- 15.Haudenshield J S, Palukaitis P. Diversity among isolates of squash mosaic virus. J Gen Virol. 1998;79:2331–2341. doi: 10.1099/0022-1317-79-10-2331. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 17.Hogle J M, Chow M, Filman D J. Three-dimensional structure of poliovirus at 2.9 Å resolution. Science. 1985;229:1358–1365. doi: 10.1126/science.2994218. [DOI] [PubMed] [Google Scholar]
- 18.Hosur M V, Schmidt T, Tucker R C, Johnson J E, Gallagher T M, Selling B H, Rueckert R R. Structure of an insect virus at 3.0 Å resolution. Proteins. 1987;2:167–176. doi: 10.1002/prot.340020302. [DOI] [PubMed] [Google Scholar]
- 19.Hu J S, Pang S Z, Nagpala P G, Siemieniak D R, Slightom J L, Gonsalves D. The coat protein genes of squash mosaic virus: cloning, sequence analysis, and expression in tobacco protoplasts. Arch Virol. 1993;130:17–31. doi: 10.1007/BF01318993. [DOI] [PubMed] [Google Scholar]
- 20.Huang C C, Petterson E F, Klein T E, Ferrin T E, Langridge R. Conic: a fast renderer for space-filling molecules with shadows. J Mol Graphics. 1991;9:230–236. doi: 10.1016/0263-7855(91)80016-s. [DOI] [PubMed] [Google Scholar]
- 21.Johnson J E. Averaging of electron density maps. Acta Crystallogr. 1978;B34:576–577. [Google Scholar]
- 22.Jones T A, Zou J Y, Cowan S W, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 23.Jones T A, Kjeldgaard M. Electron density map interpretation. Methods Enzymol. 1997;277:173–208. doi: 10.1016/s0076-6879(97)77012-5. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch W, Sander S. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 25.Kim S. Auto-indexing oscillation photographs. J Appl Crystallogr. 1989;22:53–60. [Google Scholar]
- 26.Kraulis P J. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 27.Kridl J C, Bruening G. Comparison of capsids and nucleocapsids from cowpea mosaic virus-infected cowpea protoplasts and seedlings. Virology. 1983;129:369–380. doi: 10.1016/0042-6822(83)90176-9. [DOI] [PubMed] [Google Scholar]
- 28.Lapchic L G, Clark A J, Lomonossoff G P, Shanks M. Red clover mottle virus from Ukraine is an isolate of RCMV strain S. Eur J Plant Pathol. 1998;104:409–412. [Google Scholar]
- 29.Laskowski R W, MacArthur M W, Moss D S, Thornton J M. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:345–364. [Google Scholar]
- 30.Lekkerkerker A, Wellink J, Juan P, van Lent J, Goldbach R, van Kammen A B. Distinct functional domains in the cowpea mosaic virus movement protein. J Virol. 1996;70:5658–5661. doi: 10.1128/jvi.70.8.5658-5661.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lomonossoff G P, Johnson J E. The synthesis and structure of comovirus capsids. Prog Biophys Mol Biol. 1991;55:107–137. doi: 10.1016/0079-6107(91)90003-b. [DOI] [PubMed] [Google Scholar]
- 32.MacFarlane S A, Shanks M, Davies J W, Zlotnick A, Lomonossoff G P. Analysis of the nucleotide sequence of bean pod mottle virus middle component RNA. Virology. 1991;183:405–409. doi: 10.1016/0042-6822(91)90155-5. [DOI] [PubMed] [Google Scholar]
- 33.McPherson A. The preparation and analysis of protein crystals. New York, N.Y: John Wiley and Sons; 1982. [Google Scholar]
- 34.Oxelfelt P. Biological and physico-chemical characteristics of three strains of red clover mottle virus. Virology. 1976;74:73–80. doi: 10.1016/0042-6822(76)90129-x. [DOI] [PubMed] [Google Scholar]
- 35.Oxelfelt P, Abdelmoeti M. Genetic complementation between natural strains of red clover mottle virus. Intervirology. 1978;10:78–86. doi: 10.1159/000148971. [DOI] [PubMed] [Google Scholar]
- 36.Oxelfelt P, Shanks M, Widmark A K, Lomonossoff G P. Identification and characterization of pseudo-recombinants of red clover mottle comovirus. J Gen Virol. 1992;73:2121–2124. doi: 10.1099/0022-1317-73-8-2121. [DOI] [PubMed] [Google Scholar]
- 37.Rossmann M G. Processing oscillation diffraction data for very large unit cells with an automatic convolution technique and profile fitting. J Appl Crystallogr. 1979;12:225–238. [Google Scholar]
