Abstract
Introduction
Neuroinflammation evaluation after acute ischemic stroke is a promising option for selecting an appropriate post-stroke treatment strategy. To assess neuroinflammation in vivo, translocator protein PET (TSPO PET) can be used. However, the gold standard TSPO PET quantification method includes a 90 min scan and continuous arterial blood sampling, which is challenging to perform on a routine basis. In this work, we determine what information is required for a simplified quantification approach using a machine learning algorithm.
Materials and Methods
We analyzed data from 18 patients with ischemic stroke who received 0–90 min [18F]GE-180 PET as well as T1-weigted (T1w), FLAIR, and arterial spin labeling (ASL) MRI scans. During PET scans, five manual venous blood samples at 5, 15, 30, 60, and 85 min post injection (p.i.) were drawn, and plasma activity concentration was measured. Total distribution volume (VT) was calculated using Logan plot with the full dynamic PET and an image-derived input function (IDIF) from the carotid arteries. IDIF was scaled by a calibration factor derived from all the measured plasma activity concentrations. The calculated VT values were used for training a random forest regressor. As input features for the model, we used three late PET frames (60–70, 70–80, and 80–90 min p.i.), the ASL image reflecting perfusion, the voxel coordinates, the lesion mask, and the five plasma activity concentrations. The algorithm was validated with the leave-one-out approach. To estimate the impact of the individual features on the algorithm’s performance, we used Shapley Additive Explanations (SHAP). Having determined that the three late PET frames and the plasma activity concentrations were the most important features, we tested a simplified quantification approach consisting of dividing a late PET frame by a plasma activity concentration. All the combinations of frames/samples were compared by means of concordance correlation coefficient and Bland-Altman plots.
Results
When using all the input features, the algorithm predicted VT values with high accuracy (87.8 ± 8.3%) for both lesion and non-lesion voxels. The SHAP values demonstrated high impact of the late PET frames (60–70, 70–80, and 80–90 min p.i.) and plasma activity concentrations on the VT prediction, while the influence of the ASL-derived perfusion, voxel coordinates, and the lesion mask was low. Among all the combinations of the late PET frames and plasma activity concentrations, the 70–80 min p.i. frame divided by the 30 min p.i. plasma sample produced the closest VT estimate in the ischemic lesion.
Conclusion
Reliable TSPO PET quantification is achievable by using a single late PET frame divided by a late blood sample activity concentration.
Keywords: Quantitative PET, TSPO, Ischemic stroke, GE180, Image-derived input function, Machine learning
1. Introduction
Stroke is the second leading cause of death [1] and the third main cause of long-term disability worldwide (WHO 2020 report). Hence, it is important to limit the extent of neuronal damage and to improve recovery from stroke. However, such post-stroke therapy is currently available only in the acute phase of vascular occlusion, while treatment at later time points is limited to rehabilitative training. Therefore, novel therapies are needed to improve neurological recovery after brain injury.
Microglia, the brain resident innate immune cells, are an excellent target candidate for such a therapy, as these were shown to contribute to long-term neuronal repair after stroke [2], but also were linked to neurodegeneration and excessive synaptic pruning [3]. Microglia activation is strongly correlated to the 18 kDa translocator protein (TSPO) expression level on the outer membrane of microglial mitochondria [4]. There is still limited understanding of the inflammatory response after stroke. It is known that microglia activation occurs after acute ischemic stroke and that it may persist for several months [5]. Microglia may infiltrate connecting white matter tracts, as shown by means of [11C]PK11195 positron emission tomography (PET) [6], [7], thus causing a secondary neurodegeneration. However, this has not been investigated systematically. Moreover, there are no studies with [18F]-labelled second-generation PET tracers, such as flutriciclamide ([18F]GE-180), which has higher binding potential [8], improved signal-to-noise ratio, and lower nonspecific binding in healthy tissue [9] compared to the first-generation tracers as shown in animal studies. [18F]GE-180 was already successfully used by multiple authors in clinical studies to assess activated microglia in various disease, e.g. in glioma [10], [11], Alzheimer’s disease [12], multiple sclerosis [13], 4-repeat taupathies [14], [15], and fibrillar amyloidosis [15].
The gold standard approach for TSPO binding quantification is based on the reversible two-tissue compartment model to calculate the volume of distribution (VT) [16]. As shown by Fan et al. [16], the two-tissue compartment model can be replaced by the Logan plot analysis [17] for VT estimation, which was successfully implemented by several authors [13], [14], [18]. While Logan plot produces more robust estimates due to reduced number of fit parameters, it still requires a long 90 min TSPO PET scan and, in case of invasive Logan plot, continuous arterial blood sampling and estimation of tracer metabolite content in the arterial plasma. This approach is challenging for hospital staff to perform on a routine basis and reduces patients’ comfort. In our previous work [19], we showed that a simplified TSPO binding estimation based on late 60–90 min post injection (p.i.) [18F]GE-180 PET is possible in a mouse photothrombotic stroke model when using cerebellar white matter as a pseudo-reference tissue.
In this study, we aimed at establishing a similar simplified approach for human data, now including information additional to the late PET frames, that would quantify TSPO binding with high precision without the need for a reference tissue. A machine learning-based (ML-based) prediction algorithm was used to assess the importance of the additional features (shown in Fig. 2 and listed in Section 2.9) in estimating the TSPO binding. In particular, we tested whether brain perfusion information provided by arterial spin labeling (ASL) magnetic resonance imaging (MRI) that could be used to replace the early PET data, as shown in a recent study [20], improves the performance of the estimation algorithm. After determination of the essential features for a robust [18F]GE-180 quantification, we established a simplified procedure, which comprised scaling a late PET frame by activity concentration of a late plasma sample, and compared its performance to the ML-based algorithm.
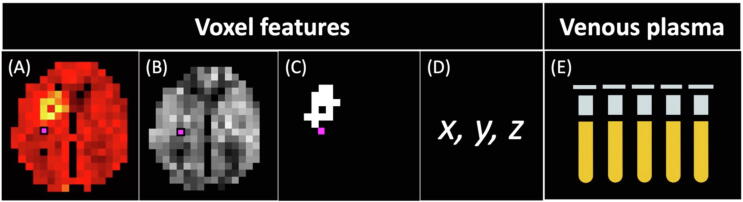
Figure 2.
Input features for random forest. Voxel features: (A) 60–90 min p.i. [18F]GE-180 PET split in three 10 min frames, (B) brain perfusion from ASL, (C) lesion in voxel (True/False), (D) voxel coordinates in MNI space. Features from venous plasma: (E) plasma activity concentration at 5, 15, 30, 60, and 85 min p.i.
