Abstract
Apicomplexan parasites constitute over 6000 species infecting a wide range of hosts. These include important pathogens such as those causing malaria and toxoplasmosis. Their evolutionary emergence coincided with the dawn of animals. Mitochondrial genomes of apicomplexan parasites have undergone dramatic reduction in their coding capacity with genes for only 3 proteins and ribosomal RNA genes present in scrambled fragments originating from both strands. Different branches of the apicomplexans have undergone rearrangements of these genes, with Toxoplasma having massive variations in gene arrangements spread over multiple copies. The vast evolutionary distance between the parasite and host mitochondria has been exploited for developing antiparasitic drugs, especially for treatment of malaria, wherein inhibition of the parasite mitochondrial respiratory chain is selectively targeted with little toxicity to the host mitochondria. We describe additional unique characteristics of the parasite mitochondria that are being investigated and provide greater insights into these deep branching eukaryotic pathogens.
Keywords: Myzozoa, Plasmodium, Toxoplasma, ATP synthase, Electron transport complexes, Antiparasitic drugs
Introduction
The emergence of eukaryotes from an exclusively prokaryotic biosphere was a momentous event that charted a divergent course for life on Earth (82). This event coincided with syntrophic adoption of mitochondria. The complexity of eukaryotes could not have existed and evolved but for the energy economy supported by mitochondrial physiology (64). Given the strong evidence for the monophyletic origin of mitochondria (37–39), it is probable that all extant mitochondria can trace their origin to that singular primordial event at the dawn of eukaryotes. Yet, evolution has resulted in a tremendous variety of mitochondria, their genomes, their regulation, and their functions, which are as diverse as the organisms they reside in, from Paramecium to the Pope. Each clade of eukaryotes appears to adjust its mitochondrial function to fit the niche in which it exists (39). Apicomplexan parasites demonstrate these evolutionary adjustments in an illuminating manner. These obligatory intracellular parasites consist of thousands of species, all making their living off a vast array of host animals (2; 3). Even corals, the deepest branching animals, have intracellular apicomplexans, suggesting that the parasitic nature of apicomplexans is as ancient as the origin of Metazoa (61; 62). The discovery of mitochondrial DNA (mtDNA) in malaria parasites as tandemly arrayed molecules with a unit length of 6 kb (51; 120; 125–127) led to the realization that a separate circular DNA of 35 kb in the parasite, believed then to be a mtDNA (34; 135), was in fact a remnant of a chloroplast genome residing in a separate organelle, now termed an apicoplast (for apicomplexan plastid). The apicoplast consists of a 4-membrane structure and thus is proposed to have originated from a secondary endosymbiotic event common to all apicomplexans, in which an algal organism was engulfed by the progenitor of both the dinoflagellates and apicomplexans (56; 57). Most extant apicomplexans have three distinct genomes: the nuclear, the mitochondrial and the apicoplast. However, the proposed secondary endosymbiotic event would have initially involved coexistence of five genomes—the progenitor’s nuclear and mitochondrial DNAs plus the algal nuclear, plastid and mitochondrial DNAs—before being reduced to three (Fig. 1). The massive quantity of gene transfers and reductions that followed has resulted in apicomplexan genomes carrying a mixture of genes inherited from 5 distinct genomes (Fig. 1). This complex provenance of apicomplexans, as well as natural selection to fit a tremendous variety of parasitic niches, has resulted in many biological characteristics that are distinct from most model eukaryotes. In this review, we aim to describe such distinct aspects of the mitochondrion in apicomplexan parasites. Our goal is not to provide a comprehensive view of mitochondria in all apicomplexans but to describe their unique properties. We refer the reader to several excellent reviews that cover additional details of these organelles (11; 41–43; 79; 85; 124; 128).
Figure 1. Schematic representation of the secondary endosymbiotic event involving the progenitor of myzozoans with two genomes and an alga with three genomes.
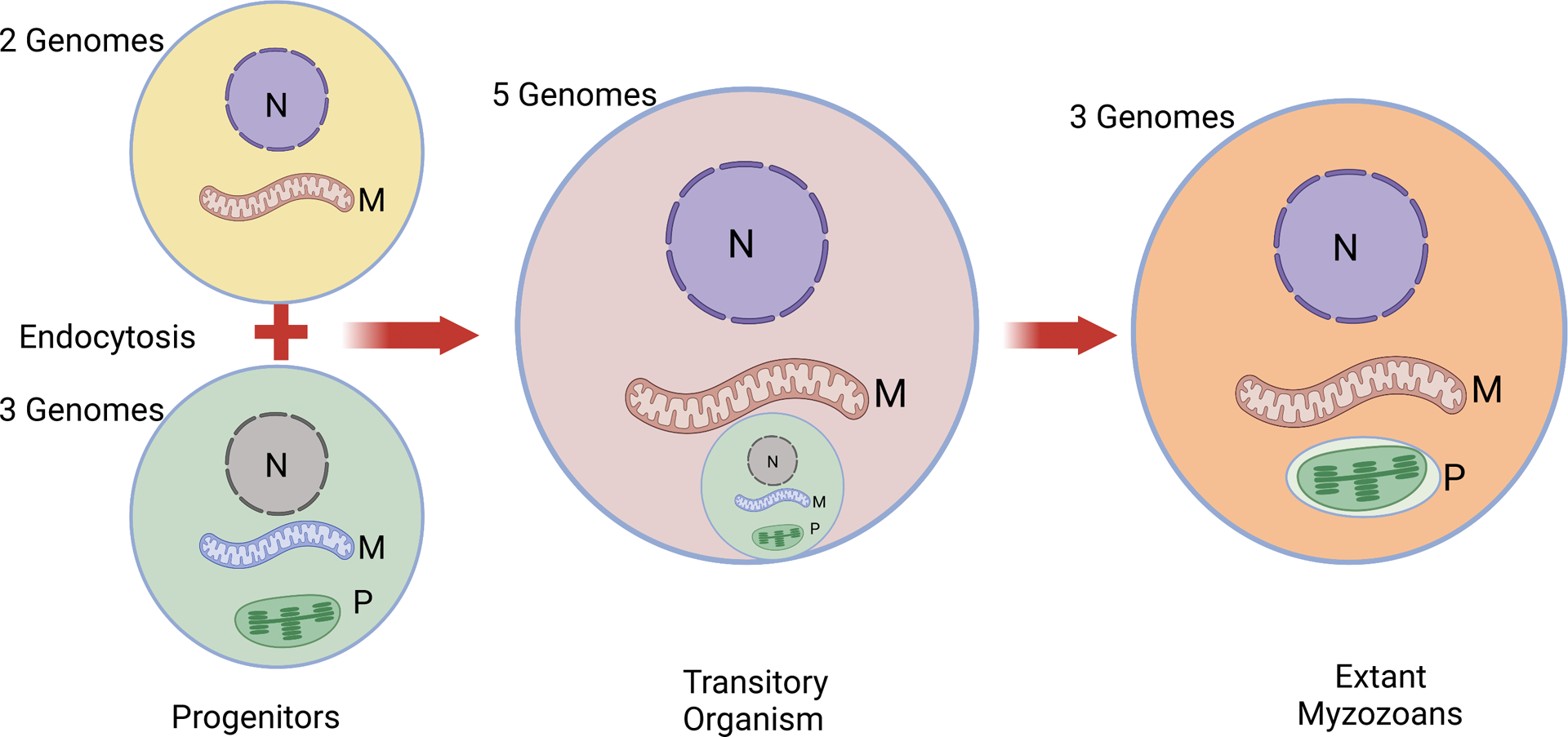
Engulfment of the alga by the progenitor of Myzozoa resulted in a transitory organism with five genomes. Massive gene transfers and losses led to the elimination of the algal nucleus and mitochondrion, thus resulting in the three genomes seen in extant Myzozoa, which remain a conglomerate of five genomes.
Downsizing and Diversification
Apicomplexans belong to the superphylum Alveolata, which diverged about 850 million years ago into two main branches: Ciliates and Myzozoa. Whereas the progenitor of alveolates likely contained a mtDNA of about 50 kb coding for 40 or so genes, and most ciliates continue to have a similar sized mtDNA, the Myzozoa underwent a drastic reduction in their capacity and now code for just 3 proteins (cytochrome oxidase subunits 1 and 3 (Cox1 and Cox 3) and cytochrome b (Cytb)) and fragmentary ribosomal RNA (rRNA) pieces (86; 128). The divergence of Myzozoa coincided with the secondary endosymbiotic acquisition of plastids (11; 49). Among the three main branches of Myzozoa, two (dinoflagellates and chromerids) consist of mostly free-living species and have plastids with photosynthesis capacity, whereas members of the third (apicomplexans) contain non-photosynthetic plastids and parasitize animals. The large reduction in the number of genes encoded by mtDNA in Myzozoa, therefore, is unlikely to be due to their dependence on an external source for their energy needs (23). One possibility could be that the progenitor of the Myzozoa, after acquiring a plastid, became more dependent on the photosynthetic supply of energy and less on the mitochondrial contribution. All through the radiation of the metazoan clade, the reduction in the gene content of their mtDNAs also appears to be accompanied by large scale rearrangements of genes as well as of the configurations of the genomes (Fig. 2). Even among the Apicomplexans, the synteny of the genes is highly divergent and the configuration of the genomes varies among different genera, and sometimes even within the same genus (Fig. 2). Unlike the three common protein coding genes, rRNA gene fragments are highly variable among different apicomplexans. The evolutionary forces that must have driven this high degree of divergence are not clear but do suggest events that occurred at branch points in the apicomplexan phylogenetic tree coincided with massive gene rearrangements within the mtDNA. Of interest, the extent of divergence seen for the mtDNA is not observed in the apicoplast genomes of the extant apicomplexans (99; 129). This suggests differential evolutionary pressures on the two apicomplexan cytoplasmic genomes.
Figure 2. Phylogenetic tree illustrating the evolutionary relationship between the subclasses of apicomplexans (branch lengths are not drawn to scale).
