Abstract
Equine infectious anemia virus (EIAV) is genetically one of the simplest lentiviruses in that the viral genome encodes only three accessory genes, tat, rev, and S2. Although serological analyses demonstrate the expression of the S2 protein in persistently infected horses, the role of this viral gene remains undefined. We recently reported that the S2 gene is not essential for EIAV replication in primary equine macrophages, as EIAV mutants lacking the S2 gene replicate to levels similar to those of the parental virus (F. Li, B. A. Puffer, and R. C. Montelaro, J. Virol. 72:8344–8348, 1998). We now describe in vivo studies that examine the evolution and role of the S2 gene in ponies experimentally infected with EIAV. The results of these studies reveal for the first time that the S2 gene is highly conserved during persistent infection and that deletion of the S2 gene reduces viral virulence and virus replication levels compared to those of the parental virus containing a functional S2 gene. These data indicate that the EIAV S2 gene is in fact an important determinant of viral replication and pathogenic properties in vivo, despite the evident lack of S2 influence on viral replication levels in vitro. Thus, these observations suggest in vivo functions of EIAV S2 that are not adequately reflected in simple infections of cultured cells, including natural target macrophages.
The relative genetic simplicity of the equine infectious anemia virus (EIAV) genome and the availability of an animal model for investigating EIAV pathogenesis provide a useful model system in which to examine the contribution of specific viral genes in lentivirus replication, persistence, and pathogenesis (16). In addition to the gag, pol, and env genes, which are present in the genomes of all retroviruses, EIAV contains only three auxiliary genes, the tat and rev genes, which are common to all lentiviruses, and the novel S2 gene, which is of unknown function and does not evidently correspond to any known lentivirus accessory gene. The S2 gene is located in the pol-env intergenic region immediately following the second exon of Tat and overlapping the amino terminus of the Env protein. The S2 open reading frame is common to all published EIAV proviral sequences and contains three potential functional motifs: GLFG (putative nucleoporin motif), PXXP (putative SH3 domain binding motif), and RRKQETKK (putative nuclear localization sequence) (9). Previous studies demonstrate the presence of serum antibodies to S2-specific peptides (20) in EIAV-infected horses, indicating that the S2 protein is expressed in vivo, presumably performing a function in viral replication. However, recent studies from our lab demonstrate that the S2 gene is not essential for virus replication in vitro (9); molecular cloned viruses with wild-type or mutated S2 genes displayed similar growth kinetics in cultured equine macrophages, the natural target of EIAV. Thus, the role of S2 in EIAV replication remains to be defined.
In light of previous studies with lentiviral accessory genes, such as simian immunodeficiency virus (SIV) nef (6), it is apparent that the relevance of a specific gene function may differ substantially in vitro in cell culture and in vivo in infected animals. To date, there has been no published assessment of S2 function during EIAV replication in experimentally infected horses. Thus, the current study was designed to examine the genetic evolution of the S2 gene during sequential disease cycles in experimentally infected ponies and to evaluate directly the role of the S2 gene in determining viral replication properties in vivo.
