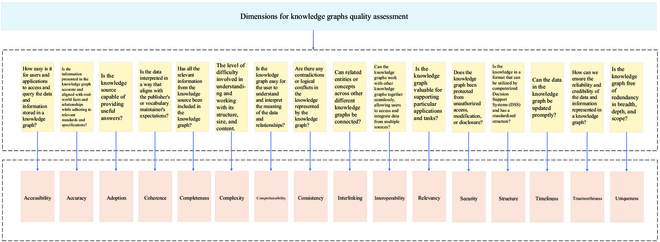
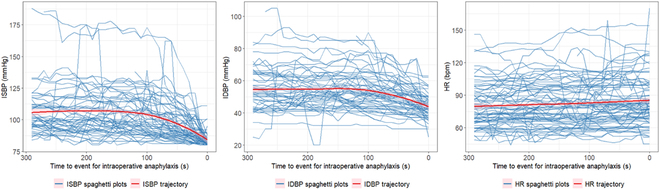
Abstract
The 5th annual Beijing Health Data Science Summit, organized by the National Institute of Health Data Science at Peking University, recently concluded with resounding success. This year, the summit aimed to foster collaboration among researchers, practitioners, and stakeholders in the field of health data science to advance the use of data for better health outcomes.
One significant highlight of this year’s summit was the introduction of the Abstract Competition, organized by Health Data Science, a Science Partner Journal, which focused on the use of cutting-edge data science methodologies, particularly the application of artificial intelligence in the healthcare scenarios. The competition provided a platform for researchers to showcase their groundbreaking work and innovations.
In total, the summit received 61 abstract submissions. Following a rigorous evaluation process by the Abstract Review Committee, eight exceptional abstracts were selected to compete in the final round and give presentations in the Abstract Competition.
The winners of the Abstract Competition are as follows:
-
•
First Prize: “Interpretable Machine Learning for Predicting Outcomes of Childhood Kawasaki Disease: Electronic Health Record Analysis” presented by researchers from the Chinese Academy of Medical Sciences, Peking Union Medical College, and Chongqing Medical University (presenter Yifan Duan).
-
•
Second Prize: “Survival Disparities among Mobility Patterns of Patients with Cancer: A Population-Based Study” presented by a team from Peking University (presenter Fengyu Wen).
-
•
Third Prize: “Deep Learning-Based Real-Time Predictive Model for the Development of Acute Stroke” presented by researchers from Beijing Tiantan Hospital (presenter Lan Lan).
We extend our heartfelt gratitude to the esteemed panel of judges whose expertise and dedication ensured the fairness and quality of the competition. The judging panel included Jiebo Luo from the University of Rochester (chair), Shenda Hong from Peking University, Xiaozhong Liu from Worcester Polytechnic Institute, Liu Yang from Hong Kong Baptist University, Ma Jianzhu from Tsinghua University, Ting Ma from Harbin Institute of Technology, and Jian Tang from Mila–Quebec Artificial Intelligence Institute. We wish to convey our deep appreciation to Zixuan He and Haoyang Hong for their invaluable assistance in the meticulous planning and execution of the event.
As the 2023 Beijing Health Data Science Summit comes to a close, we look forward to welcoming all participants to join us in 2024. Together, we will continue to advance the frontiers of health data science and work toward a healthier future for all.