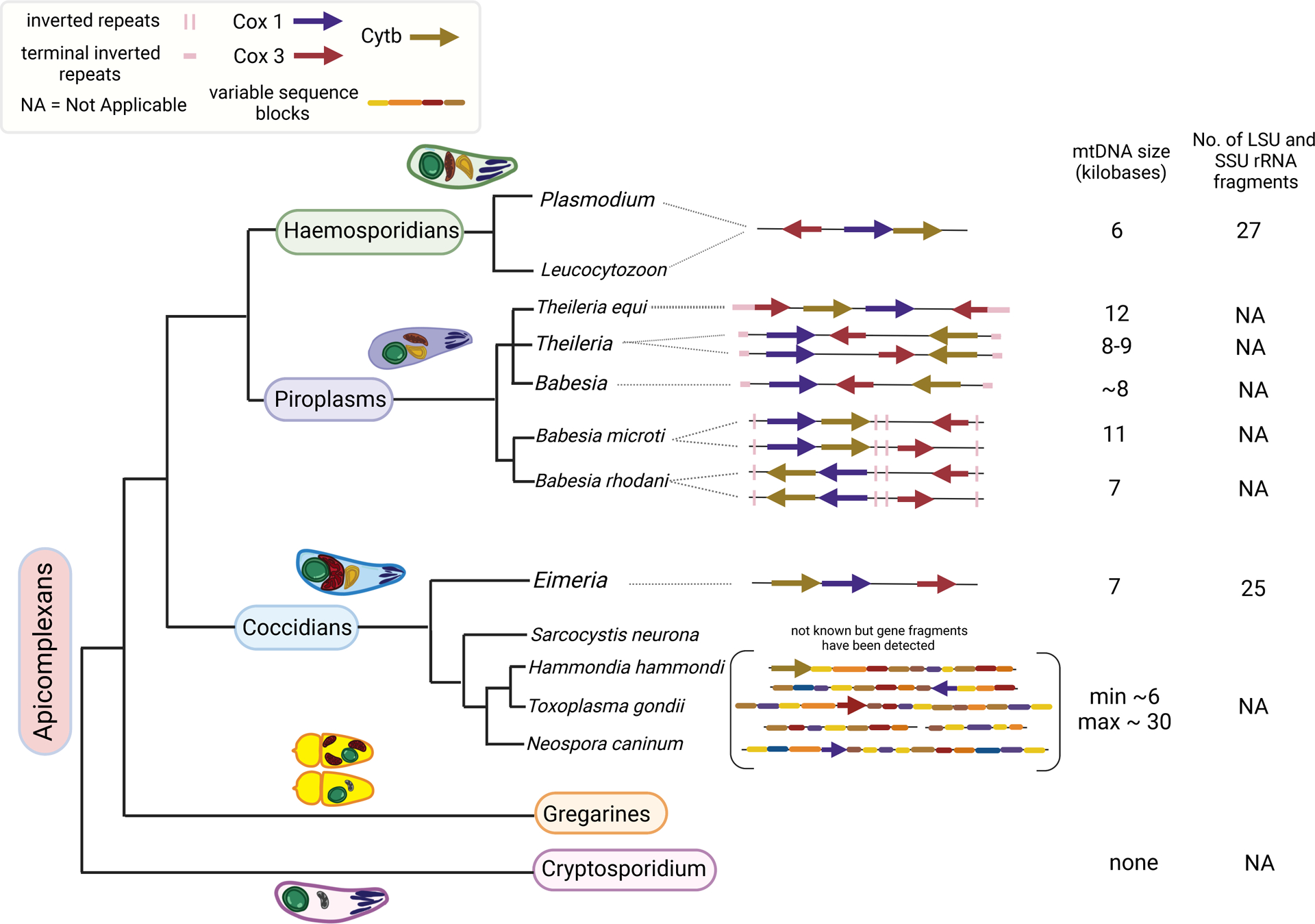
The synteny of protein-coding genes and their directionality in the apicomplexan mitochondrial genome of organisms in each subclass is shown as arrows. The size of the mitochondrial DNA as well as the known number of fragmentary mitochondrial large subunit and small subunit rRNA transcripts in each subclass are indicated to the right of the tree. This figure is modified from Figure 1 from Berná et al. (ref. 10).
Apicomplexan Mitochondrial Genomes
All apicomplexan mtDNAs encode 3 proteins and rRNA fragments but their sizes and gene arrangements vary greatly among different genera, and in some cases within a genus (Fig. 2). The size of the mtDNAs range from 6 kb (e.g., Plasmodium spp.) to 12 kb (Theileria equi) (43). In general, apicomplexan mtDNAs are present as linear molecules but in the case of Plasmodium spp., they appear to form a few circular DNA molecules, which are likely to be molecules undergoing a rolling circle mode of replication (105; 134). The tandem head-to-tail arrays with 6 kb unit length are the likely outcome of this mode of replication. Gene arrangements in Plasmodium spp. are highly conserved, with a sequence identity of >90% over an evolutionary distance among different species estimated to span millions of years based on the divergence of nuclear DNA sequences (43). For instance, the overall G+C content of the nuclear DNA varies from ~21% in P. falciparum to ~34% in P. vivax, two malaria parasites infecting humans, but both their mtDNAs have ~31% G+C content (88). It is also remarkable that, while the codon usage frequency for the nuclear genes between these species is quite divergent, it remains identical for their mtDNA-encoded genes (88).
Compared to Plasmodium spp., Theileria and Babesia mtDNAs are divergent among different species. T. equi mtDNA is a 12 kb linear molecule with terminal repeats, while other Theileria spp. have 8 to 9 kb mtDNA with genes arranged differently from those in the T. equi mtDNA. The size differences are also seen in Babesia species with B. microti having an 11 kb mtDNA compared to ~8 kb mtDNA in other species (43).
Toxoplasma gondii, and related coccidial parasites, Hammondia and Neospora, contain the most highly diverse and dramatically rearranged mtDNA among all apicomplexans (10; 96). For many years, the authentic mtDNA sequence from Toxoplasma could not be established. One complicating reason was the presence of multiple sequences of what appeared to be portions of mtDNA interspersed all throughout the nuclear genome of the parasite (101). Recent application of long-read DNA sequencing technology has revealed a startling complexity in the mitochondrial genome of T. gondii (96). Multiple molecules ranging in size from 320 to 23,600 base pairs were detected. Further analysis of all the sequences revealed that these molecules were not linear concatemers of a unit molecule but did contain 26 distinct sequence blocks that were arranged in varying configurations. Overall, coding sequences for Cox1, Cox3 and Cytb could be discerned but were arranged differently in different molecules (96). Fragments of rRNAs could also be detected but attempts to assemble them into small and large subunit rRNAs have not been carried out (96). It is a wonder how this complex arrangement of genes can result in the generation of mitochondrial complexes necessary for the survival of the organism. It is also interesting to note that the non-cyst forming coccidial parasites of Eimeria species do not have this bizarre arrangement of their mtDNA but appear to consist of tandem arrays of 7 kb molecules (44; 71).
Mitochondrial rRNA
Since the emergence of mitochondria from its alpha-proteobacterial progenitor, mitochondrial rRNAs appear to have undergone dramatic changes (24; 29; 53). This is reflected in the highly divergent structures of mitoribosomes, particularly in the increase in the ratio of protein to RNA content, suggesting the transfer of the structural role of organization and scaffolding from rRNAs to proteins in the mitoribosome (12; 106). Although, the structure of the mitoribosome of an apicomplexan parasite has not been elucidated, analysis of mitoribosomes of unicellular organisms such as Trypanosoma brucei, Tetrahymena thermophilia, and Chlamydomonas reinhardtii show a reduction in rRNA size and incorporation of new and enlarged mitoribosomal proteins. Mitoribosomal proteins form a shell that surrounds the rRNA core of the mitoribosome, stabilizing and orienting the critical regions of their fragmented rRNAs appropriately (106; 123; 132). With the much larger number of rRNA fragments encoded by apicomplexan mtDNA (see below) and the apparent absence of some of the conserved rRNA domains, the apicomplexan mitoribosomal structure, once determined, is likely to reveal many divergent and surprising features.
In contrast to most eukaryotes where mitochondrial rRNA subunits are encoded by continuous DNA sequences that are transcribed into long continuous polyribonucleotide chains, DNA sequences encoding mitochondrial rRNA in apicomplexans are fragmented and arranged in a scrambled manner originating from both strands of mtDNA (29; 125; 127). rRNA fragments have been identified in the mitochondria of all apicomplexans examined including Plasmodium spp. (28; 29; 125; 127), Babesia spp. (43; 45), Theileria spp. (45; 52), Eimeria spp. (44), and Toxoplasma gondii (96). However, investigations on these mitochondrial rRNA fragments have mostly been conducted on Plasmodium spp. parasites (29). Just as the mtDNA of each apicomplexan varies in size and organization, these mitochondrial rRNA fragments also vary in size, arrangement, and length across various apicomplexans. Eimeria spp. and Plasmodium spp. have a mtDNA of ~ 6–7 kb in length. However, most species of Eimeria have identified 14 large subunit (LSU) and 11 small subunit (SSU) rRNA fragments (69; 72), while P. falciparum has 15 LSU and 12 SSU rRNA fragments identified (29; 30). Furthermore, the DNA sequences encoding mitochondrial rRNA in Plasmodium spp and Theileria spp are arranged out of order and interspersed between protein-coding genes on either strand of the mtDNA (29; 30; 125; 127). The lengths of the mitochondrial rRNA fragments also differ vastly within each apicomplexan parasite (29; 72; 96).
Through both intra- and inter-molecular complementary base pairing, rRNA fragments in apicomplexans can be predicted to form standard RNA secondary structures, such as hairpin loops, that are consistent with the structures formed by their continuous rRNA sequence counterparts (29; 50; 68; 125). Although these fragmented rRNAs are not linked covalently, their catalytic core is preserved so they are likely to retain their function. Systematic transcript mapping of mitochondrial rRNA transcripts in P. falciparum demonstrated that some are conserved across apicomplexans and can be mapped to conserved sequences in the SSU and LSU rRNAs of E. coli (29; 30). However, there are some portions of the rRNA in E. coli that could not be identified amongst the pool of rRNA transcripts in P. falciparum. One possibility is that these missing rRNA pieces may be imported from the cytoplasm, possibly through the machinery required for tRNA import. It is also possible that they have been lost due to the evolutionary trend of reducing mitoribosomal rRNA size and transferring its function to mitoribosomal proteins (53; 132).
The process by which rRNA fragments are processed and assembled with ribosomal proteins into a functional mitoribosome remains to be elucidated. The mitoribosome of P. falciparum has been shown to be associated with the inner mitochondrial membrane, and the essentiality of the mitoribosome in P. falciparum and T. gondii has been demonstrated (54; 70; 113). Based on amino acid sequence similarity to mitoribosomal proteins of other organisms, 43 proteins have been predicted to be P. falciparum mitoribosomal proteins, six of which have been shown to be essential for mitochondrial functions (25; 54). Recently, two proteins with RNA-binding domains that are abundant in apicomplexans, PfRAP01 and PfRAP21, were identified and characterized to be essential nuclear-encoded proteins that are targeted to the mitochondria where they specifically bind mitochondrial rRNAs in P. falciparum (46; 47). PfRAP21 is involved in the control of mitochondrial rRNA expression, while both RAP proteins play a role in RNA processing, translation, and expression of mitoribosomal subunits (46; 47).
It is of interest that mitochondrial rRNAs in P. falciparum and Theileria parva have been shown to possess oligo(A) tails up to 21 nucleotides in length, which are added post-transcriptionally to their 3’ end (29; 36; 97). The length of the oligo(A) tails is transcript-specific, so it is thought their addition may serve a protective role in preventing exonucleolytic degradation of these rRNA fragments to allow for proper assembly, structure, and function of the mitoribosome. However, the roles of oligo-adenylation of specific mitochondrial rRNA transcripts in apicomplexans remain uncertain. Several investigations have revealed that the genes encoding the 16S and 12S rRNA in humans encode small open reading frames (ORFs) which are translated into “mitochondrial-derived peptides” that are involved in important cellular processes (66; 67; 90; 139). These peptides, namely Humanin, SHLPs, and MOTS-c, have been shown to have significant physiological roles and demonstrate that mitochondrial rRNA could encode peptides with important functions. Although mitochondrial rRNA-encoded peptides have not been identified in apicomplexan parasites, a possible peptide-encoding functions of small rRNA fragments could be an important topic for exploration. In addition, several small mitochondrial RNA transcripts in P. falciparum have been observed with no homology to known rRNA sequences and with no known functions (29). It would be of interest to assess the potential presence of small ORFs within these RNA molecules that may have the capacity to encode peptides.
