Abstract
The HOXA9 transcription factor serves as a molecular orchestrator in cancer stemness, epithelial-mesenchymal transition (EMT), metastasis, and generation of the tumor microenvironment in hematological and solid malignancies. However, the multiple modes of regulation, multifaceted functions, and context-dependent interactions responsible for the dual role of HOXA9 as an oncogene or tumor suppressor in cancer remain obscure. Hence, unravelling its molecular complexities, binding partners, and interacting signaling molecules enables us to comprehend HOXA9-mediated transcriptional programs and molecular crosstalk. However, it is imperative to understand its central role in fundamental biological processes such as embryogenesis, foetus implantation, hematopoiesis, endothelial cell proliferation, and tissue homeostasis before designing targeted therapies. Indeed, it presents an enormous challenge for clinicians to selectively target its oncogenic functions or restore tumor-suppressive role without altering normal cellular functions. In addition to its implications in cancer, the present review also focuses on the clinical applications of HOXA9 in recurrence and drug resistance, which may provide a broader understanding beyond oncology, open new avenues for clinicians for accurate diagnoses, and develop personalized treatment strategies. Furthermore, we have also discussed the existing therapeutic options and accompanying challenges in HOXA9-targeted therapies in different cancer types.
Graphical Abstract
Keywords: HOXA9, Transcription factor, Molecular alterations, Cancer progression, Clinical application, Therapeutic targeting
Introduction
Homeobox (HOX) genes, originally discovered in Drosophila melanogaster, are evolutionally conserved genes formed as a result of successive duplication and subsequent divergence from primordial HOX cluster genes [1, 2]. In mammals, the 39 HOX genes are organized into 4 different HOX clusters, each located at different chromosomal coordinates [3]. Following translation, HOX genes primarily function as transcription factors with a key role in regulating signaling molecules [4]. These signaling molecules govern crucial biological events that are essential for maintaining normal physiology and tissue homeostasis. The expression pattern of HOX genes plays a pivotal role during embryogenesis in determining the body axis, skeletal morphology, and organ development in a tissue-specific manner. These transcription factors possess an intrinsic ability to bind to target genes through their DNA-binding homeodomain, thereby facilitating the activation of those genes. During embryogenesis, HOX genes are transcribed in a temporally and spatially collinear manner, wherein the position of each gene within the HOX cluster corresponds to sequential activation along the anterior-posterior axis [5]. This activation occurs from the 3’ end to the 5’ end of the HOX cluster, originating sequential developmental patterns [5–7].
The HOXA9 gene has been extensively studied among the 39 HOX genes, particularly in the context of hematological as well as solid malignancies [8, 9]. Under normal physiological conditions, HOXA9 has been recognized as a crucial regulator of various processes, including morphogenesis [10], embryo implantation [11], hematopoiesis [12], and endothelial cell regulation [13]. Numerous studies have revealed a wide range of genetic and epigenetic variations occurring at the HOXA9 locus, shedding light on their biological implications during carcinogenesis. Genetic alterations, such as gene translocations and gene fusions at the HOXA9 locus, have been observed to contribute to leukemic transformation in acute myeloid leukemia (AML) [14]. On the other hand, epigenetic factors play a role in the deregulation of HOXA9 in various solid tumors. These include chromatin modification [15], promoter DNA methylation [16–18], gene-body methylation [19], microRNAs (miRNAs) [20–23], and long noncoding RNAs (lncRNAs) [24]. Aberrant expression of HOXA9 triggers an impaired immune response, resulting in sustained activation of downstream signaling pathways. This disruption of normal physiology creates a microenvironment conducive to tumor growth.
This review aims to specifically explore the functional aspects of HOXA9 in normal physiologic conditions and its involvement in oncogenesis across various cancer types. Additionally, we have summarized the clinical significance of HOXA9, its role in drug resistance and recurrence, and important strategies to therapeutically target HOXA9 in cancer.
Structural and genetic complexity of the HOXA9 locus
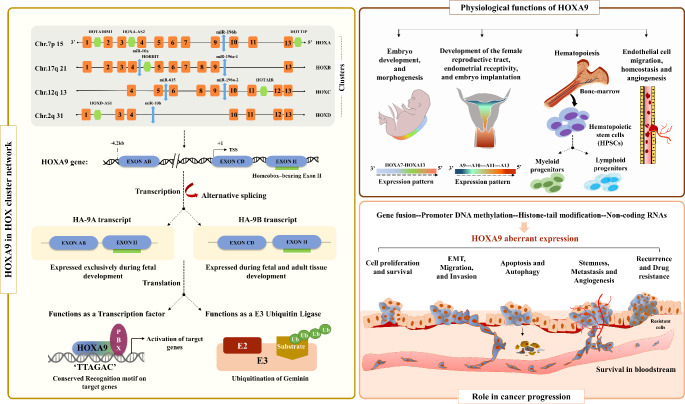
Understanding the structural complexity of the HOXA9 gene is crucial for studying its regulatory and functional roles. Throughout the evolutionary process, the 5’ end gene known as Abdominal B (AbdB) in Drosophila underwent duplication, resulting in the emergence of five AbdB-like genes positioned at the 5’ side of all four clusters in mammals. They exhibit similarities in the gene sequence and position in the HOX cluster; hence, each of the duplicated HOX genes can be aligned between the four clusters. Within the HOXA cluster, AbdB-like genes undergo duplication to form HOXA9, HOXA10, HOXA11, and HOXA13 genes in mammals [25].
As a member of the HOXA cluster, HOXA9 is located on the short arm of chromosome 7. Kim et al. sequenced the entire 7.2 kb sequence of the human HOXA9 gene. They identified three different exons, of which exon AB and exon CD are located at the 5’ end, while exon II is located at the 3’ end [26]. Due to alternative splicing at exon CD, humans have two types of HOXA9-coding transcripts known as HA-9A and HA-9B. Both transcripts share a common 3’ exon II that contains the highly conserved ‘homeobox’ sequence for DNA binding. However, they differ in their 5’ exons. HA-9A proteins are exclusively present during foetal development. In contrast, HA-9B proteins, also known as canonical HOXA9, are found in both embryonic and adult tissues [26]. It is interesting to note that the HOXA9 gene has numerous transcripts in various disorders. These transcripts are generated as a result of alternative promoter usage and alternative splicing of the HOXA9 gene. Popovic et al. extensively reviewed the transcriptional complexity of the HOXA9 gene, with particular emphasis on various human transcripts [27]. Alternative splicing at the exon CD splice site results in the generation of a premature stop codon prior to the homeobox region. This leads to the production of a protein lacking a homeodomain, which has been observed in both mice and humans [28, 29]. In contrast to the canonical isoform of HOXA9, this truncated isoform HOXA9T has been found both in the nucleus and cytoplasm of various tissues and is abundantly expressed in the embryonic genital tract, kidney, limb, and tail [27, 29]. The presence of a truncated isoform has been reported in endometrial adenocarcinoma cells. In these cells, both truncated isoforms and the canonical isoforms coexpress and compete for binding with several cofactors, including the CREB-binding protein (CBP) [29]. The truncated isoform interferes with the functions of the canonical isoform, causing molecular pathway disruption in various diseases [29]. However, a study on human MLL-rearranged leukemia demonstrated that both isoforms cooperate to promote leukemogenesis [30]. There is still much to be understood regarding the isoform-mediated molecular mechanisms involved in disease progression.
Functional role of HOXA9 under normal physiological conditions
Embryo development and morphogenesis
HOX genes are well known for their role in determining anterior-posterior segmental identity, contributing to the formation of skeletal structures, and playing a key role in organogenesis [10, 31]. In mammals, HOXA9 is involved in the morphogenesis of the embryonic skeletal system and the specification of the posterior part of the embryo [10]. The HOXA9 and HOXD9 paralogous genes are specifically involved in the axial skeletal patterning of the lumbosacral region and forelimb morphogenesis at the stylopodal stage. This was demonstrated by developing HOXA9-/HOXD9- double mutants in mice [32]. Mutations in all HOXA cluster genes can lead to the development of defects in the cardiac, respiratory, and urinogenital systems [31]. Frameshift mutations in six HOX genes, namely, HOXA9, HOXA10, HOXA11, HOXD9, HOXD10, and HOXD11 have been associated with severe limb malformation. Moreover, HOXA9,10,11 negative-mutant mice displayed a reduced length of the ulna and radius, implying their major role in stylopod development [33].
Female reproductive tract development, endometrial receptivity, and embryo implantation
The posterior or 5’ end HOX genes (HOXA9 to HOXA13), belonging to the AbdB family, are involved in the development of the mammalian female reproductive system [34]. During embryonic development, 5’ HOX genes, including HOXA9, exhibit robust expression in the paramesonephric duct. During postnatal differentiation, these follow a spatial axis, indicating their specific patterns of expression along the developing tissues or organs. HOXA cluster genes actively govern crucial physiological processes in the female reproductive tract throughout the adult stage [34, 35]. It is interesting to note that HOXA9 is expressed in the fallopian tube. Hence, aberrant expression of HOX genes leads to developmental abnormalities in the uterus, impaired growth of the endometrium, and a reduced implantation rate [34, 36]. Ovarian steroids such as progesterone and estrogen have the ability to induce the activation of HOXA9, HOXA10, and HOXA11 in a collinear manner [37]. Hormonal regulation of these HOX genes indicates the functional interaction between HOX factors and hormonal nuclear receptor families during implantation [37–40]. Considering the conserved sequences and overlapping expression pattern similar to HOXA10, Xu et al. extensively studied the role of HOXA9 and other HOX genes that are highly expressed in the endometrium during menstruation, and their expression is correlated with implantation time [11]. Interestingly, they found a significant reduction in the implantation rate upon shRNA-mediated knockdown of maternal HOX genes in mouse models, indicating that HOX genes are also essential for endometrial receptivity [11].
Hematopoiesis
Over the past decades, studies have deciphered the role of HOX genes in hematopoiesis [41]. The mammalian HOXA9 gene plays a remarkable role in hematopoiesis, and its expression is closely associated with blood formation [41, 42]. HOXA9 is abundantly expressed in hematopoietic stem and progenitor cells (HSPCs), and its expression diminishes upon differentiation [43, 44]. Researchers investigated the deleterious influence of HOXA9 on primitive blood cells by developing HOXA9-/- mutant mice. Interestingly, the mice showed a significant reduction (30–40%) in the leukocyte and lymphocyte populations (P value < 0.05). In addition, they not only observed a reduction in the myeloid and erythroid progenitors but also witnessed the reduced size of the spleen and thymus [45]. A group of researchers have demonstrated that the binding of CDX4 followed by Menin-dependent H3K4 trimethylation could induce HOXA9 expression during normal hematopoiesis [12].
Initially, hemogenic precursors (HEPs) (CD31+CD34+CD45−) show the highest expression of HOXA9, and the expression level is reduced when HEPs undergo differentiation into CD45+ blood cells [46]. In other words, in vivo overexpression and knockdown studies have revealed that HOXA9 plays a major role in the differentiation of human embryonic stem cells (hESCs) by facilitating the transition of HEPs into CD45+ blood cells by regulating the NF-κB pathway [46]. A recent study revealed that HOXA9 overexpression specifically enhances myeloid potential, rather than erythroid potential, by promoting cell cycle progression through the upregulation of the NF-κB pathway [47]. Researchers have performed ChIP sequencing to investigate the regulation and mechanisms behind the role of HOXA9 in hematopoiesis and leukemia. The study made an intriguing observation that HOXA9 proteins in conjunction with cofactors form associations with enhanceosomes at several highly conserved target sites. This interaction plays a crucial role in promoting stem cell expansion and B/T-cell development. The gene expression profiling data indicated that HOXA9 either promotes or suppresses target genes based on the type of cell and chromatin content. Various upstream oncogenic alterations lead to persistent activation of HOXA9. This activated form of HOXA9 binds with MEIS1 and several other lineage-restricted transcription factors, enabling the recruitment of P300/CBP to transcriptionally activate the target genes involved in proliferation and leukemic transformation [48].
Endothelial cell proliferation, homeostasis, and angiogenesis
Endothelial cell (EC) migration is an important phase in vascular system development. On the other hand, ECs are formed during embryogenesis through the differentiation of angioblasts, a process known as vasculogenesis. This process ultimately leads to the development of new blood vessels [49]. However, angiogenesis is the process through which newly generated blood vessels differentiate to become a complex network of new blood vessels in adulthood. Vasculogenesis and angiogenesis are crucial for maintaining vascular integrity [50]. Endothelial cell migration is needed in angiogenesis and wound healing [50]. The HOXA9-mediated activation of signaling pathways involved in this process is still under investigation [51]. Patel et al. were the first to identify HOXA9 as the sole member of the HOX family to be expressed in EC. They identified a novel splice variant, HOXA9EC, screening a human cDNA library. HOXA9EC is regulated by a novel promoter and is exclusively expressed in EC. This splice variant was found to be sensitive to TNF-α, and its expression was downregulated upon TNF-α activation. This suggests that the new promoter is activated by cytokines and contributes to maintaining ECs in a quiescent state [13]. Hence, during cytokine activation, the HOXA9 transcription factor plays a role in modulating essential genes associated with EC activation [13]. During inflammation, the release of TNF-α induces E-selectin activation. E-selectin is a leukocyte adhesion molecule that enables the recruitment of leukocytes and firm adhesion of leukocytes to inflammatory foci [52]. This cytokine-mediated activation is facilitated by the binding of NF-κB to the E-selectin promoter region [53]. Later, the same group of researchers found that the constituent expression of HOXA9 prevents EC activation by inhibiting NF-κB-mediated transcriptional activation of E-selectin in human umbilical vein endothelial cells (HUVECs) [54, 55].
