SUMMARY
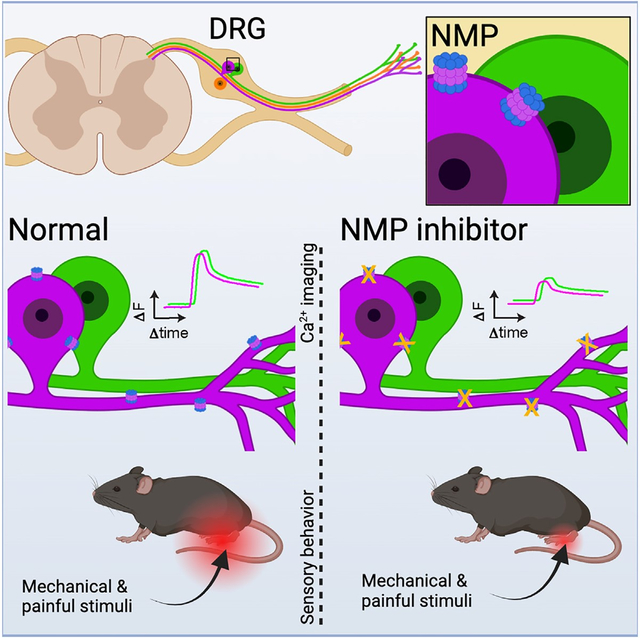
Proteasomes are critical for peripheral nervous system (PNS) function. Here, we investigate mammalian PNS proteasomes and reveal the presence of the neuronal membrane proteasome (NMP). We show that specific inhibition of the NMP on distal nerve fibers innervating the mouse hind paw leads to reduction in mechanical and pain sensitivity. Through investigating PNS NMPs, we demonstrate their presence on the somata and proximal and distal axons of a subset of dorsal root ganglion (DRG) neurons. Single-cell RNA sequencing experiments reveal that the NMP-expressing DRGs are primarily MrgprA3+ and Cysltr2+. NMP inhibition in DRG cultures leads to cell-autonomous and non-cell-autonomous changes in Ca2+ signaling induced by KCl depolarization, αβ-meATP, or the pruritogen histamine. Taken together, these data support a model whereby NMPs are expressed on a subset of somatosensory DRGs to modulate signaling between neurons of distinct sensory modalities and indicate the NMP as a potential target for controlling pain.
In brief
Proteasomes are critical for sensory neuron function. Villalón Landeros et al. uncover a specialized proteasome in somatosensory neurons called the neuronal membrane proteasome (NMP). The NMP mediates crosstalk between somatosensory neurons to modulate sensitivity to stimulation. Inhibition of the NMP reduces sensitivity to mechanical and painful stimuli in vivo.
Graphical abstract
INTRODUCTION
Optimal development and function of the peripheral nervous system (PNS) requires proteasome-dependent protein degradation.1–4 Indeed, the use of proteasome inhibitors in vivo and in vitro has revealed that proteasome activity in the PNS is essential for neurodevelopment, physiology, Ca2+ signaling, and myelination in sensory neurons.5–11 Moreover, the use of proteasome inhibitors as human cancer chemotherapies12–14 can cause painful peripheral neuropathy characterized by severe neuropathic pain, numbness, tingling, and itch sensation in distal-to-proximal extremities as well as altered proprioception.9,14–17 In contrast, proteasome inhibitors administered at low concentrations reduce pain perception without inducing neuropathy.11,18–21 Despite these extensive studies, proteasomes in PNS biology are still not well understood.
Proteasomes, which catalyze most of the protein degradation in all mammalian cells,22,23 are more complex than generally appreciated. Proteasomes exist as multi-subunit complexes in association with various regulatory particles to mediate ubiquitin-dependent and ubiquitin-independent protein degradation.22,24–27 Moreover, within neurons, proteasomes are localized throughout the cell from the nucleus to the cytosol, axons, dendrites, and synapses.28 Thus far, the majority of our understanding of the relationship between proteasomes and cellular biology has relied on the use of highly potent and specific cell-permeable pan-proteasome inhibitors. While these pan-proteasome inhibitors have proven to be invaluable research tools and therapeutics, they fail to discriminate between alternative proteasome complexes or differentially localized proteasomes.
Our laboratory discovered a neuronal-specific proteasome complex in the central nervous system (CNS).29–31 This proteasome complex is tightly associated with the neuronal plasma membrane in a manner that allows for the degradation of intracellular neuronal proteins into signaling peptides that are released directly into the extracellular space. Remarkably, these extracellular peptides were determined to modulate the activity of naive CNS neurons, in part through N-methyl-D-aspartate receptor-dependent Ca2+ signaling. These data are extraordinary and reveal the NMP as a mediator of communication between neurons (inter-neuronal) important for modulating nervous system signaling.
