Abstract
We report that endoplasmic reticulum α-glucosidase inhibitors have antiviral effects on dengue (DEN) virus. We found that glucosidase inhibition strongly affects productive folding pathways of the envelope glycoproteins prM (the intracellular glycosylated precursor of M [membrane protein]) and E (envelope protein): the proper folding of prM bearing unprocessed N-linked oligosaccharide is inefficient, and this causes delayed formation of prME heterodimer. The complexes formed between incompletely folded prM and E appear to be unstable, leading to a nonproductive pathway. Inhibition of α-glucosidase-mediated N-linked oligosaccharide trimming may thus prevent the assembly of DEN virus by affecting the early stages of envelope glycoprotein processing.
The dengue (DEN) virion is composed of three structural proteins designated C (core protein), M (membrane protein), and E (envelope glycoprotein) (6). The genomic plus-stranded RNA molecule is translated as a single polyprotein precursor which is cotranslationally processed by host cell- and virus-encoded proteinases to give three structural proteins, C (capsid protein), prM (the intracellular glycosylated precursor of M), and E, and at least seven nonstructural (NS) proteins, NS1 to NS5 (5, 26). The order of these individual proteins in the polyprotein precursor is C-prM-E-NS1-NS2A-NS2B-NS3-NS4A-NS4B-NS5.
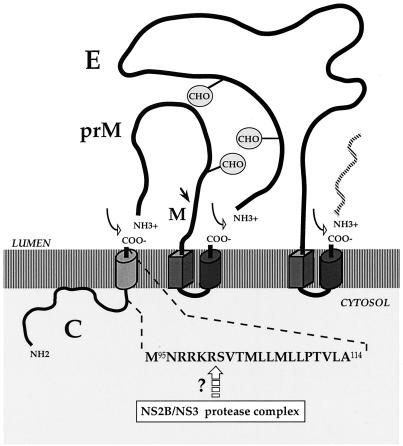
The first steps of DEN virus assembly take place in association with the membranes of the endoplasmic reticulum (ER) (5, 26). The internal signal sequence at the junction of C and prM directs the translocation of prM across the ER (5, 26) (Fig. 1). The virus-encoded NS2B-NS3 protease complex which catalyzes cleavage of the COOH terminus of the C protein on the cytoplasmic side of the ER may promote signal peptidase cleavage at the junction of C and prM (2, 19, 29, 34, 35) (Fig. 1). Indeed, the signal peptidase cleavage site at the NH2 terminus of prM in the C-prM precursor may remain in a cryptic conformation unless the cytoplasmic region is cleaved (2, 19, 33). The junction at prM-E is processed by signal peptidases on the luminal side of the ER membrane (5, 26, 29) (Fig. 1). Type I transmembrane glycoproteins prM and E appear inside the lumen of the ER (5, 26). The DEN virion is first assembled as an immature particle which contains prM noncovalently associated with E in a heterodimeric complex (5, 26). In the prME heterodimer, prM prevents an irreversible conformational change of E during virus secretion through acidified sorting compartments (15). Proteolysis of prM leads to the formation of homodimeric forms of E in virus particles before their release from the cell (25, 26).
FIG. 1.
Production of structural proteins of DEN-1 virus. At the top is a schematic representation of the orientation in the ER membranes of C, prM, and E (5, 6, 26). Signal peptides are shown as cylinders, and stop-transfer sequences are shown as rectangular blocks. The proteolytic cleavage sites are indicated by arrows (open arrow, signal peptidase cleavage site; dark arrow, furin cleavage site) (5, 6, 26, 27). The circled CHOs are potential sites for N-glycosylation (9, 17). At the bottom is shown the amino acid sequence of residues 95 to 114 of C of FGA/89, numbered from the N terminus of C (9). Below the sequence, the open arrow indicates the putative cleavage site that reacts with the NS2B/NS3 protease complex (2, 19, 28, 33, 35). Amino acids 101 to 114 are thought to constitute a transmembrane domain (9; P. Desprès, V. Deubel, and F. Pénin, unpublished results). The model is not to scale.
There is evidence that the N-linked oligosaccharide processing events in the ER are important for the secretion of some enveloped viruses (21). Recently, it has been shown that life cycles of enveloped viruses such as human immunodeficiency virus and human hepatitis B virus are greatly altered in cells in which α-glucosidase-mediated N-linked oligosaccharide trimming is inhibited (3, 4, 11, 20, 21). The initial steps of N-linked oligosaccharide processing on the glycoprotein in the ER involve the sequential trimming of the glucose residues on oligosaccharide precursor Glc3Man9GlcNAc2 after it is transferred from the dolichol diphosphate to the growing polypeptide backbone (30). ER α-glucosidases I and II are involved in the first steps in the trimming pathway. α-Glucosidase I removes the terminal α(1,2)-linked glucose from Glc3Man9GlcNAc2, and α-glucosidase II removes the second and possibly the third α(1,3)-linked glucose residues (13).
Castanospermine (CST) and deoxynojirimycin (DNJ) are ER α-glucosidase inhibitors, and both potently inhibit the early stages of glycoprotein processing. In this study, we investigated whether these α-glucosidase inhibitors affect DEN virus production in a mouse neuronal model of DEN virus infection.
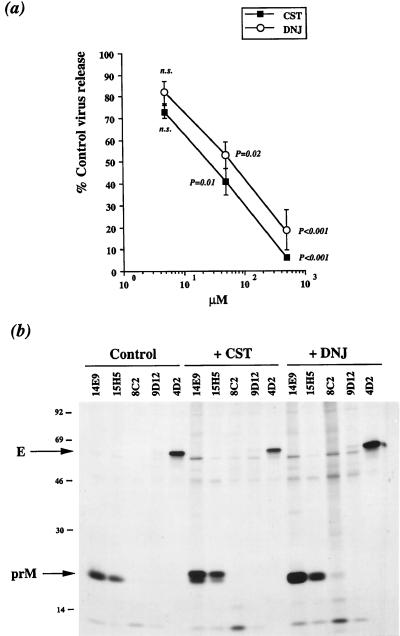
The derivation and characterization of FGA/89, a wild-type human isolate of DEN type 1 virus (DEN-1 virus), has previously been described (9). FGA/89 glycoprotein prM has a single N-linked glycosylation site at position Asn68 whereas glycoprotein E has two potential sites, Asn67 and Asn153 (Fig. 1) (9). Both N-glycosylation sites of the DEN-1 virus glycoprotein E appear to be utilized during the N-glycosylation process (12, 17). Previous studies showed that the rate of FGA/89 protein synthesis in mouse neuroblastoma Neuro 2a cells reached a maximum 20 h after infection and then decreased by 25 h when apoptotic cell death occurred (7, 8). To study the effect of the α-glucosidase inhibitors on DEN virus release, FGA/89-infected Neuro 2a cells were incubated with CST or DNJ for 6 h after 19 h of infection. Infective particles produced in the presence of various concentrations of CST or DNJ were harvested and titrated by focus immunoassay on the mosquito cell line AP61 (9). Treatment with CST resulted in a dose-dependent reduction in the amount of infectious virus, down to 5% of the control value at 500 μM (Fig. 2a). Similarly, the response to DNJ was dose dependent, and 500 μM DNJ reduced the production of infective particles to 20% (Fig. 2a). Thus, the amount of infectious FGA/89 virus released into the supernatants was significantly reduced by treatment with 500 μM CST or DNJ for 6 h.