Mitochondrial Proteomes in Apicomplexans
Except for the three mitochondrial electron transport chain (mtETC) subunits encoded by the mtDNA, all other mitochondrial proteins are encoded by the nucleus and imported into the mitochondrion. A variety of approaches have been used to generate a predicted compilation of mitochondrial proteomes in P. falciparum and T. gondii. Bioinformatic (18) and manual (17) annotations initially generated a list of about 400 proteins likely to be imported into the Plasmodium mitochondrion. Since the mitochondrion contains multiple sub-compartments with multiple import mechanisms, comprehensive lists are inevitably difficult to complete. Current manual curation suggests a proteome size of about 500 proteins associated with the P. falciparum mitochondrion; the T. gondii proteome may be slightly larger given its more complex metabolism. Experimental validations of the predicted mitochondrial proteome have been limited but are gradually accumulating. A mitochondrially-targeted proximity biotinylation followed by enrichment and mass spectrometry approach was used to identify uncharacterized P. falciparum proteins putatively targeted to the mitochondrion (63). A list of 122 putative mitochondrial proteins was generated with many proteins that were unannotated and likely to be essential for parasite survival (63). However, biotinylated proteins identified in this study also included apparently cytoplasmic proteins, thereby pointing to the limitations of this approach.
Proteomic evidence suggesting a unique architecture of apicomplexan respiratory chain complexes was reported in T. gondii using mitochondrially-targeted proximity biotinylation approaches (111). In addition to five canonical subunits of the cytochrome c oxidase (Complex IV) (Cox1, Cox2, Cox3, Cox5b, and Cox6b), this analysis identified 11 apicomplexan-specific Complex IV subunits (111). Complexome profiling, in which label-free quantitative mass spectrometry is paired with microscale fractionation of large complexes in blue native gels, has the power to identify novel protein subunits that co-migrate with such complexes (107; 136). This approach has recently been applied to P. falciparum mitochondria-enriched samples and led to identification of novel components of mitochondrial respiratory chain complexes (27). Orthologues of all 11 previously reported Complex IV subunits were identified in the P. falciparum complexome profile, confirming the unique Complex IV architecture in these organisms (27). Five uncharacterized, mostly myzozoan-specific, putative subunits also co-migrated with Complex IV, which were termed respiratory chain Complex 4 associated proteins 1–5 (C4AP1–5). In most eukaryotes, the Complex IV subunit Cox2 is encoded by the mtDNA. Uniquely, in apicomplexans and other myzozoans, the gene is split into two nuclear-encoded genes, Cox2a and Cox2b (131). Both of these proteins were detected in the P. falciparum complexome profile, confirming their mitochondrial targeting (27).
Canonically, at least four subunit proteins compose succinate dehydrogenase (Complex II): succinate dehydrogenase subunit A (SDHA), SDHB, SDHC, and SDHD. Only SDHA and SDHB are experimentally validated in Plasmodium (121; 122), but candidates for SDHC (PF3D7_0611100) and SDHD (PF3D7_1010300) have been proposed (91). In the Plasmodium respiratory chain complexome profile, SDHA and SDHB were observed to be co-migrating together in a ~530 kDa complex, but the previously proposed SDHC and SDHD proteins were not found to co-migrate with this complex (27). However, the authors identified five putative subunits that all shared a common dominant band and assigned one as a candidate for PfSDHC (PF3D7_1448900). This assignment was based upon the conserved “DY” motif found in SDHC in many species (93). The other putative Complex II subunits were termed respiratory chain complex 2 associated proteins 1–4 (C2AP1–4). One of these proteins, PF3D7_0808450, is restricted only to myzozoans and was previously reported as a mitochondrion-localized protein essential for malaria transmission (59).
Complexome profiling has also been carried out on T. gondii mitochondria (78). In this study, 60 proteins were identified and assigned to Complexes II, IV, and the F1Fo-ATP synthase (Complex V). The details of recent work investigating F1Fo-ATP synthase complexes in apicomplexans will be discussed in detail below. Sixteen of these 60 respiratory chain complex proteins had not been previously identified. The study also determined the composition of T. gondii Complex III, a known drug target. In doing so, two new homologous subunits and two parasite-specific subunits were identified. All four of these proteins were found to be essential for Complex III stability and parasite viability. In addition, depletion of these four subunits led to collapse of the mitochondrial membrane potential, consistent with their assignment as Complex III subunits (78). Similar to P. falciparum, the T. gondii Complex II subunit SDHB was also observed to migrate as a ~500 kDa complex (78), a much larger size than observed in yeast and mammalian Complex II (~130 kDa) (109; 110). This size was confirmed using endogenous tagging of TgSDHB (TGGT1_215280) and subsequent blue native PAGE analysis, making it a larger Complex II than reported in any other well studied organisms outside of apicomplexans. Seven other proteins co-migrated with TgSDHB, all of which are annotated as hypothetical proteins that bear no obvious protein features indicating function, highlighting the unique nature of Complex II in apicomplexans.
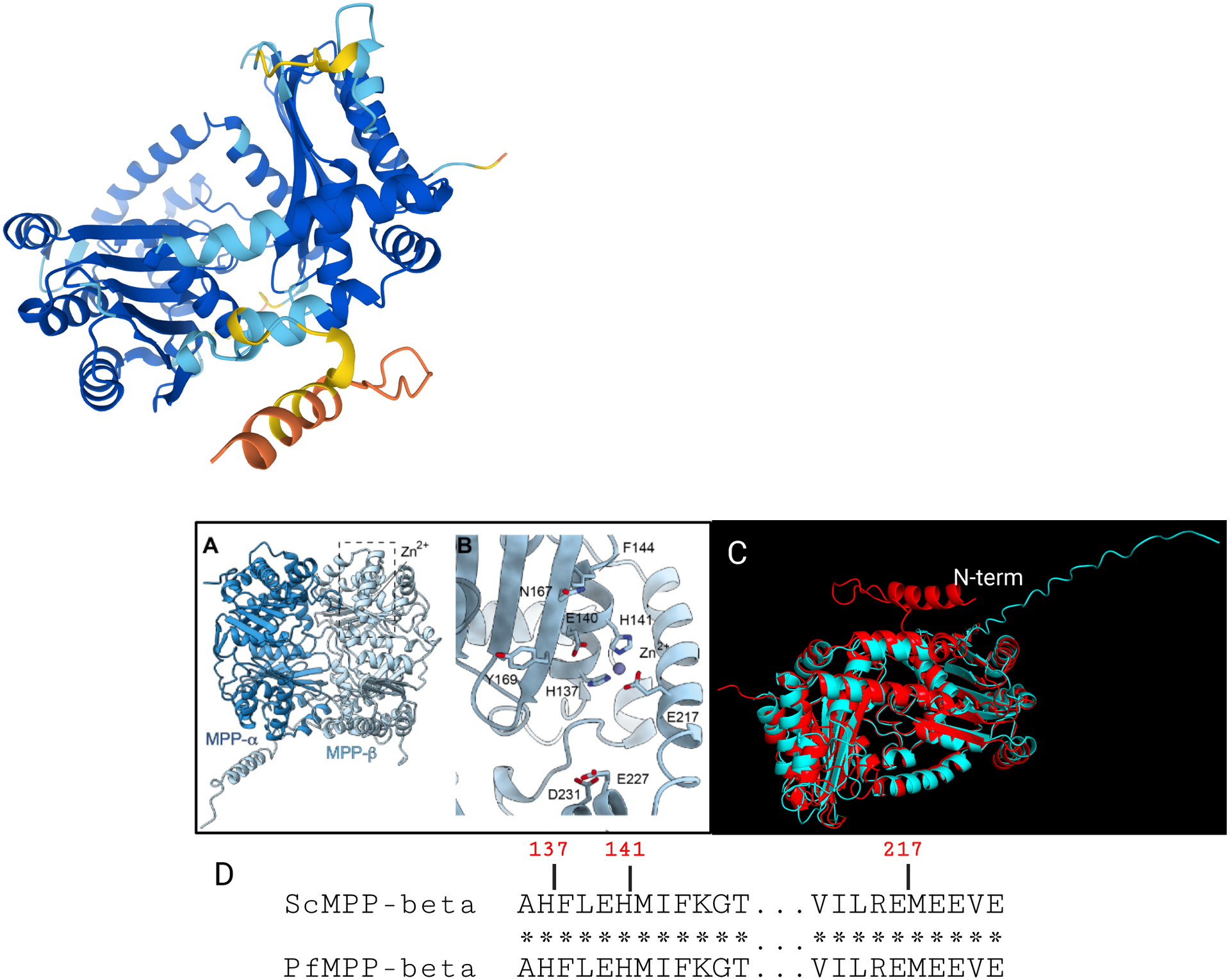
Nucleus-encoded mitochondrial matrix proteins often have N-terminal targeting peptides that are cleaved during import by mitochondrial processing peptidases (MPP) (33; 60). In yeast and mammals, MPPs are heterodimers of subunits α and β, which are located in the mitochondrial matrix (33). In addition to these matrix MPPs, the cytochrome bc1 complex (Complex III) in eukaryotes contains Core1 and Core2 subunits with high amino acid sequence homology to MPP subunits α and β (137). In contrast, plant mitochondria do not have matrix resident MPPs. It has therefore been suggested that, in plants, Complex III has two functions: ubiquinol oxidation and processing of mitochondrially targeted proteins (15; 16). The P. falciparum genome encodes annotated α and β MPP subunits that were clearly shown to be part of Complex III (27). This suggests a unique aspect of the malaria parasite mitochondrion in which MPPs are part of Complex III as in plants, playing a role in its structural assembly. In addition to co-migration of all canonical components of Complex III, four additional proteins co-migrated with the complex in the P. falciparum mitochondrial complexome profile (27). Three of these putative subunits are represented almost exclusively in apicomplexa, lacking no obvious sequence homology with characterized subunits in other phyla.
Another approach of spatial proteomics called, “hyperplexed localization of organelle proteins by isotope tagging” (hyperLOPIT) was used to determine the steady-state subcellular localization of thousands of T. gondii proteins (9). The hyperLOPIT technique capitalizes on specific abundance-distribution contours that organelles and other subcellular structures form after being subjected to density-gradient centrifugation (21; 95). This study identified 193 mitochondrial membrane-associated proteins and 274 soluble mitochondrial proteins, further enriching the proteomic landscape of Toxoplasma mitochondria (9).