2. Materials and methods
2.1. Study design
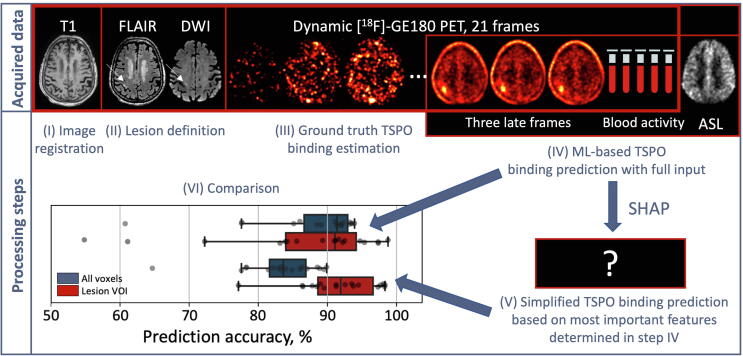
An overview of acquired data and processing steps is shown in Fig. 1. The study included 18 subjects after acute ischemic stroke. MRI was performed for all the subjects in 6.5 days (median) after stroke onset followed by 90 min [18F]GE-180 PET on the same day. During the PET acquisition, five manual venous blood samples were drawn. For each subject, we defined a TSPO binding parameter in each voxel using Logan plot based on the 90 min PET and an image-derived input function (IF). Using the calculated parameter values as ground truth, we trained a ML-based algorithm with various input features that included three late 10 min PET time frames, brain perfusion data from ASL-MRI, and plasma activity concentration from the five timepoints (see full feature list in Section 2.9). The first aim was to define which features have the highest influence on the predicted binding estimates. For this, we calculated the Shapley additive explanations (SHAP). Having determined the most important features for the algorithm, we established a simplified TSPO quantification method based on this reduced feature set and compared it to the performance of the ML-based algorithm with full input (shown in Fig. 1).
Figure 1.
Study design. (I) Image registration was performed based on T1w image. (II) FLAIR and DWI images were used to define ischemic lesion. (III) Ground truth TSPO binding estimate was derived from dynamic [18F]GE-180 PET and five venous blood samples. (IV) Input features for the ML-based TSPO binding prediction algorithm were obtained from three late PET frames, blood data, and ASL-MRI. Most important features were determined using SHAP, and (V) a simplified TSPO binding estimation method was established. Lastly, (VI) the performance of (IV) and (V) was compared.
2.2. Human data
In this study we analyzed the data from 18 ischemic stroke patients (4 females and 14 males; mean age 69.4 ± 10.3 years). Following inclusion criteria were used: age ≥50 years, acute ischemic stroke with diffusion-weighted imaging-positive (DWI-positive) lesion. Prior infectious disease, contraindications for PET and/or MRI were defined as exclusion criteria. The study was approved by the local ethics committee (IRB no. 19-428) and the German radiation protection committee (Z5-22464/2019-163-G). The research was conducted in accordance with the principles of the Declaration of Helsinki, and all subjects gave written informed consent.
2.3. MR imaging
MR imaging was performed on a Siemens MAGNETOM Prisma scanner with 64 channel head/neck coil (Siemens Healthineers, Erlangen, Germany) at the Institute of Stroke and Dementia Research of the University Hospital, LMU Munich. The MR imaging acquisition protocol included T1-weighted imaging (T1w), fluid-attenuated inversion recovery (FLAIR) imaging, DWI, and ASL.
T1w images were acquired with a multi-echo magnetization prepared rapid acquisition with Gradient Echoes using the following parameters: voxel size 0.8 × 0.8 × 0.8 mm3, repetition time (TR) 2560 ms, inversion time (TI) 1100 ms, echo times (TE) 1.68, 3.29, 4.90, 6.51 ms, flip angle 7 degrees. The root mean square across all TE was used for further processing. For FLAIR imaging, the following parameters were used: voxel size 1.0 × 1.0 × 1.0 mm3, TR 5000 ms, TI 1800 ms, TE 393 ms. DWI parameters were: voxel size 1.7 × 1.7 × 1.7 mm3, TR 3220 ms, TE 74.00 ms, multi-band acceleration factor 3, b-values 0, 1000 and 3000 s/mm2, in total 90 diffusion-encoding directions. For perfusion estimation, we used Hadamard-encoded pseudo-continuous ASL with voxel size 3.3 × 3.3 × 3.3 mm3, TR = 7 s, TE = 22.3 ms, labeling delay (LD) 0.224, 0.257, 0.305, 0.374, 0.482, 0.683, 1.175 s and postlabeling delay (PLD) 0.5 s.
2.4. PET imaging
[18F]GE-180 PET scans were acquired using a Siemens Biograph 64 PET/CT scanner (Siemens Healthineers, Erlangen, Germany) at the Department of Nuclear Medicine of the University Hospital, LMU Munich. For patient positioning, a head band was used to fix the head to both reduce possible motion and improve patient’s comfort. A low-dose CT scan was acquired before PET imaging for attenuation correction. The tracer injection was performed at the scan start. The mean injected activity was 173 ± 25 MBq. List-mode data were acquired from 0 to 90 min p.i. The images were reconstructed using the OSEM3D algorithm (4iterations,21subsets) (Siemens Healthineers, Erlangen, Germany). A 336 × 336 × 109 matrix was used, resulting in a voxel-size of 1 × 1 × 2 mm3. The list-mode data were binned into 21 frames (6 × 10 s, 4 × 30 s, 1 × 2 min, 3 × 5 min, 7 × 10 min). Standard corrections for scattered and random coincidences, decay, and dead time were performed. A post-reconstruction Gaussian filter of 5 mm full width at half maximum was applied.
2.5. Image pre-processing
Pre-processing of the ASL-MR images was performed using toolboxes from the FMRIB software library (FSL) [21]. First, the ASL data were corrected for motion using mcflirt. Next, distortion was corrected via topup based on ASL calibration scans with alternating phase-encoding polarity. The data were then decoded with the inverse Hadamard encoding matrix and mean perfusion-weighted images were generated for each PLD. The processing results and the pre-processed ASL data were taken as input for the perfusion analysis using oxford_asl.
For every subject, the ischemic lesion was manually segmented on the FLAIR image. The DWI mean image (trace-weighted) at b = 1000 s/mm2 was used to ascertain that the selected lesion was (sub)acute. All segmentations were supervised by an experienced neuroradiologist.
Next, the FLAIR, arterial blood equilibrium magnetization image (M0), and PET images were rigidly registered to the corresponding T1w image (dissimilarity function: normalized mutual information [22], interpolation method: trilinear) using the PMOD FuseIt tool (version 4.2, PMOD Technologies, Zurich, Switzerland). The resulting FLAIR transformation was applied to the lesion segmentation, while the M0 transformation was applied to the perfusion image.
T1w MR images were registered to the Montreal Neurological Institute (MNI) space using 3 Probability Maps Normalization method (SPM8) with the 1 mm SPM8 brain template consisting of gray, white matter, and CSF probability maps [23]. For this, we used the PMOD Neuro tool (version 4.2, PMOD Technologies, Zurich, Switzerland). For every subject, this elastic transformation was applied to the corresponding PET, FLAIR, perfusion image, and the lesion segmentation in the T1w space.