FIG. 2.
DEN virus production by glucosidase inhibitor-treated Neuro 2a cells. Neuro 2a cells were infected with strain FGA/89 of DEN-1 virus at a multiplicity of infection of 400 (7). (a) Three sets of infected cell monolayers were incubated with various doses of CST or DNJ for 6 h beginning at 19 h postinfection. Virus yield 25 h postinfection was determined by focus immunoassay titration (9). Virus titer is expressed as the virus titer in CST- or DNJ-treated cells as a percentage of that in mock-treated cells. Data were compared between groups with the Fisher and Yates t test. P < 0.05 was considered significant. n.s., not significant. (b) Viral protein synthesis in glucosidase inhibitor-treated cells. Infected Neuro 2a cells were incubated without (Control) or with 500 μM CST (+CST) or DNJ (+DNJ) for 3 h beginning 19 h postinfection. Proteins from infected cells were labeled for 1 h at 21 h postinfection. DEN virus structural proteins from cell lysate samples were immunoprecipitated with anti-prM MAb 14E9 or 15H5 or with anti-E MAb 8C2, 9D12, or 4D2 (7, 9). Samples were analyzed by SDS-PAGE under nonreducing conditions.
We investigated the mechanisms by which α-glucosidase inhibitors affect the production of infectious FGA/89 virus. First, we examined whether the failure of virus release was linked to the effects of CST and DNJ on Neuro 2a cell viability. More than 95% of Neuro 2a cells incubated for 6 h with 500 μM CST or DNJ still excluded trypan blue. No obvious morphological changes were observed between drug-treated and mock-treated cells, as determined by fluorescent staining of the actin cytoskeleton (data not shown). These observations suggest that neither α-glucosidase inhibitor had a significant effect on the integrity of Neuro 2a cells.
We further investigated whether the overall rate of host protein synthesis was altered in Neuro 2a cells treated with α-glucosidase inhibitor. To measure total protein synthesis, two sets of Neuro 2a cell monolayers treated with 500 μM CST or DNJ for 2 h were labeled for 1 h with Tran35S-label (100 μCi/ml; ICN Pharmaceuticals Inc.) in the presence of the drugs as previously described (7, 9, 10). The cells were lysed with SUM buffer (1% sodium dodecyl sulfate [SDS], 2 M urea, 2% β-mercaptoethanol, 25 mM glucose, 5 mM EDTA, 20 mM Tris-Cl [pH 6.8]), and samples were boiled at 65°C for 15 min. Radioactive proteins were determined by scintillation counting of trichloroacetic acid-precipitated samples (10). The level of overall protein synthesis in CST- or DNJ-treated cells was about 80 or 70% of normal, respectively. Therefore, α-glucosidase inhibitor treatment resulted in a limited shutoff of host protein synthesis.
The lower rate of intracellular protein synthesis observed in the presence of α-glucosidase inhibitor may be sufficient to affect DEN-1 virus replication in Neuro 2a cells. We investigated this possibility by using a radioimmunoprecipitation (RIP) assay. To study the synthesis of viral proteins, FGA/89-infected Neuro 2a cells were treated for 3 h with 500 μM CST or DNJ after 19 h of infection. Infected cells were labeled for 1 h with Tran35S-label (200 μCi/ml) at 21 h postinfection. Cells were lysed with RIP buffer as previously described (7, 9, 10). Samples were analyzed by SDS–15% polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions. A panel of prM- and E-specific monoclonal antibodies (MAbs) directed against DEN-1 virus strain FGA/89 was used to study the synthesis of viral proteins in infected cells in the presence or absence of α-glucosidase inhibitor (7, 9). RIP analysis showed that prM (apparent molecular mass of 17 kDa) was immunoprecipitated with the anti-prM MAbs 14E9 and 15H5 (Fig. 2b) (7). The anti-E MAbs 8C2, 9D12, and 4D2 react with the E protein of cells infected with FGA/89 (7, 9). The epitopes bound by MAbs 8C2 and 9D12 are reactive in their linear conformation, whereas MAb 4D2 binds to a conformational epitope on the mature FGA/89 virion (7, 9). RIP analysis showed that MAbs 8C2 and 9D12 immunoprecipitated a small amount of labeled protein E (Fig. 2b). In contrast, a single species of protein E (apparent molecular mass of 63 kDa) was clearly detected in samples immunoprecipitated with the conformation-sensitive MAb 4D2 (Fig. 2b). This suggests that most of the protein E molecules were correctly folded. Levels of production of glycoproteins prM and E were similar in glucosidase inhibitor-treated Neuro 2a cells and untreated cells, suggesting that CST and DNJ did not affect viral protein synthesis (Fig. 2b, compare lanes +CST and +DNJ with Control). However, anti-prM MAbs and the anti-E MAb 4D2 recognized structural proteins in glucosidase inhibitor-containing samples, suggesting that the reactivities of these MAbs were unaffected (Fig. 2b). More labeled prM was present in CST- and DNJ-containing samples, and to a lesser extent, more labeled protein E was present in DNJ-containing samples, than in untreated cells (Fig. 2b). This may be due to the accumulation of newly synthesized structural proteins in glucosidase inhibitor-treated cells because the rate of virus release was low.
