Abstract
The herpes simplex virus 1 UL3 and UL4 open reading frames are expressed late in infection and are not essential for viral replication in cultured cells in vitro. An earlier report showed that the UL4 protein colocalizes with the products of the α22/US1.5 genes in small nuclear dense bodies. Here we report that the UL3 protein also colocalized in these small nuclear dense bodies and the localization of UL3 and UL4 proteins in these bodies required the presence of α22/US1.5 genes. In cells infected with a mutant lacking intact α22/US1.5 genes, UL3 was diffused throughout the nucleus even though the overall accumulation of the γ2 UL3 protein was decreased. The results suggest that ICP22 acts both as a regulator of UL3 accumulation and as the structural component and anchor of these small dense nuclear bodies.
Of the 84 open reading frames of herpes simplex virus 1 (HSV-1) known to be expressed in infected cells, more than half are dispensable for viral replication at least in some cell lines maintained in culture (18, 19). These open reading frames appear to be essential for viral survival in nature, since viruses lacking these genes have not been isolated and in experimental animal systems mutants lacking these genes tend to be attenuated. In many instances the functions of these genes are difficult to study, since they appear to have no obvious phenotype in infected cells. This report is an extension of recently published studies on two such genes, UL3 and UL4 (8, 12).
Both UL3 and UL4 are dispensable in all cell lines tested to date (3; N. S. Markovitz and B. Roizman, unpublished data). Neither gene appears to play a role in pathogenicity or latent infections caused by HSV-1 (9; J. D. Baines and B. Roizman, unpublished data). The UL3 gene products are expressed as multiple isoforms: two isoforms result from alternate translation initiation of the UL3 open reading frame; the phospho-isoforms result from posttranslational processing by UL13 viral protein kinase and by at least one additional, as yet unidentified protein kinase (7, 12). The HSV-2 UL3 phosphoprotein is present in multiple isoforms and localizes in the nucleus after the onset of DNA synthesis (22). Both UL3 and UL4 are late, or γ2 proteins whose synthesis depends on the onset of viral DNA replication. HSV-1 and HSV-2 UL4 proteins have been described elsewhere (5, 8, 23). Recently this laboratory reported that UL4 colocalizes with the products of the α22/US1.5 genes in the nucleus at 17 h after infection (8).
The α22/US1.5 gene is located at the left terminus of the unique short sequence (US) when the genome is presented in its prototype orientation (4, 15, 21). α22 encodes the 420-amino-acid ICP22. The US1.5 protein is expressed from a shorter mRNA and consists of approximately 60% of carboxyl-terminal amino acid sequence of the ICP22 (4). The shared portion of the α22/US1.5 gene products is required for efficient viral replication in rodent and rabbit cell lines (14), and its presence enhances the expression of a subset of γ2 genes (14, 16, 17, 20). Although we refer to ICP22 in the colocalization studies, our studies do not differentiate between ICP22 and the US1.5 protein.
The studies described in this report utilized the F strain of HSV-1 [HSV-1(F)]. In the recombinant viruses R8105 and R4660, a sequence encoding an epitope reacting with an available monoclonal antibody was inserted in frame into the coding sequences of UL3 and UL4 genes, respectively (8, 12). The monoclonal antibody CH28-2 (Goodwin Cancer Research Institute, Plantation, Fla.) reacts with an epitope mapped to the glycoprotein B of human cytomegalovirus and does not react with HSV-1 proteins (11). In recombinant virus R325, the carboxyl-terminal 220 amino acids of the α22/US1.5 gene had been deleted (15). In the recombinant R4968, the deletion in α22 has been restored (13). The origin and maintenance of the HEp-2 and rabbit skin cells have been described elsewhere (8, 12). The antisera used in these studies have also been reported elsewhere. Briefly, rabbit antisera were raised against a chimeric protein containing amino acids 44 to 235 of the HSV-1 UL3 protein and, separately, against the entire HSV-1 UL4 protein (8, 12). Rabbit antiserum (R77) to ICP22 was made against the amino-terminal domain of ICP22 and does not react with the US1.5 protein (2). The monoclonal antibody to ICP4 (H640) described elsewhere (1) was purchased from Goodwin Cancer Research Institute.