In a later study, Zhang et al.,2012 demonstrated the potential of HOXA9EC in reducing endothelial dysfunction induced by high glucose [56]. This leads to the downregulation of crucial endothelial genes such as eNOS (endothelial nitric oxide synthase), VEGFR2 (vascular endothelial growth factor receptor 2), and VE-cadherin. Furthermore, the overexpression of HOXA9EC could serve as a viable strategy to mitigate endothelial dysfunction [56].
It is interesting to note that HOXA9 is crucial for endothelial cell migration and tube formation. This was demonstrated through siRNA-mediated transfection of HOXA9 into HUVECs. The study elaborated on the proangiogenic nature of HOXA9 and revealed that EphB4 receptor tyrosine kinase is a novel downstream target of HOXA9. Depletion of HOXA9 resulted in the downregulation of EphB4 expression, which in turn led to reduced migration and impaired tube formation in endothelial cells [57, 58]. Additionally, the ChIP assay provided evidence that HOXA9 directly interacts with the EphB4 promoter, thereby facilitating its transcriptional activation. Furthermore, the study also showed that HOXA9 stimulates EphB4 by binding to its TAAT motif located − 1365 bp upstream of the TSS in the promoter region. This was confirmed through site-directed mutagenesis and deletion constructs [58].
Regulation of HOXA9 in cancer
The structural complexity of HOXA9 plays a significant role in regulating multiple aspects of cancer. Notably, the regulation of HOXA9 in different cancer types is highly context-dependent. Surprisingly, the mode of regulation can vary not only between cancer types but also within subtypes of the same malignancy. Previous reports have indicated that HOXA9 can be regulated by both genetic and epigenetic factors in various types of cancer.
HOXA9 is mainly modulated through genetic fusion in leukemia. The chromosomal translocation t (7; 11) (p15; p15) results in the fusion of HOXA9 with nucleoporin 98 kDa (NUP98), leading to the formation of a chimeric protein called NUP98-HOXA9 (NHA9), which is observed in various malignancies [59]. The study used oligonucleotide microarray analysis to investigate the effects of the NUP98-HOXA9 fusion on hematopoietic cell proliferation, differentiation, and leukemic transformation [14]. ChIP-seq analysis revealed that MLL1 occupancy with the NHA9 protein is crucial for the activation of downstream target genes involved in leukemogenesis [60, 61]. Amplification of HOXA9 was evident in angiosarcoma, contributing significantly to abnormal vascular cell proliferation and blood vessel growth [62]. In addition to gene fusions, HOXA9 has also been regulated in multiple ways by means of epigenetic factors in AML. Aryal et al. performed an extensive literature search and summarized the molecular regulators of HOXA9 in AML. The study also outlined the different kinds of drugs that were used to target the oncogenic nature of HOXA9 in AML [63].
HOXA9 is subjected to complex regulation involving multiple epigenetic factors in certain cancer types. It is epigenetically regulated by promoter DNA methylation, miRNAs, lncRNAs, and histone complexes in different malignancies (Table 1).
Table 1.
Epigenetic regulation of HOXA9 in cancer
| Cancer | Expression of HOXA9 | Molecular mechanism | Clinical significance | References |
|---|---|---|---|---|
| HOXA9 is regulated by promoter DNA methylation | ||||
| Lung adenocarcinoma | ↓ | HOXA9 methylation, High frequency of TP53 mutations, and the upregulation of the OncomiR miR-9 | Associated with poor prognosis | [116] |
| Non-Small Cell Lung Cancer | ↓ | Transcriptional downregulation of HOXA9 | Associated with poor recurrence-free survival | [107, 163] |
| High-Grade Non-Invasive Bladder Cancer | ↓ | Increased methylation in recurrent tissues | Associated with recurrence and disease-specific mortality | [109] |
| Head and neck squamous cell carcinoma | - | HOXA9 promoter hypermethylation | Correlates with metastasis and advanced tumor stage | [17, 137] |
| HOXA9 is regulated by miRNAs | ||||
| Non-Small Cell Lung Cancer | ↓ | miR-182-5p sponges HOXA9; miR-196b sponges HOXA9 |
Contributes to tumor initiation and progression; Promotes invasiveness |
[66, 164] |
| Acute myeloid leukaemia | ↑ | miR-182/miR-196b downregulation; Reduced sponging effect of miR-182/ miR-196b/ with HOXA9 | Promotes leukemogenesis | [165, 166] |
| Oral squamous cell carcinoma | ↑ | miR-139-5p downregulation; Reduced sponging effect of miR-139-5p with HOXA9 | Promotes proliferation, invasiveness, and migration | [167] |
| Osteosarcoma | ↑ | miR-1294 downregulation; Reduced sponging effect of miR-1294 with HOXA9 | Promotes proliferation and invasion | [21] |
| Epithelial ovarian cancer | ↓ | Enhanced sponging effect of miR-196b on HOXA9 | Promotes ovarian cancer cell invasiveness; Associated with recurrence | [22] |
| Colorectal cancer | ↑ | miR-133b downregulation; Reduced sponging effect of miR-133b with HOXA9 | Promotes cancer metastasis | [103] |
| Glioma | ↑ | miR-647/miR-638 downregulation; Reduced sponging effect with HOXA9 | Promotes cell proliferation, colony formation and invasion | [67, 68] |
| Uveal melanoma | ↓ | Enhanced sponging effect of miR-652 on HOXA9; Promoted HIF-1α signaling | Promotes cell growth and migration | [168] |
| HOXA9 is regulated by other ncRNAs | ||||
| Prostate cancer | ↑ | TWIST1/WDR5/HOTTIP induces HOXA9 expression | Promotes migration, invasion, and metastasis | [104] |
| Pancreatic cancer | ↑ | LncRNA HOTTIP induces HOXA9 expression | Promotes proliferation and migration | [169] |
| Triple-negative breast cancer | ↓ | LncRNA MIR503HG regulates miR-224-5p/HOXA9 axis | Promotes proliferation, migration, and invasion | [24] |
| Colorectal adenocarcinoma | ↑ | LncRNA PCED1B-AS1 regulates miR-633/HOXA9 axis | Promotes cell proliferation and reduces apoptosis | [170] |
| Gastric cancer | ↑ | CirRNA circ_0026359 regulates miR-140-3p/HOXA9 axis | Associated with poor prognosis | [100, 171] |
| Osteosarcoma | ↑ | LncRNA DLX6-AS1 regulates miR-641/HOXA9 axis | Promotes proliferation, migration, and invasion | [172] |
| Laryngeal squamous cell carcinoma | ↑ | LncRNA KCNQ1OT1 directly binds with HOXA9 | Promotes proliferation, invasion, and metastasis | [173] |
| HOXA9 is regulated by histone-complexes | ||||
| Breast cancer | ↓ | TET1 demethylates HOXA9 promoter | Promotes tumor growth and metastasis | [99] |
| Acute myeloid leukaemia | ↑ | G9a methyl transferase promotes HOXA9 transcription | Promotes expansion and differentiation of AML cells | [174] |
Note: ↑- Upregulated; ↓-Downregulated
Indeed, HOXA9 has been found to be regulated by several miRNAs during hematopoiesis and cell differentiation [64, 65]. Deregulation of upstream regulators frequently results in abnormal expression of HOXA9, resulting in tumor aggressiveness. HOXA9 is regulated by various miRNAs in solid cancers (Table 1), including miR-196b, which is embedded within the HOX cluster [22, 66]. Furthermore, overexpression of HOXA9 in glioma is mainly due to the downregulation of miR-638 and miR-647. The reduced sponging effect on HOXA9 triggers its oncogenic potential, causing enhanced cell proliferation, colony formation, and invasion [67, 68]. In addition, evidence supports that other ncRNAs, such as lncRNAs and circRNAs, are often involved in the molecular sponging mechanism to regulate HOXA9 in cancer (Table 1).
Downregulation of HOXA9 due to promoter-DNA methylation serves as a tumor suppressor and prognostic marker in solid tumors [9]. In addition to promoter DNA methylation, increased methylation at the first exon of HOXA9 has also been correlated with gene repression in cervical cancer (CC). Restoring the expression of HOXA9 in CC cell lines resulted in reduced cell proliferation and migration [19]. Remarkably, a unique interaction exists between HOXA9 and HOXA10 promoters in breast cancer (BC). The CpGs of HOXA10 function as enhancers of the HOXA9 gene, causing long-range chromatin interactions [18].
Understanding the precise processes underpinning HOXA9 regulation in different cancer types is critical for designing targeted therapeutics and improving patient outcomes.
Functional implications of HOXA9 in cancer progression
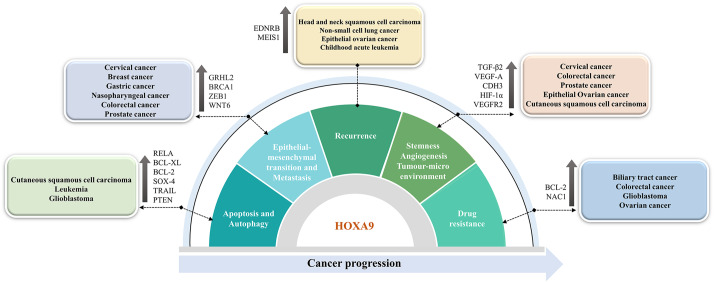
Deregulation of HOXA9 has been extensively studied in hematological as well as solid malignancies, including gastrointestinal cancers, skin-related cancers, head and neck cancer types, and gynecological cancers [69, 70] (Fig. 1). However, the role of HOXA9 as an oncogene or tumor suppressor gene particularly depends on tumor heterogeneity. Therefore, it is crucial to investigate the molecular mechanisms underlying the regulation of HOXA9 and its implication in cancer progression. This section will discuss HOXA9-mediated cancer-associated molecular events in detail, which could provide valuable insights for the design and development of targeted therapies for cancer.
Fig. 1.
Role of aberrantly expressed HOXA9 in the acquisition of cancer hallmarks in different malignancies
HOXA9 in the tumor microenvironment, stemness, and angiogenesis
The dynamics of the tumor microenvironment (TME) exhibit the characteristics that encompass tumor growth, angiogenesis, invasion, and metastatic dissemination. The niche around the tumor mass often consists of a variety of cell types, such as inflammatory immune cells, adipocytes, endothelial cells, and fibroblasts, in the stroma, which conjugate with cancer cells to release signals that favor the migration and invasiveness of the tumor mass and thus help to establish the TME [71, 72]. In other words, aggressive growth is triggered by the growth factors and chemokines secreted by the inflammatory cells of the TME, which helps to gain cancer stemness and sustain cancer hallmarks, which in turn leads to the transition into distant metastasis [71]. Once the TME is generated, the hypoxic environment often induces the formation of abnormal vasculature favorable for the growth of cancer cells [73]. Hence, there always exists an intricate connection between the TME, angiogenesis, and stemness.
Researchers have deciphered the crosstalk between HOX transcription factors and the TME in PCa [74]. Another study on PCa demonstrated that the HOXA2, HOXA9, and HOXA10 genes play a crucial role in promoting tumor growth and progression. Aberrantly expressed HOXA2, HOXA9, and HOXA10 were found to be involved in the recruitment and infiltration of immune cells, including dendritic cells, macrophages, and mastocytes, during the progression of PCa [75]. In contrast, HOXA9 was found to be involved in the modulation of the NF-κB pathway, particularly during viral infection in CC. The study discovered a novel isoform of HOXA9 that lacks the homeodomain and is highly expressed in nontumorigenic HaCaT cells compared to CC cell lines and tumor samples. Restoring HOXA9 expression in CC cell lines resulted in the elevated expression of antigen-presenting cells (APCs) and regulators of the immune response, including IL17RD, FADD, CHEK2, TRAIL, IL-3/IL-5 pathways, and IFN-gamma signaling molecules [76].
Ko et al. in 2012 reported that elevated expression of HOXA9 in epithelial ovarian cancer (EOC) contributes to angiogenesis and the generation of a microenvironment for tumor growth [77]. This is due to the HOXA9-mediated transcriptional activation of TGF-β2, followed by subsequent activation of CXCL12, IL-6, and VEGF-A in peritoneal fibroblasts. Hence, HOXA9 promotes peritoneal fibroblasts and mesenchymal stem cells (MSCs) to acquire the properties of cancer-associated fibroblasts (CAFs) [78]. Through in vitro knockdown studies, the study also proved that the tumor-inducing property of HOXA9 is mediated only through the induction of TGF-β2 in EOC cells. HOXA9 further induces the immunosuppressive phenotype of peritoneal macrophages by inducing the expression of TGF-β2 and chemokine (C-C motif) ligand 2 [79]. Once a suitable environment is generated, floating cancer cells tend to implant in peritoneal mesothelial cells by means of the cell adhesion molecule P-cadherin to form aggregates and avoid anoikis [80]. HOXA9 induces the expression of the target gene CDH3, which encodes P-cadherin, enabling ovarian cancer (OVC) cells to adapt to the peritoneal environment and acquire an aggressive phenotype [81].