The correct folding or secretion of some viral glycoproteins is impaired in cells in which α-glucosidase-mediated N-linked oligosaccharide trimming is prevented (21). We first determined the kinetics of the folding and heterodimerization of prM and E in Neuro 2a cells by pulse-chase analysis. FGA/89-infected cells were pulse-labeled with 500 μCi of Tran35S-label per ml for 10 min at 20 h postinfection and chased as previously described (10). The formation of aberrant disulfide bonds in cysteine-containing proteins during cell lysis was prevented by washing cell monolayers in ice-cold phosphate-buffered saline containing 50 mM iodoacetamide, a sulfhydryl-alkylating agent. Cells were lysed with ice-cold 1% Triton X-100 in TNI buffer (10 mM Tris, 150 mM NaCl, 10 mM iodoacetamide [pH 8.0]) containing a set of protease inhibitors (Complete; Roche Molecular Biochemicals). Cell extracts were treated with normal mouse serum and protein A-Sepharose beads and pelleted. The supernatants were incubated with MAbs and protein A-Sepharose. The folding of protein E and its assembly with prM were assessed by using antibodies that recognized correctly folded protein E (MAb 4D2) and the prME heterodimer (MAb 15H5) (7, 9). The complexes were washed five times with ice-cold 0.1% Triton X-100 in TNI buffer at 4°C, and radiolabeled antigens were released by adding Laemmli sample loading buffer and boiling.
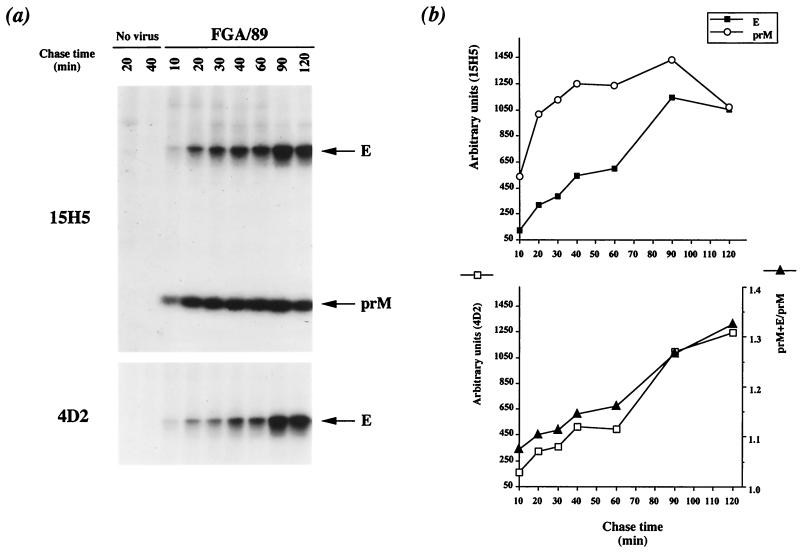
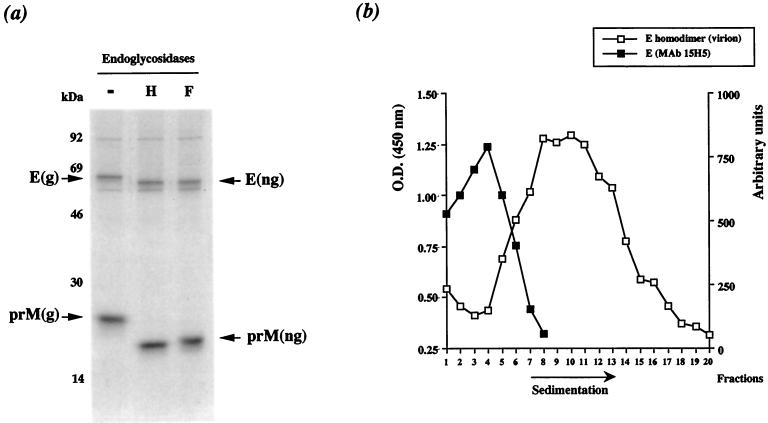
Pulse-labeled prM glycoprotein was immunoprecipitated with the anti-prM MAb 15H5 (Fig. 3a, 15H5). The amount of prM increased linearly during the first 40 min of chase and remained constant until 90 min. The time required for half-maximal prM formation (t1/2) was 15 to 20 min, indicating that this process is slow (Fig. 3b, top). Several reports provide evidence that prM production from flavivirus structural polyprotein is regulated (2, 19, 28, 29, 33, 35). The disulfide-dependent folding of prM was analyzed by SDS-PAGE under nonreducing and reducing conditions. The reduced form of protein prM migrated more slowly than did its nonreduced counterpart, indicating that correctly folded prM included disulfide bonds, consistent with the presence of six cysteine residues (data not shown). RIP analysis with the anti-prM MAb 15H5 showed that pulse-labeled prM and E interacted to form a heterodimer in FGA/89-infected Neuro 2a cells after 20 min of chase (Fig. 3a, 15H5). The molecular mass of the FGA/89 glycoprotein prM as assessed by SDS-PAGE was reduced by approximately 2 kDa by N-glycosidase F (PNGase F; New England BioLabs) digestion, consistent with the presence of one N-linked oligosaccharide at position Asn68 (Fig. 4a) (9). However, the molecular mass of FGA/89 glycoprotein E was reduced by approximately 4 kDa by PNGase F digestion, consistent with the presence of two N-linked oligosaccharides (Fig. 4a) (9). Both DEN virus envelope glycoproteins were completely susceptible to endo-β-N-acetylglucosaminidase H (endo H; Roche Molecular Biochemicals) after 30 min of chase, suggesting that prM and E associated in a compartment upstream from the medial Golgi compartment (Fig. 4a). The amount of E that coprecipitated with prM reached a maximum at 90 min of chase and then declined slightly (Fig. 3b, top). The t1/2 of the association between prM and E was 60 min, suggesting that formation of the prME heterodimer is slow (Fig. 3b, bottom). As prM formation is also a slow process (t1/2 = 20 min), it may be the rate-limiting step for prME heterodimerization.
FIG. 3.
Kinetics of FGA/89 envelope glycoprotein processing in Neuro 2a cells. Neuro 2a cells infected with DEN-1 virus (FGA/89) or mock-infected cells (No virus) were pulse-labeled for 10 min at 20 h postinfection and chased for various times. (a) Cell lysates were immunoprecipitated with anti-prM (15H5) or anti-E (4D2) MAb. (b) Samples on gels were analyzed with a PhosphorImager (Molecular Dynamics). Quantitation of the prM and E coimmunoprecipitated by MAb 15H5 (top) and the prM+E/prM ratio were used to define the kinetics of association of the prM and E proteins (bottom). Quantitation of the E protein precipitated by MAb 4D2 is shown at the bottom.
FIG. 4.