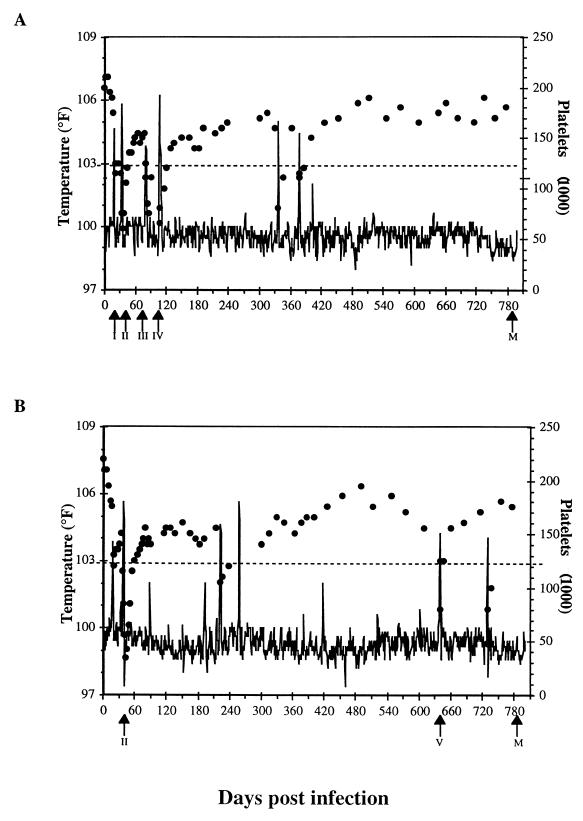
To characterize the genetic evolution of the EIAV S2 gene in experimentally infected ponies, we analyzed the S2 gene sequences of viral genomic RNA in plasma samples in two ponies infected with our reference pathogenic EIAVPV biological clone. Samples were taken during selected sequential febrile episodes characteristic of the initial chronic equine infectious anemia and during the later asymptomatic infection associated with the progression to long-term inapparent carriage (Fig. 1). Procedures for experimental infection, RNA purification, reverse transcriptase PCR (RT-PCR), cloning, and sequence analyses have been extensively described elsewhere for analyses of the evolution of the viral envelope gene in these experimentally infected ponies (8). In brief, seronegative Shetland ponies 564 and 567 were inoculated intravenously with 103 50% tissue culture infective doses of EIAVPV. Both pony 564 and pony 567 experienced five disease cycles, as measured by fever and platelet reduction. For plasma samples taken during febrile episodes and associated viremias, RNA purification and RT-PCR were performed directly on plasma samples, several independent RT-PCR products were cloned into either the low-copy-number vector pLG338 or the pGEM5ZF(+) T-A vector (Promega), and the positive clones were sequenced with a Taq Dye Deoxy Terminator Cycle Sequence kit (Applied Biosystems) and an ABI Prism 373 DNA sequencer (Applied Biosystems). Because of the low levels of viral RNA in plasma of inapparent carriers infected with EIAV, the long PCR protocols were unsuccessful in amplifying viral RNA from the plasma samples taken during asymptomatic infection. Therefore, equine macrophages were isolated from each inapparent carrier at 800 days postinfection (dpi) and cultured as described by Raabe et al. (18). Following 8 days in culture, supernatants of each macrophage culture were successfully used for RNA extraction, RT-PCR, and cloning and sequencing.
FIG. 1.
Clinical profiles of EIAVPV-infected pony 564 (A) and pony 567 (B). Pony 564 and pony 567 were both experimentally infected with 103 TCID50 of the reference pathogenic biological clone EIAVPV. Rectal temperatures (degrees Fahrenheit) and platelet counts were monitored daily to detect EIAV-associated febrile episodes and thrombocytopenia. Daily rectal temperature values are shown as a solid line, while platelet counts per microliter of whole blood are indicated by solid circles. Arrows below the x axis indicate the days on which the plasma samples were collected for EIAV genomic RNA extraction, cloning, and sequencing.
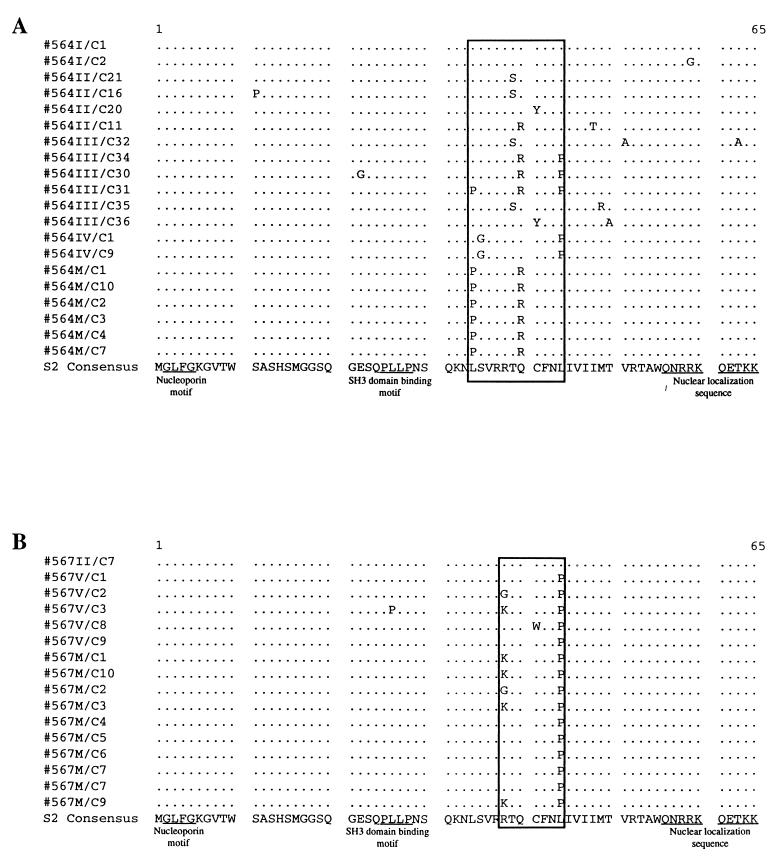
In total, the nucleotide sequences of the S2 gene from 20 clones spanning febrile episodes I, II, III, IV, and V and inapparent carrier stage of pony 564 and 16 clones spanning the febrile episodes II and V and inapparent stage of pony 567 were analyzed. The results of these sequencing analyses are summarized in Fig. 2 as translated amino acid sequences and are compared to the parental EIAVPV consensus S2 sequence. Examination of these sequence data revealed that the S2 amino acid sequences were highly conserved in vivo during the 27-month observation period. All S2 sequences during the sequential febrile episodes and inapparent stage were full length with an intact open reading frame and only minor localized amino acid variations. The three putative S2 functional motifs (GLFG, PXXP, and RRKQETKK) were all highly conserved.
