Abstract
Background
Cyclin-dependent kinases 4 and 6 (CDK4/6) inhibitors have shown significant activity against several solid tumors by reducing the phosphorylation of the canonical CDK4/6 substrate retinoblastoma (Rb) protein, while the anti-tumor effect of CDK4/6 inhibitors on Rb-deficient tumors is not clear. Most small cell lung cancers (SCLCs) are Rb-deficient and show very modest response to immune checkpoint blockade (ICB) despite recent advances in the use of immunotherapy. Here, we aimed to investigate the direct effect of CDK4/6 inhibition on SCLC cells and determine its efficacy in combination therapy for SCLC.
Methods
The immediate impact of CDK4/6 inhibitor abemaciclib on cell cycle, cell viability and apoptosis in four SCLC cell lines was initially checked. To explore the effect of abemaciclib on double-strand DNA (ds-DNA) damage induction and the combination impact of abemaciclib coupled with radiotherapy (RT), western blot, immunofluorescence (IF) and quantitative real-time polymerase chain reaction (qRT-PCR) were performed. An Rb-deficient immunocompetent murine SCLC model was established to evaluate efficacy of abemaciclib in combination therapy. Histological staining, flow cytometry analysis and RNA sequencing were performed to analyze alteration of infiltrating immune cells in tumor microenvironment (TME).
Results
Here, we demonstrated that abemaciclib induced increased ds-DNA damage in Rb-deficient SCLC cells. Combination of abemaciclib and RT induced more cytosolic ds-DNA, and activated the STING pathway synergistically. We further showed that combining low doses of abemaciclib with low-dose RT (LDRT) plus anti-programmed cell death protein-1 (anti-PD-1) antibody substantially potentiated CD8+ T cell infiltration and significantly inhibited tumor growth and prolonged survival in an Rb-deficient immunocompetent murine SCLC model.
Conclusions
Our results define previously uncertain DNA damage-inducing properties of CDK4/6 inhibitor abemaciclib in Rb-deficient SCLCs, and demonstrate that low doses of abemaciclib combined with LDRT inflame the TME and enhance the efficacy of anti-PD-1 immunotherapy in SCLC model, which represents a potential novel therapeutic strategy for SCLC.
Keywords: Small cell lung cancer (SCLC), abemaciclib, low-dose radiotherapy (LDRT), immune checkpoint blockade (ICB), STING pathway
Highlight box.
Key findings
• Cyclin-dependent kinases 4 and 6 (CDK4/6) inhibitor abemaciclib induced increased double-strand DNA (ds-DNA) damage in both retinoblastoma (Rb)-deficient and Rb-proficient small cell lung cancer (SCLC) cells. Abemaciclib combined with radiotherapy activated the STING pathway synergistically in SCLC cells.
• Low doses of abemaciclib further enhanced the anti-tumor immune responses of low-dose radiotherapy plus anti-programmed cell death protein-1 (anti-PD-1) double-therapy in an Rb-deficient immunocompetent SCLC mouse model.
What is known and what is new?
• CDK4/6 inhibitors have shown significant anti-tumor activity against multiple Rb-proficient tumors. Most SCLCs are Rb-deficient and show very modest response to immune checkpoint blockade despite recent advances in the use of immunotherapy.
• In this study, we demonstrated that abemaciclib could induce ds-DNA damage in SCLC cells regardless of Rb expression status. We further showed that abemaciclib coupled with radiotherapy exhibited a synergistic effect on STING pathway activation and enhanced the anti-tumor immune responses of anti-PD-1 antibody in SCLC mouse model.
What is the implication, and what should change now?
• The combination of low doses of abemaciclib with low-dose radiotherapy could be a promising approach to augment the efficacy of immune checkpoint blockade for SCLC and deserves further investigation.
Introduction
Small cell lung cancer (SCLC), characterized by its rapid growth and early metastasis, is the most aggressive subtype of lung cancer. Chemotherapy is the first-line treatment option for patients with extensive stage (ES) in the past decades, until recent introduction of immune checkpoint blockade (ICB) therapy (1-3). However, overall survival and progression-free survival improved by the combined therapy remain modest. Despite having a relatively high tumor mutation burden, SCLC is often accompanied by a highly immunosuppressive tumor microenvironment (TME) with low levels of T-cell infiltration (4,5), leading to ICB being ineffective for most patients with SCLC (6,7). Therefore, there is a critical need to identify therapeutic strategies that can durably activate anti-tumor responses of ICB in SCLC.
Radiotherapy (RT) is currently used as an adjuvant therapy in both limited stage (LS)- and ES-SCLC (8,9). Recently, comprehensive studies have highlighted the potential of low-dose RT (LDRT) as an immune adjuvant in both preclinical and clinical studies (10-15). LDRT, typically administered at a dose of 0.5–3 Gy per fraction for a total of 1–20 Gy, has demonstrated its ability to transform TME from an immunosuppressive state to an inflammatory phenotype through various mechanisms (10-12). We have previously shown that LDRT delivered in five fractions of 15 Gy (15 Gy/5 F) combined with anti-programmed cell death protein-1 (anti-PD-1) antibody elicited significant tumor control and improved survival in a SCLC mouse model (16).
Cyclin-dependent kinases 4 and 6 (CDK4/6) inhibitors are currently approved by Food and Drug Administration (FDA) as therapy for advanced hormone receptor-positive breast cancer (17). Recently, CDK4/6 inhibitor trilaciclib has been approved by FDA for the reduction of chemotherapy-induced bone marrow suppression in patients with SCLC (18,19). Typically, by targeting cyclin D-CDK4/6 complex, CDK4/6 inhibitors can reduce phosphorylation of the tumor suppressor protein retinoblastoma (Rb) and sequester activity of the E2F transcription factors, thus restraining cellular transition from G1 phase to S phase and inhibiting tumor cell proliferation (17). Meanwhile, CDK4 and CDK6 bind and phosphorylate numerous non-Rb substrates (20,21), and recent preclinical research findings have highlighted the potential of CDK4/6 inhibitors to exhibit enhanced anti-tumor effects when combined with ICB, RT or chemotherapy through non-Rb targeted effects (17). It has been reported that CDK4/6 inhibition could directly enhance CD8+ T cell activation and induction of effector function (22-24) and promoted memory T cell differentiation (24-26). Besides, CDK4/6 inhibitors have also shown the ability to inhibit DNA damage response (DDR) of tumor cells and enhance the efficacy of RT (27,28) or chemotherapy (29,30) in diverse tumor cell lines. The presence of these non-Rb targets provides a possibility to apply CDK4/6 inhibitors to the treatment of Rb-deficient (Rb−) tumors. In fact, majority of SCLCs exhibit Rb gene (RB1) mutations and are Rb− (31), and the direct anti-tumor biological effects of CDK4/6 inhibition on Rb− SCLCs have remained uncharted thus far.
While abemaciclib, palbociclib, and ribociclib all target both CDK4 and CDK6 proteins, there are variations in their relative potency for these kinases (32), and abemaciclib, in particular, exhibits a broader spectrum of action by inhibiting multiple other kinases (33-35). Single-agent efficacy has been observed only with abemaciclib in the treatment of hormone receptor-positive breast cancer patients (36), suggesting that the overall anti-tumor activity of abemaciclib may result from additional mechanisms beyond cell cycle arrest.
Here, we found that abemaciclib induced a higher level of double-strand DNA (ds-DNA) damage in both Rb− and Rb-proficient (Rb+) SCLC cell lines in vitro, and that abemaciclib coupled with RT exhibited a synergistic effect on STING pathway activation. We further demonstrated that abemaciclib in combination with LDRT increased tumor-infiltrating T cells in an Rb− immunocompetent murine SCLC model, sensitizing SCLC to the anti-PD-1 immunotherapy. Our results suggest that abemaciclib combined with LDRT is a potentially effective strategy to enhance the efficacy of ICB and is worthy of further investigation. This manuscript is written following the ARRIVE reporting checklist (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-33/rc).