The regions that were not present in the perfusion image (cerebellum, superior sagittal sinus), the straight sinus, and the brain ventricles were excluded from the analysis. For this purpose, we created a mask by (I) taking the intersection of all the voxels enclosed in the ASL image, (II) manually segmenting the straight sinus, the region with high unspecific [18F]GE-180 signal, on an 80–90 min p.i. PET image averaged among all the subjects, (III) subtracting the straight sinus VOI, and (IV) subtracting the brain ventricles using the definitions from the N30R83 maximum probability atlas [24], [25].
2.6. Blood data
During the 90 min PET scan, we drew five venous blood samples for blood tracer activity estimation at 5, 15, 30, 60, and 90 min p.i. Approximately 3 ml of blood were taken per sampling. 1 ml of whole blood and 1 ml of plasma were drawn from each sample, and their activity was measured with a Hidex Automatic Gamma Counter (Hidex Oy, Turku, Finland) cross-calibrated with the PET scanner. Decay correction to the injection time was applied. The blood activity values were metabolite-corrected using the population-based [18F]GE-180 parent fraction values from [18]. No parent fraction was estimated in this work.
2.7. Image-derived input function (IDIF)
We implemented a semi-automated procedure for IDIF extraction based on recommendations from [26]. The frame for segmentation was selected manually for each subject, which was the frame where the carotid arteries were clearly visible, but no significant uptake was observed in the sagittal sinus yet. The automated procedure (Python 3.7) included cropping of the selected frame so that only the carotid arteries were included. To minimize the partial volume effect, five hottest voxels from each 15 consecutive axial planes were added to the ROI, starting from below the circle of Willis, resulting in a total of 75 voxels. All automatically generated carotid VOIs were visually inspected. From the created carotid VOI, we extracted the time-activity curve (TAC). The TAC was then fitted using a three-exponential model [27]. To further reduce the partial volume effect in the carotid arteries, we multiplied the fitted TAC by a calibration factor, which was calculated as the value that minimized the sum of the mean squared difference between the plasma activities from manual sampling at the five timepoints and the corresponding TAC values. Multiple samples were needed to account for possible noise in the measured blood samples as well as due to a possible shape difference between the IDIF and the arterial IF [28]. Next, we multiplied the resulting curve by the parent fraction curve reported in [18].
2.8. Kinetic modeling
For generating ground truth VT values for the ML algorithm, we employed the Logan plot analysis, a method previously validated for the use in [18F]GE-180 quantification [16]. Logan plot was applied on interpolated image data with larger voxels to reduce noise. In addition to regional information, voxelwise modeling offers the possibility to use the voxel coordinate as input feature for the ML algorithm, contrary to a VOI-based analysis. We performed the fitting using the frames starting from 30 min p.i., i.e., the 30–40, 40–50, 50–60, 60–70, 70–80, and 80–90 min p.i. frame. VT was defined as the slope of the linear fit [17]; the intercept was not used. PET, perfusion images, and the lesion segmentations were interpolated to the same size of 8.5 × 8.5 × 8.5 mm3. For each subject, we calculated an individual VT map using an in-house Python implementation of the Logan plot algorithm with the IDIF. The VT maps were used as the ground truth for the ML algorithm. Additionally, VT values were calculated for lesion VOIs based on non-interpolated 1 × 1 × 1 mm3 images.
2.9. Machine learning algorithm for VT prediction
To make voxelwise predictions for the VT value, we used the random forest (RF) algorithm [29], a machine learning method that combines multiple randomly generated decision trees, or estimators. The algorithm was selected due to its accuracy, relative simplicity, and resistance to overfitting. The Scikit-learn implementation of RF was used (Python 3.7, Scikit-learn 0.22.1). For the algorithm, we selected input features that we assumed to be potentially relevant for the prediction. These features (Fig. 2) were:
-
•
[18F]GE-180 voxel activity from the 60–70, 70–80, and 80–90 min frames (kBq/cc) (Fig. 2(A)),
-
•
Perfusion value in the voxel from ASL MRI (ml/100g/min) (Fig. 2(B)),
-
•
Lesion in the voxel from the manual segmentation (True/False) (Fig. 2(C)),
-
•
Coordinates (x, y, z) of the voxel in the MNI space to account for possible regional differences in tracer extraction (Fig. 2(D)),
-
•
Plasma activity at 5, 15, 30, 60, and 85 min (kBq/ml) (Fig. 2(E)).
The RF was run with 100 estimators. To prevent overfitting, we performed regularization by setting the minimum number of samples required to be at a leaf node to 5, i.e., in an individual tree a split point was considered only if at least 5 training samples in each of the two branches remained after the split. When training and validating the algorithm, we used the leave-one-out approach, i.e., for the training we used the input features belonging to all the patients except the validation patient. We used 1605 input features per subject.
As a measure of the algorithm’s performance, we used accuracy that for an individual subject was calculated as follows:
where is the ground truth , is the predicted , N is the number of voxels in the image.
2.10. Model interpretation by SHAP
The impact of each individual feature was assessed by using SHAP (Python 3.7, SHAP 0.40.0). SHAP are designed to make machine learning models interpretable. Full description of the SHAP methodology is beyond the scope of this work and can be read in the original paper [30]. Briefly, a SHAP value is the average of the marginal contributions across all the feature permutations and can be calculated for each training sample.
2.11. Simplified TSPO quantification using most important features determined by SHAP
SHAP demonstrated that late PET frames and plasma activity concentrations have highest impact on the model predictions, while the lesion mask and voxel coordinates were shown to be the least important features (see Section 3.2). Based on this, we built simplified models with a reduced feature set. First model (RF1) used the three late PET frames, the five plasma activity concentrations, and the brain perfusion; second model (RF2) used the three late PET frames and the five plasma activity concentrations only. Additionally, guided by the idea that VT is a ratio of tissue to blood tracer concentration, we investigated an even simpler approach, in which we divided a late PET frame by a plasma activity concentration as a proxy for VT. Since the three late PET frames are correlated, and the same is valid for the five plasma activity concentrations, all possible combinations of PET frames and plasma samples were considered in order to define which of these features yield the closest approximation for the VT.
2.12. Reduced-features model comparison and statistical tests
To compare the VT prediction algorithms with reduced feature set to the one with the full input as well as to each other, we performed Wilcoxon signed-rank test (Python 3.7, Pingouin 0.3.8) on accuracy values from all the study subjects for each method pair. Wilcoxon signed-rank test was used instead of the paired t-test, since the distributions of the accuracies were not normal. The test was performed for both the mean accuracies derived from 8.5 × 8.5 × 8.5 mm3 voxels (lesion and non-lesion) and the mean accuracies in the non-resliced lesion VOI (i.e., directly derived from the 1 × 1 × 1 mm3 PET image in the MNI space). A significance threshold of 0.05 was used. p-values were corrected using Benjamini/Yekutieli false discovery rate correction [31]. Additionally, we calculated the Pearson correlation coefficient between the ground truth VT and VT predicted by the reduced-features methods in the lesion and performed linear fitting. To assess the concordance between the predicted values and the ground truth, we calculated the Lin’s concordance correlation coefficient (CCC). Following McBride interpretation of CCC [32], we considered CCC < 0.90 as poor concordance, 0.90 < CCC < 0.95 as moderate concordance, 0.95 < CCC < 0.99 as substantial concordance, and CCC > 0.99 as perfect concordance. To evaluate the agreement between the predicted and the ground truth VT, we performed Bland-Altman analysis [33]. Next, we determined whether there was a constant and/or proportional bias in VT estimates. To identify the existence of a constant bias, we compared mean difference between the ground truth and the predicted VT to zero by means of a single-sample t-test. A proportional bias was considered to be significant if there was a Pearson correlation between the mean of the two methods and their difference. A significance threshold of 0.01 was used in both cases.