Here, we explored proteasomes in the PNS and revealed the presence of the neuronal membrane proteasome (NMP) in the somata as well as proximal and distal nerve endings of somatosensory neurons. Selective inhibition of NMP in the hindlimb paw reduced behavioral responsiveness to punctate and sharp mechanical stimuli and noxious cold stimuli within 30 min to 2 h without any evidence of acute neuropathy. Investigating the mechanisms underlying these observations, we determined that the NMP in the PNS was enriched in a specific subpopulation of nociceptors that are positive for neurochemical markers (IB4+, CGRP+, and NF-H+) that label small, medium, and large somatosensory neurons. We also observed co-localization of NMP with the P2X3 purinoreceptor, which is known to mediate ATP-dependent nociceptor signaling. Consistent with a role for the NMP in modulating inter-neuronal signaling in the PNS, we determined that inhibition of NMP led to reduced depolarization or ATP-mediated Ca2+ signaling in vitro in a non-cell-autonomous manner. Furthermore, single-cell RNA-sequencing (scRNA-seq) analysis of NMP-expressing primary dorsal root ganglion (DRG) neurons revealed the NMP is almost (>95%) exclusively expressed in MrgprA3+ and Cysltr2+ somatosensory neurons. Based on these observations, we hypothesized that the NMP operates from these pruriceptive neurons to modulate the sensitivity of the neighboring somatosensory neurons. We investigated this by selectively stimulating NMP-containing neurons using the pruritogen histamine in the presence or absence of NMP inhibitor. We found that histamine-responsive and -non-responsive neurons, when treated with NMP inhibitor, had an enhanced sensitivity to depolarization. Taken together, these data support our model that the NMP is expressed in a subset of somatosensory DRG neurons to modulate signaling between neurons of distinct sensory modalities. While our results provide insights into understanding proteasome function in the PNS, they also reveal the NMP as a potential new therapeutic target for pain management.
RESULTS
Proteasomes localize to the plasma membranes of peripheral sensory neurons
The localization of proteasomes is closely related to their mechanism of action in regulating mammalian cellular and tissue biology.28 To assess proteasome localization in the PNS, we used previously established protocols29,30,32 to perform immunogold electron microscopy (immuno-EM) analysis of mouse tissue dissected from the lumbar spinal cord (layers L4–L6 [L4–L6]), the L5 DRG, and the sciatic nerve trunk (Figure 1A).33–37 We used previously validated antibodies29,30 raised against the β2 and β5 proteasome subunits, which are common to all constitutive proteasomes (Figure S1A).22,38 Proteasome subunits are not present in a cell separate from the intact proteasome complex and, thus, signal from these antibodies reflect the localization of proteasomes.22 For controls, we used antibodies against the membrane proteins Na+/K+-ATPase and Kv1.3 and the cytosolic protein βIII-tubulin.39,40 Considering the average length of an antibody (~15 nm) and the size of the gold particles (~6 nm), we classified a protein as being on the membrane if its corresponding gold particle was within a 33-nm radius from the membrane (Figure S1B). Using these criteria, we quantified the localization of immunogold-labeled proteasome particles in spinal cord neuropil, DRG neuron soma, and distal nerve axons (Figures 1B–1D, S1C, and S1D). Within neurons, we observed that 20%–40% of all gold particles were found on or near the plasma membrane compared to the cytosol (Figures 1B–1D, bar graphs). We also analyzed the distribution of gold particles around the membranes by measuring the perpendicular distance of the particles from the closest membrane within a 240-nm distance and found that over 25% of gold particles labeling proteasomes localized to within 33 nm from the membrane (Figures 1B–1D, violin plots). Similar results were observed using antibodies raised against different epitopes of β2 and β5 subunits (Figure S1E). The spatial localization of proteasome gold particles resembled that of gold particles labeling for Na+/K+-ATPase (Figure S1F) and Kv1.3 membrane ion channel (Figure S1G) but contrasted with that of βIII-tubulin, which was >90% cytoplasmic (Figure S1H). These experiments provided evidence of the existence of a proteasome complex that localizes to the membranes of PNS neurons and were similar to the NMP found in CNS neurons.30
Figure 1. The NMP is expressed in PNS neuron soma, axon, and nerve endings.