In the first of a series of immunofluorescence studies (data not shown), we noted that the pattern of UL3 fluorescence was similar to that of small dense nuclear bodies containing both ICP22 and UL4 (8). Specifically, an earlier report showed that UL4 protein colocalized with ICP22 in small dense nuclear structures (8). To verify this conclusion, rabbit skin cells grown in wells on glass slides were exposed to approximately 10 PFU of HSV-1(F) or of recombinant virus per cell and incubated at 37°C. The cells were fixed at 17 h after infection in methanol and processed as described elsewhere (8). The results were as follows.
(i) The UL3 protein localized in small dense nuclear bodies (Fig. 1A to C). Furthermore, this experiment showed that identical images were obtained with antibody to the tag (fluorescein isothiocyanate [FITC]) and the authentic antibody to the UL3 protein (Texas red). Thus, as was the case for UL4 (8), the virus carrying the tagged UL3 could be used as a substitute for the untagged virus in double-label studies.
FIG. 1.
Colocalization of ICP22 and UL4 protein with UL3 protein. Rabbit skin cells were exposed to HSV-1 recombinant virus R8105 (A to I) or R4660 (J to L) at a ratio of 10 PFU/cell. At 17 h after infection, the cells were fixed with cold ethanol and reacted with the indicated antibodies. Primary antibodies were diluted 1:500 to 1:1,000. Goat anti-rabbit immunoglobulin conjugated to Texas red (Molecular Probes, Eugene, Oreg.) was used to reveal the antigen reacting to the rabbit antisera against UL3, UL4, and ICP22, while anti-mouse immunoglobulin conjugated to FITC (Sigma) was used to visualize the mouse monoclonal antibody CH28-2. The slides were examined under a Zeiss confocal fluorescence microscope, and laser-scanned digitized images of the cells stained with fluorescent antibody were acquired with software provided with the Zeiss confocal microscope. Ab., antibody.
(ii) The UL3 proteins colocalized with ICP22 in cells infected with R8105 (UL3 epitope tagged) and reacted with the antibody against ICP22 (R77) and the anti-tag monoclonal antibody (Fig. 1D to F). In this instance the superimposition shown in Fig. 1F was not nearly as complete as that seen in Fig. 1C. It is noteworthy that in some of the dense bodies the UL3 protein was either more diffuse or more abundant than ICP22.
(iii) The amount of UL4 antigen was significantly lower than that of the UL3 protein in cells infected with R8105 (UL3 epitope tagged) and reacted with the rabbit polyclonal antibody to UL4 and to the anti-tag monoclonal antibody. Nevertheless, the UL3 and UL4 proteins colocalized, especially in dense bodies in which the UL4 protein was readily detectable (Fig. 1G to I). The reverse experiment yielded results consistent with that conclusion. Specifically, in cells infected with R4660 (tagged UL4) and reacted with the polyclonal rabbit antibody to UL3 and the anti-tag monoclonal antibody, the signal for the tagged UL4 protein was stronger than that obtained in Fig. 1G with the anti-UL4 antibody. In this instance the two images produced by the two antibodies were superimposable.
(iv) We have selected rabbit skin cells for these studies on the basis of the observation that the nuclear structures containing UL3 and UL4 were larger or more readily detectable. Studies on infected HEp-2 cells yielded results identical to those described above (data not shown).
In earlier reports we described the properties of the antibodies to the UL3, UL4, and ICP22 proteins (2, 8, 12). Relevant to the studies described here, the antibodies made against UL3 and UL4 are specific for each protein and do not cross-react. In both immunofluorescence and immunoblot assays, UL3 and UL4 proteins were not visualized when the anti-UL4 and UL3 antibodies, respectively, were used (Fig. 2) (8, 12), and UL3 and UL4 do not have protein sequence homology. We conclude from these studies that ICP22, UL3, and UL4 proteins colocalize in nuclear structures of infected cells.