3. Results
3.1. Random forest predicts VT values with high accuracy
The random forest algorithm was run for all the 18 study subjects. Mean VT prediction accuracy in an 8.5 × 8.5 × 8.5 mm3 voxel across all the subjects was 87.8 ± 8.3%. An example of RF predictions for all the voxels from one of the subjects plotted against the ground truth VT is shown in Fig. 3(A). The corresponding ground truth VT map and predicted VT map are depicted in Fig. 3(B, C), respectively.
Figure 3.
(A) Random forest VT predictions plotted against the ground truth for an example subject. Red dots show lesion voxels, blue dots represent non-lesion voxels. The line of identity is shown in black. (B) Ground truth VT map for the example subject. (C) Corresponding predicted VT map.
3.2. SHAP values demonstrate high impact of late PET frames and plasma samples on the RF predictions, but low influence of ASL-derived perfusion
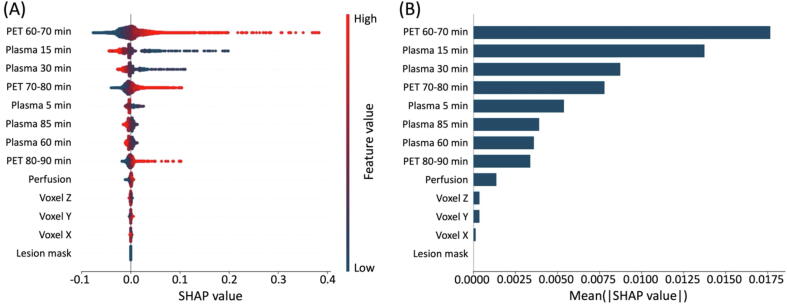
For each training sample, we calculated a set of SHAP values, which are represented as individual dots in Fig. 4(A). We then calculated the mean of the absolute SHAP values for each feature (Fig. 4(B)). It was shown that the three late PET frames as well as the five plasma activity concentrations have the highest impact on the model prediction, with the influence of the 60–70 min p.i. frame and the 15 min plasma sample being the strongest. The perfusion value from the ASL image, voxel dimensions, and lesion mask had little impact on the predictions.
Figure 4.
RF feature importance demonstrated by SHAP values. (A) SHAP values for each single voxel of each subject. The color shows the value of the feature. (B) Average absolute SHAP values representing average impact on the model output. The features are sorted according to their importance.
3.3. Late PET frames divided by late plasma activity concentration demonstrate accuracy similar to RF
It was demonstrated that RF1 and RF2 had the same accuracy when run on the whole image (p = 1) as well as on the lesion VOI (p = 0.82) only (Fig. 5). Among the scaled PET values, the best accuracy was achieved with the 15 and 30 min p.i. plasma samples when run on all the voxels and with the 30 min p.i. plasma sample when run on the lesion VOI (Fig. 5, see Supplementary Table 1, 2 for individual p-values). RF2 significantly outperformed all the scaling methods when run on all the voxels (except 30 min plasma, p > 0.27) while no differences between RF2 and all the scaling methods were observed for the lesion VOI (except 5 min plasma, p < 0.006) (Fig. 5).
Figure 5.
Performance of simplified VT prediction methods based on [18F]GE-180 PET uptake values scaled by a single plasma activity concentration versus the RF algorithm. Blue boxes show the accuracy on all the 8.5 mm voxels, red boxes represent the accuracy for the lesion VOI with no reslicing. Dots represent individual subjects.
3.4. 70–80 min p.i. PET frame divided by single 30 min p.i. plasma activity concentration yields best VT prediction in ischemic lesions
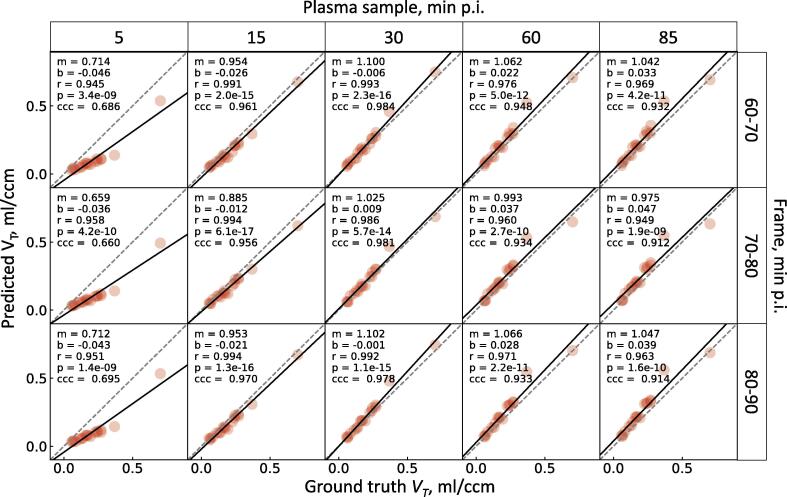
All the combinations of the PET frames and the plasma samples showed very strong correlation with the ground truth VT (r >= 0.94, p < 10−8, Fig. 6). 15 and 30 min plasma activity-scaled frames showed substantial concordance, 60 and 85 min scaled frames showed moderate concordance, while for 5 min scaled frames poor concordance was observed. The highest concordance was achieved when using 60–70 and 70–80 min frames scaled by 30 min plasma sample (CCC = 0.984, CCC = 0.981, respectively) (Fig. 6). The Bland-Altman plots for all the combinations showed high degree of agreement between the ground truth and the predicted VT, since most of the data points lie within the limits of agreement (from 16 to 18 subject out of 18 subjects, see Fig. 7, Supplementary Table 3). A significant constant bias was observed for all the combinations apart from 60–70 and 70–80 min p.i. PET and 30 min p.i. plasma sample (p < 0.01, mean difference against zero by single-sample t-test, see Supplementary Table 3). We also observed a significant proportional bias for the following combinations: 5 min p.i. plasma sample with 60–70, 70–80, 80–90 min p.i. PET frame; 15 min p.i. plasma sample with 70–80 min p.i. PET frame; 30 min p.i. plasma sample with 60–70, 80–90 min p.i. PET frame (Supplementary Table 3). A combination of 30 min p.i. plasma sample and 70–80 min p.i. PET frame is therefore the only one devoid of both forms of bias.