Methods
Cell lines and chemical compounds
Human SCLC cell lines H82, SHP77 and DMS53 were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). Murine SCLC cell line PRM, with Tp53 deletion, Rb1 deletion, and c-Myc overexpression, was provided by Feifei Na, Sichuan University, Chengdu (37). H82, SHP77 and PRM were cultured in RPMI 1640 media (Gibco, 11875500, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, 10099-141) and 5% penicillin-streptomycin solution (Hyclone, SV30010, Logan, UT, USA) at 37 ℃ with 5% CO2. DMS53 were cultured in Waymouth’s MB 752/1 media (Gibco, 11220035) supplemented with 10% FBS and 5% penicillin-streptomycin solution at 37 ℃ with 5% CO2. Prior to any experiments, all cell lines were authenticated using short tandem repeat profiling (DNA fingerprinting) and regularly tested for mycoplasma species. Abemaciclib (HY-16297A) was purchased from MedChemExpress (MCE, Shanghai, China). Mouse anti-PD-1 antibody (A2122) was purchased from Selleck (Houston, TX, USA).
Western blotting
Protein lysates were extracted using RIPA lysis buffer supplemented with protease and phosphatase inhibitors (MCE, HY-K0021 and K0010). After quantification and normalization with a bicinchoninic acid (BCA) protein assay kit (EpiZyme, ZJ102, Shanghai, China), 20 µg of total protein was subjected to electrophoresis on 6–12.5% polyacrylamide gels (EpiZyme, PG210, PG211, PG212, and PG213). The separated proteins were then transferred onto polyvinylidene fluoride (PVDF) membranes and blocked with protein-free rapid blocking buffer (EpiZyme, PS108P) for 30 min at room temperature. Subsequently, the membranes were incubated with primary antibodies at 4 ℃ overnight. Secondary antibodies were detected using the e-BLOT Touch Imager (e-BLOT, Shanghai, China). Western blotting images were representative of three independent experiments. Relative protein expression levels were quantified using ImageJ (v.1.53). The antibodies used for western blotting are listed in Table S1.
Cell cycle detection
Cells (1×106) were cultured in 6-well plates and treated with dimethyl sulfoxide (DMSO) or 1 µM abemaciclib for 24 h. After treatment, the cells were collected and washed twice with cold phosphate-buffered saline (PBS). Subsequently, cells were fixed with 70% cold ethanol at 4 ℃ overnight. The fixed cells were then stained with propidium iodide (PI)/RNase staining solution (Beyotime, C1052, Shanghai, China) for 30 min at room temperature. Three independent experiments were performed. Flow cytometry data were acquired using the CytoFLEX Flow Cytometer (Beckman, Brea, CA, USA) and analyzed with FlowJo (v.10.4).
Cell viability assay
Cells (n=5,000) were plated into 96-well plates and treated with DMSO or 1 µM abemaciclib for five days. After treatment, 10 µL of Cell Counting Kit 8 (CCK8) reagent (Beyotime, C0037, Shanghai, China) was added to each well, and the absorbance value [optical density (OD)] was measured at 450 nm after 3 h of incubation.
Apoptosis detection
Cells (5×105) cultured in 6-well plates treated with DMSO or 1 µM abemaciclib for 24 h were collected and incubated with Annexin V-PI staining solution (Beyotime, C1062S, Shanghai, China) for 15 min at 4 ℃ after washing with cold PBS twice. Three independent experiments were performed. Data were obtained on a CytoFLEX Flow Cytometer (Beckman) and analyzed with CytExpert (v.2.4).
Cell irradiation
Cells were plated and irradiated with 10 Gy of X-ray using an RS-2000 X-ray irradiator (Rad Source Technologies, Alpharetta, GA, USA) operated at 160 kV, 20 mA and a dose rate of 1.836 Gy/min (0.3 mm copper filtration) with source-surface distance (SSD) of 30 cm. The single irradiation of 10 Gy in vitro, which is biologically effective dose of 15 Gy/5 F, was performed to match the dose used in vivo. Abemaciclib was added 2 h after irradiation when combined with RT.
Immunofluorescence (IF)
Cells (1×105) cultured on 6-well chamber slides after treatment were washed with PBS twice and fixed with 4% paraformaldehyde for 20 min at room temperature. After penetration with 0.3% Triton X-100 for 20 min and washed with PBS for three times, cells were blocked with 5% bovine serum albumin at room temperature for 1 h. Then cells were incubated with primary antibodies at 4 ℃ overnight. After washing with PBS for 3 times, secondary antibodies were applied at room temperature for 1 h. Finally, cells were stained with DAPI-Fluoromount-G (Southern Biotech, 0100-20, Tuscaloosa, AL, USA) and visualized with IX 73 DP80 fluorescent microscope (Olympus, Tokyo, Japan). Three random fields were selected under the microscope to determine the number of γH2AX foci or the relative fluorescence intensity (RFI) of ds-DNA. RFI of ds-DNA was calculated using ImageJ (v.1.53). The antibodies used for IF are listed in the Table S2.
RNA extraction and quantitative real-time polymerase chain reaction (qRT-PCR)
RNA extractions were performed using the Total RNA Isolation Kit (Vazyme, RC112-01, Nanjing, China). cDNA was synthesized from RNA by reverse transcription PCR using HiScript III RT SuperMix (Vazyme, R323-01). Triplicate PCR reactions were performed using ChamQ SYBR qPCR Master Mix (Vazyme, Q711-02) and run on CFX96 Real-Time System (Bio-Rad, Hercules, CA, USA). Relative expression of each targeted gene including IFN-β, CCL5 and CXCL10 was calculated and normalized using 2−ΔΔCt method. GAPDH gene expression was used for the relative quantification. The primers are listed in the Table S3.
Mice
Six-week-old male C57BL/6J mice were purchased from the Gempharmatech Co., Ltd. (JCYK Bioscience, Jiangsu, China) and maintained under specific pathogen-free conditions in Sichuan University with autoclaved food, bedding, and water. Animals were housed at room temperature (23±2 ℃) at a humidity of 30–70% on a 12-h light/12-h dark cycle (6:00–18:00). Mice were inoculated with 1×105 PRM cells, subcutaneously (s.c.) in the right hind limb. When the tumor size reached approximately 150–200 mm3, the mice were randomly assigned to different groups such that each group had similar mean tumor volume and mean weight. Sample sizes were five to six per group, consistent with our previous study (16). Abemaciclib was administered by oral gavage (75 mg/kg) every 3 days. Radiation was delivered in 5 F of 15 Gy on first 5 days using the RS-2000 X-ray irradiator operated at 160 kV, 20 mA and a dose rate of 1.836 Gy/min (0.3 mm copper filtration) with SSD of 30 cm. Before radiation treatment, each mouse was anesthetized and shielded using a lead box such that only the irradiated tumor was exposed. Anti-PD-1 antibody was administered intraperitoneally (200 µg/mouse) every 3 days. Abemaciclib was administered 2 h after radiation, and anti-PD-1 was administered 4 h after abemaciclib administration. Mice from control group were also administered with vehicle of abemaciclib by oral gavage and PBS intraperitoneally every 3 days. Tumor volumes were recorded every 3 days using calipers and were calculated as length × width2 × 0.5. Mice were euthanized when any one of the implanted tumors reached a volume of 2,000 mm3. Survival was calculated from the date of treatment initiation to death. Weight was recorded every 3 days. All the mouse experiments were performed in compliance with the Guide for the Care and Use of Laboratory Animals of Sichuan University and protocol of this study was registered and approved by the Institutional Animal Care and Use Committee of Sichuan University (No. 20221229002).
Histological staining
Tumor samples resected at day 8 were fixed in 10% neutral buffered formalin embedded in formalin-fixed paraffin-embedded (FFPE). FFPE samples were sectioned at 4 µm thickness. Immunohistochemistry (IHC) staining was performed using the Dako REAL EnVision Detection Kit (Dako, K5007, Copenhagen, Denmark). IHC images were acquired with SLIDEVIEW VS200 Slide Scanner (Olympus, Tokyo, Japan). Multiple immunofluorescence (mIF) staining was performed using the Opal 6Plex Manual Detection Kit (Akoya, NEL861001KT, Marlborough, MA, USA). mIF images were acquired using the Akoya Vectra POLARIS multispectral microscope (Akoya, MA, USA). The antibodies used for IHC and mIF are shown in the Table S2.