Mitochondrial Contributions to Metabolism
Since apicomplexans are phylogenetically distant from mammals and other model organisms, it is not surprising that their mitochondria exhibit significant differences from those of model organisms. They are also a large phylum containing diverse parasitic species existing in numerous host species and environments. Hence, there is a wide range of bioenergetic capabilities and metabolism associated with apicomplexan mitochondria and related organelles (86). Expression of mitochondrial activities also frequently varies significantly between parasite life cycle stages. Several metabolic pathways common in more familiar model organisms are not found in apicomplexan mitochondria, such as beta oxidation of fatty acids and portions of amino acid and steroid biosynthesis. Fig. 3 provides a schematic description of mitochondrial functions in P. falciparum, a parasite for which the most extensive information is available. Apicomplexan parasites of vertebrates, such as Plasmodium and Toxoplasma have a nearly complete mtETC (lacking only Complex I) and an ATP synthase complex, providing robust oxidative phosphorylation, at least in some life stages. The metabolic partner of the oxidative phosphorylation (OxPhos) ATP generation pathway is the oxidative TCA cycle that provides reducing equivalents to the mtETC. In Plasmodium and Toxoplasma, a repurposed branched-chain alpha-keto acid dehydrogenase (BCKDH) complex in the mitochondrion, rather than a direct ortholog of the pyruvate dehydrogenase found in model organisms, provides acetyl-CoA to the cycle (100), and a set of five ubiquinone-dependent dehydrogenases provide reducing equivalents to the mtETC. Among the dehydrogenases, only dihydroorotate dehydrogenase is essential in asexual blood stage Plasmodium, as a key enzyme in the pyrimidine biosynthesis pathway, and critical for virulence in Toxoplasma, even though the later parasite has a salvage pathway for pyrimidines (31; 40; 102). In these parasites 2-oxoglutarate (from glutamine), in addition to glycolytic pyruvate, is a major contributor to carbon flux through the TCA cycle (55; 80; 81). In Toxoplasma, the presence of a γ-aminobutyric acid shunt pathway allows for additional input to the TCA cycle of succinate derived from glutamine. Two of the TCA cycle enzymes are novel relative to those in more familiar organisms, malate-quinone oxidoreductase in place of NADH-dependent malate dehydrogenase, rendering this step essentially irreversible, and in place of the more familiar metal free class II fumarase, a class I fumarate hydratase containing an iron-sulfur cluster [Fe-S], rendering the parasite enzyme sensitive to reactive oxygen species. The very low oxidation-reduction potential maintained in the parasite mitochondrion should, however, help protect Fe-S clusters in fumarate hydratase, aconitase and other Fe-S containing proteins (92). Combined genetic and metabolomic experiments have found that acetyl-CoA from the Plasmodium mitochondrion is required for acetylation of histones and other cytoplasmic and nuclear proteins, which is provided by BCKDH, or by alpha-ketoglutarate dehydrogenase if BCKDH is ablated (22). Since Plasmodium spp., unlike animals and Toxoplasma, lack ATP-citrate lyase, which cleaves citrate exported from the mitochondrion to acetyl-CoA and oxaloacetate, it is presently unclear how acetyl-CoA made in the Plasmodium mitochondrion reaches other compartments.
Figure 3. A schematic of mitochondrial functions in P. falciparum with associated drug targets.
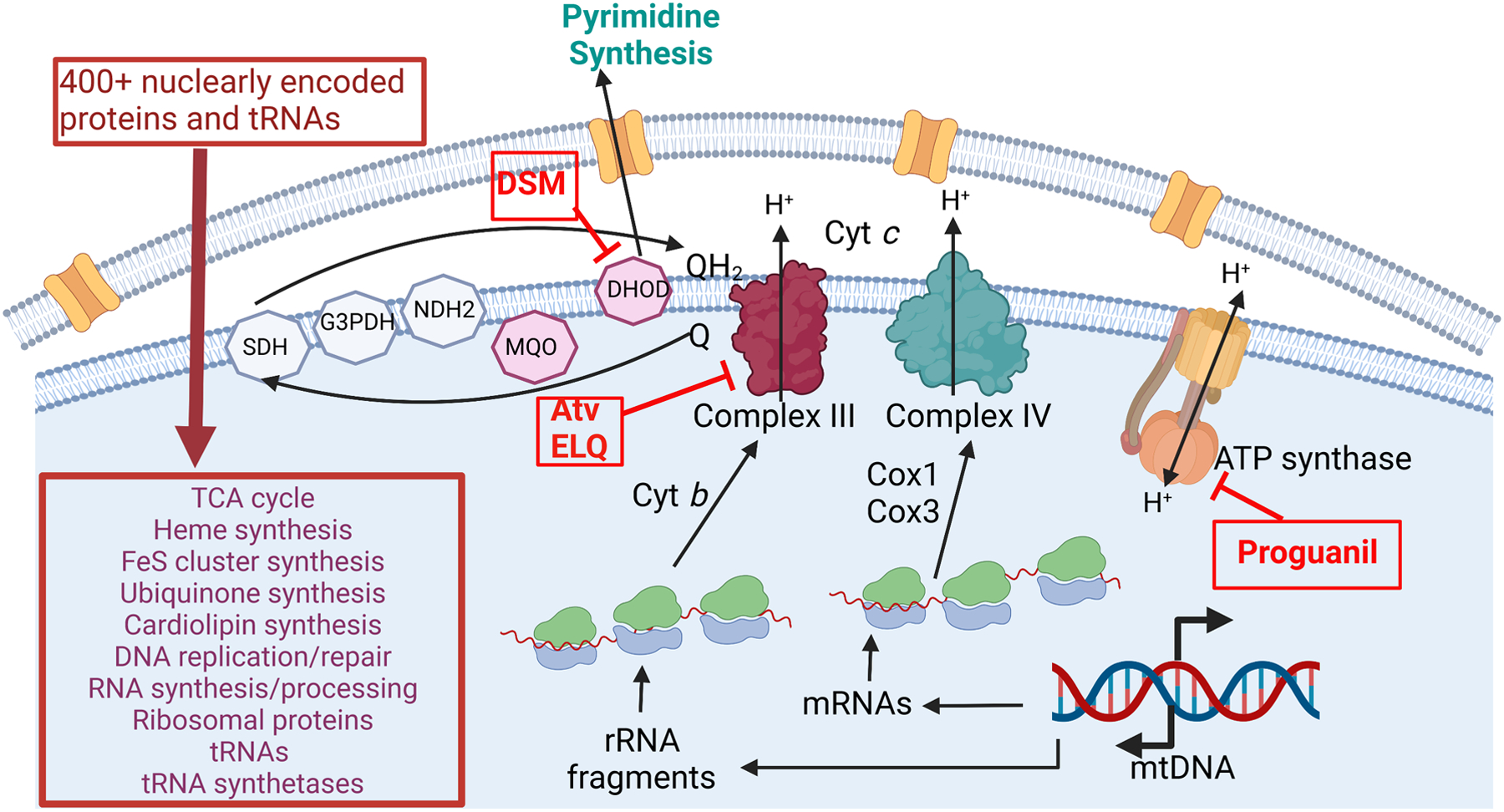
Maroon boxes depict the estimated 400+ nuclearly encoded proteins targeted to the mitochondrion and their associated biological processes. Drugs that target respiratory chain proteins are shown in red. Reduced ubiquinone (QH2) is generated by 5 dehydrogenases, of which SDH, MQO, G3PDH and NDH2 were found to be not essential in blood stages but DHOD is essential for its role in pyrimidine biosynthesis and is the target of DSM antimalarial compounds. QH2- is oxidized by Complex III, a step that is inhibited by atovaquone, ELQs and other inhibitors. Complex III transfers electrons to cytochrome c, which is then oxidized by Complex IV. Both Complexes III and IV pump H+ across the inner membrane, generating protonmotive force. Only 3 components of these complexes are encoded by the mtDNA: Cyt b, Cox1, and Cox3. These are translated by unusual ribosomes assembled from fragmented rRNAs encoded by mtDNA and ribosomal proteins encoded on the nuclear genome and imported into the mitochondrion. ATP synthase does not seem to be a significant source of ATP in blood stages but can work in reverse by hydrolyzing ATP to pump H+. This step is proposed to be targeted by proguanil, a component of antimalarial drug Malarone. rRNA= ribosomal RNA. Cyt b=cytochrome b. Cox1/3= cytochrome c oxidase 1/3. SDH = Succinate Dehydrogenase. G3PDH= glycerol-3-phosphate dehydrogenase. NDH2= type II NADH dehydrogenase. MQO= malate quinone oxidoreductase. DHOD= dihydroorotate dehydrogenase. ELQs = Endochin-Like Quinolones.
In the most virulent malaria parasite species P. falciparum, expression of the enzymes of most mitochondrial pathways including oxidative phosphorylation, is greatly reduced in the asexual blood stage, and the enzymes of the TCA cycle are dispensable (55). Nevertheless, the mtETC and ATP synthase complexes remain essential in this stage, as well as in the insect and liver stages. Expression of these complexes is greatly increased in the sexual blood stage gametocytes (27), although this stage is not fast growing and was thought to be less metabolically active. Stable isotope metabolomic experiments, however, suggest that gametocytes actually exhibit increased utilization of glucose with increased glycolytic flux directed through the TCA cycle (80). This increased expression of mitochondrial enzymes may be in preparation for the increased oxidative phosphorylation activity needed in the following insect stages. In Toxoplasma, on the other hand, the fast-growing, invasive tachyzoite stage has relatively robust oxidative phosphorylation activity, but upon stress from the immune system or treatment with certain drugs, including inhibitors of the mtETC, Toxoplasma undergoes stage conversion to the slow-growing bradyzoite cyst form with decreased reliance on mitochondrial oxidative phosphorylation and TCA cycle activities (20; 103).
Many Cryptosporidium parasites of the mammalian lower intestine and gregarine parasites of invertebrates have extremely reduced mitochondria related organelles called mitosomes that lack internal structure, as well as a TCA cycle, mtETC complexes and ATP synthase, but do contain a small number of ubiquinone-dependent dehydrogenases, such as NADH dehydrogenase and malate-quinone oxidoreductase, and an ubiquinol-utilizing alternative oxidase (1; 73; 86; 138). It is not clear how such a reduced mitosome would energize its membrane to provide for protein import, which, at a minimum, is required to carry out the common essential function of all mitochondria and mitosomes, i.e. iron-sulfur cluster ([Fe-S]) biogenesis. One possibility is a combination of external proton release concomitant to oxidation of ubiquinol and electrogenic exchange of ADP3- for ATP4- via the mitochondrial adenine nucleotide transporter.
Mitochondrial Targets for Antiparasitic Drugs
Early investigations using preparations enriched in mitochondria from malaria parasites showed that an antimalarial hydroxynaphthoquinone under development, subsequently named atovaquone, inhibited Complex III of the parasites (32). An examination of the sequence of P. falciparum cytochrome b, the central subunit of Complex III, revealed subtle but significant differences in the ubiquinol oxidation (Qo) and ubiquinone reduction (Qi) sites of the parasite cytochrome b compared to its host counterpart, which were proposed as the basis for selective toxicity of antimalarial drugs such as atovaquone (127). Atovaquone treatment also caused a collapse of the membrane potential across the inner membrane of parasite mitochondria (116). Subsequent studies revealed that mutations surrounding the Qo site in the parasite cytochrome b were responsible for resistance to atovaquone (58; 83; 87; 115). Structural studies using heterologous Complex III have supported the Qo site as the area of interaction with atovaquone (13). Because cytochrome b is largely conserved among mitochondriate apicomplexan parasites, atovaquone has been shown to be also effective against parasites such as Toxoplasma and Babesia (4; 19). While atovaquone is a potent drug, resistance to it arises rapidly when administered as a monotherapy (76). Therefore, a synergistic combination of atovaquone with a biguanide, proguanil, has been used for malaria prophylaxis and treatment (75; 77). The synergy between atovaquone and proguanil results from the ability of proguanil to inhibit a secondary pathway for generating membrane potential across the inner mitochondrial membrane in malaria parasites (117). Another series of compounds called endochin-like quinolones (ELQs) have been extensively investigated with one of them, ELQ-331, being designated a clinical candidate (89; 98; 118). ELQ-331 is a prodrug that is converted to ELQ-300, which has been shown to work at the Qi site of the parasite cytochrome b (89; 98). Other ELQ compounds have been shown to inhibit the growth of Toxoplasma and Babesia (26; 65). Medicinal chemistry exploration of the ELQ compounds has revealed that subtle differences in their structure can result in compounds that target either the Qi or Qo site (118). A combination of ELQ-300 and atovaquone to treat rodent malaria was found to be highly effective without engendering the emergence of resistant parasites (119). Such a combination holds promise for a malaria treatment with a greatly reduced chance of succumbing to the development of drug resistance. Long-lasting implants of atovaquone or ELQ-331 in mice have been shown to provide protection lasting months against experimental malaria (5; 114). Such a strategy has the potential for mass drug administration to control and eliminate malaria in endemic regions.