Figure 6.
Relationship between the ground truth VT and the predicted VT in the lesion VOI of all the study subjects. The predicted VT is the lesion uptake in a late frame divided by a plasma activity concentration. All combinations of late frames and plasma samples are shown. The solid black line shows the linear fit, the dashed line is the line of identity. m, b are the slope and the intercept of the fit line, r is the Pearson’s r, p is the linear correlation p-value, CCC is the Lin’s concordance correlation coefficient.
Figure 7.
Bland-Altman plots for the ground truth VT versus predicted VT in the lesion VOI of all the study subjects. The predicted VT is the lesion uptake in a late frame divided by a plasma activity concentration. All combinations of late frames and plasma samples are shown. The solid line indicates the mean difference between the calculation methods, the two dashed lines depict the limits of agreement (95% confidence interval of the mean difference). SD – standard deviation.
4. Discussion
In this study, we established a ML-based algorithm that predicts VT with high accuracy (87.8 ± 8.3%) compared to the ground truth VT calculated using the Logan plot with an IDIF. By means of SHAP values, we were able to show that the excellent performance of the algorithm was attributed to the late PET frames (60–70, 70–80, and 80–90 min p.i.) and plasma activity concentrations (5, 15, 30, 60, 85 min p.i.) only, while the ASL-derived brain perfusion, voxel coordinates, and the manually defined lesion mask had little influence on the performance (Fig. 4). The low importance of the ASL-derived values was also demonstrated by the Wilcoxon signed-rank test between the accuracies of the RF with and without the perfusion values (Fig. 5, RF1 versus RF2). This finding suggests possible [18F]GE-180 signal independence of the blood-brain barrier (BBB), which was previously demonstrated by Kaiser et al. [10] for a group of glioma patients. Contrary, Scott et al. [20] reported significant correlation between the non-displaceable binding potential (BPND) and a simplified binding estimate calculated based on a shortened PET acquisition and ASL-derived brain perfusion for a mixed group of cognitively normal and Alzheimer’s disease subjects. However, in their work the authors used a tracer with different kinetics ([18F]florbetapir) and applied a different kinetic model (simplified reference tissue model). Another study [34] reported non-specific binding of [18F]GE-180 in ischemic lesions with BBB damage as well as high contribution of the vascular signal to the overall uptake for a group of stroke patients; however, the authors selected a 15–30 min p.i. frame for evaluations, where tissue TACs do not reach the plateau yet [16], [35].
There is an ongoing discussion on how much the [18F]GE-180 signal is reflecting BBB damage versus specific binding. In several studies of glioma patients [10], [11], [36], [37], it was shown that [18F]GE-180 PET uptake can be clearly visualized on the PET image in areas outside of the gadolinium enhancement area on MRI and even in gliomas without any visible contrast enhancement on MRI. Additionally, Albert et al. [38], [39] showed an example of a glioma patient with a clear BBB breakdown visible on contrast MRI, but without significant [18F]GE-180 signal in the lesion. A recent longitudinal study [40] demonstrated a significant [18F]GE-180 uptake both 2 weeks before and up to 5.5 months after one could see prominent gadolinium enhancement in progressive multifocal leukoencephalopathy. Moreover, there is no correlation between [18F]GE-180 signal intensity and BBB disruption as assessed using contrast-enhanced T1-weighted MRI relative to native T1-weighted MRI in a voxelwise analysis [10]. Additionally, very similar binding patterns in in vitro autoradiography and ex vivo autoradiography were observed in animal models [38]. These finding, contrary to claims mentioned in [41], [42] clearly point to a specific component resulting in sustained [18F]GE-180 binding. Undoubtedly, the disruption of the BBB has an influence on the [18F]GE-180 uptake [41], [42], but this is the case for all TSPO tracers to a certain degree. To our knowledge, there is no study showing that the uptake of any other TSPO tracer is not influenced by BBB disruption, as well as there is no study demonstrating that the accumulation of [18F]GE-180 beyond the BBB is merely a non-specific signal. More studies are required to better understand the details of [18F]GE-180 binding in lesions with damaged BBB.
Nevertheless, [18F]GE-180 is a tracer with proven high clinical value and, contrary to [41], should not be considered a failed TSPO radioligand. For instance, [18F]GE-180 shows the tumor extent better than MRI alone [10], [11], and high [18F]GE-180 signal was not only shown to be associated with higher tumor grade [36], but it also has a prognostic value for recurrent glioma patients belonging to otherwise homogeneous molecular subgroups [43]. Additionally, [18F]GE-180 allows the representation of inflammation development over time in progressive multifocal leukoencephalopathy [40]. With that said, the clinical value of [18F]GE-180 in other conditions, especially for more radiosensitive groups of patients, such as children [44], can only be proven after careful validation for this specific patient group.
By using the Wilcoxon signed-rank test, we demonstrated that RF trained with all the three late PET frames and the five plasma activity concentrations (RF2, Fig. 5) predicts VT in all the image voxels with accuracy higher than any combination of a late PET frame scaled by a plasma activity concentration (insignificantly higher for 30 min p.i. plasma, significantly higher for the rest); however, in the lesion VOI there was no significant difference in accuracy between RF2 and other scaling methods (except for 5 min p.i. plasma). This result suggests that using longer scanning time and more than one blood sample might result in an improved VT prediction, which is, however, not the case for the ischemic stroke lesion region and therefore supports the use of a single blood sample for TSPO quantification in ischemic stroke patients. Late PET frames scaled by 30 min p.i. plasma activity concentration demonstrated the best accuracy for the lesion VOI.
The highest concordance was obtained when using 60–70 or 70–80 min p.i. frame with the 30 min p.i. plasma sample (Fig. 6). Even though the Bland-Altman plots indicated high agreement between the ground truth and all the scaling methods (Fig. 7), only the combination of 70–80 min p.i. frame and 30 min p.i. plasma sample demonstrated the lack of both constant and proportional bias (Supplementary Table 3). This result supports the use of the 30 min p.i. plasma sample for simplified TSPO quantification. However, the 60 min blood sample, which can be drawn right before the static scan and is therefore more convenient for both the patient and the personnel, also produces similarly accurate VT estimates. According to the results of this study, both 70–80 and 60–70 p.i. PET frames can be used for simplified VT estimation, which supports the use of the 60–80 min summed frame that provides better count statistics.
The two-tissue compartment model is considered to be the gold standard TSPO quantification method [16]; however, in this work we used Logan plot instead. Fan et al. [16] reported strong correlations between two-tissue compartment-derived and Logan plot-derived VT for all the brain regions investigated in their work, but the estimates had some bias. Nevertheless, the authors endorsed the use of Logan plot for voxelwise analyses. The two-tissue model is less suitable for this task as it produces high parameter errors with noisy data; in contrast, Logan plot generates robust VT estimates as it has less parameters to be fitted.