Processing of FGA/89 envelope glycoproteins in Neuro 2 cells. (a) Cell lysates chased for 30 min were immunoprecipitated with MAb 15H5. Labeled antigens were eluted from complexes in SDS and β-mercaptoethanol by boiling, and the denatured samples were divided into three aliquots for endo H (H), PNGase F (F), or mock (−) digestion as previously described (36). Glycosylated (g) and nonglycosylated (ng) forms of prM and E are indicated. (b) Pulse-labeled prME complexes chased for 90 min were subjected to sedimentation analysis on sucrose gradients. Samples were immunoprecipitated from gradient fractions with MAb 15H5 and separated by SDS-PAGE. Protein E that coprecipitated with prM was quantitated with a PhosphorImager (black boxes). The sedimentation of detergent-solubilized glycoprotein E homodimer from DEN-1 virions was determined by treating highly purified virus with 1% Triton X-100 in TNI buffer at 20°C for 1 h (1). Denatured virions were run in the parallel sucrose gradient, and protein E homodimer was detected by enzyme-linked immunosorbent assay using an anti-E specific antibody (open boxes). O.D., optical density.
DEN virus prME heterodimers are transported as oligomeric complexes through the cell secretory pathway (1, 5, 6, 15, 19, 26, 27, 31). We examined the oligomeric assembly of DEN virus envelope glycoproteins in FGA/89-infected Neuro 2a cell lysates chased for 90 min by velocity gradient centrifugation. Cells were lysed with 1% Triton X-100 in TNI buffer, and samples were directly applied to 5 to 20% (wt/vol) continuous sucrose gradients in TNI buffer containing 0.1% Triton X-100 (1). The gradients were centrifuged for 16 h at 35,000 rpm in an SW41 rotor at 4°C, and the distribution of prME complexes in gradient fractions was determined by immunoprecipitation with the anti-prM MAb, 15H5. Most mature prME complexes sedimented more slowly than DEN virus E homodimer (predicted size, about 130 kDa), suggesting that the assembly of FGA/89 glycoprotein heterodimer complexes was an inefficient process in Neuro 2a cells (Fig. 4b). However, it is possible that multimeric prME complexes were labile in the presence of 1% Triton X-100 (31).
The folding of FGA/89 glycoprotein E was monitored with the anti-E MAb 4D2. The amount of labeled E glycoprotein was low during the first 60 min of chase and then increased rapidly until 90 min (Fig. 3a, 4D2). The t1/2 of formation of the conformation-dependent 4D2 epitope was more than 60 min, indicating that FGA/89 glycoprotein E folded slowly (Fig. 3b, bottom).
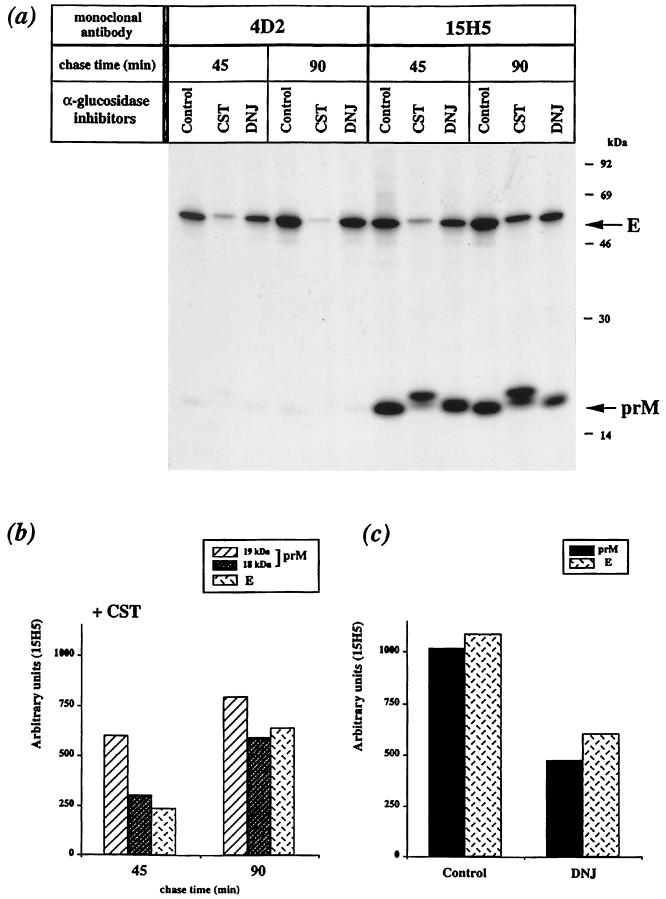
Thus, we assessed the effects of CST and DNJ on the processing of newly synthesized structural proteins in DEN virus-infected cells. FGA/89-infected Neuro 2a cells were pulse-labeled for 10 min and chased for 45 min (t1/2 for formation of prME heterodimer) or 90 min in the presence of α-glucosidase inhibitor. CST or DNJ, each at 500 μM, was included in the starvation and chase media. In the presence of the α-glucosidase inhibitor CST, the prM and E glycoproteins migrated more slowly in SDS-PAGE than did their counterparts in mock-treated cells (Fig. 5a, compare lanes CST and Control). It is likely that the presence of triglucosylated N-linked oligosaccharides on the N-glycan sites causes changes in protein conformation. Thus, as the samples were analyzed by SDS-PAGE under nonreducing conditions, the different electrophoretic mobilities may result from different conformations stabilized by disulfide bonds. The mobilities of prM and E in DNJ-containing samples were intermediate between those of their counterparts from mock- and CST-treated samples. This suggests that one or two glucose residues remained on core oligosaccharides due to incomplete inhibition of glucose trimming (Fig. 5a, compare lanes DNJ with CST and Control).
FIG. 5.
Processing of prM and E in FGA/89-infected Neuro 2a cells treated with α-glucosidase inhibitor. Infected cells were pulse-labeled 20 h postinfection and chased in the presence of the α-glucosidase inhibitor (500 μM) or mock treated (Control). (a) Samples were immunoprecipitated with anti-DEN virus antibody. Proteins prM and E are indicated. (b and c) prM and E coimmunoprecipitated by MAb 15H5 are quantified as described in the legend to Fig. 3.
In CST-treated cells, the amount of the E glycoprotein immunoprecipitated by the conformation-sensitive MAb 4D2 was about 30% of that in controls at 45 min of chase and only 15% at the end of the chase (Fig. 5a, compare lanes CST and Control, 4D2). Thus, the E-related form carrying triglucosylated N-linked oligosaccharides appeared to be misfolded, leading to a nonproductive pathway. This result demonstrates that glucose trimming contributes to the correct folding of FGA/89 glycoprotein E. Conversely, the formation of the conformation-dependent 4D2 epitope was unaffected by the presence of DNJ (Fig. 5a, compare lanes DNJ and Control, 4D2).