Plasmodium spp. are unable to salvage pyrimidines for their nucleic acid synthesis and must generate pyrimidines de novo (112). As mentioned earlier, an essential role of the parasite mtETC is to regenerate ubiquinone which is used as the electron acceptor for mitochondrially localized dihydroorotate dehydrogenase (DHODH), a critical enzyme in pyrimidine biosynthesis (102). Thus, DHODH has been shown to be an attractive target for antimalarial drug discovery (8). Several compounds have been shown to be potent inhibitors of parasite growth and a few have undergone clinical trials (74; 104). As with atovaquone, resistance to DHODH inhibitors also arise rapidly (133) and thus will require partner drugs to mitigate this risk.
Unique Features of Apicomplexan ATP synthase
F-type ATP synthases are large rotary molecular machines that use electrochemical potential across a membrane to synthesize ATP, playing a central role in the energy economy of a wide range of organisms (14; 130). They are constituted from two subcomplexes: the soluble F1 portion and the membrane -associated Fo portion. When the complete genome sequences of malaria parasites were assembled, genes encoding most of the F1 subcomplex could be detected but surprisingly highly conserved and essential genes encoding the Fo subcomplex subunits, with the exception of the subunit-c and OSCP, could not be detected through similarity searches (35); this was also observed in other apicomplexan genomes (84). In P. falciparum ATP synthase was found to form dimers and genes encoding its subunit were found to be refractory to disruption (7). More recent work, however, revealed the apicomplexan F1Fo ATP synthase to be highly divergent from its counterpart in other classes of organisms. Proteomic analyses of isolated ATP synthase from T. gondii carried out by two independent groups revealed the presence of multiple proteins in addition to the canonical F1 subunits and the subunit-c (48; 108). Nearly all these new proteins were conserved in other apicomplexan parasites as well as other Myzozoans, except Cryptosporidium and gregarine spp. that lack mtDNA (48; 108). Remarkably, these newly discovered ATP synthase subunits did not have apparent orthologous proteins in ciliates (6; 123). These studies indicated that the Myzozoa branch of the Alveolata clade possess a central bioenergetic complex that had a divergent evolutionary provenance. These myzozoa-specific subunits of the ATP synthase are divergent even from their closest sister clade of ciliates. One possible origin of these subunits could be from the ATP synthase that was part of the mitochondrion or the plastid belonging to the alga that was engulfed by the progenitor of myzozoans at the transitory stage (Fig. 1A). We acknowledge that there is no phylogenetic evidence supporting this proposition, but that could be due to an absence of sequence information from the algal progenitor. Chimeric origins of critical pathways are known to exist among unicellular parasites.
The unusual nature of the ATP synthase of these organisms became more apparent with the determination of a high-resolution cryogenic electron microscopic (cryo-EM) structure of the T. gondii complex (Fig. 4) (94). The complex was observed both as dimers and hexamers (trimers of dimers). There were 32 different proteins in each monomer, 15 of which could be discerned as structural equivalents of canonical subunits seen in ATP synthases of model organisms. Remarkably, 17 subunits were Apicomplexa-specific (identified as ATPTG1–17, for ATP synthase subunit from T. gondii). While 14 such ATPTG were observed in previous proteomic studies, 3 were identified directly from the cryo-EM structure (ATPTG1, ATPTG7 and ATPTG16). A gentler extraction revealed the dimers come together to form trimers, thus constituting hexamers, which were more akin to their native configuration in the parasite mitochondria. In the T. gondii ATP synthase dimer, the angle between the monomers is a low 19°--very different from the 100° angle seen in yeast and mammalian complexes. Because dimerization of ATP synthase is responsible for inducing curvature in the inner mitochondrial membrane, this narrow angle in the T. gondii dimer would induce reduced curvature compared to the yeast and mammalian membranes. Cryo-electron tomography of mitochondrial membranes showed bulbous vesicles that were decorated with icosahedral arrangements of ATP synthase hexamers to form pentagonal pyramids localized to the regions of curvatures in the membranes, indicating that hexamer packing is likely the primary determinant of cristae morphology in T. gondii. Overall, this work (94) constitutes the first structural elucidation of any mitochondrial complex from an apicomplexan parasite and reveals in remarkable detail the divergence and intricacies of a critical molecular machine. Further studies using cryo-EM and cryo-electron tomography to investigate mitochondrial complexes and their arrangements in situ are likely to reveal further amazing details.
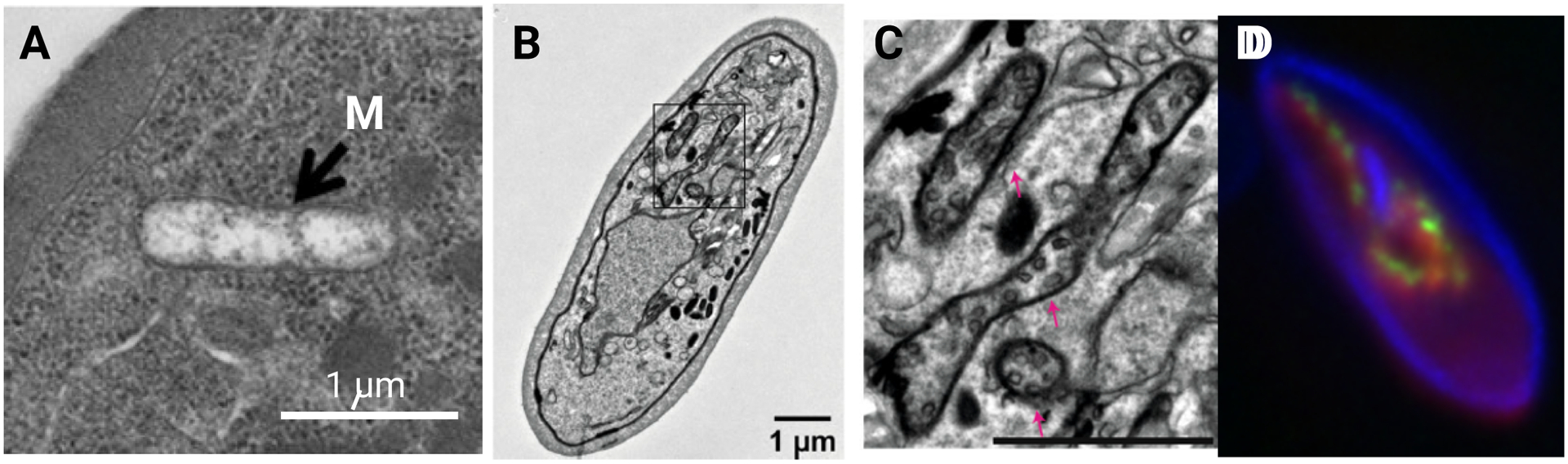
Figure 4. Cryo-EM structure of the ATP synthase complex from T. gondii.
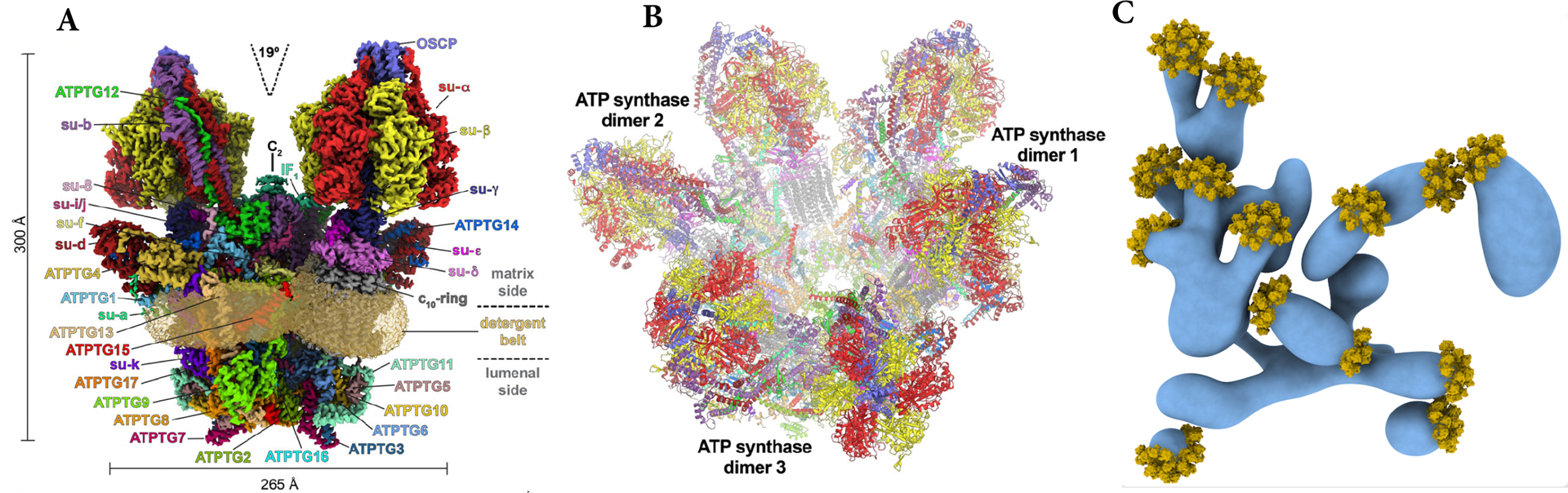
(A) A dimer of T. gondii ATP synthase. There are 32 subunit proteins in each monomer. Fifteen of these have structural equivalents in other ATP synthases and are identified with prefix “su-“; 17 other subunits are unique to T. gondii and are identified as ATPTG1 to ATPTG17. The monomers form a 19° angle relative to each other in the dimer, compared to 100° angle seen in the yeast and mammalian ATP synthase. (B) In native state, T. gondii ATP synthases form hexamers (trimers of dimers). (C) The hexamers associate at the bulbous mitochondrial inner membranes in pentameric pyramids that provide the curvature to the cristae. The figure is adapted from images in Muhleip et al. (ref. 94).