Flow cytometry analysis
Tumors were resected and digested thoroughly with RPMI 1640 containing 1.0 mg/mL collagenase I (Gibco, 17100-017), 0.5 mg/mL collagenase IV (Gibco, 17104-019), 2% FBS (Gibco, 10099-141), and 50 µg/mL deoxyribonuclease I (Sigma, D5025-15KU, Saint Louis, MO, USA) for 37 ℃ at 30 min, then filtered through a 70-µm mesh (Biofil, Guangzhou, China). Cells were resuspended in ACK lysis buffer (Leagene, CS0003, Beijing, China) for 5 min at 4 ℃ to lyse red blood cells and then resuspended in PBS; 1×106 cells/100 µL were incubated with purified anti-CD16/CD32 mAb (BioLegend, 101301, San Diego, CA, USA) for 10 min at 4 ℃ to block Fc receptors and then cells were stained with conjugated antibody cocktail for 30 min at 4 ℃. Antibodies used for flow cytometry analysis are listed in Table S4. The cells were washed twice with PBS and resuspended for flow cytometry. Flow cytometry analysis was performed using the Celesta and LSRFortessa machines (BD Bioscience, Franklin Lakes, NJ, USA), and the data were analyzed using FlowJo (v.10.4). Gating strategies were displayed in Figure S1.
RNA sequencing and analysis
RNA extractions were conducted from in vitro cultured cells and in vivo tumor samples using the TRIzol reagent (Invitrogen, 15596-018, Carlsbad, CA, USA) according to the manufacturer’s protocol. The purity of RNA samples was assessed using the NanoPhotometer (IMPLEN, Munich, Germany). Further, the concentration and integrity of RNA samples were detected by Agilent 2100 RNA Nano 6000 assay Kit (Agilent Technologies, Santa Clara, CA, USA). After the samples were qualified, libraries were constructed using the VAHTS Universal V6 RNA-seq Library Prep Kit for Illumina (NR 604-01/02, San Diego, CA, USA). Subsequently, sequencing was carried out on the Illumina platform, employing the PE150 sequencing strategy to generate 150-bp paired-end sequencing reads. The reads were aligned to the human reference genome (GRCh38) and the mouse reference genome (GRCm38) using HISAT2 (v.2.2.1). The read counts of each gene were obtained using HTSeq-count (v.0.9.1). Transcripts per million (TPM) were quantified from read counts, followed by binary log transformation using log1p function. The raw data are available in the National Genomics Data Center (NGDC) database with the accession number PRJCA022531.
Differential gene expression analysis was performed using the R package limma (v.3.50.0). For the Gene Set Enrichment Analysis (GSEA) of both in vitro and in vivo data, the R package clusterProfiler (v.4.2.2) was employed, utilizing the hallmark gene sets sourced from the MSigDB database. GSEA focused on systematically evaluating the enrichment of predefined gene sets within a ranked list of genes. Initially, genes were ordered based on their differential expression log-fold change with base 2 (log2FC) values, ranging from large positive values to large negative values. This ranked list was then compared with hallmark gene sets.
For the enrichment analysis of upregulated genes of in vivo data, genes with log2FC >2 and adjusted P value <0.05 were defined as upregulated genes. Enrichment analysis of upregulated genes based on biological process terms in the Gene Ontology (GO) database was performed also using the R package clusterProfiler (v.4.2.2). For the heatmap visualization of representative genes, the expression levels were z-score-normalized and plotted using the R package pheatmap (v.1.0.12).
Statistical analysis
The statistical significance of differences was analyzed using with GraphPad Prism (v.8.0.2) with P value less than 0.05 considered statistically significant (*, P<0.05; **, P<0.01; ***, P<0.001; ns, >0.05). Data from tumor growth curves were represented as mean ± standard error of the mean (SEM) and others as mean ± standard deviation (SD). The details of the specific statistical analysis used in the individual experiments can be found in the legends of Figures 1-3 and Figures S2,S3.
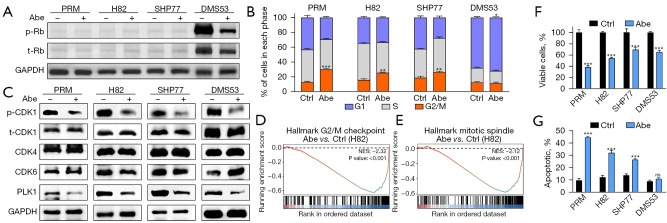
Figure 1.
The anti-tumor effects of abemaciclib in SCLC. (A) Western blots showing the protein expression of p- and t-Rb in SCLC cells treated with 1 µM abemaciclib for 24 h. (B) Cell cycle states of SCLC cells treated with 1 µM abemaciclib for 24 h as detected by PI-based flow cytometry. (C) Western blots showing the protein expression of p- and t-CDK1, CDK4, CDK6 and PLK1 in SCLC cells treated with 1 µM abemaciclib for 24 h. (D,E) In H82 cells treated with abemaciclib, gene sets for G2/M checkpoint and mitotic spindle were significantly downregulated. The y-axis represents enrichment score and the x-axis are genes (vertical black lines) represented in gene sets. The colored band at the bottom represents the degree of correlation of genes with the abemaciclib treatment (red for positive and blue for negative correlation). (F) Cell viability of SCLC cells treated with 1 µM abemaciclib for 5 days as measured by CCK8 cell viability assay. (G) Apoptotic proportion of SCLC cells treated with 1 µM abemaciclib for 24 h as detected by annexin V-PI-based flow cytometry. The statistical analysis was performed using two-tailed unpaired Student’s t-test. P values are shown and error bars indicate mean ± SD. *, P<0.05; **, P<0.01; ***, P<0.001; ns, no significance. Abe, abemaciclib; p-, phospho-; t-, total-; Rb, retinoblastoma; ctrl, control; NES, normalized enrichment score; SCLC, small cell lung cancer; PI, propidium iodide; PLK1, polo-like kinase 1; CCK8, Cell Counting Kit 8; SD, standard deviation.
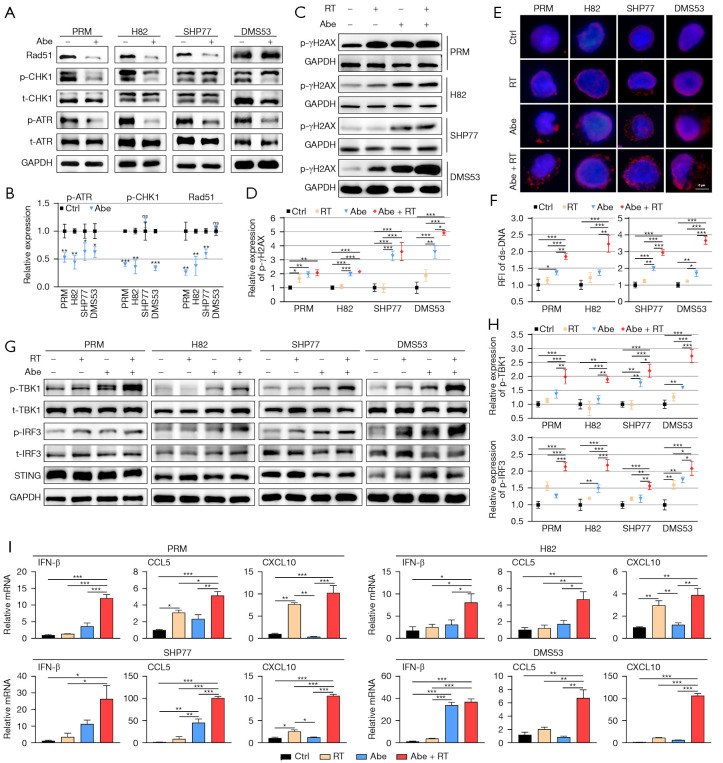
Figure 2.