Furthermore, the TME often consists of cancer stem cells (CSCs) that possess self-renewal, differentiation, and treatment-resistance capabilities [82]. Targeting CSCs is essential to prevent recurrence, overcome therapy resistance, and inhibit metastasis. Interestingly, HOXA9 is overexpressed in colorectal cancer (CRC) patients with lymph node metastasis, and it contributes to stem cell overpopulation, which could favor cancer progression [83, 84]. Indeed, it was demonstrated that HOXA9 is regulated by retinoic acid (RA) signaling, and an anticancer effect is exerted with all-trans retinoic acid (ATRA) treatment through the repression of HOXA9 in CRC [83, 85].
In addition, a study on cutaneous squamous cell carcinoma (CSCC) reported that HOXA9 plays a crucial role in reducing the hypoxic response, angiogenesis, and tumor progression. Under normal physiological conditions, HOXA9 epigenetically regulates the key factor for the hypoxia response, HIF-1α. However, during CSCC tumor progression, miR-365 inhibits HOXA9 expression, leading to the upregulation of HIF-1α and downstream genes (GLUT1, HK2, and PDK1) involved in the glycolytic pathway. In cells expressing HOXA9, it exerts a tumor-suppressive role by restricting the uptake of glucose and suppressing glycolysis in cancer cells. Therefore, the miR-365-HOXA9-HIF-1α regulatory axis could serve as a suitable target for intervention therapy [86, 87]. Furthermore, HOXA9 has been considered a master switch that directly regulates endothelial-committed target genes such as eNOs (endothelial nitric oxide synthase), CDH5 (VE-cadherin), and VEGFR2 to maintain the vasculature and angiogenesis [88]. The study also revealed that HOXA9-deficient mice exhibited impaired postnatal neovascularization [74, 88]. Hence, the expression of HOXA9 in endothelial progenitor cells is crucial for the maintenance of the tumor microenvironment and for the commitment to the endothelial cell lineage.
HOXA9 in the cell cycle, apoptosis, and autophagy
HOX transcription factors play a critical role in regulating cell proliferation during embryogenesis and morphogenesis. HOX transcription factors regulate the cell cycle machinery through protein‒protein interactions. Dysregulation of HOX transcription factors disrupts the cell cycle machinery, leading to cancer progression [89].
HOXA9 was identified as a potential regulator of the apoptosis and autophagy pathways in CSCC through the transcriptional control of RELA, a crucial element of the NF-κB pathway. In cells depleted of HOXA9, RELA was upregulated, leading to transcriptional activation of the antiapoptotic factor Bcl-XL and the autophagic molecules ATG2, ATG3, and ATG12. Accordingly, the study revealed that regulation of the NF-κB pathway by HOXA9 resulted in increased apoptosis and decreased autophagy in CSCC [90]. However, persistent activation of HOXA9 is needed for cell proliferation and survival. In leukemia, HOXA9-mediated maintenance of BCL2 expression is essential for HOXA9-dependent immortalization of hematopoietic cells and survival of myeloid progenitor cells. Further research is warranted to elucidate the molecular interactions between HOXA9 and Bcl-2 family members to develop effective therapeutic options [91]. Recently, Miyamoto et al. showed that HOXA9 plays a multifunctional role in regulating antiapoptotic pathways in leukemia. The activation of HOXA9 target genes such as BCL2 and SOX4 was attributed to the combined activation of MYC and HOXA9 caused by MLL fusion. The enhanced activation of these molecules not only repressed apoptosis but also promoted leukemogenesis, indicating that HOXA9 could be a potential regulator of apoptotic pathways [92].
Researchers have identified transcriptional cooperation between HOXA9 and JAK3/STAT5 in T-cell acute lymphoblastic leukaemia [93]. Furthermore, HOXA9 acts as a transcription factor for the oncogenic kinase enzyme PIM1, leading to the transcriptional activation of PIM1. The binding of HOXA9 to PIM1 facilitates the phosphorylation of the pro-apoptotic protein BAD, resulting in its inactivation. Hence, HOXA9-mediated activation of PIM1 kinase often results in enhanced proliferation and anti-apoptosis in leukaemia [94]. In glioblastoma (GBM), HOXA9 inhibits apoptosis by regulating tumor necrosis factor-related apoptosis-including ligand (TRAIL). The study also showed that increasing HOXA9 levels influence PTEN, a PI3K pathway antagonist, which in turn influences proliferation and apoptosis. As a result, reversing HOXA9 activation via PI3K inhibition may have therapeutic implications in human GBM [95].
HOXA9 in EMT and metastasis
HOXA9 demonstrates a dual role as a tumor suppressor and oncogene, with its function being context-dependent (Table 2). The expression of HOXA9 was found to be reduced in CC, and its restoration resulted in decreased cell proliferation and migration along with an increase in epithelial phenotype [19]. HOXA9 elevation led to a significant upregulation of the novel suppressor of EMT, GRHL2 (Grainyhead-like 2), compared to the scrambled control [19, 96, 97]. The study showed that CC cells expressing HOXA9 and infected with E6 or E7-HPV 18 oncoproteins demonstrated decreased cell proliferation, motility, colony formation, and metabolism. In fact, HOXA9 was found to be upregulated in E6- and E7-depleted cells [19]. Therefore, finding the molecular link in this phenomenon paves the way to inhibit CC progression.
Table 2.
Oncogenic and tumor suppressive function of HOXA9 in cancer
| Cancer | Functional implications upon aberrant expression | References |
|---|---|---|
| HOXA9 as an oncogene | ||
| Colorectal cancer | Contributes to stem cell overpopulation, | [83, 84, 103] |
| Pancreatic cancer | Enhances the stem cell properties | [175] |
| Ovarian cancer | Contributes to the angiogenesis and generation of a microenvironment; Induces aggressive phenotype | [77, 81] |
| Leukemia | Promotes cell proliferation and survival; Reduces apoptosis | [92, 165] |
| Gastric cancer | Associated with tumor-node-metastasis (TNM) staging | [100] |
| Nasopharyngeal cancer | Associated with tumor-node-metastasis (TNM) staging | [101] |
| Prostate cancer | Induction of metastatic phenotype | [104] |
| Osteosarcoma | Promotes proliferation and invasion | [21] |
| Oral squamous cell carcinoma | Promotes proliferation, invasiveness, and migration | [167] |
| Glioma | Promotes cell proliferation, colony formation and invasion; Inhibits apoptosis | [67, 95, 105] |
| HOXA9 as a tumor suppressive gene | ||
| Cutaneous squamous cell carcinoma | Downregulation of HOXA9 induces hypoxic response, angiogenesis, and tumor progression | [87, 90] |
| Cervical cancer | Downregulation of HOXA9 induces cell proliferation, migration and EMT | [19] |
| Breast cancer | Reduced levels of HOXA9 induces metastasis and aggressiveness | [98, 99] |
| Non-small cell lung cancer | Downregulation is correlated with disease recurrence; Promotes invasiveness | [66, 107, 163, 164] |
| High-Grade Non-Invasive Bladder Cancer | Promotes recurrence | [109] |
| Uveal melanoma | Promotes cell growth and migration | [168] |
| Lung adenocarcinoma | Associated with poor prognosis | [116] |
Reduced levels of HOXA9 transcripts have been associated with metastasis and aggressiveness in breast cancer (BC). The tumor suppressor gene BRCA1 was expressed more frequently after HOXA9 restoration, which in turn prevented the malignant behavior of BC cells [98]. In addition, researchers have performed an extensive study on BC and found that inhibiting the HMGA2-TET1-HOXA9 pathway aided tumor development, intravasation, invasion, and metastasis in BC [99]. In contrast, HOXA9 was significantly upregulated in colonic adenoma, gastric cancer (GC), nasopharyngeal cancer (NPC) and CRC and was associated with tumor-node-metastasis (TNM) staging and positive lymph node metastasis [84, 100–102]. Especially in CRC, HOXA9 upregulation was the consequence of miR-133b downregulation. Interestingly, the study found that elevated levels of HOXA9 significantly contribute to tumor invasion and metastasis by regulating the expression of ZEB1. Restoration of miR-133b and downregulation of HOXA9 significantly reduced the expression of ZEB1 and increased the expression of CDH1 in CRC cells. Hence, this molecular pathway therefore serves as a possible therapeutic target for CRC treatment [103]. In 2017, Malek et al. extensively investigated the molecular mechanism of PCa metastasis and aggressiveness. A major EMT transcription factor, TWIST1, induces HOXA9 expression either by transcriptional activation or by epigenetic reprogramming of the HOXA9 locus. TWIST1 binds with a complex of proteins associated with SET1 (COMPASS)-like complex to induce chromatin modification at the HOXA9 promoter via H3K4me3 in PCa cells. The induction of the metastatic phenotype was due to HOXA9 overactivation caused by the binding of the TWIST1-WDR5 complex to the E-box consensus sequence of the HOXA9 promoter. The study demonstrated that pharmacological inhibition of HOXA9 using 10 nM HXR9 peptide effectively reduced PCa cell migration and invasion and prevented metastasis induced by TWIST1-HOXA9 [104].
Upregulation of WNT6, a crucial regulator of the Wnt/β-catenin pathway, was associated with poor clinical outcomes in glioma patients. By performing in vitro and in vivo studies, researchers found that glioma aggressiveness might be due to HOXA9/WNT6-mediated activation of the canonical Wnt/β-catenin pathway. There exists a molecular link between HOXA9 and WNT6 where the binding of HOXA9 to the promoter region of WNT6 induces its expression. Thus, targeting the WNT6-HOXA9 pathway may be a promising therapeutic approach to inhibit the growth of glioma [105].
HOXA9 in recurrence
Although there have been considerable advances in therapeutic regimens, the recurrence rate of many types of cancer is still very high. Contemplating HOXA9 as a clinical biomarker not only facilitates the assessment of disease recurrence and cancer progression but also helps in treatment decisions in recurrent cancer patients. Interestingly, few recent studies have shown the role of HOXA9 in recurrence in different cancer types. In head and neck squamous cell carcinoma (HNSCC), the study showed a correlation between EDNRB and HOXA9 methylation in surgical margin imprints. Multivariate analysis revealed that the regions with a high risk of locoregional recurrence showed higher methylation upon surgery and hence were considered valuable predictive biomarkers for locoregional recurrence in HNSCC (HR, 3.31; P value = 0.012) [106].
According to a study on non-small cell lung carcinoma (NSCLC), HOXA9 may be a useful marker for anticipating disease recurrence. Approximately 70% of NSCLC patients had HOXA9 promoter hypermethylation and downregulation, which was associated with poor recurrence-free survival (P value = 0.01) [107].
A study on nonmuscle invasive bladder cancer (NMIBC) reported that hypermethylation status of HOXA9, ISL1 (ISL LIM homeobox 1), and ALDH1A3 (aldehyde dehydrogenase 1 family, member A3) and decreased expression of these genes were associated with aggressive clinical characteristics [108]. In a similar vein, Kitchen et al. in 2015 revealed that ISL1/HOXA9 methylation might be used as a predictive biomarker for tumor recurrence in high-grade noninvasive bladder cancer within a year of cancer diagnosis. Indeed, ISL1/HOXA9 gene methylation levels were greater in progressive and recurrent tumors than in nonrecurrent tumors (P value < 0.05), and these methylation levels were also related to disease-specific mortality [109]. Especially in patient urine samples of bladder cancer, the study showed that methylation levels of five markers (HOXA9, POU4F2, TWIST1, VIM, and ZNF154) were significantly linked to tumor recurrence. Univariate cox-regression analysis proposed these as markers for the early detection of recurrence in bladder cancer [110].
In contrast, HOXA9 and its co-factor MEIS1 upregulation were inversely correlated with relapse in pediatric acute leukemia [111]. In addition to tissue samples, researchers have investigated the prognostic significance of the methylation status of HOXA9 in circulating tumor-specific DNA (ctDNA) in the blood samples of EOC patients during chemotherapy [112–114]. Studies have shown that HOXA9 methylation could be used as a potential ctDNA biomarker to predict prognosis and recurrence and assess treatment resistance in different cancer types [112, 113, 115, 116]. Cai et al. conducted a comprehensive meta-analysis and confirmed that HOXA9 methylation has potential as a reliable biomarker for predicting the prognosis of malignant tumors [9]. Interestingly, the study observed that the increase in methylation of HOXA9 in patient blood samples after one treatment cycle was associated with reduced overall survival, especially in patients with disease recurrence [113].
HOXA9 in drug resistance
Despite advanced clinical approaches to target cancer cells, they still undergo metastasis and acquire an aggressive phenotype. The treatment ineffectiveness is due to the ability of cancer cells to resist the treatment [117]. Therapeutic regimens are typically designed to promote the immune response against growing tumors. This may involve administering specific drugs at the maximum tolerated dose (MTD) [118, 119]. While this method of treatment initially helps in depleting cancer cells, it often leads to impaired immune surveillance and drug resistance over time [120, 121]. Drug resistance is acquired due to the dynamic interaction between the host immune system and cancer cells. Initially, cancer cells respond effectively to therapies, but eventually, they tend to reoccur over a period of time [121].
A study on BRCA-mutated OVC has indicated that HOXA9 promoter methylation in the plasma of patients could serve as a promising prognostic biomarker. HOXA9 methylation in ctDNA was associated with the worst outcomes in PARP inhibitor-treated, platinum-resistant BRCA-mutated OVC. Patients with higher HOXA9 methylation in ctDNA had shorter median progression-free survival (PFS) than patients with lower methylation levels (P value < 0.05) after three treatment cycles [112].