Unresolved Aspects of Apicomplexan Mitochondria
The unusual features of apicomplexan mitochondria summarized here raise many additional questions. These involve the machinery used by the parasites to manage the separate genetic systems relegated to the mitochondria. The remarkable conservation of the mtDNA in Plasmodium spp. would suggest a robust system of DNA replication, repair and recombination. While some of the components of this system have been identified, much remains to be elucidated. For instance, drug resistance mutations that arise in the mtDNAs of both Plasmodium and Toxoplasma quicky achieve fixation in the multiple copies of the genome such that wild-type sequences are no longer detected; such mutations are also very stable with no reversion to the wild type. This suggests a “copy-correction” machinery that remains unexplored. Plasmodium mtDNA molecules in linear arrays with intermediates suggesting strand invasions were observed in an early study. This would suggest extensive gene conversion events among the multi-copy mtDNA molecules, requiring a variety of enzymes that would mediate this process. At this point none of these mediators have been identified or investigated. Transcription regulation and processing of RNA encoded by mtDNA have also been underexplored. About 40 distinct RNA molecules are generated from the 6 kb mtDNA of Plasmodium spp. Enzymes involved in precise processing of these molecules are not known. The finding that a large number of proteins with RNA binding motifs amplified in Apicomplexa (RAP proteins) appear to be localized to the parasite mitochondrion suggest their potential role in post-transcriptional processing of mitochondrial RNA molecules. The most intriguing aspect of mitochondrial functions in apicomplexans is the manner in which the 3 mtETC proteins encoded by the mtDNA are translated. The structure of the mitochondrial ribosomes remains unresolved as is the process by which such ribosomes are assembled from multiple rRNA fragments and a large number of mitoribosomal proteins imported from the cytoplasm. With the availability of powerful new technologies, we anticipate many of these questions will be addressed in coming years.
Acknowledgements
We thank our colleagues for lively discussions. We thank Alexey Amunts and Lilach Sheiner for providing images included in Figure 4. The work in our laboratory is supported by grants R01 AI028398 and R01 AI100569 from NIH.
Literature Cited
- 1.Abrahamsen MS, Templeton TJ, Enomoto S, Abrahante JE, Zhu G, et al. 2004. Complete genome sequence of the apicomplexan, Cryptosporidium parvum. Science 304:441–5 [DOI] [PubMed] [Google Scholar]
- 2.Adl SM, Bass D, Lane CE, Lukes J, Schoch CL, et al. 2019. Revisions to the Classification, Nomenclature, and Diversity of Eukaryotes. J Eukaryot Microbiol 66:4–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adl SM, Simpson AG, Lane CE, Lukes J, Bass D, et al. 2012. The revised classification of eukaryotes. J Eukaryot Microbiol 59:429–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araujo FG, Huskinson J, Remington JS. 1991. Remarkable in vitro and in vivo activities of the hydroxynaphthoquinone 566C80 against tachyzoites and tissue cysts of Toxoplasma gondii. Antimicrob Agents Chemother 35:293–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bakshi RP, Tatham LM, Savage AC, Tripathi AK, Mlambo G, et al. 2018. Long-acting injectable atovaquone nanomedicines for malaria prophylaxis. Nature communications 9:315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balabaskaran Nina P, Dudkina NV, Kane LA, van Eyk JE, Boekema EJ, et al. 2010. Highly divergent mitochondrial ATP synthase complexes in Tetrahymena thermophila. PLoS Biol 8:e1000418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balabaskaran Nina P, Morrisey JM, Ganesan SM, Ke H, Pershing AM, et al. 2011. ATP synthase complex of Plasmodium falciparum: dimeric assembly in mitochondrial membranes and resistance to genetic disruption. J Biol Chem 286:41312–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baldwin J, Michnoff CH, Malmquist NA, White J, Roth MG, et al. 2005. High-throughput screening for potent and selective inhibitors of plasmodium falciparum dihydroorotate dehydrogenase. J Biol Chem 280:21847–53 [DOI] [PubMed] [Google Scholar]
- 9.Barylyuk K, Koreny L, Ke H, Butterworth S, Crook OM, et al. 2020. A Comprehensive Subcellular Atlas of the Toxoplasma Proteome via hyperLOPIT Provides Spatial Context for Protein Functions. Cell Host Microbe 28:752–66 e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berna L, Marquez P, Cabrera A, Greif G, Francia ME, Robello C. 2021. Reevaluation of the Toxoplasma gondii and Neospora caninum genomes reveals misassembly, karyotype differences, and chromosomal rearrangements. Genome Res 31:823–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berna L, Rego N, Francia ME. 2021. The Elusive Mitochondrial Genomes of Apicomplexa: Where Are We Now? Front Microbiol 12:751775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bieri P, Greber BJ, Ban N. 2018. High-resolution structures of mitochondrial ribosomes and their functional implications. Curr Opin Struct Biol 49:44–53 [DOI] [PubMed] [Google Scholar]
- 13.Birth D, Kao WC, Hunte C. 2014. Structural analysis of atovaquone-inhibited cytochrome bc1 complex reveals the molecular basis of antimalarial drug action. Nature communications 5:4029. [DOI] [PubMed] [Google Scholar]
- 14.Boyer PD. 1997. The ATP synthase--a splendid molecular machine. Annu Rev Biochem 66:717–49 [DOI] [PubMed] [Google Scholar]
- 15.Braun HP. 2021. The two roles of complex III in plants. eLife 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Braun HP, Schmitz UK. 1995. Are the ‘core’ proteins of the mitochondrial bc1 complex evolutionary relics of a processing protease? Trends Biochem Sci 20:171–5 [DOI] [PubMed] [Google Scholar]
- 17.Bushell E, Gomes AR, Sanderson T, Anar B, Girling G, et al. 2017. Functional Profiling of a Plasmodium Genome Reveals an Abundance of Essential Genes. Cell 170:260–72 e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, et al. 2002. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature 419:512–9 [DOI] [PubMed] [Google Scholar]
- 19.Chiu JE, Renard I, Pal AC, Singh P, Vydyam P, et al. 2021. Effective Therapy Targeting Cytochrome bc(1) Prevents Babesia Erythrocytic Development and Protects from Lethal Infection. Antimicrob Agents Chemother 65:e0066221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christiansen C, Maus D, Hoppenz E, Murillo-Leon M, Hoffmann T, et al. 2022. In vitro maturation of Toxoplasma gondii bradyzoites in human myotubes and their metabolomic characterization. Nature communications 13:1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christoforou A, Mulvey CM, Breckels LM, Geladaki A, Hurrell T, et al. 2016. A draft map of the mouse pluripotent stem cell spatial proteome. Nature communications 7:8992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cobbold SA, Santos JM, Ochoa A, Perlman DH, Llinas M. 2016. Proteome-wide analysis reveals widespread lysine acetylation of major protein complexes in the malaria parasite. Scientific reports 6:19722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danne JC, Gornik SG, Macrae JI, McConville MJ, Waller RF. 2013. Alveolate mitochondrial metabolic evolution: dinoflagellates force reassessment of the role of parasitism as a driver of change in apicomplexans. Mol Biol Evol 30:123–39 [DOI] [PubMed] [Google Scholar]
- 24.Dass S, Mather MW, Ke H. 2020. Divergent Mitochondrial Ribosomes in Unicellular Parasitic Protozoans. Trends Parasitol 36:318–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dass S, Mather MW, Morrisey JM, Ling L, Vaidya AB, Ke H. 2022. Transcriptional changes in Plasmodium falciparum upon conditional knock down of mitochondrial ribosomal proteins RSM22 and L23. PLoS One 17:e0274993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doggett JS, Nilsen A, Forquer I, Wegmann KW, Jones-Brando L, et al. 2012. Endochin-like quinolones are highly efficacious against acute and latent experimental toxoplasmosis. Proc Natl Acad Sci U S A 109:15936–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Evers F, Cabrera-Orefice A, Elurbe DM, Kea-Te Lindert M, Boltryk SD, et al. 2021. Composition and stage dynamics of mitochondrial complexes in Plasmodium falciparum. Nature communications 12:3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feagin JE, Gardner MJ, Williamson DH, Wilson RJ. 1991. The putative mitochondrial genome of Plasmodium falciparum. J Protozool 38:243–5 [DOI] [PubMed] [Google Scholar]
- 29.Feagin JE, Harrell MI, Lee JC, Coe KJ, Sands BH, et al. 2012. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum. PLoS One 7:e38320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feagin JE, Mericle BL, Werner E, Morris M. 1997. Identification of additional rRNA fragments encoded by the Plasmodium falciparum 6 kb element. Nucleic Acids Res 25:438–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fox BA, Bzik DJ. 2002. De novo pyrimidine biosynthesis is required for virulence of Toxoplasma gondii. Nature 415:926–9 [DOI] [PubMed] [Google Scholar]
- 32.Fry M, Pudney M. 1992. Site of action of the antimalarial hydroxynaphthoquinone, 2-[trans-4-(4’-chlorophenyl) cyclohexyl]-3-hydroxy-1,4-naphthoquinone (566C80). Biochem Pharmacol 43:1545–53 [DOI] [PubMed] [Google Scholar]
- 33.Gakh O, Cavadini P, Isaya G. 2002. Mitochondrial processing peptidases. Biochim Biophys Acta 1592:63–77 [DOI] [PubMed] [Google Scholar]
- 34.Gardner MJ, Bates PA, Ling IT, Moore DJ, McCready S, et al. 1988. Mitochondrial DNA of the human malarial parasite Plasmodium falciparum. Mol Biochem Parasitol 31:11–7 [DOI] [PubMed] [Google Scholar]
- 35.Gardner MJ, Hall N, Fung E, White O, Berriman M, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419:498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillespie DE, Salazar NA, Rehkopf DH, Feagin JE. 1999. The fragmented mitochondrial ribosomal RNAs of Plasmodium falciparum have short A tails. Nucleic Acids Res 27:2416–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gray MW. 1988. Organelle origins and ribosomal RNA. Biochem Cell Biol 66:325–48 [DOI] [PubMed] [Google Scholar]
- 38.Gray MW. 1993. Origin and evolution of organelle genomes. Curr Opin Genet Dev 3:884–90 [DOI] [PubMed] [Google Scholar]
- 39.Gray MW, Lang BF, Burger G. 2004. Mitochondria of Protists. Annu Rev Genet 38:477–524 [DOI] [PubMed] [Google Scholar]
- 40.Gutteridge WE, Dave D, Richards WH. 1979. Conversion of dihydroorotate to orotate in parasitic protozoa. Biochim Biophys Acta 582:390–401 [DOI] [PubMed] [Google Scholar]
- 41.Habib S, Vaishya S, Gupta K. 2016. Translation in Organelles of Apicomplexan Parasites. Trends Parasitol 32:939–52 [DOI] [PubMed] [Google Scholar]