Combination of abemaciclib and RT induced ds-DNA damage and activated the STING pathway in SCLC. (A) Western blots showing the protein expression of p- and t-ATR, p- and t-CHK1 and Rad51 in SCLC cells treated with 1 µM abemaciclib for 24 h. (B) Quantification of p-ATR, p-CHK1 and Rad51 in SCLC cells. (C) Western blots showing the protein expression of p-γH2AX in SCLC cells treated with RT (10 Gy), abemaciclib (1 µM) and their combination for 24 h. (D) Quantification of p-γH2AX in SCLC cells. (E) Representative images of ds-DNA staining (red) in SCLC cells treated with RT (10 Gy), abemaciclib (1 µM) and their combination for 24 h. (F) RFI of ds-DNA in SCLC cells. (G) Western blots showing the protein expression of p- and t-TBK1, p- and t-IRF3 and STING in SCLC cells treated with RT (10 Gy), abemaciclib (1 µM) and their combination for 24 h. (H) Quantification of p-TBK1 and p-IRF3 in SCLC cells. (I) Quantitative mRNA expression of IFN-β, CCL5 and CXCL10 after treatment with RT (10 Gy), abemaciclib (1 µM) and their combination for 24 h in SCLC cells as measured by qRT-PCR. The statistical analysis was performed using two-tailed unpaired Student’s t-test (B) and one-way ANOVA with Tukey’s multiple comparisons test (D,F,H,I). P values are shown and error bars indicate mean ± SD. *, P<0.05; **, P<0.01; ***, P<0.001; ns, no significance. Abe, abemaciclib; p-, phospho-; t-, total-; ctrl, control; RT, radiotherapy; RFI, relative fluorescence intensity; ds-DNA, double-strand DNA; SCLC, small cell lung cancer; qRT-PCR, quantitative real-time polymerase chain reaction; ANOVA, analysis of variance; SD, standard deviation.
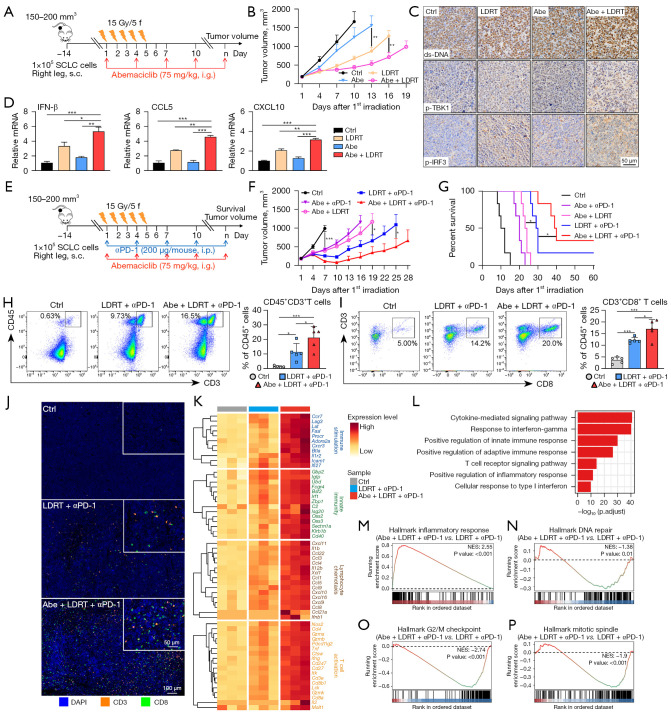
Figure 3.
Abemaciclib combined with LDRT enhanced anti-tumor immune responses of anti-PD-1 immunotherapy in Rb− SCLC mouse model. (A,E) Schematic of PRM tumor-bearing mouse model experiment design and treatment plan. (B) Tumor growth curves of mice with different treatments (n=5–6). (C) Representative IHC of ds-DNA, p-IRF3 and p-TBK1 in tumors resected at day 8. (D) Quantitative mRNA expression of IFN-β, CCL5 and CXCL10 in tumors resected at day 8 as measured by qRT-PCR. (F,G) Tumor growth curves (F) and Kaplan-Meier survival curves (G) of mice with different treatments (n=6). (H,I) Representative flow cytometry plots (left) and the proportion (right) of CD45+CD3+ (H) and CD3+CD8+ (I) T cells in tumors resected at day 8 (n=5). (J) Representative images of mIF staining indicating CD3+ and CD8+ T cells in tumors resected at day 8. (K) Heatmap showing the expression of immune-related genes from tumors resected at day 8. (L) Representative enriched GO terms in biological processes upregulated in tumors treated with Abe + LDRT + αPD-1 compared with LDRT + αPD-1. (M-P) Gene sets significantly upregulated (M) and downregulated (N-P) in tumors treated with Abe + LDRT + αPD-1 compared with LDRT + αPD-1. The y-axis represents enrichment score and the x-axis are genes (vertical black lines) represented in gene sets. The colored band at the bottom represents the degree of correlation of genes with the triple treatment (red for positive and blue for negative correlation). The statistical analysis was performed using two-way ANOVA (B,F), one-way ANOVA (D,H,I) with Tukey’s multiple comparisons test and log-rank test (G). P values are shown and error bars indicate mean ± SEM (B,F) or ± SD (D,H,I). *, P<0.05; **, P<0.01; ***, P<0.001. SCLC, small cell lung cancer; s.c., subcutaneously; i.g., oral gavage; i.p., intraperitoneal injection; ctrl, control; Abe, abemaciclib; LDRT, low-dose radiotherapy; p-, phospho-; ds-DNA, double-strand DNA; αPD-1, anti-programmed cell death protein-1 antibody; NES, normalized enrichment score; Rb−, retinoblastoma-deficient; IHC, immunohistochemistry; qRT-PCR, quantitative real-time polymerase chain reaction; mIF, multiple immunofluorescence; GO, Gene Ontology; ANOVA, analysis of variance; SEM, standard error of mean; SD, standard deviation.
Results
Abemaciclib exhibited distinct effects on cell cycle and apoptosis in Rb− SCLC cells in contrast with Rb+ SCLC cells
RB1 mutations and loss of Rb expression are recognized as a molecular hallmark of SCLC (31), while there are still about 6% Rb+ subset (38). Here we used both Rb− and Rb+ SCLC cell lines for in vitro experiments to evaluate the direct effects of CDK4/6 inhibition. Western blotting showed that murine SCLC cell line PRM and human SCLC cell lines H82 and SHP77 were Rb− (Figure 1A). Human SCLC cell line DMS53 was Rb+ and phospho (p)-Rb was suppressed after administration of 1 µM abemaciclib for 24 h (Figure 1A).
By reducing p-Rb, CDK4/6 inhibitors can induce G1 cell cycle arrest in Rb+ tumor cells (17). However, the effect of CDK4/6 inhibition on cell cycle in Rb− SCLC cells has not been reported. Therefore, we first examined alteration of cell cycle using the PI staining. As expected, we observed a slight G1 cell cycle arrest in Rb+ cell line DMS53 after treatment with abemaciclib (Figure 1B and Figure S4A). Predictably, treatment with 1 µM abemaciclib for 24 h did not result in G1 cell cycle arrest in Rb− SCLC cell lines; however, G2/M cell cycle arrest was observed after treatment with abemaciclib (Figure 1B and Figure S4A). Loss of Rb protein expression and concomitant TP53 alterations disrupt the G1/S checkpoint and consequently most of SCLCs rely on G2/M checkpoint (39). Western blotting showed that phosphorylation of CDK1 was suppressed (Figure 1C), indicating increased cells entering the M phase and potential changes in G2/M checkpoint. A previous study reported that treatment with abemaciclib downregulated genes relating to G2/M checkpoint in a breast carcinoma mouse model (24). Consistently, based on RNA sequencing data, GSEA showed that pathway related to G2/M checkpoint was downregulated in H82 after treatment with abemaciclib (Figure 1D). Another previous report showed that abemaciclib inhibited the expression of polo-like kinase 1 (PLK1) (40), which is a regulator of G2/M phase transition (41) and mitotic kinase signaling (42) and has been previously demonstrated to be upregulated in Rb− triple-negative breast cancer clinical specimens (43). Here we also demonstrated downregulated expression of PLK1 after treatment with abemaciclib in all four SCLC cell lines (Figure 1C). PLK1 inhibition is associated with mitosis dysfunction (44), and our data showed that abemaciclib downregulated genes related to mitosis in H82 according to GSEA (Figure 1E).