The amount of the E-related form that coprecipitated with prM from CST-containing samples was about 25% of that of the control at 45 min of chase (Fig. 5a, compare lanes CST with Control, 15H5). It appeared, therefore, that CST decreased the formation efficiency of prME heterodimer in FGA/89-infected Neuro 2a cells. The anti-prM MAb 15H5 immunoprecipitated two closely related forms of prM from CST-treated cells with apparent molecular masses of 18 and 19 kDa; both migrated more slowly than prM from mock-treated samples (Fig. 5a, compare lanes CST with Control, 15H5). The intensity of the prM-related 19-kDa band increased slightly during the chase period (Fig. 5b). Conversely, the intensity of the prM-related 18-kDa band increased twofold, and there was a parallel increase in the amount of the E-related form associated with prM in a heterodimeric complex (Fig. 5b). Therefore, the formation of prME heterodimer appeared to require the prM-related 18-kDa form in CST-treated cells. The position within the NH2 terminus of prM at which signal peptidase cleavage occurs may be affected by CST, causing slower conversion of this site from a cryptic to an accessible conformation on the luminal side of the ER membrane. If this is so, the prM-related 19-kDa form may be a prM precursor containing the signal sequence at its NH2 terminus (see below). However, the association between prM and E does not appear to necessitate the trimming of the glucosylated N-linked oligosaccharides, as these proteins can be recovered as a heterodimeric complex in CST-treated cells.
In the presence of DNJ, the amount of prME heterodimer declined during the chase (Fig. 5a, compare 45 and 90 min, 15H5). At 90 min of chase, the amount of complex formed between the prM and E-related forms in DNJ-containing samples was about 50% of that in controls (Fig. 5c). Thus, the presence of partially deglucosylated N-linked oligosaccharides at the N-glycan sites may interfere with the stability of DEN virus envelope glycoprotein complexes which are probably misassociated heterodimers leading to a nonproductive pathway.
Therefore, inhibition of α-glucosidase-mediated N-linked oligosaccharide trimming seemed to impair the correct folding and optimal assembly of FGA/89 envelope glycoproteins in mouse neuroblastoma cells. We investigated the impact of glucose trimming on DEN virus envelope glycoprotein processing by expressing the FGA/89 cDNA coding for prM and E in Neuro 2a cells, using the ecdysone-inducible gene expression system (22). Transient expression in Neuro 2a cells indicated that the prM gene in cis greatly increases the production of correctly folded E protein (data not shown), supporting the notion that prM may have a chaperone-like role in the folding of E in the ER (1, 18). The cell clone N2aprM+E was established. It carries the cDNA comprising the FGA/89 prM+E region (Met95 to Ala773) and the genes encoding the modified ecdysone receptor VgEcR and the retinoic X receptor. The 20 amino acids (Met95 to Ala114) at the COOH terminus of protein C are required for translocation of the prM protein (26, 29, 34) (Fig. 1). In the presence of the ecdysone analog ponasterone A, the retinoic X receptor and VgEcR heterodimerize and then transactivate the ecdysone response element-containing promoter. The ecdysone response elements are upstream from a minimal heat shock promoter that drives the expression of the FGA/89 prM and E genes. After 15 h of induction with ponasterone A (5 μM), recombinant FGA/89 envelope glycoproteins were readily detected in N2aprM+E cells (data not shown).
The electrophoretic mobilities of the recombinant prM and E proteins under nonreducing conditions were identical to those of their counterparts from FGA/89-infected Neuro 2a cells (data not shown). Furthermore, treatment with PNGase F indicated that a single N-linked oligosaccharide was present on recombinant prM and that the two N-linked oligosaccharides were present on recombinant E (data not shown). These results suggest that the newly synthesized prM and E proteins in N2aprM+E cells were correctly processed and folded.
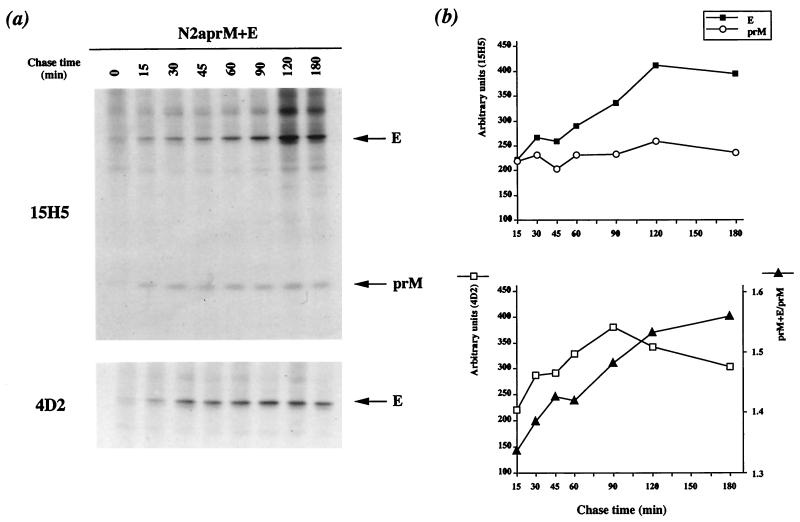
The kinetics of recombinant DEN virus envelope glycoprotein processing were examined by pulse-chase experiments. Induced N2aprM+E cells were pulse-labeled for 10 min after 20 h of induction and chased for 180 min. Recombinant prM glycoprotein was clearly detected 15 min after the pulse (Fig. 6a, 15H5), and the amount of prM was constant until the end of the chase (Fig. 6b, top). Signal peptidase cleavage of the prM signal sequence is modulated by the presence upstream of protein C (2, 19, 28, 34). The t1/2 of formation of prM was shorter in N2aprM+E cells than that of its counterpart in FGA/89-infected Neuro 2a cells, supporting the notion that the absence of the C protein enhances the efficiency of prM processing.
FIG. 6.
Kinetics of recombinant DEN virus envelope glycoprotein processing in N2aprM+E cells. N2aprM+E cells were briefly pulse-labeled 20 h after addition of ponasterone A (5 μM) and chased for various times. (a) Cell lysates were immunoprecipitated with anti-DEN virus antibody; (b) samples on gels were quantified as described in the legend to Fig. 3.