- 42.Hayward JA, van Dooren GG. 2019. Same same, but different: Uncovering unique features of the mitochondrial respiratory chain of apicomplexans. Mol Biochem Parasitol 232:111204. [DOI] [PubMed] [Google Scholar]
- 43.Hikosaka K, Kita K, Tanabe K. 2013. Diversity of mitochondrial genome structure in the phylum Apicomplexa. Mol Biochem Parasitol 188:26–33 [DOI] [PubMed] [Google Scholar]
- 44.Hikosaka K, Nakai Y, Watanabe Y, Tachibana S, Arisue N, et al. 2011. Concatenated mitochondrial DNA of the coccidian parasite Eimeria tenella. Mitochondrion 11:273–8 [DOI] [PubMed] [Google Scholar]
- 45.Hikosaka K, Watanabe Y, Tsuji N, Kita K, Kishine H, et al. 2010. Divergence of the mitochondrial genome structure in the apicomplexan parasites, Babesia and Theileria. Mol Biol Evol 27:1107–16 [DOI] [PubMed] [Google Scholar]
- 46.Hollin T, Abel S, Falla A, Pasaje CFA, Bhatia A, et al. 2022. Functional genomics of RAP proteins and their role in mitoribosome regulation in Plasmodium falciparum. Nature communications 13:1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollin T, Jaroszewski L, Stajich JE, Godzik A, Le Roch KG. 2021. Identification and phylogenetic analysis of RNA binding domain abundant in apicomplexans or RAP proteins. Microb Genom 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huet D, Rajendran E, van Dooren GG, Lourido S. 2018. Identification of cryptic subunits from an apicomplexan ATP synthase. eLife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Janouskovec J, Paskerova GG, Miroliubova TS, Mikhailov KV, Birley T, et al. 2019. Apicomplexan-like parasites are polyphyletic and widely but selectively dependent on cryptic plastid organelles. eLife 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ji YE, Mericle BL, Rehkopf DH, Anderson JD, Feagin JE. 1996. The Plasmodium falciparum 6 kb element is polycistronically transcribed. Mol Biochem Parasitol 81:211–23 [DOI] [PubMed] [Google Scholar]
- 51.Joseph JT, Aldritt SM, Unnasch T, Puijalon O, Wirth DF. 1989. Characterization of a conserved extrachromosomal element isolated from the avian malarial parasite Plasmodium gallinaceum. Mol Cell Biol 9:3621–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kairo A, Fairlamb AH, Gobright E, Nene V. 1994. A 7.1 kb linear DNA molecule of Theileria parva has scrambled rDNA sequences and open reading frames for mitochondrially encoded proteins. Embo J 13:898–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kamikawa R, Inagaki Y, Sako Y. 2007. Fragmentation of mitochondrial large subunit rRNA in the dinoflagellate Alexandrium catenella and the evolution of rRNA structure in alveolate mitochondria. Protist 158:239–45 [DOI] [PubMed] [Google Scholar]
- 54.Ke H, Dass S, Morrisey JM, Mather MW, Vaidya AB. 2018. The mitochondrial ribosomal protein L13 is critical for the structural and functional integrity of the mitochondrion in Plasmodium falciparum. J Biol Chem 293:8128–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ke H, Lewis IA, Morrisey JM, McLean KJ, Ganesan SM, et al. 2015. Genetic Investigation of Tricarboxylic Acid Metabolism during the Plasmodium falciparum Life Cycle. Cell reports 11:164–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keeling PJ. 2013. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Annu Rev Plant Biol 64:583–607 [DOI] [PubMed] [Google Scholar]
- 57.Keeling PJ, Burki F. 2019. Progress towards the Tree of Eukaryotes. Curr Biol 29:R808–R17 [DOI] [PubMed] [Google Scholar]
- 58.Kessl JJ, Lange BB, Merbitz-Zahradnik T, Zwicker K, Hill P, et al. 2003. Molecular basis for atovaquone binding to the cytochrome bc1 complex. J Biol Chem 278:31312–8 [DOI] [PubMed] [Google Scholar]
- 59.Klug D, Mair GR, Frischknecht F, Douglas RG. 2016. A small mitochondrial protein present in myzozoans is essential for malaria transmission. Open Biol 6:160034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kunova N, Havalova H, Ondrovicova G, Stojkovicova B, Bauer JA, et al. 2022. Mitochondrial Processing Peptidases-Structure, Function and the Role in Human Diseases. Int J Mol Sci 23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kwong WK, Del Campo J, Mathur V, Vermeij MJA, Keeling PJ. 2019. A widespread coral-infecting apicomplexan with chlorophyll biosynthesis genes. Nature 568:103–7 [DOI] [PubMed] [Google Scholar]
- 62.Kwong WK, Irwin NAT, Mathur V, Na I, Okamoto N, et al. 2021. Taxonomy of the Apicomplexan Symbionts of Coral, including Corallicolida ord. nov., Reassignment of the Genus Gemmocystis, and Description of New Species Corallicola aquarius gen. nov. sp. nov. and Anthozoaphila gnarlus gen. nov. sp. nov. J Eukaryot Microbiol:e12852. [DOI] [PubMed] [Google Scholar]
- 63.Lamb IM, Rios KT, Shukla A, Ahiya AI, Morrisey J, et al. 2022. Mitochondrially targeted proximity biotinylation and proteomic analysis in Plasmodium falciparum. PLoS One 17:e0273357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lane N, Martin W. 2010. The energetics of genome complexity. Nature 467:929–34 [DOI] [PubMed] [Google Scholar]
- 65.Lawres LA, Garg A, Kumar V, Bruzual I, Forquer IP, et al. 2016. Radical cure of experimental babesiosis in immunodeficient mice using a combination of an endochin-like quinolone and atovaquone. J Exp Med 213:1307–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee C, Kim KH, Cohen P. 2016. MOTS-c: A novel mitochondrial-derived peptide regulating muscle and fat metabolism. Free Radic Biol Med 100:182–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee C, Zeng J, Drew BG, Sallam T, Martin-Montalvo A, et al. 2015. The mitochondrial-derived peptide MOTS-c promotes metabolic homeostasis and reduces obesity and insulin resistance. Cell Metab 21:443–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J, Maga JA, Cermakian N, Cedergren R, Feagin JE. 2001. Identification and characterization of a Plasmodium falciparum RNA polymerase gene with similarity to mitochondrial RNA polymerases. Mol Biochem Parasitol 113:261–9 [DOI] [PubMed] [Google Scholar]
- 69.Lin RQ, Qiu LL, Liu GH, Wu XY, Weng YB, et al. 2011. Characterization of the complete mitochondrial genomes of five Eimeria species from domestic chickens. Gene 480:28–33 [DOI] [PubMed] [Google Scholar]
- 70.Ling L, Mulaka M, Munro J, Dass S, Mather MW, et al. 2020. Genetic ablation of the mitoribosome in the malaria parasite Plasmodium falciparum sensitizes it to antimalarials that target mitochondrial functions. J Biol Chem 295:7235–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu GH, Hou J, Weng YB, Song HQ, Li S, et al. 2012. The complete mitochondrial genome sequence of Eimeria mitis (Apicomplexa: Coccidia). Mitochondrial DNA 23:341–3 [DOI] [PubMed] [Google Scholar]
- 72.Liu GH, Tian SQ, Cui P, Fang SF, Wang CR, Zhu XQ. 2015. The complete mitochondrial genomes of five Eimeria species infecting domestic rabbits. Exp Parasitol 159:67–71 [DOI] [PubMed] [Google Scholar]
- 73.Liu S, Roellig DM, Guo Y, Li N, Frace MA, et al. 2016. Evolution of mitosome metabolism and invasion-related proteins in Cryptosporidium. BMC Genomics 17:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Llanos-Cuentas A, Casapia M, Chuquiyauri R, Hinojosa JC, Kerr N, et al. 2018. Antimalarial activity of single-dose DSM265, a novel plasmodium dihydroorotate dehydrogenase inhibitor, in patients with uncomplicated Plasmodium falciparum or Plasmodium vivax malaria infection: a proof-of-concept, open-label, phase 2a study. Lancet Infect Dis 18:874–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Looareesuwan S, Chulay JD, Canfield CJ, Hutchinson DB. 1999. Malarone (atovaquone and proguanil hydrochloride): a review of its clinical development for treatment of malaria. Malarone Clinical Trials Study Group. Am J Trop Med Hyg 60:533–41 [DOI] [PubMed] [Google Scholar]
- 76.Looareesuwan S, Viravan C, Webster HK, Kyle DE, Hutchinson DB, Canfield CJ. 1996. Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am J Trop Med Hyg 54:62–6 [DOI] [PubMed] [Google Scholar]
- 77.Looareesuwan S, Wilairatana P, Glanarongran R, Indravijit KA, Supeeranontha L, et al. 1999. Atovaquone and proguanil hydrochloride followed by primaquine for treatment of Plasmodium vivax malaria in Thailand. Trans R Soc Trop Med Hyg 93:637–40 [DOI] [PubMed] [Google Scholar]
- 78.Maclean AE, Bridges HR, Silva MF, Ding S, Ovciarikova J, et al. 2021. Complexome profile of Toxoplasma gondii mitochondria identifies divergent subunits of respiratory chain complexes including new subunits of cytochrome bc1 complex. PLoS Pathog 17:e1009301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Maclean AE, Hayward JA, Huet D, van Dooren GG, Sheiner L. 2022. The mystery of massive mitochondrial complexes: the apicomplexan respiratory chain. Trends Parasitol [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacRae JI, Dixon MW, Dearnley MK, Chua HH, Chambers JM, et al. 2013. Mitochondrial metabolism of sexual and asexual blood stages of the malaria parasite Plasmodium falciparum. BMC Biol 11:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.MacRae JI, Sheiner L, Nahid A, Tonkin C, Striepen B, McConville MJ. 2012. Mitochondrial metabolism of glucose and glutamine is required for intracellular growth of Toxoplasma gondii. Cell Host Microbe 12:682–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin W, Muller M. 1998. The hydrogen hypothesis for the first eukaryote. Nature 392:37–41 [DOI] [PubMed] [Google Scholar]
- 83.Mather MW, Darrouzet E, Valkova-Valchanova M, Cooley JW, McIntosh MT, et al. 2005. Uncovering the molecular mode of action of the antimalarial drug atovaquone using a bacterial system. J Biol Chem 280:27458–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mather MW, Henry KW, Vaidya AB. 2007. Mitochondrial drug targets in apicomplexan parasites. Curr Drug Targets 8:49–60 [DOI] [PubMed] [Google Scholar]
- 85.Mather MW, Vaidya AB. 2008. Mitochondria in malaria and related parasites: ancient, diverse and streamlined. J Bioenerg Biomembr 40:425–33 [DOI] [PubMed] [Google Scholar]
- 86.Mathur V, Wakeman KC, Keeling PJ. 2021. Parallel functional reduction in the mitochondria of apicomplexan parasites. Curr Biol 31:2920–8 e4 [DOI] [PubMed] [Google Scholar]
- 87.McFadden DC, Tomavo S, Berry EA, Boothroyd JC. 2000. Characterization of cytochrome b from Toxoplasma gondii and Q(o) domain mutations as a mechanism of atovaquone-resistance. Mol Biochem Parasitol 108:1–12 [DOI] [PubMed] [Google Scholar]
- 88.McIntosh MT, Srivastava R, Vaidya AB. 1998. Divergent evolutionary constraints on mitochondrial and nuclear genomes of malaria parasites. Mol Biochem Parasitol 95:69–80 [DOI] [PubMed] [Google Scholar]