Next, to confirm the sensitivity of SCLC to abemaciclib, we performed CCK8 cell viability assay. Treatment with 1 µM abemaciclib for 5 days induced significant growth inhibition in both Rb− and Rb+ cell lines (Figure 1F). The inhibition of G2/M checkpoint and premature entering into M phase could result in mitotic catastrophe and apoptosis in SCLC cells (1). So we next investigated cell death using annexin V-PI staining and our data showed that treatment with 1 µM abemaciclib for 24 h significantly increased the apoptotic fraction in Rb− SCLC cell lines relative to control, which had not been observed in Rb+ cell line DMS53 (Figure 1G and Figure S4B). Preclinical investigations have revealed that the primary response in Rb+ tumors following CDK4/6 inhibition is the suppression of growth, aligning with the mechanism of action in inducing G1 cell cycle arrest, rather than triggering cell death (45). Our results showed that while abemaciclib effectively inhibited the growth of both Rb− and Rb+ SCLC cells, it specifically induced apoptosis in Rb− SCLC cells during early stages of administration.
Our findings indicate that abemaciclib has the potential to induce growth inhibition in SCLC cells while its impact on cell cycle and apoptosis appears to differ between Rb− and Rb+ subsets.
Abemaciclib combined with RT induced increased ds-DNA damage and activated the STING pathway synergistically in SCLC cells
Previous studies have demonstrated that CDK4/6 inhibitors could induce ds-DNA damage in multiple Rb+ tumor cell lines (27-30,46,47), which was correlative to homologous recombination (HR) dysfunction after CDK4/6 inhibition (29,47,48). However, it is unknown whether CDK4/6 inhibition could induce more ds-DNA damage in Rb− SCLC cells. Here, we first evaluated the expression of key DNA repair proteins such as p-ATR, p-Chk1 and Rad51 after CDK4/6 inhibition in SCLC cell lines. Our data showed that after treatment with 1 µM abemaciclib for 24 h, p-ATR was observed to be reduced in all four cell lines; p-Chk1 was observed to be reduced in PRM, H82 and DMS53 and Rad51 was reduced in PRM, H82 and SHP77 (Figure 2A,2B), suggesting that abemaciclib may have a negative regulatory effect on DDR.
A recent study reported that inhibition of CDK4/6 combined with RT led to more DNA damage in glioblastoma cells (28). We next investigated that whether abemaciclib coupled with RT could induce more ds-DNA damage in SCLC cells. Western blotting showed that the phosphorylation level of γH2AX, which is indicative of DNA double-strand break (DSB) repair (49), significantly increased in both Rb− and Rb+ SCLC cells when treated with 1 µM abemaciclib combined with or without RT for 24 h (Figure 2C,2D). Consistently, IF staining of γH2AX showed that abemaciclib led to increased γH2AX foci formation in all four SCLC cell lines (Figure S2A,S2B). Next, to determine whether the accumulation of γH2AX resulted in increased level of ds-DNA in cytoplasm, we performed IF staining and found that the combination of abemaciclib and RT induced more cytosolic ds-DNA than single treatment alone in SCLC cells (Figure 2E,2F).
It was reported that presence of cytosolic ds-DNA can activate the STING pathway, an innate antitumor immune response, in SCLC and other tumors (50-53). Thus, we performed western blotting to investigate whether combination of abemaciclib and RT could activate STING pathway more potently compared with single treatment alone. Our results showed that compared with single treatment, abemaciclib combined with RT activated STING pathway more remarkably in SCLC cells, as indicated by increased p-TBK1 and p-IRF3 (Figure 2G,2H). Moreover, mRNA expression of downstream cytokines (IFN-β) and chemokines (CCL5, CXCL10) of STING pathway was enhanced in SCLC cells treated with abemaciclib plus RT compared with single treatment (Figure 2I).
Altogether, our results demonstrated that combination of abemaciclib and RT led to increased ds-DNA damage and activated the STING pathway synergistically in SCLC cells.
Abemaciclib further enhanced anti-tumor immune responses of LDRT plus anti-PD-1 double-therapy in Rb− SCLC mouse model
Subcutaneous PRM tumor-bearing immunocompetent C57BL/6J mouse model was established to evaluate the anti-tumor effect of abemaciclib in combination therapy of SCLC in vivo. As we have demonstrated that abemaciclib combined with RT induced increased ds-DNA and activated STING pathway more remarkably in vitro, our focus was on the combination effect of abemaciclib plus LDRT first. Mice were treated with abemaciclib (75 mg/kg) and LDRT (15 Gy/5 F) as single agents or in combination (Figure 3A). In contrast to the utilization of standard daily administration of CDK4/6 inhibitors in Rb+ tumors for inducing tumor cell cycle arrest and inhibiting tumor cell proliferation, abemaciclib here was used for its DNA damage-inducing properties and was administrated every 3 days. Our data showed that the combination of abemaciclib and LDRT inhibited tumor growth significantly compared with single agent treatment (Figure 3B). Body weight loss was not observed (Figure S3A). IHC staining of tumors resected at day 8 revealed that treatment with abemaciclib in combination with LDRT increased the level of ds-DNA, p-TBK1 and p-IRF3 in tumor tissues (Figure 3C). Moreover, the mRNA expression level of IFN-β, CCL5 and CXCL10 significantly increased in tumors treated with combined therapy compared with abemaciclib or LDRT alone (Figure 3D), consisting with the results of in vitro experiments.
Previous studies have demonstrated that activation of the STING pathway and the subsequent release of downstream cytokines and chemokines promoted the infiltration of CD8+ T cells into the TME in SCLC and other tumors (50-53). Our results in vitro and in vivo showed that the combination of abemaciclib and RT induced more ds-DNA damage and activated the STING pathway synergistically, which might induce a more inflamed TME and potentiate the anti-tumor immune responses induced by anti-PD-1 antibody. So we next investigated whether abemaciclib could further enhance anti-tumor effects of the LDRT plus anti-PD-1 double-therapy, which was demonstrated to inhibit tumor growth and extend mouse survival significantly in the PRM mouse model according to our previous study (16). Mice were treated with abemaciclib (75 mg/kg), LDRT (15 Gy/5 F) and anti-PD-1 antibody (200 µg/mouse) in double or triple combination (Figure 3E). Our results showed that triple-therapy with abemaciclib, LDRT and anti-PD-1 significantly inhibited tumor growth (Figure 3F) and extended animal survival (Figure 3G) compared with other combined double-therapy. Body weight loss was not observed in any of the groups (Figure S3B). At day 8, tumors from a cohort of mice (control, LDRT plus anti-PD-1, and abemaciclib plus LDRT plus anti-PD-1) were resected to analyze alteration of tumor-infiltrating immune cells. Multicolor flow cytometry showed that triple-therapy significantly increased infiltration of CD45+CD3+ total T cells (Figure 3H), CD3+CD8+ cytotoxic T cells (Figure 3I) and CD44highCD62Llow memory/effector T cells (Figure S3C) in tumors compared with double-therapy. mIF staining demonstrated that tumors treated with triple-therapy exhibited more CD3+ and CD8+ T cell infiltration compared with LDRT plus anti-PD-1 double-therapy (Figure 3J). Next, we performed RNA sequencing of tumors that were collected from each treatment group described above. The expression of immune-related genes (e.g., genes involved in immune stimulation, innate immunity, cytokines, and T cell activation) was strongly upregulated after triple-therapy (Figure 3K). Consistently, enrichment analysis showed that pathways related to immune response were upregulated in triple-therapy group (Figure 3L,3M). Downregulated gene sets included DNA repair (Figure 3N), G2/M checkpoint (Figure 3O) and mitotic spindle (Figure 3P), consistent with results in vitro.
In summary, these results together demonstrated that abemaciclib could further enhance the anti-tumor immune responses of LDRT plus anti-PD-1 double-therapy via reshaping a more inflamed TME in vivo.
Discussion
Considering that Rb is the canonical CDK4/6 substrate, the overwhelming majority of CDK4/6 inhibitor research in cancer has been on Rb+ tumors. Notably, a recent study reported that Rb+ SCLC cells are sensitive to growth inhibition by CDK4/6 inhibition, and that a SCLC patient with Rb expression benefited from the treatment with abemaciclib monotherapy (54). Our study here focused on the dominant Rb− SCLC subset and showed that abemaciclib induced G2/M cell cycle arrest and apoptosis in Rb− SCLC cells, contrasting with the results observed in Rb+ SCLC cells. Further studies are needed to clarify the molecular mechanisms underlying the direct biological effects of CDK4/6 inhibition on Rb− SCLCs. Of note, one previous research has demonstrated synergistic effects of combined CDK4/6 and PARP inhibition in Rb− breast cancer cells by inducing more DNA damage, whereas CDK4/6 inhibition alone had little impact (55). In our study, though abemaciclib demonstrated significant growth inhibition in Rb− SCLC cells in vitro, its efficacy as monotherapy in Rb− PRM mouse model was notably limited, in contrast with its effect when combined with RT and ICB. Therefore, for Rb− tumors, including SCLC, CDK4/6 inhibitors should be considered as a part of combination therapy rather than administered as a standalone treatment.