The clinical significance of HOXA9 methylation was investigated in patients with biliary tract cancer (BTC). The patients receiving erlotinib and bevacizumab drug treatment showed an increase in HOXA9 methylation levels, which negatively correlated with patient survival [122]. In contrast, researchers have suggested that chemotherapy resistance in high-grade serous ovarian cancer (HGSOC) samples is due to the upregulation of HOXA9 [123]. In CRC, nucleus accumbens‑associated protein 1 (NAC1) contributes to drug resistance by inducing the expression of HOXA9 [124]. Researchers conducted a comprehensive study on GBM and identified HOXA9 as a key factor in promoting cancer stemness, aggressiveness, and drug resistance. The study validated that HOXA9 promotes the malignant transformation of immortalized astrocytes and induces temozolomide drug resistance via Bcl-2 upregulation. Targeting Bcl-2 with ABT-737 significantly reversed temozolomide resistance in HOXA9-expressing cells [125].
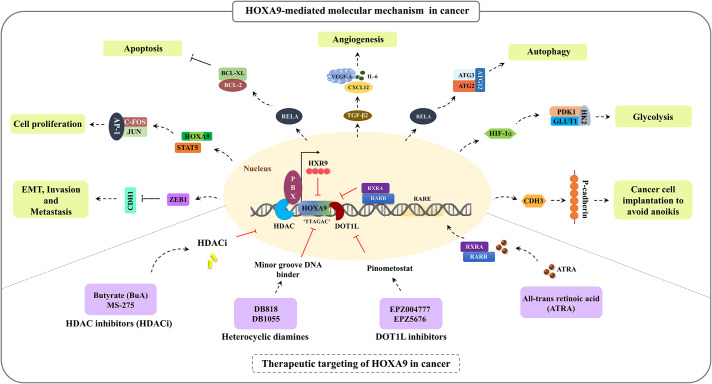
Figure 2 summarizes the molecular mechanism and functional implications of HOXA9 during cancer progression.
Fig. 2.
HOXA9-mediated molecular mechanism and its functional implications during cancer progression
Multifunctional role of HOXA9 in cancer
HOX proteins are well-known transcription factors that play a fundamental role in several cellular processes by binding to the promoter region of target genes with their DNA-binding homeodomain [4]. HOXA9 directly targets insulin-like growth factor 1 (IGF1) by binding to specific regions on its promoter, including the first intronic and DNase-hypersensitive region. This interaction leads to the stimulation of autocrine signaling, which is needed for hematopoietic transformation [126]. However, dysregulation of HOXA9 often leads to its binding to the promoter of oncogenes, resulting in cancer development. This section provides a comprehensive overview of the extensive role of HOXA9 as a transcription factor under normal physiological conditions and during the progression of cancer (Table 3).
Table 3.
Role of HOXA9 as a transcription factor
| HOXA9 as a transcription factor in normal physiological conditions | ||
|---|---|---|
| Normal physiological function | HOXA9 targets | References |
| Hematopoietic cell differentiation | IGF1 | [126] |
| Endothelial cell differentiation | SELE | [176] |
| Lymphoid and B-cell development | FLT3 | [177] |
| Myeloid differentiation | CYBB | [178] |
| Endothelial tube formation | EPHB4 | [58] |
| HOXA9 as a transcription factor in cancer | ||
| Cancer | HOXA9 targets | References |
| Ovarian cancer | TGF-β2, CDH3 | [78, 80] |
| Leukemia | BCL2, SOX4, CDX4, PIM1 | [91, 92, 94, 179] |
| Cervical cancer | GRHL2, IL17RD, FADD, CHEK2, TRAIL, IL-3/IL-5 | [19, 76] |
| Glioma | TRAIL, PTEN, HOTAIR,WNT6 | [95, 105, 180] |
| Breast cancer | BRCA1 | [98] |
| Cutaneous squamous cell carcinoma | HIF-1α, RELA | [87, 90] |
Interestingly, Shi et al., 2001 demonstrated that HOXA9 acts as a strong transcriptional repressor in the TGF-β pathway. Smad-4 interacts with HOXA9 and displaces it from its DNA-binding site, thereby facilitating the transcriptional activation of the OPN (osteopontin) promoter upon TGF-β stimulation [127].
Furthermore, Ohno et al. (2013) reported the nontranscriptional activity of HOXA9. It was discovered that HOXA9 acts as an E3 ubiquitin ligase in hematopoietic cells, leading to the degradation of the DNA replication inhibitor Geminin through ubiquitination [128]. HOXA9 transduction results in the stimulation of the activity of hematopoietic stem cells (HSCs) and progenitor cells, enhancing their activity [128]. However, its complex function in controlling a wide range of biological phenomena is mostly attributable to the tissue-specific expression of the protein and its collaboration with cofactors and binding partners.
HOXA9 is frequently associated with three-amino loop extension (TALE) homeodomain-containing cofactors, namely, MEIS1 and PBX [129, 130]. HOXA9 cooperates with PBX with a highly conserved motif called hexapeptide (HX), and the additional binding of MEIS induces the remodelling of the trimeric complex. In a study utilizing Bimolecular Fluorescence Complementation (BiFC) in cell lines, researchers discovered that the interaction between HOXA9-TALE protein is dependent on the specific activity of the HX motif and paralogue-specific motif of the HD domain [131]. The association of HOXA9 with PBX3 cofactors has been widely reported in leukemia and in a few solid cancer types [100, 132, 133]. The coexpression and association of HOXA9 with PBX3 enhance their binding to the promoter region of downstream target genes, potentially leading to a worse prognosis in cancer. It has been reported that poor prognosis in GC patients with upregulated HOXA9/PBX3 expression was found to be associated with lymph node metastasis and TNM stage (P value < 0.05) [100]. High coincidental expression and association of HOXA9 and PBX3 have demonstrated a synergistic impact on leukemic cell transformation and leukemogenesis. The study showed a strong association between HOXA9 and PBX3, specifically in patients with different subtypes of AML, including MLL-AML and cytologically abnormal AML (CA-AML) [132, 133]. Researchers have proposed the development of the HXR9 peptide as a potential strategy to disrupt the interaction between HOXA9 and PBX3, aiming to mitigate leukemogenesis [132].
Knockdown of HOXA9/TALE in cytologically normal AML (CN-AML) led to a reduction in aggressiveness and increased sensitivity of cancer cells to cytarabine chemotherapy [134]. In AML, the NPM + mutation has been found to induce high expression of HOXA9-PBX3, resulting in increased di- and trimethylation at the H3K79 locus. The study additionally demonstrated the use of a DOTL1 inhibitor (EPZ5676) to target HOXA9/PBX3, which effectively inhibited the survival of leukemic cells by inducing cell apoptosis [135]. Therefore, TALE factors can be considered potential pathological cofactors of HOXA9 in driving carcinogenesis.
Clinical applications of HOXA9 in cancer
HOXA9 serves as a potential biomarker for cancer diagnosis and prognosis. Determining the correlation between its expression patterns and disease progression aids clinicians in predicting patient outcomes, ultimately leading to suitable therapeutic interventions. In this section, we have summarized the clinical significance of HOXA9 in different cancer types.
HOXA9 as a diagnostic and prognostic marker
Accurate and early diagnosis of the condition is crucial for reducing the severity of disease progression and improving prognosis. HOXA9 and its binding partner MEIS1 have been identified as diagnostic markers for early cancer detection in paediatric acute leukaemia. Their expression has been observed to be inversely correlated with overall survival [111]. A recent study has demonstrated the upregulation of HOXA9/MEIS1 in adult acute leukaemia, suggesting its potential as a diagnostic marker for this condition. However, no significant association was observed between disease-free survival (DFS) and overall survival (OS) in the study population [136]. Indeed, several studies have demonstrated that HOXA9 methylation can serve as a valuable diagnostic and prognostic marker in various types of solid and hematological malignancies. It has also been suggested as a promising therapeutic target for these cancers [9, 69]. Methylation at the HOXA9 locus has emerged as a widely used biomarker in HNSC [17, 137, 138], BC [18], OVC [16, 112, 113], CC [19], lung cancer [115, 116, 139–141], PCa [75] and bladder cancer [109, 110, 142]. Hence, regular monitoring of HOXA9 methylation levels can provide valuable insights for making accurate clinical decisions.
HOXA9 as a marker for clinical staging application
Few studies have reported the emerging role of HOXA9 in differentiating clinical stages, which not only allows the detection of cancer at the early stage but also helps in guiding therapeutic decisions, which eventually aids in better clinical outcomes. In nasopharyngeal cancer (NPC), high expression of HOXA9 has been associated with advanced tumor stage (T), implying its role in cancer progression (P value < 0.05). Multivariate analysis revealed a worse prognosis in patients with HOXA9 overexpression [101]. In addition, HOXA9 overexpression has been significantly associated with higher TNM stage and positive lymph node metastasis in CRC [84].
In contrast, HOXA9 has been found to be downregulated in CC [18]. Downregulation of HOXA9 in CC patients (N = 154) has been significantly associated with clinical outcomes, including TNM stage, pathological grade, and differentiation (P value < 0.05) [143]. Furthermore, increased HOXA9 methylation levels were observed in more aggressive tumors, and these methylation levels strongly correlated with tumor number, size, grade, and stage in NMIBC [108]. In GC, the upregulation of HOXA9 expression was associated with cell differentiation and clinical staging, particularly in lymph node metastasis [100].
Therapeutic targeting of HOXA9 in cancer
Although aberrant expression of HOXA9 has been implicated in many cancer types, pharmacologically targeting HOXA9 in various cancer types presents a significant challenge owing to the complexity of the structure of HOXA9, its interaction with several binding partners, and its involvement in normal physiological processes. Nevertheless, a few studies have proposed the pharmacological targeting of HOXA9 in leukemia. In this section, we will discuss the emerging drugs that have undergone preclinical and clinical trials for the treatment of hematological and solid malignancies. However, the development of suitable drugs and targeted therapies specifically aimed at HOXA9 in solid malignancies is still in the early stages of research.
Lambert et al., 2019 conducted a comprehensive review on various strategies for targeting HOXA9 in AML. The overexpression of HOXA9 can be effectively targeted either by suppressing its expression through epigenetic modulation or by inhibiting its activity at the protein level [144]. Heterocyclic diamines, namely, DB818 and DB1055, were proposed as inhibitors of the HOXA9 transcription factor in leukemia, which function as minor groove DNA ligands on HOXA9, effectively competing with the HOXA9/DNA interaction [145]. Later, it was proposed that DB818 efficiently downregulated the expression of HOXA9 target genes such as the MYB, MYC, and BCL2 genes in AML [146].
DOT1L, a histone H3-lysine 79 (H3K79) methyltransferase enzyme, plays a crucial role in cell proliferation and cell survival in MLL. It facilitates methylation at the H3K79 position, leading to the overexpression of HOXA9 and MEIS1 [147]. Initially, researchers developed DOT1L inhibitors namely EPZ004777 and EPZ5676, generally known as pinometostat. These inhibitors specifically target and inhibit the activity of the DOT1L enzyme. Treatment with DOT1L inhibitors resulted in the downregulation of HOXA9 and induced cell death in AML cells carrying MLL translocations and NPM1 mutations [135, 148, 149]. Promising results were obtained from successful preclinical studies in xenograft mouse models and phase 1 clinical trials at safer doses, leading to improved clinical outcomes [150–152].
Subsequently, another inhibitor of DOT1L called SYC-522 was developed that shows potential in treating AML. The study revealed that treatment with SYC-522 resulted in a significant decrease in the expression levels of key leukaemia driver genes, including HOXA9 and MEIS1. Furthermore, it also led to a reduction in the expression of cell cycle-related and antiapoptotic genes. SYC-522-mediated inhibition of DOT1L was associated with increased chemosensitivity and improved clinical outcome in AML. Therefore, the combination of a DOT1L inhibitor and chemotherapy holds promise as a reliable approach for the treatment of AML [153]. Several therapeutic drugs, such as LSD1 inhibitors, HDAC class I inhibitors, and WDR5/MLL inhibitors, have demonstrated success in reducing the expression of HOXA9 in leukemia [88, 154–157].
Morgan et al. provided a comprehensive review on the therapeutic targeting of HOXA9 in solid tumors [158]. A synthetic peptide called HXR9 has been developed to specifically target the interaction between HOX and PBX. This peptide has shown promise in reducing the aggressive behavior of cancer cells in a few cancer types [159–161]. In OVC, the disruption of HOX/PBX complexes using the HXR9 peptide has led to HOXA9-mediated transcriptional alteration and induction of apoptosis in cancer cells [161]. In meningioma, targeting HOXA9 with HXR9 has shown promise as a reliable therapeutic option. The synthetic peptide HXR9 has been shown to effectively reduce the proliferation rate of meningioma cells and inhibit tumor aggressiveness. This is achieved by disrupting the HOXA9/PBX interaction, interfering with DNA binding, and subsequently altering the expression of target genes [160]. The HXR9 peptide has demonstrated its effectiveness in inducing apoptosis specifically in premalignant and oral squamous cell carcinoma (OSCC) cells, while having minimal impact on normal oral keratinocytes [162].