- 38.Rossmann M G, Johnson J E. Icosahedral RNA virus structure. Annu Rev Biochem. 1989;58:533–573. doi: 10.1146/annurev.bi.58.070189.002533. [DOI] [PubMed] [Google Scholar]
- 39.Rossmann M G, Leslie A G W, Abdel-Meguid S S, Tsukihara T. Processing and post-refinement of oscillation camera data. J Appl Crystallogr. 1979;12:570–581. [Google Scholar]
- 40.Rossmann M G, McKenna R, Tong L, Xia D, Dai J-B, Wu H, Choi H K. Molecular replacement real-space averaging. J Appl Crystallogr. 1992;25:166–180. [Google Scholar]
- 41.Shanks M, Stanley J, Lomonossoff G P. The primary structure of red clover mottle virus middle component RNA. Virology. 1986;155:687–706. doi: 10.1016/0042-6822(86)90229-1. [DOI] [PubMed] [Google Scholar]
- 42.Shanks M, Tomenius K, Clapham D, Huskisson N S, Barker P J, Wilson I G, Maule A J, Lomonossoff G P. Identification and subcellular localization of a putative cell-to-cell transport protein from red clover mottle virus. Virology. 1989;173:400–407. doi: 10.1016/0042-6822(89)90552-7. [DOI] [PubMed] [Google Scholar]
- 43.Shanks M, Lomonossoff G P. The primary structure of the 24K protease from red clover mottle virus: implications for the mode of action of comoviral proteases. J Gen Virol. 1990;71:735–738. doi: 10.1099/0022-1317-71-3-735. [DOI] [PubMed] [Google Scholar]
- 44.Shanks M, Lomonossoff G P. The nucleotide sequence of red clover mottle virus bottom component RNA. J Gen Virol. 1992;73:2473–2477. doi: 10.1099/0022-1317-73-9-2473. [DOI] [PubMed] [Google Scholar]
- 45.Shanks M, Dessens J T, Lomonossoff G P. The 24-kDa proteinases of comoviruses are virus-specific in cis as well as in trans. J Gen Virol. 1996;77:2365–2369. doi: 10.1099/0022-1317-77-9-2365. [DOI] [PubMed] [Google Scholar]
- 46.Shindo N, Vicente A C P, Krengiel R, Oliveira D E. Nucleotide sequence analysis of an Andean potato mottle virus middle component RNA cDNA clone: comparison of the encoded proteins with those of other comoviruses. Intervirology. 1993;36:169–180. doi: 10.1159/000150337. [DOI] [PubMed] [Google Scholar]
- 47.Sinha R C. Red clover mottle virus. Ann Appl Biol. 1960;48:742–748. [Google Scholar]
- 48.Stauffacher C V, Usha R, Harrington M, Schmidt T, Hosur M, Johnson J E. The structure of cowpea mosaic virus at 3.5 Å resolution. In: Moras D, Drenth J, Strandberg B, Suck D, Wilson K, editors. Crystallography in molecular biology. New York, N.Y: Plenum Publishing Corporation; 1987. pp. 293–308. [Google Scholar]
- 49.Taylor K M, Spall V E, Butler P J G, Lomonossoff G P. The cleavable C-terminus of the small coat protein of cowpea mosaic virus is involved in RNA encapsidation. Virology. 1999;255:129–137. doi: 10.1006/viro.1998.9567. [DOI] [PubMed] [Google Scholar]
- 50.Tong L, Rossmann M G. The locked rotation function. Acta Crystallogr. 1990;A46:783–792. doi: 10.1107/s0108767390005530. [DOI] [PubMed] [Google Scholar]
- 51.Tong L, Rossmann M G. Rotation function calculation with GLRF program. Methods Enzymol. 1997;276:594–611. [PubMed] [Google Scholar]
- 52.van Wezenbeek P, Verver J, Harmsen J, Vos P, van Kammen A. Primary structure and gene organization of the middle-component RNA of cowpea mosaic virus. EMBO J. 1983;2:941–946. doi: 10.1002/j.1460-2075.1983.tb01525.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Virudachalam R, Harrington M, Johnson J E, Markley J L. 1H, 13C and 31P nuclear magnetic resonance studies of cowpea mosaic virus: detection and exchange of polyamines and dynamics of the RNA. Virology. 1985;141:43–50. doi: 10.1016/0042-6822(85)90181-3. [DOI] [PubMed] [Google Scholar]
- 54.Wilson A J C. Determination of absolute from relative X-ray intensity data. Nature. 1942;150:151. [Google Scholar]
- 55.Zimmern D. The 5′ end group of tobacco mosaic virus RNA is m7G5′ppp5′Gp. Nucleic Acids Res. 1975;2:1189–1201. doi: 10.1093/nar/2.7.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]