FIG. 2.
Analysis of S2 evolution in experimentally infected ponies. The region spanning the S2 gene was sequenced from EIAVPV stock and plasma viral RNA obtained during febrile episodes and during long-term asymptomatic infection of EIAVPV-infected pony 564 and pony 567 (see Fig. 1). The deduced amino acid sequences were aligned and compared to S2 consensus derived from EIAVPV. A dot indicates an amino acid identical to the consensus sequence.
In contrast to these highly conserved S2 motifs, a relatively high frequency of amino acid substitutions was evident in one segment of S2 located between the putative SH3 domain binding motif and the nuclear localization motifs. In certain cases, identical substitutions were found to occur or reoccur at specific amino acids in this variable region during successive febrile episodes and/or during inapparent carrier infection. Examples include a leucine (L)-to-proline (P) change at amino acid 34, an arginine (R)-to-lysine (K) or -glycine (G) change at amino acid 38, a threonine (T)-to-serine (S) change at amino acid 39, a glutamine (Q)-to-arginine (R) change at amino acid 40, and another leucine (L)-to-proline (P) change at amino acid 44. Because the initiation codon for the envelope gene is 23 bp downstream of the S2 start codon, resulting in the overlapping of most of the S2 open reading frame with the envelope open reading frame, it was possible that the observed variations reflected selection for changes from the overlapping envelope gene during disease progression. Among the five most frequent substitutions observed in the S2 gene in pony 564 and pony 567, however, only the conservative threonine (T)-to-serine (S) change at amino acid 39 in pony 564 is coincident with a substitution occurring in the overlapping region of the envelope gene. The remaining substitutions are specific for the S2 amino acid sequence without altering envelope residues.
Previous sequencing studies from this lab and others of proviral DNA isolated from various cell-adapted strains of EIAV have indicated a high degree of S2 gene sequence conservation among different viral isolates, even after long-term passage in cell culture (5, 8, 13, 19). The current studies, however, are the first published analyses of the evolution of the S2 gene during persistent infection and the first analysis that is based on sequencing of EIAV genomic RNA, ensuring that the S2 sequences are from replication-competent genomes. The highly conserved nature of the EIAV S2 gene established by these sequencing analyses is suggestive of a critical functional role in virus replication that is preserved despite the frequent and random nucleotide variation intrinsic to lentivirus replication. More specifically, it is important to note that the minor amount of amino acid sequence variation observed in the S2 gene was localized to one segment of the S2 gene outside of the three putative functional motifs. The conservation of these putative motifs may imply their activity in virus replication, although the role of the EIAV S2 gene in the EIAV replication cycle remains to be determined.