There is an enormous gap in targeting the oncogenic nature of HOXA9 in solid tumors. Further investigations are needed to understand the complexity of the regulation of HOXA9 and address off-target effects that may arise when designing and delivering drugs targeting this pathway.
Conclusion and future prospects
The HOXA9 transcription factor plays a crucial role in various biological processes, including embryonic development, embryo implantation, endothelial cell differentiation, and hematopoiesis. Aberrant expression of HOXA9 triggers it to be oncogenic, leading to the activation of cancer-associated signaling pathways. Its dysregulated expression has been observed in both hematological and solid malignancies, exhibiting context-dependent roles as either an oncogene or tumor suppressor.
This review provides a comprehensive summary of the regulation and functional role of HOXA9 in cancer progression. Several studies have highlighted the significance of HOXA9 as a biomarker for cancer prognosis and clinical staging, emphasizing its versatility in regulating several targets, modulating numerous signaling pathways, and promoting therapy resistance and recurrence. Despite extensive research conducted on HOXA9, there are still some notable research gaps in our understanding of its precise role in cancer.
Despite being deregulated in various cancer types, the full understanding of HOXA9-mediated molecular events driving cancer progression remains incomplete. Further research is needed to unravel the precise mechanisms through which HOXA9 promotes tumorigenesis and influences disease outcomes in the context of different cancers.
More studies are required to understand the regulation of HOXA9 by other HOX transcription factors, HOX cluster-embedded lncRNAs and miRNAs. Identifying the binding partners and cofactors of HOXA9 is crucial for unravelling downstream molecular pathways during cancer progression. Gaining a deeper understanding of the functional consequences of HOXA9 dysregulation would provide valuable insights for developing therapeutic interventions.
Further studies are needed to fully evaluate the clinical significance of HOXA9 in cancer, which is pivotal for the development of targeted therapies.
Further investigation is essential to investigate the prognostic and diagnostic roles of HOXA9 in larger patient cohorts across various cancer types.
It is important to explore the feasibility of using HOXA9 as a circulating biomarker in easily accessible bodily fluids such as serum, saliva, plasma, and blood. This approach would not only enhance affordability for patients but also enable clinicians to expedite the diagnosis of disease severity.
Targeting HOXA9 poses a significant challenge due to its transcriptional complexity, generation of multiple isoforms, tissue-specific expression, involvement in normal physiological functions, and roles in therapy resistance and recurrence. By carefully considering its context-specific dual role, it is crucial to design drugs that specifically and selectively inhibit its activity, minimizing any potential off-target effects.
It is worth noting that understanding the molecular behavior of HOXA9 in cancer requires further investigation. Addressing the existing research gaps is imperative for translating findings into effective therapies.
Acknowledgements
We thank the Wellcome Trust DBT India Alliance, Government of India (Grant No. IA/CPHI/18/1/503927), Joint CSIR-UGC NET Senior Research Fellowship, Government of India (File No. 09/1165(0011)/2020-EMR-I) and ICMR SRF, Government of India (2019/4115/CMB/BMS) and Manipal School of Life Sciences, MAHE, Manipal for infrastructure support.
Abbreviations
- EMT
epithelial-mesenchymal transition
- HOX
Homeobox
- AML
acute myeloid leukemia
- miRNAs
microRNAs
- lncRNAs
long noncoding RNAs
- CBP
CREB-binding protein
- AbdB
Abdominal B
- HSC
hematopoietic stem cells
- HSPCs
hematopoietic stem and progenitor cells
- HEP
hemogenic precursors
- EC
Endothelial cell
- eNOS
endothelial nitric oxide synthase
- VEGFR2
vascular endothelial growth factor receptor 2
- CC
cervical cancer
- CSCs
cancer stem cells
- CRC
colorectal cancer
- ATRA
all-trans retinoic acid
- EOC
epithelial ovarian cancer
- MSCs
mesenchymal stem cells
- CAFs
cancer-associated fibroblasts
- OVC
ovarian cancer
- CSCC
cutaneous squamous cell carcinoma
- HDAC
histone deacetylases
- PCa
prostate cancer
- GBM
glioblastoma
- TRAIL
tumor necrosis factor-related apoptosis-including ligand
- BC
breast cancer
- GC
gastric cancer
- TNM
tumor-node-metastasis
- NSCLC
non-small cell lung carcinoma
- NMIBC
nonmuscle invasive bladder cancer
- ctDNA
circulating tumor-specific DNA
- MTD
maximum tolerated dose
- IGF1
Insulin-like growth factor 1
- HX
hexapeptide
- TALE
three-amino loop extension
- BiFC
Bimolecular Fluorescence Complementation
- HGSOC
high-grade serous ovarian cancer
- NAC1
nucleus accumbens‑associated protein 1
- CA
AML-cytologically abnormal-AML
- CN
AML-cytologically normal-AML
- PML
paediatric acute leukaemia
- OS
overall survival
- DFS
disease-free survival
- NPC
nasopharyngeal cancer
Author Contributions
U.S.S performed the literature search and wrote the manuscript; D.A and U.S.S designed the figures and tables. F.A. edited the manuscript. D.A, S.P.K, and R.R edited the manuscript and did the critical revision. All the authors have read and approved the final manuscript.
Funding
Open access funding provided by Manipal Academy of Higher Education, Manipal. Wellcome Trust DBT India Alliance, Government of India (Grant No. IA/CPHI/18/1/503927).
Open access funding provided by Manipal Academy of Higher Education, Manipal
Data Availability
Not Applicable.
Not applicable.
Declarations
Ethical approval
Not Applicable.
Consent for publication
All the authors have read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Consent for participation
Not Applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Ferrier DE, Holland PW. Ancient origin of the hox gene cluster. Nature Reviews Genetics. 2001;2(1):33–38. doi: 10.1038/35047605. [DOI] [PubMed] [Google Scholar]
- 2.Holland PWH. Evolution of homeobox genes. Wiley Interdisciplinary Reviews Developmental Biology. 2013;2(1):31–45. doi: 10.1002/wdev.78. [DOI] [PubMed] [Google Scholar]
- 3.Paço A, Freitas R. HOX genes as transcriptional and epigenetic regulators during tumorigenesis and their value as therapeutic targets. Epigenomics. 2019;11(13):1539–1552. doi: 10.2217/epi-2019-0090. [DOI] [PubMed] [Google Scholar]
- 4.Svingen T, Tonissen KF. Hox transcription factors and their elusive mammalian gene targets. Heredity. 2006;97(2):88–96. doi: 10.1038/sj.hdy.6800847. [DOI] [PubMed] [Google Scholar]
- 5.Tschopp P, Tarchini B, Spitz F, Zakany J, Duboule D. Uncoupling time and space in the collinear regulation of hox genes. PLoS Genetics. 2009;5(3):e1000398. doi: 10.1371/journal.pgen.1000398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaunt, S. J., & Strachan, L. (1996). Temporal colinearity in expression of anterior hox genes in developing chick embryos. Developmental Dynamics: An Official Publication of the American Association of Anatomists, 207(3), 270–280. 10.1002/(SICI)1097-0177(199611)207:3<270::AID-AJA4>3.0.CO;2-E. [DOI] [PubMed]
- 7.Durston AJ. Vertebrate hox temporal collinearity: Does it exist and what is it’s function? Cell Cycle (Georgetown Tex) 2019;18(5):523–530. doi: 10.1080/15384101.2019.1577652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins CT, Hess JL. Role of HOXA9 in Leukemia: Dysregulation, cofactors and essential targets. Oncogene. 2016;35(9):1090–1098. doi: 10.1038/onc.2015.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cai H, Ke ZB, Dong RN, Chen H, Lin F, Zheng WC, Xu N. The prognostic value of homeobox A9 (HOXA9) methylation in solid tumors: A systematic review and meta-analysis. Translational cancer Research. 2021;10(10):4347–4354. doi: 10.21037/tcr-21-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wellik DM. Hox patterning of the vertebrate axial skeleton. Developmental Dynamics: An Official Publication of the American Association of Anatomists. 2007;236(9):2454–2463. doi: 10.1002/dvdy.21286. [DOI] [PubMed] [Google Scholar]
- 11.Xu B, Geerts D, Bu Z, Ai J, Jin L, Li Y, Zhu G. Regulation of endometrial receptivity by the highly expressed HOXA9, HOXA11 and HOXD10 HOX-class homeobox genes. Human Reproduction (Oxford England) 2014;29(4):781–790. doi: 10.1093/humrep/deu004. [DOI] [PubMed] [Google Scholar]
- 12.Yan J, Chen YX, Desmond A, Silva A, Yang Y, Wang H, Hua X. Cdx4 and menin coregulate Hoxa9 expression in hematopoietic cells. PloS One. 2006;1(1):e47. doi: 10.1371/journal.pone.0000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel CV, Sharangpani R, Bandyopadhyay S, DiCorleto PE. Endothelial cells express a novel, Tumor necrosis factor-alpha-regulated variant of HOXA9. The Journal of Biological Chemistry. 1999;274(3):1415–1422. doi: 10.1074/jbc.274.3.1415. [DOI] [PubMed] [Google Scholar]
- 14.Takeda A, Goolsby C, Yaseen NR. NUP98-HOXA9 induces long-term proliferation and blocks differentiation of primary human CD34 + hematopoietic cells. Cancer Research. 2006;66(13):6628–6637. doi: 10.1158/0008-5472.CAN-06-0458. [DOI] [PubMed] [Google Scholar]
- 15.Agrawal-Singh S, Bagri J, Giotopoulos G, Azazi DMA, Horton SJ, Lopez CK, Huntly BJP. HOXA9 forms a repressive complex with nuclear matrix-associated protein SAFB to maintain acute Myeloid Leukemia. Blood. 2023;141(14):1737–1754. doi: 10.1182/blood.2022016528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faaborg L, Jakobsen A, Waldstrøm M, Petersen CB, Andersen RF, Steffensen KD. HOXA9-methylated DNA as a diagnostic biomarker of ovarian malignancy. Biomarkers in Medicine. 2021;15(15):1309–1317. doi: 10.2217/bmm-2021-0144. [DOI] [PubMed] [Google Scholar]
- 17.Zhou C, Li J, Li Q, Liu H, Ye D, Wu Z, Deng H. The clinical significance of HOXA9 promoter hypermethylation in head and neck squamous cell carcinoma. Journal of Clinical Laboratory Analysis. 2019;33(5):e22873. doi: 10.1002/jcla.22873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park SM, Choi EY, Bae M, Choi JK, Kim YJ. A long-range interactive DNA methylation marker panel for the promoters of HOXA9 and HOXA10 predicts survival in Breast cancer patients. Clinical Epigenetics. 2017;9:73. doi: 10.1186/s13148-017-0373-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alvarado-Ruiz L, Martinez-Silva MG, Torres-Reyes LA, Pina-Sanchez P, Ortiz-Lazareno P, Bravo-Cuellar A, Jave-Suarez LF. HOXA9 is underexpressed in Cervical Cancer cells and its restoration decreases Proliferation, Migration and expression of epithelial-to-mesenchymal transition genes. Asian Pacific Journal of cancer Prevention: APJCP. 2016;17(3):1037–1047. doi: 10.7314/apjcp.2016.17.3.1037. [DOI] [PubMed] [Google Scholar]
- 20.Xu Q, Zhang Q, Dong M, Yu Y. MicroRNA-638 inhibits the progression of Breast cancer through targeting HOXA9 and suppressing Wnt/β-cadherin pathway. World Journal of Surgical Oncology. 2021;19(1):247. doi: 10.1186/s12957-021-02363-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang ZF, Li GR, Cao CN, Xu Q, Wang GD, Jiang XF. MicroRNA-1294 targets HOXA9 and has a Tumor suppressive role in osteosarcoma. European Review for Medical and Pharmacological Sciences. 2018;22(24):8582–8588. doi: 10.26355/eurrev_201812_16621. [DOI] [PubMed] [Google Scholar]
- 22.Chong GO, Jeon HS, Han HS, Son JW, Lee YH, Hong DG, Cho YL. Overexpression of microRNA-196b accelerates invasiveness of Cancer cells in recurrent epithelial Ovarian Cancer through regulation of Homeobox A9. Cancer Genomics & Proteomics. 2017;14(2):137–141. doi: 10.21873/cgp.20026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu C, Li B, Zhao S, Jin B, Jia R, Ge J, Xu H. MicroRNA-186-5p inhibits Proliferation and Metastasis of Esophageal Cancer by mediating HOXA9. OncoTargets and Therapy. 2019;12:8905–8914. doi: 10.2147/OTT.S227920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang SM, Pang J, Zhang KJ, Zhou ZY, Chen FY. lncRNA MIR503HG inhibits cell proliferation and promotes apoptosis in TNBC cells via the miR-224-5p/HOXA9 axis. Molecular Therapy Oncolytics. 2021;21:62–73. doi: 10.1016/j.omto.2021.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benson GV, Nguyen TH, Maas RL. The expression pattern of the murine Hoxa-10 gene and the sequence recognition of its homeodomain reveal specific properties of abdominal B-like genes. Molecular and Cellular Biology. 1995;15(3):1591–1601. doi: 10.1128/MCB.15.3.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim MH, Chang HH, Shin C, Cho M, Park D, Park HW. Genomic structure and sequence analysis of human HOXA-9. DNA and cell Biology. 1998;17(5):407–414. doi: 10.1089/dna.1998.17.407. [DOI] [PubMed] [Google Scholar]