One of the limitations of this study is the lack of arterial IF (AIF). Invasive arterial blood sampling is associated with risks for the patient and was not possible in this study given the condition of the patients that had ischemic stroke less than a week before their [18F]GE-180 PET scan. Another way to approximately estimate the blood peak would be to draw several early venous blood samples with a 15-20 s interval. It was not performed either due to the condition of the veins of several study patients, for which it took more than a minute to draw a single blood sample. Therefore, we used carotid IDIF for the Logan plot. Due to the finite frame length (10sduringthefirstminute), the blood peak magnitude could not be precisely estimated. IDIFs are known to be prone to the partial volume effect (PVE) and therefore underestimate the signal. To reduce the influence of the PVE, we used only the hottest voxels from the carotid arteries to derive the IDIF. But, as mentioned by Zanotti-Fregonara et al. [28], methods based on the use of n hottest voxels closely approximate the AIF only when several imaging parameters are a posteriori adjusted, and choosing a different reconstruction algorithm [45] or using a different tracer [26] results in reduced accuracy. This issue was addressed by scaling using multiple blood samples, a recommendation [28] based on the fact that IDIF and AIF might differ in shape. On the other hand, IDIF offers some advantages over AIF, as it does not require delay and dispersion correction as well as the cross-calibration between the continuous blood sampler and the scanner, which are common sources of error when dealing with AIF. Carotid artery IDIF was successfully validated for using with TSPO tracers by several groups [26], [46]. Using IF derived from sources that are less prone to PVE, such as ascending aorta, available when using long axial field-of-view PET [47] could improve VT estimates. Additional limitations are the lack of parent fraction correction and the fact that metabolite correction was achieved by using population-based data [18].
[18F]GE-180 is a tracer with slow kinetics [16], which makes kinetic modelling, especially voxelwise, challenging, since the tracer cannot be clearly classified as reversible or irreversible. On the other hand, a VOI-based analysis was not desirable since one of the tested input features was the coordinate in the MNI space, and VOIs do not have the same shape. The use of VOI centers as input features could have possibly reduced the performance of the ML algorithm. Using coarse voxels (8.5 × 8.5 × 8.5 mm3) was a good compromise that allowed both the coordinate estimation and robust VT calculation. Additionally, coarse voxel size was a way to achieve a better match between the PET and the perfusion image, which both have limited resolution. No reslicing to coarse voxels is required for kinetic modelling when using simplified approaches without ASL-based perfusion and coordinates (such as RF2), provided that the parameter error is acceptable.
Another limitation of the study is the sample size. The values of plasma activity concentration were identical for the voxels belonging to a single patient, i.e. only 18 unique concentration values were present in the dataset. However, the number of single samples was relatively high (1605perpatient). Since RF regressor is not able to extrapolate beyond the value range of the training samples, the accuracy was reduced in samples with extreme feature values. This can be seen on Supplementary Fig. 1 for the subject with the lowest accuracy (the first bar), who had the smallest plasma activity concentration among the studied group. To improve the accuracy on extreme validation samples, one can combine RF with linear regressor by stacking or use a regression-enhanced RF [48]. Increasing the number of training subjects is likely to improve the overall accuracy.
To our knowledge, this is the first study that validated a semi-quantitative TSPO binding estimate based on a late PET frame and a blood sample against a quantitative parameter from the full 90 min dynamic scan in a human ischemic stroke cohort for [18F]GE-180. The use of the late frame is also supported by other clinical and preclinical [18F]GE-180 studies in different patient groups as well as disease models. For instance, Vomacka et al. [13] reported a very strong correlation (Pearson’s r > 0.9) between the standardized uptake value ratio (SUVR) from static 60–90 min p.i. images and the distribution volume ratio (DVR) derived from reference-tissue Logan plot in patients with multiple sclerosis when using the frontal cortex as a reference. A high correlation between SUVR und DVR derived from the same kinetic model with the white matter as the reference region was also reported by Brendel et al. [49] in a mouse study, while another preclinical work [50] showed that scaling the late static frame by myocardial uptake can further improve the estimates. In a clinical study, Albert et al. [11] reported [18F]GE-180 TAC reaching a plateau after 60 min p.i. in the glioblastoma region and used the 60–80 min p.i. frame for further evaluations. In a preclinical model of ischemic stroke, Chaney et al. [51] performed dynamic 0–60 min p.i. [18F]GE-180 scan and selected the 50–60 min p.i. frame for the analysis as it provided the highest signal-to-background ratio.
In conclusion, our study shows that late [18F]GE-180 PET and a single late plasma sample are sufficient for a robust quantification of neuroinflammation in ischemic stroke patients. The 70–80 min p.i. frame and the 30 min p.i. plasma sample yield the closest VT approximation. This procedure replaces the 90 min [18F]GE-180 PET and does not require kinetic modeling, thus this procedure could be easily implemented in the clinical workflow.
Author Contributions
AZ: conceptualization, PET data processing, data analysis, and manuscript writing. AK: patient recruitment, MR imaging, manuscript review and editing. AH: PET imaging. SH: MR imaging. AS: lesion segmentation. MD: MR data processing. LK and NA: manuscript review and editing. SL: tracer production. MS: ASL methodology, manuscript editing. PB: funding acquisition, manuscript review and editing. MB and SZ: conceptualization, funding acquisition, manuscript review and editing. All authors contributed to the article and approved the submitted version.
Funding
This project was supported by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany's Excellence Strategy (EXC 2145 SyNergy – ID 390857198) and through FOR 2879 (project number 428668490). The procurement of the MRI scanner was supported by the DFG grant for major research instrumentation (DFG, INST 409/193-1 FUGG).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.zemedi.2022.11.008.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Statistics
Data Availability Statement
Data and code reported in this article will be shared with any appropriately qualified investigator upon email request.