To evaluate directly the role of the S2 gene in viral replication properties in vivo, we investigated the replication kinetics of EIAV with wild-type and mutated S2 genes in parallel experimental infections of ponies. For this comparison, we used our reference EIAVUK molecular clone and the S2 mutant EIAVUK2M/X, both described in detail previously (9). EIAVUK2M/X was generated by introduction of multiple termination codons into the S2 gene by a strategy that does not alter the amino acid sequence of the overlapping env gene downstream (9). Both EIAVUK and EIAVUK2M/X virus stocks were prepared by harvesting the medium from Lipofectamine (Gibco BRL)-transfected primary fetal equine kidney cells. Virus stocks of the EIAVUK and EIAVUK2M/X were characterized for RT activity and infectivity by assay procedures described by Lichtenstein et al. (10). Two groups of three seronegative ponies each were inoculated intravenously with 106.0 infectious-center doses of EIAVUK and EIAVUK2M/X, respectively. Infected ponies were monitored daily for clinical symptoms, and plasma samples for measurements of virus load were collected from each pony at regular intervals over the 6-month observation period, as previously described (10).
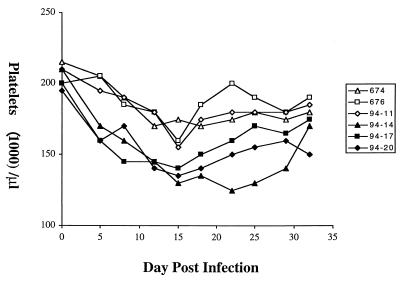
Viral pathogenicity was monitored in both sets of experimentally infected ponies by daily measurements of rectal temperature and blood platelet levels, the latter having been established as a sensitive and quantitative measure of EIAV disease severity (8, 17). Neither the EIAVUK- nor the EIAVUK2M/X-infected ponies displayed febrile episodes. The failure of the EIAVUK infection to produce a febrile episode is in contrast to the pathogenic properties originally described for the EIAVUK molecular clone (1). The lack of an obvious fever episode in these experimental infections, however, is consistent with more recent observations of additional EIAVUK infections of ponies indicating that the molecular clone is only weakly pathogenic, often without evident febrile episodes associated with the acute infection (C. J. Issel, unpublished data). In the absence of defined febrile episodes, we monitored the relative pathogenesis of the parental and S2 mutant virus infections by measuring daily levels of blood platelets during the acute stage of infection. These data (Fig. 3) demonstrated that during the first several weeks postinfection the ponies infected with the parental EIAVUK clone experienced a more rapid and more severe depletion of blood platelets than observed in the ponies infected with the S2 mutant virus at all time points tested. After 35 dpi, the blood platelet levels in both sets of experimentally infected ponies returned to normal for the remainder of the observation period (data not shown). The maximum platelet depletion associated with acute infection in the EIAVUK-infected ponies was an average of about 35% at 15 dpi, when the average platelet count dropped to 125,000 per μl compared to the normal level of about 200,000 per μl. In contrast, the ponies infected with the EIAVUK2M/X mutant virus experienced less severe platelet reductions, with an average minimum platelet count of about 175,000 per μl at 15 dpi representing only a 12% reduction in platelet levels. While neither of the observed platelet reductions qualifies by our standards (8) as clinical thrombocytopenia (defined as less than 105,000 per μl), the platelet data did clearly demonstrate a reduction of in vivo virulence by the S2 mutant virus compared to the parental virus. The absence of more-pathogenic clones of EIAV that can produce chronic disease precluded a more detailed examination of the role of the S2 gene as a determinant of viral virulence.
FIG. 3.
Viral virulence as monitored by reduction in platelet levels during acute infection. Blood platelet levels were assayed daily after infection of ponies with either the S2 mutant virus EIAVUK2M/X (ponies 674, 676, and 94-11) or the parental virus EIAVUK (ponies 94-14, 94-17, and 94-20). Platelet levels are expressed as the number of cells per microliter of whole blood. Clinical thrombocytopenia in EIAV-infected ponies has been defined as platelet levels below 105,000 per microliter, compared to normal platelet levels averaging about 200,000 cells per microliter.