- 89.Miley GP, Pou S, Winter R, Nilsen A, Li Y, et al. 2015. ELQ-300 prodrugs for enhanced delivery and single-dose cure of malaria. Antimicrob Agents Chemother 59:5555–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Miller B, Kim SJ, Kumagai H, Yen K, Cohen P. 2022. Mitochondria-derived peptides in aging and healthspan. J Clin Invest 132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mogi T, Kita K. 2009. Identification of mitochondrial Complex II subunits SDH3 and SDH4 and ATP synthase subunits a and b in Plasmodium spp. Mitochondrion 9:443–53 [DOI] [PubMed] [Google Scholar]
- 92.Mohring F, Rahbari M, Zechmann B, Rahlfs S, Przyborski JM, et al. 2017. Determination of glutathione redox potential and pH value in subcellular compartments of malaria parasites. Free Radic Biol Med 104:104–17 [DOI] [PubMed] [Google Scholar]
- 93.Morales J, Mogi T, Mineki S, Takashima E, Mineki R, et al. 2009. Novel mitochondrial complex II isolated from Trypanosoma cruzi is composed of 12 peptides including a heterodimeric Ip subunit. J Biol Chem 284:7255–6319122194 [Google Scholar]
- 94.Muhleip A, Kock Flygaard R, Ovciarikova J, Lacombe A, Fernandes P, et al. 2021. ATP synthase hexamer assemblies shape cristae of Toxoplasma mitochondria. Nature communications 12:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mulvey CM, Breckels LM, Geladaki A, Britovsek NK, Nightingale DJH, et al. 2017. Using hyperLOPIT to perform high-resolution mapping of the spatial proteome. Nat Protoc 12:1110–35 [DOI] [PubMed] [Google Scholar]
- 96.Namasivayam S, Baptista RP, Xiao W, Hall EM, Doggett JS, et al. 2021. A novel fragmented mitochondrial genome in the protist pathogen Toxoplasma gondii and related tissue coccidia. Genome Res 31:852–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Nene V, Morzaria S, Bishop R. 1998. Organisation and informational content of the Theileria parva genome. Mol Biochem Parasitol 95:1–8 [DOI] [PubMed] [Google Scholar]
- 98.Nilsen A, LaCrue AN, White KL, Forquer IP, Cross RM, et al. 2013. Quinolone-3-diarylethers: a new class of antimalarial drug. Science translational medicine 5:177ra37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Obornik M, Janouskovec J, Chrudimsky T, Lukes J. 2009. Evolution of the apicoplast and its hosts: from heterotrophy to autotrophy and back again. Int J Parasitol 39:1–12 [DOI] [PubMed] [Google Scholar]
- 100.Oppenheim RD, Creek DJ, Macrae JI, Modrzynska KK, Pino P, et al. 2014. BCKDH: the missing link in apicomplexan mitochondrial metabolism is required for full virulence of Toxoplasma gondii and Plasmodium berghei. PLoS Pathog 10:e1004263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ossorio PN, Sibley LD, Boothroyd JC. 1991. Mitochondrial-like DNA sequences flanked by direct and inverted repeats in the nuclear genome of Toxoplasma gondii. J Mol Biol 222:525–36 [DOI] [PubMed] [Google Scholar]
- 102.Painter HJ, Morrisey JM, Mather MW, Vaidya AB. 2007. Specific role of mitochondrial electron transport in blood-stage Plasmodium falciparum. Nature 446:88–91 [DOI] [PubMed] [Google Scholar]
- 103.Pamukcu S, Cerutti A, Bordat Y, Hem S, Rofidal V, Besteiro S. 2021. Differential contribution of two organelles of endosymbiotic origin to iron-sulfur cluster synthesis and overall fitness in Toxoplasma. PLoS Pathog 17:e1010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Phillips MA, Lotharius J, Marsh K, White J, Dayan A, et al. 2015. A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Science translational medicine 7:296ra111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Preiser PR, Wilson RJ, Moore PW, McCready S, Hajibagheri MA, et al. 1996. Recombination associated with replication of malarial mitochondrial DNA. Embo J 15:684–93 [PMC free article] [PubMed] [Google Scholar]
- 106.Ramrath DJF, Niemann M, Leibundgut M, Bieri P, Prange C, et al. 2018. Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Science 362 [DOI] [PubMed] [Google Scholar]
- 107.Rudashevskaya EL, Sickmann A, Markoutsa S. 2016. Global profiling of protein complexes: current approaches and their perspective in biomedical research. Expert Rev Proteomics 13:951–64 [DOI] [PubMed] [Google Scholar]
- 108.Salunke R, Mourier T, Banerjee M, Pain A, Shanmugam D. 2018. Highly diverged novel subunit composition of apicomplexan F-type ATP synthase identified from Toxoplasma gondii. PLoS Biol 16:e2006128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schagger H, Pfeiffer K. 2000. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. EMBO J 19:1777–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Schilling B, Murray J, Yoo CB, Row RH, Cusack MP, et al. 2006. Proteomic analysis of succinate dehydrogenase and ubiquinol-cytochrome c reductase (Complex II and III) isolated by immunoprecipitation from bovine and mouse heart mitochondria. Biochim Biophys Acta 1762:213–22 [DOI] [PubMed] [Google Scholar]
- 111.Seidi A, Muellner-Wong LS, Rajendran E, Tjhin ET, Dagley LF, et al. 2018. Elucidating the mitochondrial proteome of Toxoplasma gondii reveals the presence of a divergent cytochrome c oxidase. eLife 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sherman IW. 1979. Biochemistry of Plasmodium (malarial parasites). Microbiol Rev 43:453–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Shikha S, Silva MF, Sheiner L. 2022. Identification and Validation of Toxoplasma gondii Mitoribosomal Large Subunit Components. Microorganisms 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Smilkstein MJ, Pou S, Krollenbrock A, Bleyle LA, Dodean RA, et al. 2019. ELQ-331 as a prototype for extremely durable chemoprotection against malaria. Malar J 18:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Srivastava IK, Morrisey JM, Darrouzet E, Daldal F, Vaidya AB. 1999. Resistance mutations reveal the atovaquone-binding domain of cytochrome b in malaria parasites. Mol Microbiol 33:704–11 [DOI] [PubMed] [Google Scholar]
- 116.Srivastava IK, Rottenberg H, Vaidya AB. 1997. Atovaquone, a broad spectrum antiparasitic drug, collapses mitochondrial membrane potential in a malarial parasite. J Biol Chem 272:3961–6 [DOI] [PubMed] [Google Scholar]
- 117.Srivastava IK, Vaidya AB. 1999. A mechanism for the synergistic antimalarial action of atovaquone and proguanil. Antimicrob Agents Chemother 43:1334–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Stickles AM, de Almeida MJ, Morrisey JM, Sheridan KA, Forquer IP, et al. 2015. Subtle changes in endochin-like quinolone structure alter the site of inhibition within the cytochrome bc1 complex of Plasmodium falciparum. Antimicrob Agents Chemother 59:1977–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stickles AM, Smilkstein MJ, Morrisey JM, Li Y, Forquer IP, et al. 2016. Atovaquone and ELQ-300 Combination Therapy as a Novel Dual-Site Cytochrome bc1 Inhibition Strategy for Malaria. Antimicrob Agents Chemother 60:4853–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Suplick K, Morrisey J, Vaidya AB. 1990. Complex transcription from the extrachromosomal DNA encoding mitochondrial functions of Plasmodium yoelii. Mol Cell Biol 10:6381–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Takeo S, Kokaze A, Ng CS, Mizuchi D, Watanabe JI, et al. 2000. Succinate dehydrogenase in Plasmodium falciparum mitochondria: molecular characterization of the SDHA and SDHB genes for the catalytic subunits, the flavoprotein (Fp) and iron-sulfur (Ip) subunits. Mol Biochem Parasitol 107:191–205 [DOI] [PubMed] [Google Scholar]
- 122.Tanaka TQ, Hirai M, Watanabe Y, Kita K. 2012. Toward understanding the role of mitochondrial complex II in the intraerythrocytic stages of Plasmodium falciparum: gene targeting of the Fp subunit. Parasitol Int 61:726–8 [DOI] [PubMed] [Google Scholar]
- 123.Tobiasson V, Amunts A. 2020. Ciliate mitoribosome illuminates evolutionary steps of mitochondrial translation. eLife 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Usey MM, Huet D. 2022. Parasite powerhouse: A review of the Toxoplasma gondii mitochondrion. J Eukaryot Microbiol:e12906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vaidya AB, Akella R, Suplick K. 1989. Sequences similar to genes for two mitochondrial proteins and portions of ribosomal RNA in tandemly arrayed 6-kilobase-pair DNA of a malarial parasite. Mol Biochem Parasitol 35:97–107 [DOI] [PubMed] [Google Scholar]
- 126.Vaidya AB, Arasu P. 1987. Tandemly arranged gene clusters of malarial parasites that are highly conserved and transcribed. Mol Biochem Parasitol 22:249–57 [DOI] [PubMed] [Google Scholar]
- 127.Vaidya AB, Lashgari MS, Pologe LG, Morrisey J. 1993. Structural features of Plasmodium cytochrome b that may underlie susceptibility to 8-aminoquinolines and hydroxynaphthoquinones. Mol Biochem Parasitol 58:33–42 [DOI] [PubMed] [Google Scholar]
- 128.Vaidya AB, Mather MW. 2009. Mitochondrial evolution and functions in malaria parasites. Annu Rev Microbiol 63:249–67 [DOI] [PubMed] [Google Scholar]
- 129.van Dooren GG, Striepen B. 2013. The algal past and parasite present of the apicoplast. Annu Rev Microbiol 67:271–89 [DOI] [PubMed] [Google Scholar]
- 130.Walker JE. 1994. The regulation of catalysis in ATP synthase. Curr Opin Struct Biol 4:912–8 [DOI] [PubMed] [Google Scholar]
- 131.Waller RF, Keeling PJ. 2006. Alveolate and chlorophycean mitochondrial cox2 genes split twice independently. Gene 383:33–7 [DOI] [PubMed] [Google Scholar]
- 132.Waltz F, Salinas-Giege T, Englmeier R, Meichel H, Soufari H, et al. 2021. How to build a ribosome from RNA fragments in Chlamydomonas mitochondria. Nature communications 12:7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.White J, Dhingra SK, Deng X, El Mazouni F, Lee MCS, et al. 2019. Identification and Mechanistic Understanding of Dihydroorotate Dehydrogenase Point Mutations in Plasmodium falciparum that Confer in Vitro Resistance to the Clinical Candidate DSM265. ACS Infect Dis 5:90–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Williamson DH, Preiser PR, Wilson RJ. 1996. Organelle DNAs: The bit players in malaria parasite DNA replication. Parasitol Today 12:357–62 [DOI] [PubMed] [Google Scholar]
- 135.Williamson DH, Wilson RJ, Bates PA, McCready S, Perler F, Qiang BU. 1985. Nuclear and mitochondrial DNA of the primate malarial parasite Plasmodium knowlesi. Mol Biochem Parasitol 14:199–209 [DOI] [PubMed] [Google Scholar]
- 136.Wittig I, Braun HP, Schagger H. 2006. Blue native PAGE. Nat Protoc 1:418–28 [DOI] [PubMed] [Google Scholar]
- 137.Xia D, Yu CA, Kim H, Xia JZ, Kachurin AM, et al. 1997. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science 277:60–6 [DOI] [PubMed] [Google Scholar]
- 138.Xu P, Widmer G, Wang Y, Ozaki LS, Alves JM, et al. 2004. The genome of Cryptosporidium hominis. Nature 431:1107–12 [DOI] [PubMed] [Google Scholar]
- 139.Yoon TK, Lee CH, Kwon O, Kim MS. 2022. Exercise, Mitohormesis, and Mitochondrial ORF of the 12S rRNA Type-C (MOTS-c). Diabetes Metab J 46:402–13 [DOI] [PMC free article] [PubMed] [Google Scholar]