References
- 1.Campbell B.C., De Silva D.A., Macleod M.R., Coutts S.B., Schwamm L.H., Davis S.M., et al. Ischaemic stroke. Nat Rev Dis Primers. 2019;5(1):1–22. doi: 10.1038/s41572-019-0118-8. [DOI] [PubMed] [Google Scholar]
- 2.Szalay G., Martinecz B., Lénárt N., Környei Z., Orsolits B., Judák L., et al. Microglia protect against brain injury and their selective elimination dysregulates neuronal network activity after stroke. Nat Commun. 2016;7(1):1–13. doi: 10.1038/ncomms11499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong S., Beja-Glasser V.F., Nfonoyim B.M., Frouin A., Li S., Ramakrishnan S., et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352(6286):712–716. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivash L., O’Brien T.J. Imaging microglial activation with TSPO PET: lighting up neurologic diseases? J Nucl Med. 2016;57(2):165–168. doi: 10.2967/jnumed.114.141713. [DOI] [PubMed] [Google Scholar]
- 5.Wimmer I., Zrzavy T., Lassmann H. Neuroinflammatory responses in experimental and human stroke lesions. J Neuroimmunol. 2018;323:10–18. doi: 10.1016/j.jneuroim.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Shi K., Tian D.-C., Li Z.-G., Ducruet A.F., Lawton M.T., Shi F.-D. Global brain inflammation in stroke. Lancet Neurol. 2019;18(11):1058–1066. doi: 10.1016/S1474-4422(19)30078-X. [DOI] [PubMed] [Google Scholar]
- 7.Thiel A., Radlinska B.A., Paquette C., Sidel M., Soucy J.-P., Schirrmacher R., et al. The temporal dynamics of poststroke neuroinflammation: a longitudinal diffusion tensor imaging–guided PET study with 11C-PK11195 in acute subcortical stroke. J Nucl Med. 2010;51(9):1404–1412. doi: 10.2967/jnumed.110.076612. [DOI] [PubMed] [Google Scholar]
- 8.Dickens A.M., Vainio S., Marjamäki P., Johansson J., Lehtiniemi P., Rokka J., et al. Detection of microglial activation in an acute model of neuroinflammation using PET and radiotracers 11C-(R)-PK11195 and 18F-GE-180. J Nucl Med. 2014;55(3):466–472. doi: 10.2967/jnumed.113.125625. [DOI] [PubMed] [Google Scholar]
- 9.Boutin H., Murray K., Pradillo J., Maroy R., Smigova A., Gerhard A., et al. 18 F-GE-180: a novel TSPO radiotracer compared to 11 CR-PK11195 in a preclinical model of stroke. Eur J Nucl Med Mol Imaging. 2015;42(3):503–511. doi: 10.1007/s00259-014-2939-8. [DOI] [PubMed] [Google Scholar]
- 10.Kaiser L., Holzgreve A., Quach S., Ingrisch M., Unterrainer M., Dekorsy F.J., et al. Differential Spatial Distribution of TSPO or Amino Acid PET Signal and MRI Contrast Enhancement in Gliomas. Cancers. 2022;14(1):53. doi: 10.3390/cancers14010053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Albert N.L., Unterrainer M., Fleischmann D., Lindner S., Vettermann F., Brunegraf A., et al. TSPO PET for glioma imaging using the novel ligand 18F-GE-180: first results in patients with glioblastoma. Eur J Nucl Med Mol Imaging. 2017;44(13):2230–2238. doi: 10.1007/s00259-017-3799-9. [DOI] [PubMed] [Google Scholar]
- 12.Rauchmann B.-S., Brendel M., Franzmeier N., Trappmann L., Zaganjori M., Morenas-Rodriguez E., et al. MRI connectivity-based spread of microglial activation in early Alzheimer's disease. Ann Neurol. 2022 doi: 10.1002/ana.26465. [DOI] [PubMed] [Google Scholar]
- 13.Vomacka L., Albert N.L., Lindner S., Unterrainer M., Mahler C., Brendel M., et al. TSPO imaging using the novel PET ligand [18 F] GE-180: quantification approaches in patients with multiple sclerosis. EJNMMI Res. 2017;7(1):1–9. doi: 10.1186/s13550-017-0340-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palleis C., Sauerbeck J., Beyer L., Harris S., Schmitt J., Morenas-Rodriguez E., et al. In vivo assessment of neuroinflammation in 4-repeat tauopathies. Mov Disord. 2021;36(4):883–894. doi: 10.1002/mds.28395. [DOI] [PubMed] [Google Scholar]
- 15.Xiang X., Wind K., Wiedemann T., Blume T., Shi Y., Briel N., et al. Microglial activation states drive glucose uptake and FDG-PET alterations in neurodegenerative diseases. Sci Transl Med. 2021;13(615):eabe5640 doi: 10.1126/scitranslmed.abe5640. [DOI] [PubMed] [Google Scholar]
- 16.Fan Z., Calsolaro V., Atkinson R.A., Femminella G.D., Waldman A., Buckley C., et al. Flutriciclamide (18F-GE180) PET: first-in-human PET study of novel third-generation in vivo marker of human translocator protein. J Nucl Med. 2016;57(11):1753–1759. doi: 10.2967/jnumed.115.169078. [DOI] [PubMed] [Google Scholar]
- 17.Logan J., Fowler J.S., Volkow N.D., Wolf A.P., Dewey S.L., Schlyer D.J., et al. Graphical analysis of reversible radioligand binding from time—activity measurements applied to [N-11C-methyl]-(−)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab. 1990;10(5):740–747. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- 18.Buchert R., Dirks M., Schütze C., Wilke F., Mamach M., Wirries A.-K., et al. Reliable quantification of 18 F-GE-180 PET neuroinflammation studies using an individually scaled population-based input function or late tissue-to-blood ratio. Eur J Nucl Med Mol Imaging. 2020;47(12):2887–2900. doi: 10.1007/s00259-020-04810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zatcepin A., Heindl S., Schillinger U., Kaiser L., Lindner S., Bartenstein P., et al. Reduced Acquisition Time [18F] GE-180 PET Scanning Protocol Replaces Gold-Standard Dynamic Acquisition in a Mouse Ischemic Stroke Model. Front Med. 2022;9 doi: 10.3389/fmed.2022.830020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scott C.J., Jiao J., Melbourne A., Burgos N., Cash D.M., De Vita E., et al. Reduced acquisition time PET pharmacokinetic modelling using simultaneous ASL–MRI: proof of concept. J Cereb Blood Flow Metab. 2019;39(12):2419–2432. doi: 10.1177/0271678X18797343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith S.M., Jenkinson M., Woolrich M.W., Beckmann C.F., Behrens T.E., Johansen-Berg H., et al. Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage. 2004;23:S208–S219. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 22.Studholme C., Hill D.L., Hawkes D.J. An overlap invariant entropy measure of 3D medical image alignment. Pattern Recogn. 1999;32(1):71–86. [Google Scholar]
- 23.Ashburner J., Friston K.J. Unified segmentation. Neuroimage. 2005;26(3):839–851. doi: 10.1016/j.neuroimage.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 24.Hammers A., Allom R., Koepp M.J., Free S.L., Myers R., Lemieux L., et al. Three-dimensional maximum probability atlas of the human brain, with particular reference to the temporal lobe. Hum Brain Mapp. 2003;19(4):224–247. doi: 10.1002/hbm.10123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gousias I.S., Rueckert D., Heckemann R.A., Dyet L.E., Boardman J.P., Edwards A.D., et al. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. Neuroimage. 2008;40(2):672–684. doi: 10.1016/j.neuroimage.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 26.Mourik J.E., Lubberink M., Schuitemaker A., Tolboom N., van Berckel B.N., Lammertsma A.A., et al. Image-derived input functions for PET brain studies. Eur J Nucl Med Mol Imaging. 2009;36(3):463–471. doi: 10.1007/s00259-008-0986-8. [DOI] [PubMed] [Google Scholar]
- 27.Feng D., Huang S.-C., Wang X. Models for computer simulation studies of input functions for tracer kinetic modeling with positron emission tomography. Int J Biomed Comput. 1993;32(2):95–110. doi: 10.1016/0020-7101(93)90049-c. [DOI] [PubMed] [Google Scholar]
- 28.Zanotti-Fregonara P., Chen K., Liow J.-S., Fujita M., Innis R.B. Image-derived input function for brain PET studies: many challenges and few opportunities. J Cereb Blood Flow Metab. 2011;31(10):1986–1998. doi: 10.1038/jcbfm.2011.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breiman L. Random forests. Mach Learn. 2001;45(1):5–32. [Google Scholar]
- 30.Lundberg S.M., Lee S.-I. A unified approach to interpreting model predictions. Adv Neural Inf Proces Syst. 2017;30 [Google Scholar]
- 31.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;1165–88 [Google Scholar]
- 32.McBride G. A proposal for strength-of-agreement criteria for Lin’s concordance correlation coefficient. NIWA client report: HAM2005-062. 2005;45:307–10.