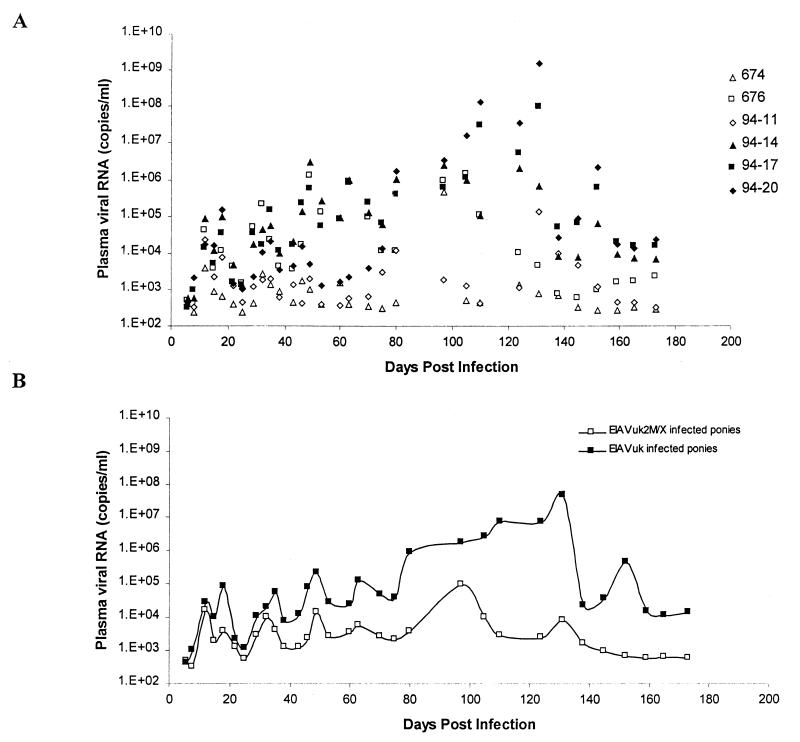
The severity of EIAV disease has been correlated closely with the levels of virus replication in experimentally infected ponies (17), as observed with other human and animal lentiviral infections (3, 11, 14). Therefore, we closely monitored the levels of virus replication in the two experimental infections to assess the effect of S2 mutation on virus replication properties. To quantify the level of virus replication, we used a recently developed nonradioactive single-tube semiquantitative RT-PCR (15) with a detection limit of 20 to 50 molecules of RNA to monitor EIAV genomic RNA levels in plasma samples taken twice a week during the first 8 weeks postinfection and weekly thereafter up to 6 months. The individual plasma RNA levels measured for each plasma sample from the six ponies are presented in Fig. 4A, and a calculated median of the RNA levels for the two groups of ponies at each time point is summarized in Fig. 4B. As detailed in Fig. 4A, all three of the EIAVUK-infected ponies displayed similar levels of plasma viral RNA at the various time points and waves of viremia, with levels of viral RNA similar to the values measured in historical panels of EIAVPV-infected ponies (17). Among the EIAVUK2M/X-infected ponies, ponies 94-11 and 674 displayed similar levels of viral RNA at most of the time points examined, and these levels were in general 101- to 104-fold lower than the RNA levels in the EIAVUK-infected ponies at the same times postinfection, with the differential in virus replication levels steadily increasing with time. In marked contrast, the third pony (676) infected with the EIAVUK2M/X mutant virus displayed viral RNA levels that were similar to those in the parental EIAVUK infections to about 110 dpi. After this time, the viral RNA levels observed in pony 676 were 101- to 104-fold less than the RNA levels observed in the parental EIAVUK infections, as seen with the other two ponies infected with the mutant EIAVUK2M/X virus.
FIG. 4.