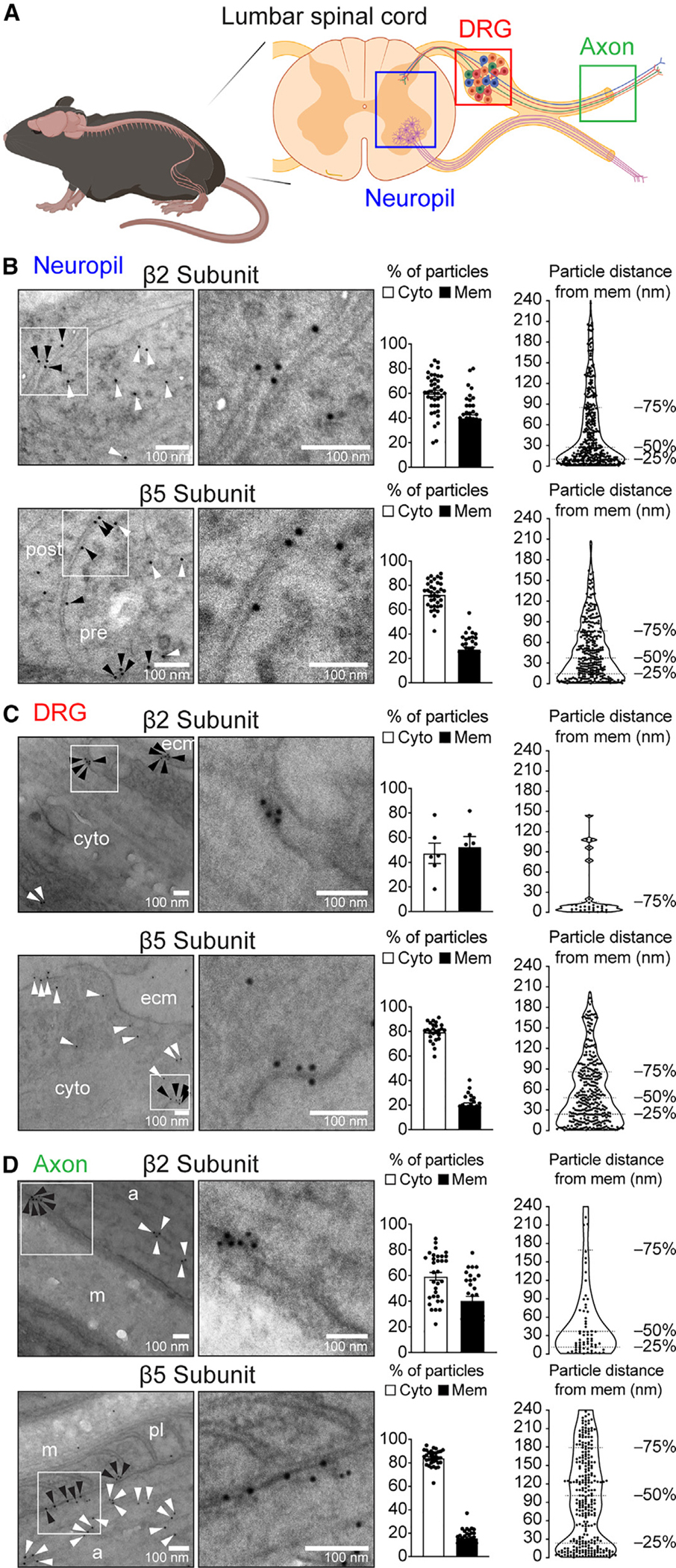
(A) Schematic of tissues used in immuno-EM experiments.
(B–D) Representative images of immunogold labeling in neuropil of the spinal cord (B), DRG (C), and axon (D). White boxes: sections shown magnified on the right. White arrowheads: gold particles localized on the cytoplasm. Black arrowheads: gold particles localized on the membrane. Bar graphs to the right show the proportion of gold particles localized on the cytoplasm (Cyto) and membrane (Mem). Violin plots show the distribution of gold particles within 240 nm of the neuronal membranes. Twelve to twenty micrographs for each antibody were analyzed for each tissue region from two mice. ecm, extracellular matrix; post, post-synaptic cell; pre, pre-synaptic cell; a, axon; m, myelin; pl, paranodal loop. Data are presented as mean ± SEM (n = 25–45 [except for C, top, where n = 6] micrographs analyzed from two independent animals).
See also Figure S1.