- 27.Popovic R, Erfurth F, Zeleznik-Le N. Transcriptional complexity of the HOXA9 locus. Blood Cells Molecules & Diseases. 2008;40(2):156–159. doi: 10.1016/j.bcmd.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fujimoto S, Araki K, Chisaka O, Araki M, Takagi K, Yamamura K. Analysis of the murine Hoxa-9 cDNA: An alternatively spliced transcript encodes a truncated protein lacking the homeodomain. Gene. 1998;209(1–2):77–85. doi: 10.1016/s0378-1119(98)00014-6. [DOI] [PubMed] [Google Scholar]
- 29.Dintilhac A, Bihan R, Guerrier D, Deschamps S, Pellerin I. A conserved nonhomeodomain Hoxa9 isoform interacting with CBP is coexpressed with the typical Hoxa9 protein during embryogenesis. Gene Expression Patterns: GEP. 2004;4(2):215–222. doi: 10.1016/j.modgep.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 30.He M, Chen P, Arnovitz S, Li Y, Huang H, Neilly MB, Li Z. Two isoforms of HOXA9 function differently but work synergistically in human MLL-rearranged Leukemia. Blood Cells Molecules & Diseases. 2012;49(2):102–106. doi: 10.1016/j.bcmd.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di-Poï N, Koch U, Radtke F, Duboule D. Additive and global functions of HoxA cluster genes in mesoderm derivatives. Developmental Biology. 2010;341(2):488–498. doi: 10.1016/j.ydbio.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 32.Fromental-Ramain C, Warot X, Lakkaraju S, Favier B, Haack H, Birling C, Chambon P. Specific and redundant functions of the paralogous Hoxa-9 and Hoxd-9 genes in forelimb and axial skeleton patterning. Development (Cambridge England) 1996;122(2):461–472. doi: 10.1242/dev.122.2.461. [DOI] [PubMed] [Google Scholar]
- 33.Raines AM, Magella B, Adam M, Potter SS. Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Developmental Biology. 2015;15:28. doi: 10.1186/s12861-015-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He B, Ni ZL, Kong SB, Lu JH, Wang HB. Homeobox genes for embryo implantation: From mouse to human. Animal Models and Experimental Medicine. 2018;1(1):14–22. doi: 10.1002/ame2.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taylor HS, Vanden Heuvel GB, Igarashi P. A conserved hox axis in the mouse and human female reproductive system: Late establishment and persistent adult expression of the Hoxa cluster genes. Biology of Reproduction. 1997;57(6):1338–1345. doi: 10.1095/biolreprod57.6.1338. [DOI] [PubMed] [Google Scholar]
- 36.Du H, Taylor HS. The role of hox genes in Female Reproductive Tract Development, adult function, and fertility. Cold Spring Harbor Perspectives in Medicine. 2015;6(1):a023002. doi: 10.1101/cshperspect.a023002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ma L, Benson GV, Lim H, Dey SK, Maas RL. Abdominal B (AbdB) Hoxa genes: Regulation in adult uterus by estrogen and progesterone and repression in müllerian duct by the synthetic estrogen diethylstilbestrol (DES) Developmental Biology. 1998;197(2):141–154. doi: 10.1006/dbio.1998.8907. [DOI] [PubMed] [Google Scholar]
- 38.Daftary GS, Taylor HS. Implantation in the human: The role of HOX genes. Seminars in Reproductive Medicine. 2000;18(3):311–320. doi: 10.1055/s-2000-12568. [DOI] [PubMed] [Google Scholar]
- 39.Eun Kwon H, Taylor HS. The role of HOX genes in human implantation. Annals of the New York Academy of Sciences. 2004;1034:1–18. doi: 10.1196/annals.1335.001. [DOI] [PubMed] [Google Scholar]
- 40.Ashary N, Laheri S, Modi D. Homeobox genes in endometrium: From development to decidualization. The International Journal of Developmental Biology. 2020;64(1-2-3):227–237. doi: 10.1387/ijdb.190120dm. [DOI] [PubMed] [Google Scholar]
- 41.Lawrence HJ, Sauvageau G, Humphries RK, Largman C. The role of HOX homeobox genes in normal and leukemic hematopoiesis. Stem Cells (Dayton Ohio) 1996;14(3):281–291. doi: 10.1002/stem.140281. [DOI] [PubMed] [Google Scholar]
- 42.Magli, M. C., Largman, C., & Lawrence, H. J. (1997). Effects of HOX homeobox genes in blood cell differentiation. Journal of Cellular Physiology, 173(2), 168–177. 10.1002/(SICI)1097-4652(199711)173:2<168::AID-JCP16>3.0.CO;2-C. [DOI] [PubMed]
- 43.Lawrence HJ, Christensen J, Fong S, Hu YL, Weissman I, Sauvageau G, Largman C. Loss of expression of the Hoxa-9 homeobox gene impairs the proliferation and repopulating ability of hematopoietic stem cells. Blood. 2005;106(12):3988–3994. doi: 10.1182/blood-2005-05-2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sauvageau G, Lansdorp PM, Eaves CJ, Hogge DE, Dragowska WH, Reid DS, Humphries RK. Differential expression of homeobox genes in functionally distinct CD34 + subpopulations of human bone marrow cells. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(25):12223–12227. doi: 10.1073/pnas.91.25.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lawrence HJ, Helgason CD, Sauvageau G, Fong S, Izon DJ, Humphries RK, Largman C. Mice bearing a targeted interruption of the homeobox gene HOXA9 have defects in myeloid, erythroid, and lymphoid hematopoiesis. Blood. 1997;89(6):1922–1930. doi: 10.1182/blood.V89.6.1922. [DOI] [PubMed] [Google Scholar]
- 46.Ramos-Mejía V, Navarro-Montero O, Ayllón V, Bueno C, Romero T, Real PJ, Menendez P. HOXA9 promotes hematopoietic commitment of human embryonic stem cells. Blood. 2014;124(20):3065–3075. doi: 10.1182/blood-2014-03-558825. [DOI] [PubMed] [Google Scholar]
- 47.Zeng J, Yi D, Sun W, Liu Y, Chang J, Zhu L, Ma F. Overexpression of HOXA9 upregulates NF-κB signaling to promote human hematopoiesis and alter the hematopoietic differentiation potentials. Cell Regeneration (London England) 2021;10(1):9. doi: 10.1186/s13619-020-00066-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang Y, Sitwala K, Bronstein J, Sanders D, Dandekar M, Collins C, Hess JL. Identification and characterization of Hoxa9 binding sites in hematopoietic cells. Blood. 2012;119(2):388–398. doi: 10.1182/blood-2011-03-341081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schmidt A, Brixius K, Bloch W. Endothelial precursor cell migration during vasculogenesis. Circulation Research. 2007;101(2):125–136. doi: 10.1161/CIRCRESAHA.107.148932. [DOI] [PubMed] [Google Scholar]
- 50.Michaelis UR. Mechanisms of endothelial cell migration. Cellular and Molecular life Sciences: CMLS. 2014;71(21):4131–4148. doi: 10.1007/s00018-014-1678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bruderer M, Alini M, Stoddart MJ. Role of HOXA9 and VEZF1 in endothelial biology. Journal of Vascular Research. 2013;50(4):265–278. doi: 10.1159/000353287. [DOI] [PubMed] [Google Scholar]
- 52.Silva M, Videira PA, Sackstein R. E-Selectin ligands in the human mononuclear Phagocyte System: Implications for Infection, inflammation, and Immunotherapy. Frontiers in Immunology. 2017;8:1878. doi: 10.3389/fimmu.2017.01878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lewis H, Kaszubska W, DeLamarter JF, Whelan J. Cooperativity between two NF-kappa B complexes, mediated by high-mobility-group protein I(Y), is essential for cytokine-induced expression of the E-selectin promoter. Molecular and Cellular Biology. 1994;14(9):5701–5709. doi: 10.1128/mcb.14.9.5701-5709.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trivedi CM, Patel RC, Patel CV. Homeobox gene HOXA9 inhibits nuclear factor-kappa B dependent activation of endothelium. Atherosclerosis. 2007;195(2):e50–60. doi: 10.1016/j.atherosclerosis.2007.04.055. [DOI] [PubMed] [Google Scholar]
- 55.Trivedi CM, Patel RC, Patel CV. Differential regulation of HOXA9 expression by nuclear factor kappa B (NF-kappaB) and HOXA9. Gene. 2008;408(1–2):187–195. doi: 10.1016/j.gene.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Zhang N, Gong L, Zhang H, Cao C. High glucose-induced dysfunction of endothelial cells can be restored by HoxA9EC. Annals of Vascular Surgery. 2012;26(7):1002–1010. doi: 10.1016/j.avsg.2012.05.011. [DOI] [PubMed] [Google Scholar]
- 57.Du E, Li X, He S, Li X, He S. The critical role of the interplays of EphrinB2/EphB4 and VEGF in the induction of angiogenesis. Molecular Biology Reports. 2020;47(6):4681–4690. doi: 10.1007/s11033-020-05470-y. [DOI] [PubMed] [Google Scholar]
- 58.Bruhl T, Urbich C, Aicher D, Acker-Palmer A, Zeiher AM, Dimmeler S. Homeobox A9 transcriptionally regulates the EphB4 receptor to modulate endothelial cell migration and tube formation. Circulation Research. 2004;94(6):743–751. doi: 10.1161/01.RES.0000120861.27064.09. [DOI] [PubMed] [Google Scholar]
- 59.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: Common themes and new biologic insights. Blood. 2011;118(24):6247–6257. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu H, Valerio DG, Eisold ME, Sinha A, Koche RP, Hu W, Armstrong SA. NUP98 Fusion proteins interact with the NSL and MLL1 complexes to Drive Leukemogenesis. Cancer cell. 2016;30(6):863–878. doi: 10.1016/j.ccell.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shima Y, Yumoto M, Katsumoto T, Kitabayashi I. MLL is essential for NUP98-HOXA9-induced Leukemia. Leukemia. 2017;31(10):2200–2210. doi: 10.1038/leu.2017.62. [DOI] [PubMed] [Google Scholar]
- 62.Xie, H. M., & Bernt, K. M. (2022). HOXA amplification defines a genetically distinct subset of Angiosarcomas. Biomolecules, 12(8), 10.3390/biom12081124. [DOI] [PMC free article] [PubMed]
- 63.Aryal S, Zhang Y, Wren S, Li C, Lu R. Molecular regulators of HOXA9 in acute Myeloid Leukemia. The FEBS Journal. 2023;290(2):321–339. doi: 10.1111/febs.16268. [DOI] [PubMed] [Google Scholar]
- 64.Hu YL, Fong S, Largman C, Shen WF. HOXA9 regulates miR-155 in hematopoietic cells. Nucleic Acids Research. 2010;38(16):5472–5478. doi: 10.1093/nar/gkq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Schneider E, Pochert N, Ruess C, MacPhee L, Escano L, Miller C, Rouhi A. MicroRNA-708 is a novel regulator of the Hoxa9 program in myeloid cells. Leukemia. 2020;34(5):1253–1265. doi: 10.1038/s41375-019-0651-1. [DOI] [PubMed] [Google Scholar]
- 66.Yu SL, Lee DC, Sohn HA, Lee SY, Jeon HS, Lee JH, Kang J. Homeobox A9 directly targeted by miR-196b regulates aggressiveness through nuclear factor-kappa B activity in non-small cell Lung cancer cells. Molecular Carcinogenesis. 2016;55(12):1915–1926. doi: 10.1002/mc.22439. [DOI] [PubMed] [Google Scholar]
- 67.Qin K, Tian G, Chen G, Zhou D, Tang K. miR-647 inhibits glioma cell proliferation, colony formation and invasion by regulating HOXA9. The Journal of gene Medicine. 2020;22(3):e3153. doi: 10.1002/jgm.3153. [DOI] [PubMed] [Google Scholar]
- 68.Zheng DH, Wang X, Lu LN, Chen DL, Chen JM, Lin FM, Xu XB. MiR-638 serves as a Tumor suppressor by targeting HOXA9 in glioma. European Review for Medical and Pharmacological Sciences. 2018;22(22):7798–7806. doi: 10.26355/eurrev_201811_16404. [DOI] [PubMed] [Google Scholar]
- 69.Talarmain L, Clarke MA, Shorthouse D, Cabrera-Cosme L, Kent DG, Fisher J, Hall BA. HOXA9 has the hallmarks of a biological switch with implications in blood cancers. Nature Communications. 2022;13(1):5829. doi: 10.1038/s41467-022-33189-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tang L, Peng L, Tan C, Liu H, Chen P, Wang H. Role of HOXA9 in solid tumors: Mechanistic insights and therapeutic potential. Cancer cell International. 2022;22(1):349. doi: 10.1186/s12935-022-02767-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nallasamy P, Nimmakayala RK, Parte S, Are AC, Batra SK, Ponnusamy MP. Tumor microenvironment enriches the stemness features: The architectural event of therapy resistance and Metastasis. Molecular cancer. 2022;21(1):225. doi: 10.1186/s12943-022-01682-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Baghban R, Roshangar L, Jahanban-Esfahlan R, Seidi K, Ebrahimi-Kalan A, Jaymand M, Zare P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Communication and Signaling: CCS. 2020;18(1):59. doi: 10.1186/s12964-020-0530-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kim HJ, Ji YR, Lee YM. Crosstalk between angiogenesis and immune regulation in the Tumor microenvironment. Archives of Pharmacal Research. 2022;45(6):401–416. doi: 10.1007/s12272-022-01389-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morgan R, Pandha HS. HOX transcription factors and the prostate Tumor microenvironment. Journal of Cancer Metastasis and Treatment. 2017;3(12):278. doi: 10.20517/2394-4722.2017.31. [DOI] [Google Scholar]
- 75.Song, Y. P., Xian, P., Luo, H., Dai, J. Y., Bai, Y., Li, Y., & Tang, X. L. (2022). Comprehensive Landscape of HOXA2, HOXA9, and HOXA10 as Potential Biomarkers for Predicting Progression and Prognosis in Prostate Cancer. Journal of immunology research, 2022, 5740971. 10.1155/2022/5740971. [DOI] [PMC free article] [PubMed]
- 76.Alvarado-Ruíz, L., Aguilar-Lemarroy, A., Alejandro, B., & Jave Suarez, L. (2015). Determination of biological processes regulated by HOXA9 in cells derived from Cervical, Uterine and its influence on the immune response. Frontiers in Immunology, 6. 10.3389/conf.fimmu.2015.05.00056.