Dynamics of viral replication in ponies inoculated intravenously with the S2 mutant virus EIAVUK2M/X (ponies 674, 676, and 94-11) or the parental virus EIAVUK (ponies 94-14, 94-17, and 94-20). (A) Individual EIAV plasma viral RNA levels measured for each pony; (B) median plasma RNA levels calculated for wild-type and mutant S2 virus-infected pony groups, respectively. Plasma viral RNA levels were analyzed by a recently developed semiquantitative RT-PCR assay with gag-specific amplification primers. The lower limit of the assay was determined to be 20 to 50 molecules of RNA per ml. Briefly, the virus pelleting and RNA extraction from plasma samples collected from experimentally infected ponies were performed as previously described (10). The Promega Access RT-PCR system (Promega) based on a single-tube reaction was employed for the semiquantitative assay (15). RT-PCR was carried out with 4 μl of plasma viral RNA sample as specified by the manufacturer, with an EIAV gag-specific primer, Gag34 (5′ GCTGACTCTTCTGTTGTATCG 3′), for both RT and PCR and an EIAV gag-specific primer, Gag11 (5′ ATGTATGCTTGCAGAGACATTG 3′), for PCR only. The first-strand cDNA was synthesized at 48°C for 45 min followed by denaturing at 94°C for 2 min. Second-strand cDNA synthesis and DNA amplification were then carried out under the following cycling conditions: 30 s at 94°C, 30 s at 55°C, and 30 s at 72°C for 35 cycles; 10 min at 72°C for 1 cycle; and holding at 4°C. RT-PCR products were separated by electrophoresis in a 2% agarose gel. Gels were stained in pH 8.0 Tris-acetate-EDTA buffer containing a 1:10,000 dilution of SYBR Green I stock solution (Molecular Probes, Eugene, Oreg.) for 45 min. The intensity in each band was quantified with a PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.) and the program ImageQuant (Molecular Dynamics). A standard curve, based on the known amount of synthetic RNA prepared by in vitro transcription with a T7 MEGAscript kit (Ambion, Austin, Tex.), was determined by linear regression analysis. The number of viral RNA molecules was then calculated by using the equation for this standard curve.
To account for the natural variation observed in EIAV infection in outbred ponies (17), we also calculated median RNA levels for each group of three ponies from the data presented in Fig. 4A. These median RNA levels are summarized for each group of infections in Fig. 4B. In this context, the general pattern of the kinetics of virus replication levels characteristic of the EIAVUK and EIAVUK2M/X infections becomes more obvious. These data revealed similar levels of virus replication in the EIAVUK and EIAVUK2M/X infections during the first 5 weeks postinfection with the average initial viremia in both pony groups being about 3 × 104 copies of viral RNA per ml of plasma. After this time, however, there was evident a progressively increasing disparity in the replication levels of the parental and mutant viruses as measured by plasma RNA levels. After about 35 dpi, the median levels of plasma viral RNA levels in the EIAVUK-infected ponies were consistently greater than the median levels of plasma viral RNA measured in the EIAVUK2M/X-infected ponies. At time points between 35 and 100 dpi, the parental virus replication levels were on average 10- to 100-fold greater than the levels of virus replication observed with the S2 mutant virus. A maximum differential between virus replication levels (6,000-fold) appeared at about 130 dpi, when the EIAVUK RNA levels averaged about 5 × 107 copies/ml and the EIAVUK2M/X RNA levels averaged about 8 × 103 copies/ml. At the terminal 6-month time point, the EIAVUK infections averaged about 1.4 × 104 copies per ml compared to the EIAVUK2M/X plasma RNA levels of about 600 copies per ml. While the latter time points are suggestive of viral set points, as reported for SIV and human immunodeficiency virus (HIV) infections (3, 11, 14), the unpredictable irregular waves of viremia associated with EIAV infections during the first year postinfection (16) preclude any definitive identification of a viral set point.
Statistical analysis by the nonparametric Mann-Whitney test of the differences of the median levels of virus replication between the two groups of infections revealed statistically relevant differences in median viral RNA levels at intermittent time points during the first 80 dpi (e.g., 8, 15, 18, 43, 75, and 80 dpi), usually associated with peaks of viremia. However, statistically significant differences (P < 0.05) for the level of virus replication between the two infection groups were consistently observed at all time points after 120 dpi.
To ensure that the S2 mutations in the EIAVUK2M/X virus were stable during the 6-month infection, we analyzed the S2 gene sequences present in plasma RNA from the three EIAVUK2M/X-infected ponies to confirm the presence of the two stop codons that were engineered into the S2 gene. Plasma samples collected at 194 dpi from each EIAVUK2M/X-infected pony were used for viral RNA isolation and RT-PCR sequence analyses of the S2 gene (data not shown). These S2 sequence analyses demonstrated a conservation of the two engineered termination codons in all of the EIAVUK2M/X S2 genes analyzed from each pony, confirming a lack of S2 reversion in these experimentally infected ponies. Therefore, the observed differences in levels of virus replication that developed late in infection cannot be attributed to in vivo changes in the S2 gene sequence of the mutant virus. While rapid in vivo reversions of many site-directed mutations of lentiviral accessory genes have been reported (6, 7), the absence of S2 reversion over the 6.5-month observation period may be attributed to the nature of the engineered mutations that require a minimum of 9 base changes to accomplish a reversion to wild-type and S2 gene expression (9).