- 33.Bland J.M., Altman D. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;327(8476):307–310. [PubMed] [Google Scholar]
- 34.Visi E, Hinz R, Punter M, Majid A, Gerhard A, Herholz K. Positron emission tomography to image cerebral neuroinflammation in ischaemic stroke: a pilot study. 2020. [PubMed]
- 35.Feeney C., Scott G., Raffel J., Roberts S., Coello C., Jolly A., et al. Kinetic analysis of the translocator protein positron emission tomography ligand [18 F] GE-180 in the human brain. Eur J Nucl Med Mol Imaging. 2016;43(12):2201–2210. doi: 10.1007/s00259-016-3444-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unterrainer M., Fleischmann D., Vettermann F., Ruf V., Kaiser L., Nelwan D., et al. TSPO PET, tumour grading and molecular genetics in histologically verified glioma: A correlative 18F-GE-180 PET study. Eur J Nucl Med Mol Imaging. 2020;47(6):1368–1380. doi: 10.1007/s00259-019-04491-5. [DOI] [PubMed] [Google Scholar]
- 37.Unterrainer M., Fleischmann D., Diekmann C., Vomacka L., Lindner S., Vettermann F., et al. Comparison of 18F-GE-180 and dynamic 18F-FET PET in high grade glioma: a double-tracer pilot study. Eur J Nucl Med Mol Imaging. 2019;46(3):580–590. doi: 10.1007/s00259-018-4166-1. [DOI] [PubMed] [Google Scholar]
- 38.Albert N.L., Unterrainer M., Kaiser L., Brendel M., Vettermann F., Holzgreve A., et al. In response to: Anatomy of 18F-GE180, a failed radioligand for the TSPO protein. Eur J Nucl Med Mol Imaging. 2020;47(10):2237–2241. doi: 10.1007/s00259-020-04885-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albert N.L., Unterrainer M., Brendel M., Kaiser L., Zweckstetter M., Cumming P., et al. In response to: The validity of 18F-GE180 as a TSPO imaging agent. Eur J Nucl Med Mol Imaging. 2019;46(6):1208–1211. doi: 10.1007/s00259-019-04294-8. [DOI] [PubMed] [Google Scholar]
- 40.Mahler C., Schumacher A.-M., Unterrainer M., Kaiser L., Höllbacher T., Lindner S., et al. TSPO PET imaging of natalizumab-associated progressive multifocal leukoencephalopathy. Brain. 2021;144(9):2683–2695. doi: 10.1093/brain/awab127. [DOI] [PubMed] [Google Scholar]
- 41.Zanotti-Fregonara P., Pascual B., Rostomily R.C., Rizzo G., Veronese M., Masdeu J.C., et al. Anatomy of 18F-GE180, a failed radioligand for the TSPO protein. Eur J Nucl Med Mol Imaging. 2020;47(10):2233–2236. doi: 10.1007/s00259-020-04732-y. [DOI] [PubMed] [Google Scholar]
- 42.Zanotti-Fregonara P., Veronese M., Pascual B., Rostomily R.C., Turkheimer F., Masdeu J.C. The validity of 18F-GE180 as a TSPO imaging agent. Eur J Nucl Med Mol Imaging. 2019;46(6):1205–1207. doi: 10.1007/s00259-019-4268-4. [DOI] [PubMed] [Google Scholar]
- 43.Quach S., Holzgreve A., Kaiser L., Unterrainer M., Dekorsy F.J., Nelwan D.V., et al. TSPO PET signal using [18F] GE180 is associated with survival in recurrent gliomas. Eur J Nucl Med Mol Imaging. 2022;1–11 doi: 10.1007/s00259-022-06006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zanotti-Fregonara P., Rizzo G., Veronese M., Innis R.B., Turkheimer F.E. 18F-GE180, a failed tracer for translocator protein, has no place in child abuse imaging. Pediatr Radiol. 2022;52(5):1015–1016. doi: 10.1007/s00247-021-05248-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parker B.J., Feng D. Graph-based Mumford-Shah segmentation of dynamic PET with application to input function estimation. IEEE Trans Nucl Sci. 2005;52(1):79–89. [Google Scholar]
- 46.Fang Y.-H.-D., McConathy J.E., Yacoubian T.A., Zhang Y., Kennedy R.E., Standaert D.G. Image Quantification for TSPO PET with a Novel Image-Derived Input Function Method. Diagnostics. 2022;12(5):1161 doi: 10.3390/diagnostics12051161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sari H., Mingels C., Alberts I., Hu J., Buesser D., Shah V., et al. First results on kinetic modelling and parametric imaging of dynamic 18F-FDG datasets from a long axial FOV PET scanner in oncological patients. Eur J Nucl Med Mol Imaging. 2022;1–13 doi: 10.1007/s00259-021-05623-6. [DOI] [PubMed] [Google Scholar]
- 48.Zhang H, Nettleton D, Zhu Z. Regression-enhanced random forests. arXiv preprint arXiv:190410416. 2019.
- 49.Brendel M., Probst F., Jaworska A., Overhoff F., Korzhova V., Albert N.L., et al. Glial activation and glucose metabolism in a transgenic amyloid mouse model: a triple-tracer PET study. J Nucl Med. 2016;57(6):954–960. doi: 10.2967/jnumed.115.167858. [DOI] [PubMed] [Google Scholar]
- 50.Deussing M., Blume T., Vomacka L., Mahler C., Focke C., Todica A., et al. Coupling between physiological TSPO expression in brain and myocardium allows stabilization of late-phase cerebral [18F] GE180 PET quantification. Neuroimage. 2018;165:83–91. doi: 10.1016/j.neuroimage.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 51.Chaney A., Cropper H.C., Johnson E.M., Lechtenberg K.J., Peterson T.C., Stevens M.Y., et al. 11C-DPA-713 versus 18F-GE-180: a preclinical comparison of translocator protein 18 kDa PET tracers to visualize acute and chronic neuroinflammation in a mouse model of ischemic stroke. J Nucl Med. 2019;60(1):122–128. doi: 10.2967/jnumed.118.209155. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Statistics
Data Availability Statement
Data and code reported in this article will be shared with any appropriately qualified investigator upon email request.