Previous studies have shown that CDK4/6 inhibitors could induce DNA damage via suppressing DDR in multiple tumor cell types (27-30,46-48), while part of these studies reported that the inhibitory effect on DDR might be Rb-dependent (29,47). Our results showed that abemaciclib downregulated the expression of DDR-related proteins and induced ds-DNA damage in SCLC cells regardless of Rb expression status. These differences can be partly explained by disparities of cell lines or CDK4/6 inhibitors utilized, and indicate the presence of DDR inhibition pathways which are not dependent on Rb. In a recent research, CDK6 was found to play a direct role in stabilizing FOXO3 and inducing ATR, with CDK4/6 inhibition demonstrating to increase the sensitivity of ovarian cancer cells to platinum therapies by downregulating FOXO3-ATR expression (30). Besides, PLK1 has recently been proved to play an important role in DSB repair during mitosis by phosphorylating DNA polymerase theta (56). Our data showed that abemaciclib suppressed the expression of PLK1, which might contribute to the ds-DNA induction in SCLC cells. These findings represent a part, probably not all, of the non-Rb-dependent DDR inhibition pathways following CDK4/6 inhibition. Furthermore, it has been reported that the level of reactive oxygen species (ROS), which can directly induce DNA damage and affect DDR (57), increased in Rb− tumor cells through a cell cycle-independent mechanism after inhibition of CDK4/6 (55,58), representing another possible Rb-independent manner to induce DNA damage. Previous studies have demonstrated the upregulation of DDR components (such as CHK1, ATR, WEE1, PARP) in SCLC, and the effectiveness of DDR inhibitors has been observed in preclinical SCLC models (39,50-53,59,60). Our findings indicate that the ability of abemaciclib to induce ds-DNA damage in SCLC provides a rationale for its further clinical exploration.
Given recent research showing that CDK4/6 inhibitors exert various biological effects on tumor cells, evaluations of their application in combination with current standard therapy in tumor types other than breast cancer are underway (61). Cytotoxic therapies, such as chemotherapy and RT, are conventional treatments for a wide range of cancer types including SCLC. Recent studies have questioned the notion that CDK4/6 inhibitors should not be combined with cytotoxic therapies. For example, CDK4/6 inhibitors applied after chemotherapy was proved to enhance therapeutic benefit in pancreatic cancer models, which was associated with impaired recovery from chemotherapy-induced DNA damage due to suppressed DNA repair by HR caused by CDK4/6 inhibition (29). Similarly, our results also showed that the combination of abemaciclib and RT had synergetic effects on ds-DNA damage induction and STING pathway activation. Besides, recently there is a growing consensus that CDK4/6 inhibitors can activate anti-tumor immune responses and could be used in combination with immunotherapy. The direct immunomodulatory effects of CDK4/6 inhibitors on tumor cells known so far include the upregulation of major histocompatibility complex (MHC) I/II expression (24), induction of immunogenic cell death (62) and upregulation of programmed death-ligand 1 (PD-L1) (63), etc. Importantly, several recent reports have started to unravel how inhibition of CDK4/6 directly influences the function of immune cells, including induction of CD8+ T cell effector function (22-24) and promotion of memory T cell differentiation (24-26). Since our data from transcriptomic analysis of tumors revealed a substantial upregulation of immune response pathways upon the addition of abemaciclib in the double-therapy of LDRT plus anti-PD-1, we believe that intricate synergies between abemaciclib and immunotherapy extend beyond the activation of STING pathway.
Regrettably, the combination of CDK4/6 inhibitors with PD-1 blockade in clinical trials for hormone receptor-positive breast cancers and non-small cell lung cancer (NSCLC) resulted in a high incidence of hepatitis and several cases of fatal pneumonitis (64-66). Consequently, these trials have been largely halted due to safety concerns. The toxicity induced by combination therapy has spurred efforts to explore alternative modes of administration strategy. Some suggested that early treatment with CDK4/6 inhibitors to promote CD8+ T cell memory formation followed by checkpoint blockade would avoid concurrent administration of these agents and their synergistic toxicity (25). Another potential strategy to mitigate toxicity is by reducing the dosage of CDK4/6 inhibitors. To maximize their direct Rb-dependent tumor growth inhibitory effects, CDK4/6 inhibitors are typically administered orally on a daily basis, both in preclinical studies and clinical practice. In our study, however, abemaciclib was used for its DNA damage-inducing properties and was administrated every 3 days, and was proved to be capable of enhancing the efficacy of LDRT plus immunotherapy in such an unconventional dosing regimen. Our findings propose a concept to utilize reduced-dose CDK4/6 inhibitors as an adjunctive therapy to immunotherapy. Whether this strategy could alleviate the toxicity of combination therapy requires further clinical validation.
Conclusions
In this study, we demonstrated that CDK4/6 inhibitor abemaciclib could lead to increased ds-DNA damage and that the combination of abemaciclib and RT synergistically activated the STING pathway in Rb− SCLC cells. We further demonstrated that abemaciclib in combination with LDRT inflamed the TME and sensitizing Rb− SCLC to the anti-PD-1 immunotherapy. Our findings indicated that the combination of abemaciclib with LDRT could be a promising approach to augment the efficacy of ICB for SCLC and deserves further investigation.
Supplementary
The article’s supplementary files as
Acknowledgments
The authors thank Li Li, Fei Chen, Chunjuan Bao and Yang Deng (Institute of Clinical Pathology, West China Hospital, Sichuan University) for processing histological staining.
Funding: This study was supported by 1·3·5 Project for Disciplines of Excellence, West China Hospital, Sichuan University (No. ZYJC21003), National Natural Science Foundation of China (No. 82073336, No. 82303773), Sichuan Provincial Research Foundation (No. 2023NSFSC0699) and Postdoctoral Fellowship Program of CPSF (No. GZB20230481).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All the mouse experiments were performed in compliance with the Guide for the Care and Use of Laboratory Animals of Sichuan University and protocol of this study was registered and approved by the Institutional Animal Care and Use Committee of Sichuan University (No. 20221229002).
Footnotes
Reporting Checklist: The authors have completed the ARRIVE reporting checklist. Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-33/rc
Data Sharing Statement: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-33/dss
Peer Review File: Available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-33/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tlcr.amegroups.com/article/view/10.21037/tlcr-24-33/coif). The authors have no conflicts of interest to declare.