FIG. 2.
Immunoblot of electrophoretically separated proteins from cell lysates of rabbit skin cells (RSC) (A) or HEp-2 cells (B) probed with antibody (Ab) to UL3 (lanes 1 to 4) or UL4 (lanes 5 to 8). Lysates of cells mock infected or infected with HSV-1(F) or the indicated recombinant virus were prepared as described elsewhere (12).
The second series of experiments was prompted by earlier observations that ICP22 enhances the expression of a subset of γ2 proteins and that modification of the phosphoprotein isoforms of UL3 is mediated by the viral UL13 protein kinase. In numerous studies, this laboratory has found a link between UL13 protein kinase and ICP22 (14, 16, 17, 20). In every case examined, the phenotype of UL13− mutants was similar to that of α22−/US1.5− mutants (13, 14, 16, 17). To unravel the effect of ICP22 on the accumulation of UL3 and UL4, electrophoretically separated lysates of mock-infected cells or cells infected with HSV-1(F), R325 (Δα22/US1.5), or the repair virus R4968 were transferred to nitrocellulose sheets and probed with antibodies to either UL3 or UL4. The results of this experiment shown in Fig. 2 indicate that, consistent with earlier results (Markovitz and Roizman, unpublished), in cells infected with the R325 (Δα22/US1.5) virus, all isoforms of UL3 protein were present but in smaller amounts than in cells infected with the wild-type virus or with the recombinant virus in which the α22 gene had been repaired (R4968) (Fig. 2, lanes 1 to 4), whereas the levels of UL4 protein showed a less dramatic decrease (Fig. 2, lanes 5 to 8).
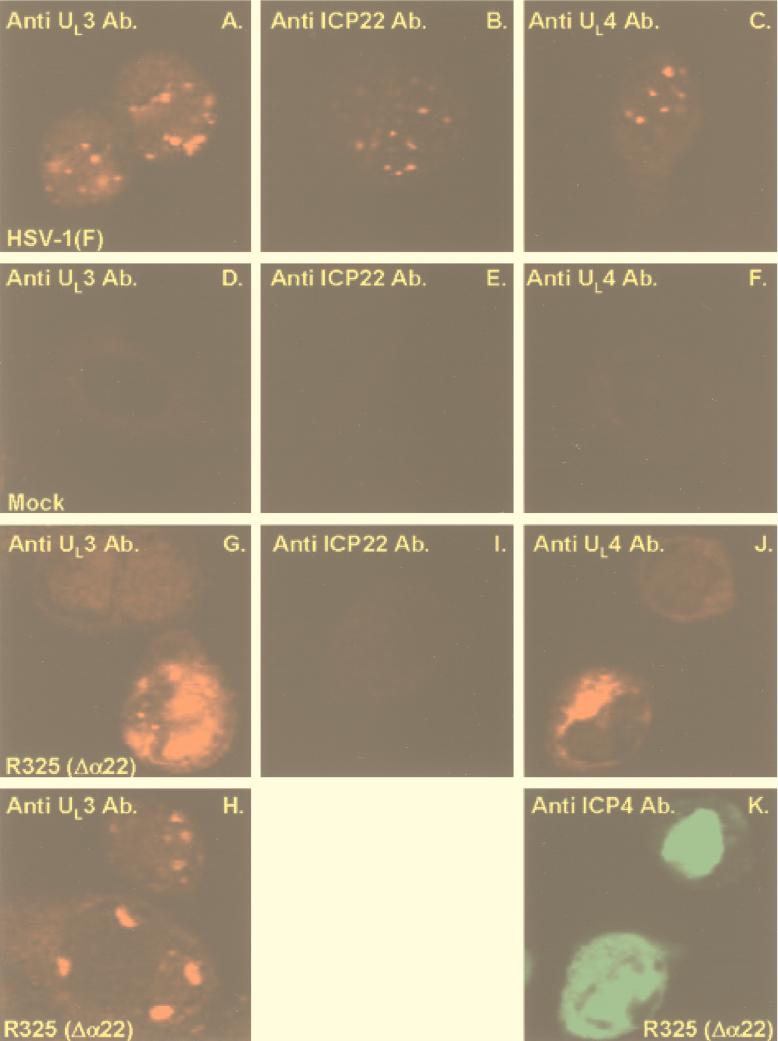
Since UL4 and UL3 proteins colocalize with ICP22 and deletion of ICP22 resulted in a decreased accumulation of UL3 and, to a lesser extent, of UL4 proteins, the question arose as to whether the localization of these proteins was affected in cells infected with the recombinant virus R325 from which the carboxyl-terminal 220 codons of the α22 gene had been deleted. In this series of experiments, rabbit skin cells were mock infected or infected with HSV-1(F) or recombinant R325 virus and reacted with single antibodies (Fig. 3A to I) or UL4 and ICP4 antibodies (Fig. 3J and K). The results were as follows.