The amounts of recombinant E protein that coprecipitated with prM increased slowly during the first 120 min of chase and then remained constant until the end of the chase (Fig. 6a, 15H5). The t1/2 of formation of the prME heterodimer was 75 to 90 min (Fig. 6b, bottom). The size distribution of recombinant prME heterodimers in samples chased for 90 min was examined by sedimentation through a sucrose gradient. The faster-sedimenting complexes cosedimented with the FGA/89 prME heterodimer, suggesting that most recombinant DEN virus envelope glycoprotein complexes are single heterodimers (data not shown).
The amount of recombinant E glycoprotein immunoprecipitated with MAb 4D2 increased linearly during the first 90 min of chase and then declined slowly until the end of the chase (Fig. 6a, 4D2). The t1/2 for formation of the conformation-dependent 4D2 epitope was less than 60 min (Fig. 6b, bottom), suggesting that the folding of recombinant E glycoprotein was faster than that of its counterpart in FGA/89-infected Neuro 2a cells (t1/2 > 60 min).
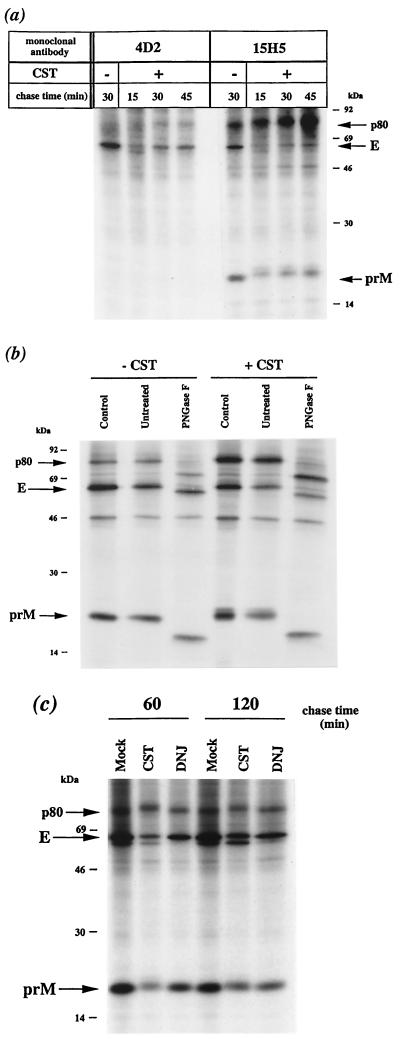
We examined the effects of trimming inhibition on recombinant DEN virus envelope glycoprotein processing. Induced N2aprM+E cells were pulse-labeled for 10 min after 20 h of induction and chased for various times in the presence of the α-glucosidase inhibitor. In the presence of CST, recombinant DEN virus envelope glycoproteins had a slightly greater electrophoretic mobility than their counterparts from mock-treated cells, suggesting that glucose trimming was efficiently inhibited in N2aprM+E cells (Fig. 7a).
FIG. 7.
Processing of recombinant prM and E in N2aprM+E cells treated with α-glucosidase inhibitor. (a) Mock-treated (−) or CST-treated (+) cells were pulse-labeled and chased as described in the legend to Fig. 5. Cell lysates were immunoprecipitated with anti-DEN virus antibody. (b) Mock-treated (−CST) or CST-treated (+CST) samples chased for 30 min were immunoprecipitated with MAb 15H5. Immunoprecipitated proteins were directly analyzed (Control) or incubated in the presence (PGNase F) or absence (Untreated) of PGNase F as described in the legend to Fig. 4. (c) α-Glucosidase inhibitor-containing samples (CST or DNJ) or mock-treated samples (Mock) were immunoprecipitated with MAb 15H5.
Detailed analysis of CST-containing samples revealed the existence of a polypeptide with a molecular mass of about 80 kDa (p80) which also reacted with the anti-prM MAb 15H5 (Fig. 7a). The samples were subjected to PNGase F digestion, and the molecular mass of the polypeptide was found to have been reduced by about 7 kDa in SDS-PAGE under reducing conditions, consistent with the cleavage of three N-linked oligosaccharides (Fig. 7b, compare lanes Untreated and PGNase F). Therefore, p80 may be the uncleaved precursor of recombinant prM and E in its glycosylated form. There was significantly more putative uncleaved precursor of recombinant DEN virus glycoproteins in CST-treated samples, suggesting that glucosidase inhibition reduced the efficiency of signal peptidase cleavage at the junction between prM and E in N2aprM+E cells (Fig. 7b, compare lanes −CST and +CST). Thus, the maintenance of triglucosylated N-linked oligosaccharides may change the position of the signal peptidase cleavage site at the NH2 terminus of the E glycoprotein, by reducing its accessibility on the luminal side of the ER membrane.
In CST-treated N2aprM+E cells, the amount of labeled recombinant E glycoprotein immunoprecipitated with MAb 4D2 increased slowly during the first 45 min of chase (Fig. 7a, compare lanes +CST with −CST, 4D2). The formation of the conformation-dependent 4D2 epitope was reduced to 50% of that in controls after 60 min of chase (data not shown). Therefore, the folding efficiency of recombinant E glycoprotein was diminished by CST as seen in DEN virus-infected cells.
In N2aprM+E cells treated with CST, the anti-prM MAb 15H5 immunoprecipitated two forms of recombinant prM which migrated with apparent molecular masses of 18 and 19 kDa, higher than the molecular mass of prM from mock-treated cells (Fig. 7a, compare lanes +CST with −CST, 15H5). The amount of the prM-related 18-kDa form increased during the first 45 min of chase (Fig. 7a, lanes +CST, 15H5), and this form was the most abundant after 60 min (Fig. 7c, lanes CST). There was a parallel increase in the amount of recombinant E protein that coprecipitated with prM, suggesting that the prM-related 18-kDa form was required for efficient assembly of prME heterodimer as observed in FGA/89-infected Neuro 2a cells (Fig. 7a, lanes +CST, 15H5).