References
- 1.Rudin CM, Brambilla E, Faivre-Finn C, et al. Small-cell lung cancer. Nat Rev Dis Primers 2021;7:3. 10.1038/s41572-020-00235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn L, Mansfield AS, Szczęsna A, et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N Engl J Med 2018;379:2220-9. 10.1056/NEJMoa1809064 [DOI] [PubMed] [Google Scholar]
- 3.Paz-Ares L, Dvorkin M, Chen Y, et al. Durvalumab plus platinum-etoposide versus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer (CASPIAN): a randomised, controlled, open-label, phase 3 trial. Lancet 2019;394:1929-39. 10.1016/S0140-6736(19)32222-6 [DOI] [PubMed] [Google Scholar]
- 4.Ishii H, Azuma K, Kawahara A, et al. Significance of programmed cell death-ligand 1 expression and its association with survival in patients with small cell lung cancer. J Thorac Oncol 2015;10:426-30. 10.1097/JTO.0000000000000414 [DOI] [PubMed] [Google Scholar]
- 5.Carvajal-Hausdorf D, Altan M, Velcheti V, et al. Expression and clinical significance of PD-L1, B7-H3, B7-H4 and TILs in human small cell lung Cancer (SCLC). J Immunother Cancer 2019;7:65. 10.1186/s40425-019-0540-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonia SJ, López-Martin JA, Bendell J, et al. Nivolumab alone and nivolumab plus ipilimumab in recurrent small-cell lung cancer (CheckMate 032): a multicentre, open-label, phase 1/2 trial. Lancet Oncol 2016;17:883-95. 10.1016/S1470-2045(16)30098-5 [DOI] [PubMed] [Google Scholar]
- 7.Ott PA, Elez E, Hiret S, et al. Pembrolizumab in Patients With Extensive-Stage Small-Cell Lung Cancer: Results From the Phase Ib KEYNOTE-028 Study. J Clin Oncol 2017;35:3823-9. 10.1200/JCO.2017.72.5069 [DOI] [PubMed] [Google Scholar]
- 8.Grønberg BH, Killingberg KT, Fløtten Ø, et al. High-dose versus standard-dose twice-daily thoracic radiotherapy for patients with limited stage small-cell lung cancer: an open-label, randomised, phase 2 trial. Lancet Oncol 2021;22:321-31. 10.1016/S1470-2045(20)30742-7 [DOI] [PubMed] [Google Scholar]
- 9.Slotman BJ, van Tinteren H, Praag JO, et al. Use of thoracic radiotherapy for extensive stage small-cell lung cancer: a phase 3 randomised controlled trial. Lancet 2015;385:36-42. 10.1016/S0140-6736(14)61085-0 [DOI] [PubMed] [Google Scholar]
- 10.Klug F, Prakash H, Huber PE, et al. Low-dose irradiation programs macrophage differentiation to an iNOS+/M1 phenotype that orchestrates effective T cell immunotherapy. Cancer Cell 2013;24:589-602. 10.1016/j.ccr.2013.09.014 [DOI] [PubMed] [Google Scholar]
- 11.Herrera FG, Ronet C, Ochoa de Olza M, et al. Low-Dose Radiotherapy Reverses Tumor Immune Desertification and Resistance to Immunotherapy. Cancer Discov 2022;12:108-33. 10.1158/2159-8290.CD-21-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barsoumian HB, Ramapriyan R, Younes AI, et al. Low-dose radiation treatment enhances systemic antitumor immune responses by overcoming the inhibitory stroma. J Immunother Cancer 2020;8:e000537. 10.1136/jitc-2020-000537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin L, Xue J, Li R, et al. Effect of Low-Dose Radiation Therapy on Abscopal Responses to Hypofractionated Radiation Therapy and Anti-PD1 in Mice and Patients With Non-Small Cell Lung Cancer. Int J Radiat Oncol Biol Phys 2020;108:212-24. 10.1016/j.ijrobp.2020.05.002 [DOI] [PubMed] [Google Scholar]
- 14.Patel RR, He K, Barsoumian HB, et al. High-dose irradiation in combination with non-ablative low-dose radiation to treat metastatic disease after progression on immunotherapy: Results of a phase II trial. Radiother Oncol 2021;162:60-7. 10.1016/j.radonc.2021.06.037 [DOI] [PubMed] [Google Scholar]
- 15.Schoenfeld JD, Giobbie-Hurder A, Ranasinghe S, et al. Durvalumab plus tremelimumab alone or in combination with low-dose or hypofractionated radiotherapy in metastatic non-small-cell lung cancer refractory to previous PD(L)-1 therapy: an open-label, multicentre, randomised, phase 2 trial. Lancet Oncol 2022;23:279-91. 10.1016/S1470-2045(21)00658-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Yu M, Na F, et al. Striking effect of low-dose radiotherapy combined with PD-1 blockade on small cell lung cancer in mice and refractory patients (Achilles Study). J Clin Oncol 2022;40:e20608. 10.1200/JCO.2022.40.16_suppl.e20608 [DOI] [Google Scholar]
- 17.Goel S, Bergholz JS, Zhao JJ. Targeting CDK4 and CDK6 in cancer. Nat Rev Cancer 2022;22:356-72. 10.1038/s41568-022-00456-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He S, Roberts PJ, Sorrentino JA, et al. Transient CDK4/6 inhibition protects hematopoietic stem cells from chemotherapy-induced exhaustion. Sci Transl Med 2017;9:eaal3986. 10.1126/scitranslmed.aal3986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weiss JM, Csoszi T, Maglakelidze M, et al. Myelopreservation with the CDK4/6 inhibitor trilaciclib in patients with small-cell lung cancer receiving first-line chemotherapy: a phase Ib/randomized phase II trial. Ann Oncol 2019;30:1613-21. 10.1093/annonc/mdz278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jirawatnotai S, Sharma S, Michowski W, et al. The cyclin D1-CDK4 oncogenic interactome enables identification of potential novel oncogenes and clinical prognosis. Cell Cycle 2014;13:2889-900. 10.4161/15384101.2014.946850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders L, Ke N, Hydbring P, et al. A systematic screen for CDK4/6 substrates links FOXM1 phosphorylation to senescence suppression in cancer cells. Cancer Cell 2011;20:620-34. 10.1016/j.ccr.2011.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng J, Wang ES, Jenkins RW, et al. CDK4/6 Inhibition Augments Antitumor Immunity by Enhancing T-cell Activation. Cancer Discov 2018;8:216-33. 10.1158/2159-8290.CD-17-0915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schaer DA, Beckmann RP, Dempsey JA, et al. The CDK4/6 Inhibitor Abemaciclib Induces a T Cell Inflamed Tumor Microenvironment and Enhances the Efficacy of PD-L1 Checkpoint Blockade. Cell Rep 2018;22:2978-94. 10.1016/j.celrep.2018.02.053 [DOI] [PubMed] [Google Scholar]
- 24.Goel S, DeCristo MJ, Watt AC, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature 2017;548:471-5. 10.1038/nature23465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heckler M, Ali LR, Clancy-Thompson E, et al. Inhibition of CDK4/6 Promotes CD8 T-cell Memory Formation. Cancer Discov 2021;11:2564-81. 10.1158/2159-8290.CD-20-1540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lelliott EJ, Kong IY, Zethoven M, et al. CDK4/6 Inhibition Promotes Antitumor Immunity through the Induction of T-cell Memory. Cancer Discov 2021;11:2582-601. 10.1158/2159-8290.CD-20-1554 [DOI] [PubMed] [Google Scholar]
- 27.Naz S, Sowers A, Choudhuri R, et al. Abemaciclib, a Selective CDK4/6 Inhibitor, Enhances the Radiosensitivity of Non-Small Cell Lung Cancer In Vitro and In Vivo. Clin Cancer Res 2018;24:3994-4005. 10.1158/1078-0432.CCR-17-3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashizume R, Zhang A, Mueller S, et al. Inhibition of DNA damage repair by the CDK4/6 inhibitor palbociclib delays irradiated intracranial atypical teratoid rhabdoid tumor and glioblastoma xenograft regrowth. Neuro Oncol 2016;18:1519-28. 10.1093/neuonc/now106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salvador-Barbero B, Álvarez-Fernández M, Zapatero-Solana E, et al. CDK4/6 Inhibitors Impair Recovery from Cytotoxic Chemotherapy in Pancreatic Adenocarcinoma. Cancer Cell 2020;37:340-353.e6. 10.1016/j.ccell.2020.01.007 [DOI] [PubMed] [Google Scholar]