FIG. 3.
Localization of UL3 and UL4 proteins and of ICP22 in infected cells. Rabbit skin cells were mock infected or exposed to 10 PFU of HSV-1(F) or R325 per cell and processed as described in the legend to Fig. 1. Anti-mouse immunoglobulin conjugated to FITC (Sigma) was used to visualize the mouse monoclonal antibody to ICP4 (K). Ab., antibody.
(i) As expected, neither UL3, UL4, or ICP22 antibodies reacted with mock-infected cells (Fig. 3D to F).
(ii) In HSV-1(F)-infected cells, polyclonal antibodies to UL3, UL4, and ICP22 reacted with their respective antigens in small dense nuclear structures (Fig. 3A to C).
(iii) In R325 mutant-infected cells, the expression patterns of UL3 and UL4 differed from that observed in HSV-1(F)-infected cells in two respects. First, we observed that fewer infected rabbit skin cells expressed detectable UL3 protein—a finding consistent with the observation that UL3 protein accumulated in smaller amounts in cells infected with R325 recombinant virus than in cells infected with wild-type virus. The fraction of cells expressing UL4 was also decreased in R325-infected cells. To verify that the difference in the proportion of cells expressing UL3 and UL4 in R325-infected cells was related to relative UL3 and UL4 levels and not to a difference in infectivity, replicate wells were reacted with antibodies to the ICP4 protein. Figure 3J and K show the same two cells. While virtually all cells were infected, as evidenced by the presence of the ICP4 protein (Fig. 3K), only some cells were positive for UL3 (data not shown) or UL4 (Fig. 3J) protein expression. Thus, the microscopic fields shown in Fig. 3J and K are identical; only one of the two infected cells shown in Fig. 3K contains UL4 protein. The second interesting finding was that in the cells positive for UL3 protein, the distribution of UL3 was more diffuse, formed irregularly shaped structures that occupied a significantly larger portion of the nucleus (Fig. 3G and H). A similar phenomenon was observed for UL4 (Fig. 3J).
(iv) Studies on infected HEp-2 cells yielded results identical to those described above.
A curious phenomenon observed in these studies is that although UL3 protein accumulated in reduced quantities in cells infected with α22−/US1.5− virus-infected cells, the quantity of UL3 protein as measured by immunofluorescence appeared to be greater in those cells expressing UL3 than that observed in cells infected with wild-type virus. One likely explanation is that in a dispersed mode, as seen in cells infected with α22−/US1.5− virus, the UL3 protein was far more accessible to its antibody than in the compressed mode in the small, dense nuclear structures of cells infected with wild-type virus. We conclude from the studies described in this report the following.