To confirm these findings biochemically, we harvested the lumbar spinal cord and proximal sciatic nerve from wild-type mice to perform surface protein biotinylation assays.30,41 These samples were then resolved by SDS-PAGE followed by immunoblotting for proteasome core subunits α1–7 (detects α subunits 1, 2, 3, 4, 5, 6, 7), as well as membrane (Na+/K+-ATPase) and cytoplasmic (β-actin) protein controls. Immunoblots revealed 20S proteasome subunits and Na+/K+-ATPase, but not β-actin, in the purified surface biotinylated protein pool from the spinal cord and proximal sciatic nerve (Figures S1I and S1J). No proteins were detected in non-biotinylated samples.
Taken together with our immuno-EM data, these results highlighted that proteasomes localize to the plasma membranes of peripheral sensory neurons and are exposed to the extracellular space in a manner similar to that of CNS NMPs.30 Such a localized proteasome, we believed, would have significance for PNS function, and we thus proceeded to investigate this observation further.
In the PNS, NMP localizes to sensory nerve terminals
Sensory neurons of the PNS innervate peripheral organs such as the skin, where they receive and transduce information from the environment to allow for sensation.42,43 To investigate whether NMPs localized on distal peripheral nerve processes, we developed a proteasome antibody feeding protocol to label NMPs in fresh hind-paw skin tissue from wild-type mice (Figure 2A). Given that antibodies are not cell permeable, this approach enables labeling of proteins, including proteasomes that are exposed to the extracellular space.30,44,45 Following antibody feeding to label surface membrane proteasomes, we then permeabilized the tissue and co-stained with antibodies against cytosolic proteins to label sensory nerve fibers (protein gene product 9.5 [PGP9.5] and neurofilament heavy [NF-H]). PGP9.5 predominantly labels the thinly myelinated and non-myelinated neurons,46,47 and NF-H predominantly labels myelinated and thinly myelinated neurons.46 Antibody-labeled samples were then prepared for confocal microscopy to measure the overlap between surface proteasome labeling, PGP9.5, and NF-H (Figures 2B and S2A). PGP9.5 and NF-H staining was similar to standard protocols48,49 and, in our experiments, unaffected by surface proteasome labeling (Figure S2B).
Figure 2. The NMP localizes to nerve endings on the skin.
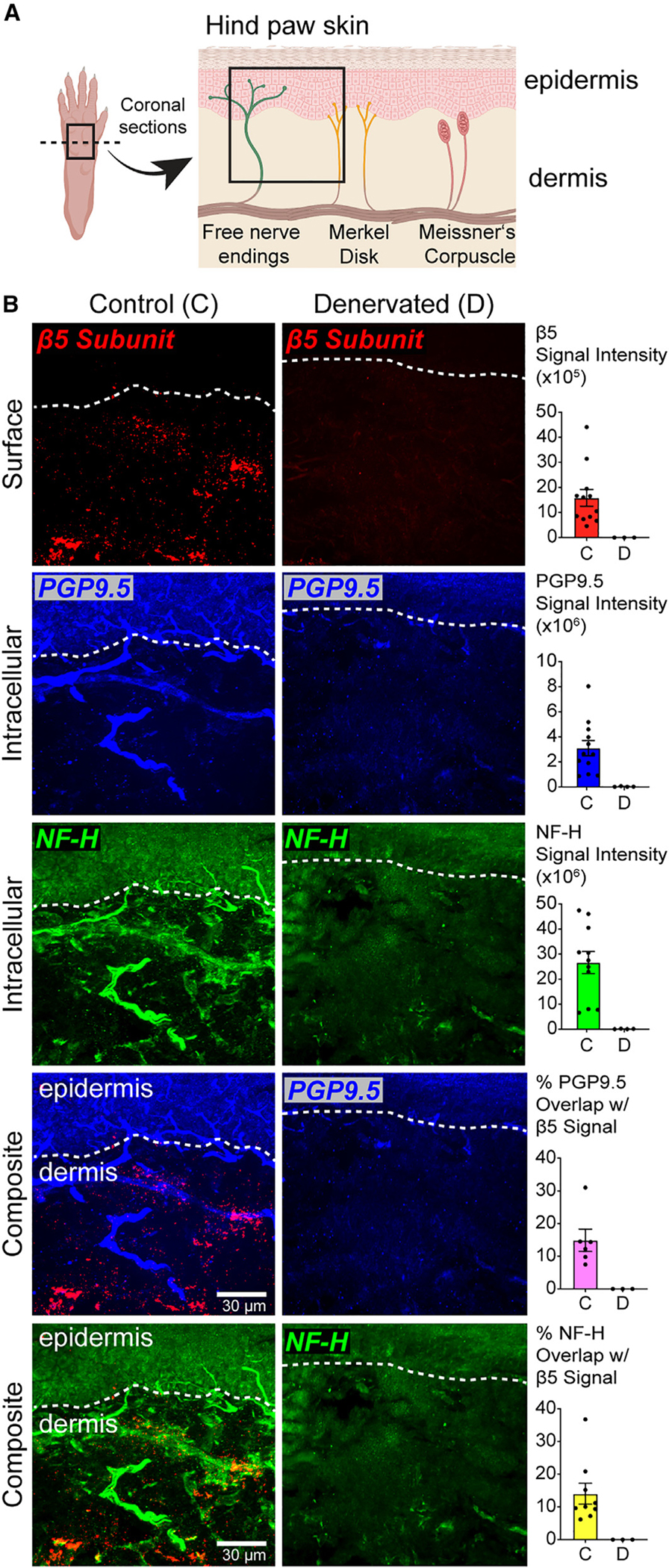
(A) Schematic of paw skin nerve endings.