- 30.Dall'Acqua A, Sonego M, Pellizzari I, et al. CDK6 protects epithelial ovarian cancer from platinum-induced death via FOXO3 regulation. EMBO Mol Med 2017;9:1415-33. 10.15252/emmm.201607012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.George J, Lim JS, Jang SJ, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47-53. 10.1038/nature14664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherr CJ, Beach D, Shapiro GI. Targeting CDK4 and CDK6: From Discovery to Therapy. Cancer Discov 2016;6:353-67. 10.1158/2159-8290.CD-15-0894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klein ME, Kovatcheva M, Davis LE, et al. CDK4/6 Inhibitors: The Mechanism of Action May Not Be as Simple as Once Thought. Cancer Cell 2018;34:9-20. 10.1016/j.ccell.2018.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.George MA, Qureshi S, Omene C, et al. Clinical and Pharmacologic Differences of CDK4/6 Inhibitors in Breast Cancer. Front Oncol 2021;11:693104. 10.3389/fonc.2021.693104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marra A, Curigliano G. Are all cyclin-dependent kinases 4/6 inhibitors created equal? NPJ Breast Cancer 2019;5:27. 10.1038/s41523-019-0121-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patnaik A, Rosen LS, Tolaney SM, et al. Efficacy and Safety of Abemaciclib, an Inhibitor of CDK4 and CDK6, for Patients with Breast Cancer, Non-Small Cell Lung Cancer, and Other Solid Tumors. Cancer Discov 2016;6:740-53. 10.1158/2159-8290.CD-16-0095 [DOI] [PubMed] [Google Scholar]
- 37.Na F, Pan X, Chen J, et al. KMT2C deficiency promotes small cell lung cancer metastasis through DNMT3A-mediated epigenetic reprogramming. Nat Cancer 2022;3:753-67. 10.1038/s43018-022-00361-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Febres-Aldana CA, Chang JC, Ptashkin R, et al. Rb Tumor Suppressor in Small Cell Lung Cancer: Combined Genomic and IHC Analysis with a Description of a Distinct Rb-Proficient Subset. Clin Cancer Res 2022;28:4702-13. 10.1158/1078-0432.CCR-22-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Foy V, Schenk MW, Baker K, et al. Targeting DNA damage in SCLC. Lung Cancer 2017;114:12-22. 10.1016/j.lungcan.2017.10.006 [DOI] [PubMed] [Google Scholar]
- 40.Pandey K, Katuwal NB, Park N, et al. Combination of Abemaciclib following Eribulin Overcomes Palbociclib-Resistant Breast Cancer by Inhibiting the G2/M Cell Cycle Phase. Cancers (Basel) 2022;14:210. 10.3390/cancers14010210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roshak AK, Capper EA, Imburgia C, et al. The human polo-like kinase, PLK, regulates cdc2/cyclin B through phosphorylation and activation of the cdc25C phosphatase. Cell Signal 2000;12:405-11. 10.1016/S0898-6568(00)00080-2 [DOI] [PubMed] [Google Scholar]
- 42.Combes G, Alharbi I, Braga LG, et al. Playing polo during mitosis: PLK1 takes the lead. Oncogene 2017;36:4819-27. 10.1038/onc.2017.113 [DOI] [PubMed] [Google Scholar]
- 43.Witkiewicz AK, Chung S, Brough R, et al. Targeting the Vulnerability of RB Tumor Suppressor Loss in Triple-Negative Breast Cancer. Cell Rep 2018;22:1185-99. 10.1016/j.celrep.2018.01.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strebhardt K. Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat Rev Drug Discov 2010;9:643-60. 10.1038/nrd3184 [DOI] [PubMed] [Google Scholar]
- 45.Torres-Guzmán R, Calsina B, Hermoso A, et al. Preclinical characterization of abemaciclib in hormone receptor positive breast cancer. Oncotarget 2017;8:69493-507. 10.18632/oncotarget.17778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liang ML, Chen CH, Liu YR, et al. Abemaciclib, A Selective CDK4/6 Inhibitor, Restricts the Growth of Pediatric Ependymomas. Cancers (Basel) 2020;12:3597. 10.3390/cancers12123597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dean JL, McClendon AK, Knudsen ES. Modification of the DNA damage response by therapeutic CDK4/6 inhibition. J Biol Chem 2012;287:29075-87. 10.1074/jbc.M112.365494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun CY, Talukder M, Cao D, et al. Gilteritinib Enhances Anti-Tumor Efficacy of CDK4/6 Inhibitor, Abemaciclib in Lung Cancer Cells. Front Pharmacol 2022;13:829759. 10.3389/fphar.2022.829759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sedelnikova OA, Horikawa I, Redon C, et al. Delayed kinetics of DNA double-strand break processing in normal and pathological aging. Aging Cell 2008;7:89-100. 10.1111/j.1474-9726.2007.00354.x [DOI] [PubMed] [Google Scholar]
- 50.Sen T, Della Corte CM, Milutinovic S, et al. Combination Treatment of the Oral CHK1 Inhibitor, SRA737, and Low-Dose Gemcitabine Enhances the Effect of Programmed Death Ligand 1 Blockade by Modulating the Immune Microenvironment in SCLC. J Thorac Oncol 2019;14:2152-63. 10.1016/j.jtho.2019.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sen T, Rodriguez BL, Chen L, et al. Targeting DNA Damage Response Promotes Antitumor Immunity through STING-Mediated T-cell Activation in Small Cell Lung Cancer. Cancer Discov 2019;9:646-61. 10.1158/2159-8290.CD-18-1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taniguchi H, Caeser R, Chavan SS, et al. WEE1 inhibition enhances the antitumor immune response to PD-L1 blockade by the concomitant activation of STING and STAT1 pathways in SCLC. Cell Rep 2022;39:110814. 10.1016/j.celrep.2022.110814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang N, Gao Y, Huang Z, et al. PARP inhibitor plus radiotherapy reshapes an inflamed tumor microenvironment that sensitizes small cell lung cancer to the anti-PD-1 immunotherapy. Cancer Lett 2022;545:215852. 10.1016/j.canlet.2022.215852 [DOI] [PubMed] [Google Scholar]
- 54.Wildey G, Shay AM, McColl KS, et al. Retinoblastoma Expression and Targeting by CDK4/6 Inhibitors in Small Cell Lung Cancer. Mol Cancer Ther 2023;22:264-73. 10.1158/1535-7163.MCT-22-0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Zhang Y, Wang N, et al. Pan-cancer analysis reveals synergistic effects of CDK4/6i and PARPi combination treatment in RB-proficient and RB-deficient breast cancer cells. Cell Death Dis 2020;11:219. 10.1038/s41419-020-2408-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gelot C, Kovacs MT, Miron S, et al. Polθ is phosphorylated by PLK1 to repair double-strand breaks in mitosis. Nature 2023;621:415-22. 10.1038/s41586-023-06506-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Srinivas US, Tan BWQ, Vellayappan BA, et al. ROS and the DNA damage response in cancer. Redox Biol 2019;25:101084. 10.1016/j.redox.2018.101084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang H, Nicolay BN, Chick JM, et al. The metabolic function of cyclin D3-CDK6 kinase in cancer cell survival. Nature 2017;546:426-30. 10.1038/nature22797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Byers LA, Wang J, Nilsson MB, et al. Proteomic profiling identifies dysregulated pathways in small cell lung cancer and novel therapeutic targets including PARP1. Cancer Discov 2012;2:798-811. 10.1158/2159-8290.CD-12-0112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sen T, Tong P, Stewart CA, et al. CHK1 Inhibition in Small-Cell Lung Cancer Produces Single-Agent Activity in Biomarker-Defined Disease Subsets and Combination Activity with Cisplatin or Olaparib. Cancer Res 2017;77:3870-84. 10.1158/0008-5472.CAN-16-3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malumbres M. CDK4/6 Inhibitors: What Is the Best Cocktail? Clin Cancer Res 2019;25:6-8. 10.1158/1078-0432.CCR-18-2177 [DOI] [PubMed] [Google Scholar]
- 62.Tong J, Tan X, Song X, et al. CDK4/6 Inhibition Suppresses p73 Phosphorylation and Activates DR5 to Potentiate Chemotherapy and Immune Checkpoint Blockade. Cancer Res 2022;82:1340-52. 10.1158/0008-5472.CAN-21-3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Bu X, Wang H, et al. Cyclin D-CDK4 kinase destabilizes PD-L1 via cullin 3-SPOP to control cancer immune surveillance. Nature 2018;553:91-5. 10.1038/nature25015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rugo HS, Kabos P, Beck JT, et al. Abemaciclib in combination with pembrolizumab for HR+, HER2- metastatic breast cancer: Phase 1b study. NPJ Breast Cancer 2022;8:118. 10.1038/s41523-022-00482-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pujol JL, Vansteenkiste J, Paz-Ares Rodríguez L, et al. Abemaciclib in Combination With Pembrolizumab for Stage IV KRAS-Mutant or Squamous NSCLC: A Phase 1b Study. JTO Clin Res Rep 2021;2:100234. 10.1016/j.jtocrr.2021.100234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jerusalem G, Prat A, Salgado R, et al. Neoadjuvant nivolumab + palbociclib + anastrozole for oestrogen receptor-positive/human epidermal growth factor receptor 2-negative primary breast cancer: Results from CheckMate 7A8. Breast 2023;72:103580. 10.1016/j.breast.2023.103580 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as