We investigated whether the lower electrophoretic mobility of the prM-related 19-kDa form in CST-treated N2aprM+E cells was due to the maintenance of the putative signal sequence. Cell lysates chased for 30 min were immunoprecipitated with MAb 15H5, and samples were digested with PNGase F. Under the reducing conditions used for this assay, both prM-related forms were observed, suggesting that the changes in electrophoretic mobility were not due to modifications in disulfide bonding (Fig. 7b, lane Control, +CST). After digestion with PNGase F, which cleaves all N-linked oligosaccharides, the two prM-related forms in CST-containing samples migrated as a single band at the same speed as the deglycosylated recombinant prM (Fig. 7b, compare lanes Untreated and PNGase F, +CST). The signal peptidase therefore appeared to have cleaved prM efficiently at the NH2 terminus in CST-treated N2aprM+E cells. The deglycosylated form of prM in CST-containing samples (Fig. 7b, lane PNGase F, +CST) had a slightly lower mobility than its counterpart in inhibitor-free samples (Fig. 7b, lane PNGase F, −CST), indicating a modification unrelated to N-glycosylation. Further investigation is required to determine whether the 18- and 19-kDa polypeptides in CST-treated cells are incompletely folded or misfolded forms of prM bearing unprocessed N-linked oligosaccharides.
In the presence of the α-glucosidase inhibitor DNJ, the yield of prME complexes in induced N2aprM+E cells was about 60% of the control value after 60 min of chase (Fig. 7c, compare lanes DNJ with Control). Thus, DNJ affected the formation of recombinant prME heterodimer as observed in FGA/89-infected Neuro 2a cells.
We found that DEN virus envelope glycoprotein processing in mouse neuroblastoma cells was strongly affected if α-glucosidase-mediated N-linked oligosaccharide trimming was inhibited. Inhibition of glucose trimming has a major effect on the productive folding pathways of DEN virus glycoproteins prM and E, indicating that N-glycosylation processing is required for folding into the correct conformation. The folding efficiency of prM appeared to be reduced by the presence of triglucosylated N-linked oligosaccharide at residue Asn68 of prM, causing a delay in formation of the prME heterodimer. Glucosidase inhibition did not prevent the heterodimeric association between prM and E, but the prME complexes appeared to be unstable, reducing the efficiency of folded prME complex assembly. It is likely that these transient heterodimeric complexes are misassociated prME complexes carrying unprocessed N-linked oligosaccharides. It has been suggested that domain II of flavivirus E glycoprotein is involved in prME heterodimerization (25), and the presence of glucosylated N-linked oligosaccharide within this region at residue Asn67 may destabilize the complexes formed between prM and E, leading to a nonproductive assembly pathway. These findings suggest that the formation of properly folded DEN virus envelope glycoprotein complexes requires a lectin chaperone pathway. The molecular chaperones calnexin and calreticulin have lectin-like affinity for monoglucosylated N-linked oligosaccharides and interact with intermediate oligomeric complexes of viral envelope glycoproteins in the ER (14, 16, 23, 24, 32). However, preliminary results suggest that neither calreticulin nor calnexin associates with DEN virus envelope glycoproteins in FGA/89-infected Neuro 2a cells (M. P. Courageot, V. Deubel, and P. Desprès, unpublished results). Glucose trimming of N-linked oligosaccharides may have a direct effect in promoting the correct folding of DEN virus envelope glycoproteins.
This study shows that N-linked oligosaccharide processing may play a crucial role in the early stages of DEN virus morphogenesis. We also found that α-glucosidase inhibitors can be used to prevent the first steps of DEN virus envelope glycoprotein processing and that the inhibition of glucose trimming has antiviral effects.
Acknowledgments
We thank Mary K. Gentry and Robert Putnak (Walter Reed Army Institute of Research, Washington, D.C.) for providing MAbs.
This investigation was supported by research grants from the CNRS Interdisciplinaire de Recherche Environnement Vie et Société, program 95N82/0134, DGA, program 99 34 031, and CAPES/COFECUB, program 254/98. M.-P. Courageot was supported by scholarship funds received from the Ministère de l'Education Nationale, de la Recherche et de la Technologie.
REFERENCES
- 1.Allison S L, Stadler K, Mandl C W, Kunz C, Heinz F X. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol. 1995;69:5816–5820. doi: 10.1128/jvi.69.9.5816-5820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amberg S M, Nestorowicz A, McCourt D W, Rice C M. NS2B-3 proteinase-mediated processing in the yellow fever virus structural region: in vitro and in vivo studies. J Virol. 1994;68:3794–3802. doi: 10.1128/jvi.68.6.3794-3802.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Block T M, Lu X, Mehta A, Blumberg B S, Tennant B, Ebling M, Korba B, Lansky D M, Jacob G S, Dwek R A. Treatment of chronic hepadnavirus infection in a woodchuck animal model with an inhibitor of protein folding and trafficking. Nat Med. 1998;4:610–614. doi: 10.1038/nm0598-610. [DOI] [PubMed] [Google Scholar]
- 4.Block T M, Lu X, Platt F M, Foster G R, Gerlich W H, Blumberg B S, Dwek R A. Secretion of human hepatitis B virus is inhibited by the imino sugar N-butyldeoxynojirimycin. Proc Natl Acad Sci USA. 1994;91:2235–2239. doi: 10.1073/pnas.91.6.2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers T J, Hahn C S, Galler R, Rice C M. Flavivirus genome organization, expression, and replication. Annu Rev Microbiol. 1990;44:649–688. doi: 10.1146/annurev.mi.44.100190.003245. [DOI] [PubMed] [Google Scholar]
- 6.Chang G J. Molecular biology of dengue viruses. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. New York, N.Y: CAB International; 1997. pp. 175–198. [Google Scholar]
- 7.Desprès P, Flamand M, Ceccaldi P E, Deubel V. Human isolates of dengue type 1 virus induce apoptosis in mouse neuroblastoma cells. J Virol. 1996;70:4090–4096. doi: 10.1128/jvi.70.6.4090-4096.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desprès P, Frenkiel M P, Ceccaldi P E, Duarte Dos Santos C, Deubel V. Apoptosis in the mouse central nervous system in response to infection with mouse-neurovirulent dengue viruses. J Virol. 1998;72:823–829. doi: 10.1128/jvi.72.1.823-829.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Desprès P, Frenkiel M P, Deubel V. Differences between cell membrane fusion activities of two dengue type-1 isolates reflect modifications of viral structure. Virology. 1993;196:209–219. doi: 10.1006/viro.1993.1469. [DOI] [PubMed] [Google Scholar]