(i) Both UL3 and UL4 are late or γ2 genes dependent on viral DNA synthesis for their expression. Efficient accumulation of UL3 protein, nascent or posttranslationally modified, requires the expression of UL13 protein kinase (12) and of the α22/US1.5 genes. In the absence of the α22/US1.5 genes, neither UL3 nor UL4 localizes in the small dense nuclear structures seen in cells infected with wild-type virus. ICP22 by itself in transfection assays forms small dense nuclear bodies (V. Galvan and B. Roizman, unpublished data). Thus, no late protein is required for the formation of the dense bodies, since ICP22 alone is sufficient. Furthermore, the UL3 and UL4 genes do not have appreciable protein sequence homology. These results indicate that colocalization of UL3 and UL4 is directed by ICP22 rather than by the interaction of UL3 and UL4 proteins.
(ii) The function of these small dense nuclear bodies is not known. Earlier in infection, ICP22 localized in structures resembling the small dense nuclear structures described here. After the onset of viral DNA, ICP22 colocalizes with ICP4, RNA polymerase II, and nascent DNA and occupies a much larger volume than the small dense nuclear structures seen very early or very late in infection do (10). Late in infection, ICP22 again localizes in small dense nuclear structures along with UL3 and UL4. The observation that ICP22 directs the localization of the newly synthesized UL3 and UL4 proteins to such structures suggests that they play a role in the viral replicative cycle late in infection.
Acknowledgments
These studies were aided by grants from the National Cancer Institute (CA47451, CA71933, and CA78766) of the United States Public Health Service.
REFERENCES
- 1.Ackerman M, Braun D K, Pereira L, Roizman B. Characterization of herpes simplex virus 1 α proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ackermann M, Sarmiento M, Roizman B. Application of antibody to synthetic peptides for characterization of the intact and truncated α22 protein specified by herpes simplex virus 1 and the R325 α22− deletion mutant. J Virol. 1985;56:207–215. doi: 10.1128/jvi.56.1.207-215.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baines J D, Roizman B. The open reading frames UL3, UL4, UL10, and UL16 are dispensable for the replication of herpes simplex virus 1 in cell culture. J Virol. 1991;65:938–944. doi: 10.1128/jvi.65.2.938-944.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carter K L, Roizman B. The promoter and transcriptional unit of a novel herpes simplex virus 1 α gene are contained in, and encode a protein in frame with, the open reading frame of the α22 gene. J Virol. 1996;70:172–178. doi: 10.1128/jvi.70.1.172-178.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eide T, Marsden H S, Leib D A, Cunningham C, Davidson A J, Langeland N, Haarr L. Identification of the UL4 protein of herpes simplex virus type 1. J Gen Virol. 1998;79:3033–3038. doi: 10.1099/0022-1317-79-12-3033. [DOI] [PubMed] [Google Scholar]
- 6.Ejercito P M, Kieff E D, Roizman B. Characteristics of herpes simplex virus strains differing in their effect on social behavior of infected cells. J Gen Virol. 1968;2:357–364. doi: 10.1099/0022-1317-2-3-357. [DOI] [PubMed] [Google Scholar]
- 7.Ghiasi H, Perng G-C, Cai S, Nesburn A B, Wechsler S L. The UL3 open reading frame of herpes simplex virus type 1 codes for a phosphoprotein. Virus Res. 1996;44:137–142. doi: 10.1016/0168-1702(96)01330-5. [DOI] [PubMed] [Google Scholar]
- 8.Jahedi S, Markovitz N S, Filatov F, Roizman B. Colocalization of the herpes simplex virus 1 UL4 protein with infected cell protein 22 in small, dense nuclear structures formed prior to onset of DNA synthesis. J Virol. 1999;73:5132–5136. doi: 10.1128/jvi.73.6.5132-5136.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jun P Y, Strelow L I, Herman R C, Marsden H S, Eide T, Haarr L, Leib D A. The UL4 gene of herpes simplex virus type 1 is dispensable for latency, reactivation and pathogenesis in mice. J Gen Virol. 1998;79:1603–1611. doi: 10.1099/0022-1317-79-7-1603. [DOI] [PubMed] [Google Scholar]