- 77.Ko SY, Barengo N, Ladanyi A, Lee JS, Marini F, Lengyel E, Naora H. HOXA9 promotes Ovarian cancer growth by stimulating cancer-associated fibroblasts. The Journal of Clinical Investigation. 2012;122(10):3603–3617. doi: 10.1172/JCI62229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ko, S. Y., & Naora, H. (2014). Adaptation of Ovarian cancer cells to the peritoneal environment: Multiple mechanisms of the developmental patterning gene HOXA9. Cancer cell & Microenvironment, 1(6), e379. [PMC free article] [PubMed]
- 79.Ko SY, Ladanyi A, Lengyel E, Naora H. Expression of the homeobox gene HOXA9 in Ovarian cancer induces peritoneal macrophages to acquire an M2 tumor-promoting phenotype. The American Journal of Pathology. 2014;184(1):271–281. doi: 10.1016/j.ajpath.2013.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Usui A, Ko SY, Barengo N, Naora H. P-cadherin promotes Ovarian cancer dissemination through Tumor cell aggregation and tumor-peritoneum interactions. Molecular cancer Research: MCR. 2014;12(4):504–513. doi: 10.1158/1541-7786.MCR-13-0489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ko SY, Naora H. HOXA9 promotes homotypic and heterotypic cell interactions that facilitate Ovarian cancer dissemination via its induction of P-cadherin. Molecular cancer. 2014;13:170. doi: 10.1186/1476-4598-13-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang JC. Cancer stem cells: Role in Tumor growth, recurrence, Metastasis, and treatment resistance. Medicine. 2016;95(1 Suppl 1):S20–S25. doi: 10.1097/MD.0000000000004766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Osmond, B., Facey, C. O. B., Zhang, C., & Boman, B. M. (2022). HOXA9 overexpression contributes to Stem Cell Overpopulation that drives Development and Growth of Colorectal Cancer. International Journal of Molecular Sciences, 23(12), 10.3390/ijms23126799. [DOI] [PMC free article] [PubMed]
- 84.Oncology letters, 15(3), 2756–2762. https://doi.org/10.3892/ol.2017.7650.
- 85.PloS one, 10(7), e0132566. https://doi.org/10.1371/journal.pone.0132566.
- 86.Jing X, Yang F, Shao C, Wei K, Xie M, Shen H, Shu Y. Role of hypoxia in cancer therapy by regulating the Tumor microenvironment. Molecular cancer. 2019;18(1):157. doi: 10.1186/s12943-019-1089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.α-mediated glycolysis through interacting with CRIP2 to repress cutaneous squamous cell carcinoma development. Nature communications, 9(1), 1480. https://doi.org/10.1038/s41467-018-03914-5. [DOI] [PMC free article] [PubMed]
- 88.Dimmeler, S. (2005). Histone deacetylase activity is essential for the expression of HoxA9 and for endothelial commitment of progenitor cells. The Journal of experimental medicine, 201(11), 1825–1835. https://doi.org/10.1084/jem.20042097. [DOI] [PMC free article] [PubMed]
- 89.Del Bene F, Wittbrodt J. Cell cycle control by homeobox genes in development and Disease. Seminars in cell & Developmental Biology. 2005;16(3):449–460. doi: 10.1016/j.semcdb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 90.Han, S., Li, X., Liang, X., & Zhou, L. (2019). HOXA9 transcriptionally promotes apoptosis and represses autophagy by targeting NF-κB in cutaneous squamous cell carcinoma. Cells, 8(11), 10.3390/cells8111360. [DOI] [PMC free article] [PubMed]
- 91.Oncotarget, 4(11), 1933–1947. https://doi.org/10.18632/oncotarget.1306.
- 92.eLife, 10. https://doi.org/10.7554/eLife.64148.
- 93.Cools, J. (2018). HOXA9 Cooperates with Activated JAK/STAT Signaling to Drive Leukemia Development. Cancer discovery, 8(5), 616–631. https://doi.org/10.1158/2159-8290.CD-17-0583. [DOI] [PubMed]
- 94.Hu YL, Passegué E, Fong S, Largman C, Lawrence HJ. Evidence that the Pim1 kinase gene is a direct target of HOXA9. Blood. 2007;109(11):4732–4738. doi: 10.1182/blood-2006-08-043356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cancer research, 70(2), 453–462. https://doi.org/10.1158/0008-5472.CAN-09-2189.
- 96.International journal of clinical and experimental pathology, 7(11), 7409–7418. [PMC free article] [PubMed]
- 97.The Journal of biological chemistry, 290(32), 19999–20008. https://doi.org/10.1074/jbc.M115.659144.
- 98.Weaver, V. M. (2010). HOXA9 regulates BRCA1 expression to modulate human breast tumor phenotype. The Journal of clinical investigation, 120(5), 1535–1550. https://doi.org/10.1172/JCI39534. [DOI] [PMC free article] [PubMed]
- 99.Proceedings of the National Academy of Sciences of the United States of America, 110(24), 9920–9925. https://doi.org/10.1073/pnas.1305172110. [DOI] [PMC free article] [PubMed]
- 100.Ma YY, Zhang Y, Mou XZ, Liu ZC, Ru GQ, Li E. High level of homeobox A9 and PBX homeobox 3 expression in gastric cancer correlates with poor prognosis. Oncology Letters. 2017;14(5):5883–5889. doi: 10.3892/ol.2017.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu T, Ji C, Sun Y, Bai W. HOXA9 expression is Associated with Advanced Tumour Stage and Prognosis in Nasopharyngeal Carcinoma. Cancer Management and Research. 2021;13:4147–4154. doi: 10.2147/CMAR.S305814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Peppelenbosch, M. P. (2019). HOXA9 mediates and marks premalignant compartment size expansion in colonic adenomas. Carcinogenesis, 40(12), 1514–1524. https://doi.org/10.1093/carcin/bgz038. [DOI] [PubMed]
- 103.Hou, B. (2017). miR-133b suppresses metastasis by targeting HOXA9 in human colorectal cancer. Oncotarget, 8(38), 63935–63948. https://doi.org/10.18632/oncotarget.19212. [DOI] [PMC free article] [PubMed]
- 104.Cancer research, 77(12), 3181–3193. https://doi.org/10.1158/0008-5472.CAN-16-2797.
- 105.Molecular oncology, 14(6), 1224–1241. https://doi.org/10.1002/1878-0261.12633.
- 106.Cancer, 121(12), 1957–1965. https://doi.org/10.1002/cncr.29303.
- 107.Kim, D.-H. (2015). HOXA9 inhibits migration of lung cancer cells and its hypermethylation is associated with recurrence in non-small cell lung cancer. Molecular carcinogenesis, 54 Suppl 1, E72-80. https://doi.org/10.1002/mc.22180. [DOI] [PubMed]
- 108.International Journal of Cancer, 133(5), 1135–1142. https://doi.org/10.1002/ijc.28121.
- 109.PloS one, 10(9), e0137003. https://doi.org/10.1371/journal.pone.0137003.
- 110.Reinert T, Borre M, Christiansen A, Hermann GG, Ørntoft TF, Dyrskjøt L. Diagnosis of Bladder cancer recurrence based on urinary levels of EOMES, HOXA9, POU4F2, TWIST1, VIM, and ZNF154 hypermethylation. PloS One. 2012;7(10):e46297. doi: 10.1371/journal.pone.0046297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adamaki M, Lambrou GI, Athanasiadou A, Vlahopoulos S, Papavassiliou AG, Moschovi M. HOXA9 and MEIS1 gene overexpression in the diagnosis of childhood acute leukemias: Significant correlation with relapse and overall survival. Leukemia Research. 2015;39(8):874–882. doi: 10.1016/j.leukres.2015.04.012. [DOI] [PubMed] [Google Scholar]
- 112.Rusan M, Andersen RF, Jakobsen A, Steffensen KD. Circulating HOXA9-methylated tumour DNA: A novel biomarker of response to poly (ADP-ribose) polymerase inhibition in BRCA-mutated epithelial Ovarian cancer. European Journal of Cancer. 2020;125:121–129. doi: 10.1016/j.ejca.2019.11.012. [DOI] [PubMed] [Google Scholar]
- 113.Faaborg, L., Andersen, R. F., Waldstrøm, M., Henriksen, J. R., Adimi, P., Jakobsen, A., & Steffensen, K. D. (2022). Prognostic impact of circulating methylated homeobox A9 DNA in patients undergoing treatment for recurrent Ovarian Cancer. Cancers, 14(7), 10.3390/cancers14071766. [DOI] [PMC free article] [PubMed]
- 114.Clinica chimica acta; international journal of clinical chemistry, 522, 152–157. https://doi.org/10.1016/j.cca.2021.08.020. [DOI] [PubMed]
- 115.Wen SWC, Andersen RF, Hansen TF, Nyhus CH, Hager H, Hilberg O, Jakobsen A. The prognostic impact of circulating homeobox A9 methylated DNA in advanced non-small cell Lung cancer. Translational Lung cancer Research. 2021;10(2):855–865. doi: 10.21037/tlcr-20-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lung cancer (Amsterdam, Netherlands), 122, 151–159. https://doi.org/10.1016/j.lungcan.2018.05.021. [DOI] [PMC free article] [PubMed]
- 117.Kossatz-Boehlert, U. (2020). Cancer Stem Cells-Origins and Biomarkers: Perspectives for Targeted Personalized Therapies. Frontiers in immunology, 11, 1280. https://doi.org/10.3389/fimmu.2020.01280. [DOI] [PMC free article] [PubMed]
- 118.Schito L, Rey S, Xu P, Man S, Cruz-Muñoz W, Kerbel RS. Metronomic chemotherapy offsets HIFα induction upon maximum-tolerated dose in metastatic cancers. EMBO Molecular Medicine. 2020;12(9):e11416. doi: 10.15252/emmm.201911416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.De Souza R, Zahedi P, Badame RM, Allen C, Piquette-Miller M. Chemotherapy dosing schedule influences drug resistance development in Ovarian cancer. Molecular cancer Therapeutics. 2011;10(7):1289–1299. doi: 10.1158/1535-7163.MCT-11-0058. [DOI] [PubMed] [Google Scholar]
- 120.Zahreddine H, Borden KLB. Mechanisms and insights into drug resistance in cancer. Frontiers in Pharmacology. 2013;4:28. doi: 10.3389/fphar.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tran AP, Al-Radhawi A, Kareva M, Wu I, Waxman J, Sontag ED. Delicate balances in Cancer Chemotherapy: Modelling Immune recruitment and emergence of systemic drug resistance. Frontiers in Immunology. 2020;11:1376. doi: 10.3389/fimmu.2020.01376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Andersen, L. B., Mahler, M. S. K., Andersen, R. F., Jensen, L. H., & Raunkilde, L. (2022). The clinical impact of methylated homeobox A9 ctDNA in patients with non-resectable biliary Tract Cancer treated with Erlotinib and Bevacizumab. Cancers, 14(19), 10.3390/cancers14194598. [DOI] [PMC free article] [PubMed]
- 123.Li XF, Zhang HB, Huo Y. High HOXA9 gene expression predicts response to chemotherapy and prognosis of high-grade serous Ovarian cancer patients. The Journal of International Medical Research. 2022;50(11):3000605221135864. doi: 10.1177/03000605221135864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ju T, Jin H, Ying R, Xie Q, Zhou C, Gao D. Overexpression of NAC1 confers drug resistance via HOXA9 in colorectal carcinoma cells. Molecular Medicine Reports. 2017;16(3):3194–3200. doi: 10.3892/mmr.2017.6986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Oncotarget, 6(10), 7657–7674. https://doi.org/10.18632/oncotarget.3150.
- 126.Steger J, Füller E, Garcia-Cuellar MP, Hetzner K, Slany RK. Insulin-like growth factor 1 is a direct HOXA9 target important for hematopoietic transformation. Leukemia. 2015;29(4):901–908. doi: 10.1038/leu.2014.287. [DOI] [PubMed] [Google Scholar]
- 127.Shi X, Bai S, Li L, Cao X. Hoxa-9 represses transforming growth factor-beta-induced osteopontin gene transcription. The Journal of Biological Chemistry. 2001;276(1):850–855. doi: 10.1074/jbc.M005955200. [DOI] [PubMed] [Google Scholar]
- 128.PloS one, 8(1), e53161. https://doi.org/10.1371/journal.pone.0053161.