Taken together, the studies described here reveal for the first time that deletion of the S2 gene by introducing stop codons from the EIAV genome reduces the level of virus replication and apparent virulence in vivo, with the differences in virus replication levels between the parental and mutant S2 viral strains becoming more pronounced as the infection progresses. This in vivo phenotype was somewhat unexpected, as we previously observed no differences in the in vitro replication properties of these same parental and S2 mutant viruses in cultured equine macrophages, even after prolonged serial passage (9). Thus, these experiments suggest that while the S2 gene is not essential for virus replication, it does influence viral replication properties in vivo, presumably via some accessory role in virus replication. In this regard, the EIAV S2 gene function may be analogous to the HIV-SIV accessory gene nef and others (including vif, vpr, and vpu in HIV type 1 and Vpx in HIV type 2 and SIV) in that, while dispensable for virus growth in most tissue culture systems, these accessory genes are critically important for viral growth in vivo (2, 22). The current studies of EIAV S2 function in viral replication also emphasize the importance of evaluating gene function under conditions of natural infection in the appropriate host, as in vitro cell culture assays may not detect critical in vivo properties of lentivirus genes. In addition, the studies with EIAV demonstrate that phenotypic differences may not be evident early in infection but can become more apparent with time as the effect is amplified through multiple rounds of virus replication.
Although the current studies clearly demonstrated the importance of the S2 gene in vivo, the functional role of S2 remains unknown. Equine macrophages in tissues are the predominant in vivo target cells for EIAV, and differentiation of the monocyte to macrophage is required to produce a productive infection (12, 18, 21). Our previous experiments addressing the importance of the S2 gene for viral replication by modulation of macrophage/monocyte lineage cells failed to disclose any obvious differences in viral replication between EIAVUK2M/X- and parental virus-infected cultures (9). However, the replication of the S2 mutant virus EIAVUK2M/X in equine blood monocyte-derived macrophages and an equine blood monocyte differentiation-maturation culture system did consistently reveal an initial delay in S2 mutant virus replication compared to the parental virus, perhaps suggesting a subtle in vitro effect that is amplified in vivo. Studies of the accessory genes of other lentiviruses define a variety of functions, a number of which affect the activation state of target cells (2, 4). Additional experiments are being directed toward defining the role of the S2 gene of EIAV in different steps of viral replication and in the effect of S2 protein expression on monocyte/macrophage activation.
Acknowledgments
We thank Scott Hammond for advice in statistical analysis and Bridget Puffer for many helpful discussions.
This work was supported by National Institutes of Health grant R01CA49296 and by funds from the Kentucky Agricultural Experiment Station and the Lucille Markey Charitable Trust.
REFERENCES
- 1.Cook R F, Leroux C, Cook S J, Berger S L, Lichtenstein D L, Ghabrial N N, Montelaro R C, Issel C J. Development and characterization of an in vitro pathogenic molecular clone of equine infectious anemia virus. J Virol. 1998;72:1383–1393. doi: 10.1128/jvi.72.2.1383-1393.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 3.Haaft P T, Verstrepen B, Überla K, Rosenwirth B, Heeney J. A pathogenic threshold of virus load defined in simian immunodeficiency virus- or simian-human immunodeficiency virus-infected macaques. J Virol. 1998;72:10281–10285. doi: 10.1128/jvi.72.12.10281-10285.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hirsch V M, Sharkey M E, Brown C R, Brichacek B, Goldstein S, Wakefield J, Byrum R, Elkins W R, Hahn B H, Lifson J D, Stevenson M. Vpx is required for dissemination and pathogenesis of SIVSMPBJ: evidence of macrophage-dependent viral amplification. Nat Med. 1998;4:1401–1408. doi: 10.1038/3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawakamin T, Sherman L, Dahlberg J, Gazit A, Yaniv A, Tronick S R, Aaronson S A. Nucleotide sequence analysis of equine infectious anemia virus proviral DNA. Virology. 1987;158:300–312. doi: 10.1016/0042-6822(87)90202-9. [DOI] [PubMed] [Google Scholar]