- 10.Leopardi R, Ward P L, Ogle W O, Roizman B. Association of herpes simplex virus regulatory protein ICP22 with transcriptional complexes containing EAP, ICP4, RNA polymerase II, and viral DNA requires posttranslational modification by the UL13 protein kinase. J Virol. 1997;71:1133–1139. doi: 10.1128/jvi.71.2.1133-1139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F, Roizman B. The promoter, transcriptional unit, and coding sequence of herpes simplex virus 1 family 35 proteins are contained within and in frame with the UL26 open reading frame. J Virol. 1991;65:206–212. doi: 10.1128/jvi.65.1.206-212.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Markovitz N S, Filatov F, Roizman B. The UL3 protein of herpes simplex virus 1 is translated predominantly from the second in-frame methionine codon and is subject to at least two posttranslational modifications. J Virol. 1999;73:8010–8018. doi: 10.1128/jvi.73.10.8010-8018.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng T I, Chang Y E, Roizman B. Infected cell protein 22 of herpes simplex virus 1 regulates the expression of virion host shutoff gene UL41. Virology. 1997;234:226–234. doi: 10.1006/viro.1997.8659. [DOI] [PubMed] [Google Scholar]
- 14.Ogle W O, Roizman B. Functional anatomy of herpes simplex virus 1 overlapping genes encoding infected-cell protein 22 and US1.5 protein. J Virol. 1999;73:4305–4315. doi: 10.1128/jvi.73.5.4305-4315.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Post L E, Roizman B. A generalized technique for deletion of specific genes in large genomes: α gene 22 of herpes simplex virus 1 is not essential for growth. Cell. 1981;25:227–232. doi: 10.1016/0092-8674(81)90247-6. [DOI] [PubMed] [Google Scholar]
- 16.Purves F C, Ogle W O, Roizman B. Processing of the herpes simplex virus regulatory protein α22 mediated by the UL13 protein kinase determines the accumulation of a subset of α and γ mRNAs and proteins in infected cells. Proc Natl Acad Sci USA. 1993;90:6701–6705. doi: 10.1073/pnas.90.14.6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Purves F C, Roizman B. The UL13 gene of herpes simplex virus 1 encodes the functions for posttranslational processing associated with phosphorylation of the regulatory protein α22. Proc Natl Acad Sci USA. 1992;89:7310–7314. doi: 10.1073/pnas.89.16.7310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roizman B. The function of herpes simplex virus genes: a primer for genetic engineering of novel vectors. Proc Natl Acad Sci USA. 1996;93:11307–11312. doi: 10.1073/pnas.93.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roizman B, Sears A E. The replication of herpes simplex viruses. In: Fields B N, Knipe D M, Howley P, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. In Fields' virology. 3rd ed. New York, N.Y: Lippincott-Raven Press; 1996. pp. 2231–2295. [Google Scholar]
- 20.Sears A E, Halliburton I W, Meignier B, Silver A, Roizman B. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watson R J, Sullivan M, vande Woude G F. Structures of two spliced herpes simplex virus type 1 immediate-early mRNAs which map at the junctions of the unique and reiterated regions of the virus DNA S component. J Virol. 1981;37:431–444. doi: 10.1128/jvi.37.1.431-444.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worrad D M, Caradonna S. The herpes simplex virus type 2 UL3 open reading frame encodes a nuclear localizing phosphoprotein. Virology. 1993;195:364–376. doi: 10.1006/viro.1993.1386. [DOI] [PubMed] [Google Scholar]
- 23.Yamada H, Jiang Y-M, Oshima S, Wada K, Goshima F, Daikoku T, Nishiyama Y. Characterization of the UL4 gene product of herpes simplex virus type 2. Arch Virol. 1998;143:1199–1207. doi: 10.1007/s007050050367. [DOI] [PubMed] [Google Scholar]