- 129.Leukemia, 13(12), 1993–1999. https://doi.org/10.1038/sj.leu.2401578.
- 130.Collins CT, Hess JL. Deregulation of the HOXA9/MEIS1 axis in acute Leukemia. Current Opinion in Hematology. 2016;23(4):354–361. doi: 10.1097/MOH.0000000000000245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Dard A, Jia Y, Reboulet J, Bleicher F, Lavau C, Merabet S. The human HOXA9 protein uses paralogue-specific residues of the homeodomain to interact with TALE-class cofactors. Scientific Reports. 2019;9(1):5664. doi: 10.1038/s41598-019-42096-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Blood, 121(8), 1422–1431. https://doi.org/10.1182/blood-2012-07-442004.
- 133.Chen, J. (2012). Upregulation of a HOXA-PBX3 homeobox-gene signature following downregulation of miR-181 is associated with adverse prognosis in patients with cytogenetically abnormal AML. Blood, 119(10), 2314–2324. https://doi.org/10.1182/blood-2011-10-386235. [DOI] [PMC free article] [PubMed]
- 134.Haematologica, 98(8), 1216–1225. https://doi.org/10.3324/haematol.2012.079012.
- 135.Xu, J. (2018). Inactivation of PBX3 and HOXA9 by downregulating H3K79 methylation represses NPM1-mutated leukemic cell survival. Theranostics, 8(16), 4359–4371. https://doi.org/10.7150/thno.26900. [DOI] [PMC free article] [PubMed]
- 136.Abdelrahman, A. M. N., Tolba, F. M., Kamal, H. M., Abdellateif, M. S., Ahmed, H. A., & Hassan, N. M. (2023). Evaluation of the HOXA9 and MEIS1 genes as a potential biomarker in adult acute Myeloid Leukemia. Egyptian Journal of Medical Human Genetics, 24(1), 10.1186/s43042-023-00391-4.
- 137.Cancer prevention research (Philadelphia, Pa.), 4(7), 1061–1072. https://doi.org/10.1158/1940-6207.CAPR-11-0006. [DOI] [PMC free article] [PubMed]
- 138.Uchida K, Veeramachaneni R, Huey B, Bhattacharya A, Schmidt BL, Albertson DG. Investigation of HOXA9 promoter methylation as a biomarker to distinguish Oral cancer patients at low risk of neck Metastasis. BMC cancer. 2014;14:353. doi: 10.1186/1471-2407-14-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Abou-Zeid A, Hashad D, Baess A, Mosaad M, Tayae E. HOXA9 gene promotor methylation and copy number variation of SOX2 and HV2 genes in cell free DNA: A potential diagnostic panel for non-small cell Lung cancer. BMC cancer. 2023;23(1):329. doi: 10.1186/s12885-023-10793-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Hilberg, O. (2021). Validating Methylated HOXA9 in Bronchial Lavage as a Diagnostic Tool in Patients Suspected of Lung Cancer. Cancers, 13(16). https://doi.org/10.3390/cancers13164223. [DOI] [PMC free article] [PubMed]
- 141.Clinical cancer research: an official journal of the American Association for Cancer Research, 20(7), 1856–1864. https://doi.org/10.1158/1078-0432.CCR-13-2109. [DOI] [PMC free article] [PubMed]
- 142.Xu, J. (2020). HOXA9, PCDH17, POU4F2, and ONECUT2 as a Urinary Biomarker Combination for the Detection of Bladder Cancer in Chinese Patients with Hematuria. European urology focus, 6(2), 284–291. https://doi.org/10.1016/j.euf.2018.09.016. [DOI] [PubMed]
- 143.Zhang, K. (n.d.) (Ed.). The expression of HOXA9 and its prognostic value in cervical cancer, 1–12.
- 144.Lambert, M., Alioui, M., Jambon, S., Depauw, S., Van Seuningen, I., & David-Cordonnier, M. H. (2019). Direct and indirect targeting of HOXA9 transcription factor in Acute Myeloid Leukemia. Cancers, 11(6), 10.3390/cancers11060837. [DOI] [PMC free article] [PubMed]
- 145.Journal of medicinal chemistry, 62(3), 1306–1329. https://doi.org/10.1021/acs.jmedchem.8b01448.
- 146.Sonoda Y, Itoh M, Tohda S. Effects of HOXA9 inhibitor DB818 on the growth of Acute myeloid Leukaemia cells. Anticancer Research. 2021;41(4):1841–1847. doi: 10.21873/anticanres.14950. [DOI] [PubMed] [Google Scholar]
- 147.Sarno F, Nebbioso A, Altucci L. DOT1L: A key target in normal chromatin remodelling and in mixed-lineage Leukaemia treatment. Epigenetics. 2020;15(5):439–453. doi: 10.1080/15592294.2019.1699991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cancer cell, 20(1), 53–65. https://doi.org/10.1016/j.ccr.2011.06.009.
- 149.Blood, 122(6), 1017–1025. https://doi.org/10.1182/blood-2013-04-497644.
- 150.Blood, 131(24), 2661–2669. https://doi.org/10.1182/blood-2017-12-818948.
- 151.Journal of controlled release: official journal of the Controlled Release Society, 220(Pt B), 758–765. https://doi.org/10.1016/j.jconrel.2015.09.023. [DOI] [PubMed]
- 152.Waters NJ. Preclinical pharmacokinetics and pharmacodynamics of Pinometostat (EPZ-5676), a first-in-Class, small molecule S-Adenosyl methionine competitive inhibitor of DOT1L. European Journal of drug Metabolism and Pharmacokinetics. 2017;42(6):891–901. doi: 10.1007/s13318-017-0404-3. [DOI] [PubMed] [Google Scholar]
- 153.Liu W, Deng L, Song Y, Redell M. DOT1L inhibition sensitizes MLL-rearranged AML to chemotherapy. PloS One. 2014;9(5):e98270. doi: 10.1371/journal.pone.0098270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Lillico R, Lawrence CK, Lakowski TM. Selective DOT1L, LSD1, and HDAC class I inhibitors reduce HOXA9 expression in MLL-AF9 Rearranged Leukemia Cells, however, Dysregulate the expression of many histone-modifying enzymes. Journal of Proteome Research. 2018;17(8):2657–2667. doi: 10.1021/acs.jproteome.8b00118. [DOI] [PubMed] [Google Scholar]
- 155.Yang C, Fang Y, Luo X, Teng D, Liu Z, Zhou Y, Liao G. Discovery of natural product-like spirooxindole derivatives as highly potent and selective LSD1/KDM1A inhibitors for AML treatment. Bioorganic Chemistry. 2022;120:105596. doi: 10.1016/j.bioorg.2022.105596. [DOI] [PubMed] [Google Scholar]
- 156.Journal of the American Chemical Society, 135(2), 669–682. https://doi.org/10.1021/ja306028q. [DOI] [PMC free article] [PubMed]
- 157.Li, D.-D., Chen, W.-L., Wang, Z.-H., Xie, Y.-Y., Xu, X.-L., Jiang, Z.-Y., … Guo, X.-K. (2016). High-affinity small molecular blockers of mixed lineage leukemia 1 (MLL1)-WDR5 interaction inhibit MLL1 complex H3K4 methyltransferase activity. European journal of medicinal chemistry, 124, 480–489. https://doi.org/10.1016/j.ejmech.2016.08.036. [DOI] [PubMed]
- 158.Morgan R, El-Tanani M, Hunter KD, Harrington KJ, Pandha HS. Targeting HOX/PBX dimers in cancer. Oncotarget. 2017;8(19):32322–32331. doi: 10.18632/oncotarget.15971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Morgan R, Pirard PM, Shears L, Sohal J, Pettengell R, Pandha HS. Antagonism of HOX/PBX dimer formation blocks the in vivo proliferation of Melanoma. Cancer Research. 2007;67(12):5806–5813. doi: 10.1158/0008-5472.CAN-06-4231. [DOI] [PubMed] [Google Scholar]
- 160.Cancer chemotherapy and pharmacology, 73(1), 53–60. https://doi.org/10.1007/s00280-013-2316-5.
- 161.Morgan R, Ploughright L, Harrington KJ, Michael A, Pandha HS. Targeting HOX and PBX transcription factors in Ovarian cancer. BMC cancer. 2010;10:89. doi: 10.1186/1471-2407-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Platais C, Radhakrishnan R, Niklander Ebensperger S, Morgan R, Lambert DW, Hunter KD. Targeting HOX-PBX interactions causes death in oral potentially malignant and squamous carcinoma cells but not normal oral keratinocytes. BMC cancer. 2018;18(1):723. doi: 10.1186/s12885-018-4622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Clinical cancer research: an official journal of the American Association for Cancer Research, 23(22), 7141–7152. https://doi.org/10.1158/1078-0432.CCR-17-1222. [DOI] [PubMed]
- 164.Chen, G. (2020). MiR-182-5p and its target HOXA9 in non-small cell lung cancer: a clinical and in-silico exploration with the combination of RT–qPCR, miRNA-seq and miRNA-chip. BMC medical genomics, 13(1), 3. https://doi.org/10.1186/s12920-019-0648-7. [DOI] [PMC free article] [PubMed]
- 165.Theranostics, 13(1), 77–94. https://doi.org/10.7150/thno.77404.
- 166.Haematologica. Italy. https://doi.org/10.3324/haematol.2019.223297.
- 167.Wang K, Jin J, Ma T, Zhai H. MiR-139-5p inhibits the tumorigenesis and progression of oral squamous carcinoma cells by targeting HOXA9. Journal of Cellular and Molecular Medicine. 2017;21(12):3730–3740. doi: 10.1111/jcmm.13282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ma, Y. (2019). miR-652 Promotes Proliferation and Migration of Uveal Melanoma Cells by Targeting HOXA9. Medical science monitor: international medical journal of experimental and clinical research, 25, 8722–8732. https://doi.org/10.12659/MSM.917099. [DOI] [PMC free article] [PubMed]
- 169.Cheng Y, Jutooru I, Chadalapaka G, Corton JC, Safe S. The long noncoding RNA HOTTIP enhances Pancreatic cancer cell proliferation, survival and migration. Oncotarget. 2015;6(13):10840–10852. doi: 10.18632/oncotarget.3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Liu J, Qian J, Mo Q, Tang L, Xu Q. Long noncoding RNA PCED1B-AS1 promotes the proliferation of colorectal adenocarcinoma through regulating the miR-633/HOXA9 axis. Bioengineered. 2022;13(3):5407–5420. doi: 10.1080/21655979.2022.2037225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Lu, S., Lu, J., Liu, L., Sun, Y., Zhao, Y., Tan, X., & Li, J. (2022). Circ_0026359 induces HOXA9 to regulate gastric cancer malignant progression through miR-140-3p. Applied Biological Chemistry, 65(1), 10.1186/s13765-022-00726-6.
- 172.Zhang N, Meng X, Mei L, Zhao C, Chen W. LncRNA DLX6-AS1 promotes Tumor proliferation and Metastasis in osteosarcoma through modulating miR-641/HOXA9 signaling pathway. Journal of Cellular Biochemistry. 2019;120(7):11478–11489. doi: 10.1002/jcb.28426. [DOI] [PubMed] [Google Scholar]
- 173.Journal of cellular and molecular medicine, 26(2), 385–398. https://doi.org/10.1111/jcmm.17091.
- 174.Rossi, F. M. (2014). The methyltransferase G9a regulates HoxA9-dependent transcription in AML. Genes & development, 28(4), 317–327. https://doi.org/10.1101/gad.236794.113. [DOI] [PMC free article] [PubMed]
- 175.Cancer letters, 410, 68–81. https://doi.org/10.1016/j.canlet.2017.09.019.
- 176.Bandyopadhyay S, Ashraf MZ, Daher P, Howe PH, DiCorleto PE. HOXA9 participates in the transcriptional activation of E-selectin in endothelial cells. Molecular and Cellular Biology. 2007;27(12):4207–4216. doi: 10.1128/MCB.00052-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Gwin K, Frank E, Bossou A, Medina KL. Hoxa9 regulates Flt3 in lymphohematopoietic progenitors. Journal of Immunology (Baltimore Md : 1950) 2010;185(11):6572–6583. doi: 10.4049/jimmunol.0904203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bei L, Lu Y, Eklund EA. HOXA9 activates transcription of the gene encoding gp91Phox during myeloid differentiation. The Journal of Biological Chemistry. 2005;280(13):12359–12370. doi: 10.1074/jbc.M408138200. [DOI] [PubMed] [Google Scholar]
- 179.Bei L, Shah C, Wang H, Huang W, Platanias LC, Eklund EA. Regulation of CDX4 gene transcription by HoxA9, HoxA10, the mll-ell oncogene and Shp2 during leukemogenesis. Oncogenesis. 2014;3(12):e135. doi: 10.1038/oncsis.2014.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Xavier-Magalh?es, A., Gon?alves, C. S., Fogli, A., Louren?o, T., Pojo, M., Pereira, B., ? Costa, B. M. (2018). The long noncoding RNA HOTAIR is transcriptionally activated by HOXA9 and is an independent prognostic marker in patients with malignant glioma. Oncotarget, 9(21), 15740–15756. https://doi.org/10.18632/oncotarget.24597. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not Applicable.
Not applicable.