- 6.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R C. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 7.Lang M S, Weeger M, Stahl-Hennig C, Coulibaly C, Hunsmann G, Muller J, Muller-Hermelink H, Fuchs D, Wachter H, Daniel M, Desrosiers R C, Fleckenstein B. Importance of vpr for infection of rhesus monkeys with simian immunodeficiency virus. J Virol. 1993;67:902–912. doi: 10.1128/jvi.67.2.902-912.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leroux C, Issel C J, Montelaro R C. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J Virol. 1997;71:9627–9639. doi: 10.1128/jvi.71.12.9627-9639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li F, Puffer B A, Montelaro R C. S2 gene of equine infectious anemia virus is dispensable for viral replication in vitro. J Virol. 1998;72:8344–8348. doi: 10.1128/jvi.72.10.8344-8348.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lichtenstein D L, Rushlow K E, Cook R F, Raabe M L, Swardson C J, Kociba G J, Issel C J, Montelaro R C. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J Virol. 1995;69:2881–2888. doi: 10.1128/jvi.69.5.2881-2888.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lifson J D, Nowak M A, Goldstein S, Rossio J L, Kinter A, Vasquez G, Wiltrout T A, Brown C, Schneider D, Wahl L, Lloyd A L, Williams J, Elkins W R, Fauci A S, Hirsch V M. The extent of early viral replication is a critical determinant of the natural history of simian immunodeficiency virus infection. J Virol. 1997;71:9508–9514. doi: 10.1128/jvi.71.12.9508-9514.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maury W. Monocyte maturation controls expression of equine infectious anemia virus. J Virol. 1994;68:6270–6279. doi: 10.1128/jvi.68.10.6270-6279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuire T C, Lacy P A, O'Rourke K I. cDNA sequence of the env gene of a pathogenic equine infectious anemia lentivirus variant. Nucleic Acids Res. 1990;18:196. doi: 10.1093/nar/18.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellors J W, Rinaldo R C, Gupta P, White R M, Todd J A, Kingsley L A. Prognosis in HIV-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 15.Miller K, Storts D R. An improved single buffer, two enzyme system for RT-PCR. J NIH Res. 1996;8:48. [Google Scholar]
- 16.Montelaro R C, Ball J M, Rushlow K E. Equine retroviruses. In: Levy J A, editor. The Retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 257–360. [Google Scholar]
- 17.Raabe M L, Issel C J, Cook S J, Cook R F, Woodson B, Montelaro R C. Immunization with a recombinant envelope protein (rgp90) of EIAV produces a spectrum of vaccine efficacy ranging from lack of clinical disease to severe enhancement. Virology. 1998;245:151–162. doi: 10.1006/viro.1998.9142. [DOI] [PubMed] [Google Scholar]
- 18.Raabe M R, Issel C J, Montelaro R C. Equine monocyte derived macrophage cultures and their applications for infectivity and neutralization studies of equine infectious anemia virus. J Virol Methods. 1998;71:87–104. doi: 10.1016/s0166-0934(97)00204-8. [DOI] [PubMed] [Google Scholar]
- 19.Rushlow K E, Olsen K, Stiegler G, Payne S L, Montelaro R C, Issel C J. Lentivirus genomic organization: the complete nucleotide sequence of the env gene region of equine infectious anemia virus. Virology. 1986;155:309–321. doi: 10.1016/0042-6822(86)90195-9. [DOI] [PubMed] [Google Scholar]
- 20.Schiltz R L, Shih D S, Rasty S, Montelaro R C, Rushlow K E. Equine infectious anemia virus gene expression: characterization of the RNA splicing pattern and the protein products encoded by open reading frames S1 and S2. J Virol. 1992;66:3455–3465. doi: 10.1128/jvi.66.6.3455-3465.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sellon D C, Walker K M, Russell K E, Perry S T, Covington P, Fuller F J. Equine infectious anemia virus replication is upregulated during differentiation of blood monocytes from acutely infected horses. J Virol. 1996;70:590–594. doi: 10.1128/jvi.70.1.590-594.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trono D. HIV accessory proteins: leading roles for the supporting cast. Cell. 1995;82:189–192. doi: 10.1016/0092-8674(95)90306-2. [DOI] [PubMed] [Google Scholar]