- 10.Desprès P, Griffin J W, Griffin D E. Effects of anti-E2 monoclonal antibody on Sindbis virus replication in AT3 cells expressing bcl-2. J Virol. 1995;69:7006–7014. doi: 10.1128/jvi.69.11.7006-7014.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer P B, Collin M, Karlsson G B, James W, Butters T D, Davis S J, Gordon S, Dwek R A, Platt F M. The α-glucosidase inhibitor N-butyldeoxynojirimycin inhibits human immunodeficiency virus entry at the level of post-CD4 binding. J Virol. 1995;69:5791–5797. doi: 10.1128/jvi.69.9.5791-5797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonseca B A, Pincus S, Shope R E, Paoletti E, Mason P W. Recombinant vaccinia viruses co-expressing dengue-1 glycoproteins prM and E induce neutralizing antibodies in mice. Vaccine. 1994;12:279–285. doi: 10.1016/0264-410x(94)90206-2. [DOI] [PubMed] [Google Scholar]
- 13.Hebert D N, Foellmer B, Helenius A. Glucose trimming and reglucosylation determine glycoprotein association with calnexin in the endoplasmic reticulum. Cell. 1995;81:425–433. doi: 10.1016/0092-8674(95)90395-x. [DOI] [PubMed] [Google Scholar]
- 14.Hebert D N, Foellmer B, Helenius A. Calnexin and calreticulin promote folding, delay oligomerization and suppress degradation of influenza hemagglutinin in microsomes. EMBO J. 1996;15:2961–2968. [PMC free article] [PubMed] [Google Scholar]
- 15.Heinz F X, Auer G, Stiasny K, Holzmann H, Mandl C, Guirakhoo F, Kunz C. The interaction of the flavivirus envelope proteins: implications for virus entry and release. Arch Virol. 1994;9:339–348. doi: 10.1007/978-3-7091-9326-6_34. [DOI] [PubMed] [Google Scholar]
- 16.Helenius A, Trombetta E S, Hebert D N, Simons J F. Calnexin, calreticulin and the folding of glycoproteins. Trends Cell Biol. 1997;7:193–210. doi: 10.1016/S0962-8924(97)01032-5. [DOI] [PubMed] [Google Scholar]
- 17.Johnson A J, Guirakhoo F, Roehrig J T. The envelope glycoproteins of dengue 1 and dengue 2 viruses grown in mosquito cells differ in their utilization of potential glycosylation sites. Virology. 1994;203:241–249. doi: 10.1006/viro.1994.1481. [DOI] [PubMed] [Google Scholar]
- 18.Konishi E, Mason P W. Proper maturation of the Japanese encephalitis virus envelope glycoprotein requires cosynthesis with the premembrane protein. J Virol. 1993;67:1672–1675. doi: 10.1128/jvi.67.3.1672-1675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lobigs M. Flavivirus premembrane protein cleavage and spike heterodimer secretion require the function of the viral proteinase NS3. Proc Natl Acad Sci USA. 1993;90:6218–6222. doi: 10.1073/pnas.90.13.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu X, Mehta A, Dadmarz M, Dwek R, Blumberg B S, Block T M. Aberrant trafficking of hepatitis B virus glycoproteins in cells in which N-glycan processing is inhibited. Proc Natl Acad Sci USA. 1997;94:2380–2385. doi: 10.1073/pnas.94.6.2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mehta A, Zitzmann N, Rudd P M, Block T M, Dwek R A. α-Glucosidase inhibitors as potential broad based anti-viral agents. FEBS Lett. 1998;430:17–22. doi: 10.1016/s0014-5793(98)00525-0. [DOI] [PubMed] [Google Scholar]
- 22.No D, Yao T P, Evans R M. Ecdysone-inducible gene expression in mammalian cells and transgenic mice. Proc Natl Acad Sci USA. 1996;93:3346–3351. doi: 10.1073/pnas.93.8.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otteken A, Moss B. Calreticulin interacts with newly synthesized human immunodeficiency virus type 1 envelope glycoprotein, suggesting a chaperone function similar to that of calnexin. J Biol Chem. 1996;271:97–103. doi: 10.1074/jbc.271.1.97. [DOI] [PubMed] [Google Scholar]
- 24.Peterson J R, Ora A, Van P N, Helenius A. Transient, lectin-like association of calreticulin with folding intermediates of cellular and viral glycoproteins. Mol Biol Cell. 1995;6:1173–1184. doi: 10.1091/mbc.6.9.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rey F A, Heinz F X, Mandl C, Kunz C, Harrison S C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 26.Rice C M. Flaviviridae: the viruses and their replication. In: Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–959. [Google Scholar]
- 27.Stadler K, Allison S L, Schalich J, Heinz F X. Proteolytic activation of tick-borne encephalitis virus by furin. J Virol. 1997;71:8475–8481. doi: 10.1128/jvi.71.11.8475-8481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stocks C E, Lobigs M. Posttranslational signal peptidase cleavage at the flavivirus C-prM junction in vitro. J Virol. 1995;69:8123–8126. doi: 10.1128/jvi.69.12.8123-8126.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stocks C E, Lobigs M. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, the signal peptide, and prM. J Virol. 1998;72:2141–2149. doi: 10.1128/jvi.72.3.2141-2149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trombetta E S, Helenius A. Lectins as chaperones in glycoprotein folding. Curr Opin Struct Biol. 1998;8:587–592. doi: 10.1016/s0959-440x(98)80148-6. [DOI] [PubMed] [Google Scholar]
- 31.Wang S, He R, Anderson R. PrM- and cell-binding domains of dengue virus E protein. J Virol. 1999;73:2547–2551. doi: 10.1128/jvi.73.3.2547-2551.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Werr M, Prange R. Role for calnexin and N-linked glycosylation in the assembly and secretion of hepatitis B virus middle envelope protein particles. J Virol. 1998;72:778–782. doi: 10.1128/jvi.72.1.778-782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamshchikov V F, Compans R W. Formation of the flavivirus envelope: role of the viral NS2B-NS3 protease. J Virol. 1995;69:1995–2003. doi: 10.1128/jvi.69.4.1995-2003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamshchikov V F, Compans R W. Regulation of the late events in flavivirus protein processing and maturation. Virology. 1993;192:38–51. doi: 10.1006/viro.1993.1006. [DOI] [PubMed] [Google Scholar]
- 35.Yamshchikov V F, Trent D W, Compans R W. Upregulation of signalase processing and induction of prM-E secretion by the flavivirus NS2B-NS3 protease: roles of protease components. J Virol. 1997;71:4364–4371. doi: 10.1128/jvi.71.6.4364-4371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zheng Z, Maidji E, Tugizov S, Pereira L. Mutations in the carboxyl-terminal hydrophobic sequence of human cytomegalovirus glycoprotein B alter transport and protein chaperone binding. J Virol. 1996;70:8029–8040. doi: 10.1128/jvi.70.11.8029-8040.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]