(B–D) (B) Representative maximum projection of z-stack images of plantar skin coronal sections showing immunostaining on nerves in control (C) and denervated (D) tissue. Bar graphs show the quantification of surface β5 (i.e., NMP) and intracellular PGP9.5 and NF-H signal intensities. For composite images, bar graphs show the quantification of percent overlap of NMP labeling on detectable nerves on the skin. Data are presented as mean ± SEM (n = 3–9 groups of images analyzed from three independent animals).
See also Figure S2.
To verify that proteasome signal in our antibody feeding protocol was specific to nerve fibers, we performed a sciatic nerve transection injury on the right paw of mice and allowed the mice to recover for 10 days to allow degeneration of the transected nerves in the skin.50,51 Skin from both the denervated paw and the naive left hind paw was sectioned coronally to preserve the epidermis and dermis containing the sensory nerve fibers. While proteasome antibody feeding and nerve staining on skin tissue from the non-lesioned side showed substantial signal in the dermis and epidermis sections of the skin, we observed little to no signal in denervated paw skin (Figures 2B and S2B). Denervation did not affect overall tissue integrity (Figure S2C).
The proteasome signal in our images that did not overlap with PGP9.5 or NF-H signal likely represented proteasomes localized to other non-labeled nerves, or our nerve staining was below detection at these proteasome-enriched sites. In addition, no observation of NMP staining in the epidermis could be a limitation of our current labeling methods or could indicate the absence of NMP on nerve sections innervating this tissue. Together, these results supported the idea that a surface-exposed proteasome at the plasma membrane of PNS neurons was also present in the most distal sections of these neurons. Based on these data and our current understanding of the NMP in the CNS,29,30 we considered that this NMP localized in nerve terminals of the paw skin might be involved in regulating sensory neuron function relevant to the sensation of touch, temperature, itch, or pain.
Selective inhibition of the NMP in the skin does not cause acute nerve damage and leads to reduced mechanical and pain sensitivity
To investigate the role of NMP in sensory neurons in vivo, we turned to the previously described NMP inhibitor, biotin-epoxomicin (BE), a potent and specific covalent proteasome inhibitor that is not plasma membrane permeable.52,53 To demonstrate BE targeting of NMP in nerve endings of the paw, we injected 5 μmL of 10 μM BE into the right hind paw of normal and denervated mice and collected paw skin tissues at 30 min, 60 min, and 120 min post injection (Figure 3A). Immunoblot analysis of these skin samples revealed, in normal skin, biotin signal and neuronal-specific NF-H, and covalently associated BE to β5 proteasome subunit as indicated by a ~1-kDa change in molecular weight of the β5 antibody signal (Figure 3B).54 Biotin, NF-H, and shifted-β5 signal were absent in denervated skin samples (Figure 3B). These data were in contrast to those of epoxomicin (~500 Da) injection experiments, which readily crosses the cell membrane and targets all proteasomes (Figure S3A).55 Taken together, we interpreted these data to mean that intraplantar injection of BE resulted in acute (<60 min) targeting of the nerve-specific subfraction of proteasome in the skin. Combined with our NMP labeling on nerve endings (Figure 2) and our understanding of BE,29,30,52 we concluded that BE is selectively inhibiting the NMP on neurons innervating the paw skin.
Figure 3. Selective inhibition of NMP in PNS results in reduced mechanical and pain nociception without nerve degeneration.
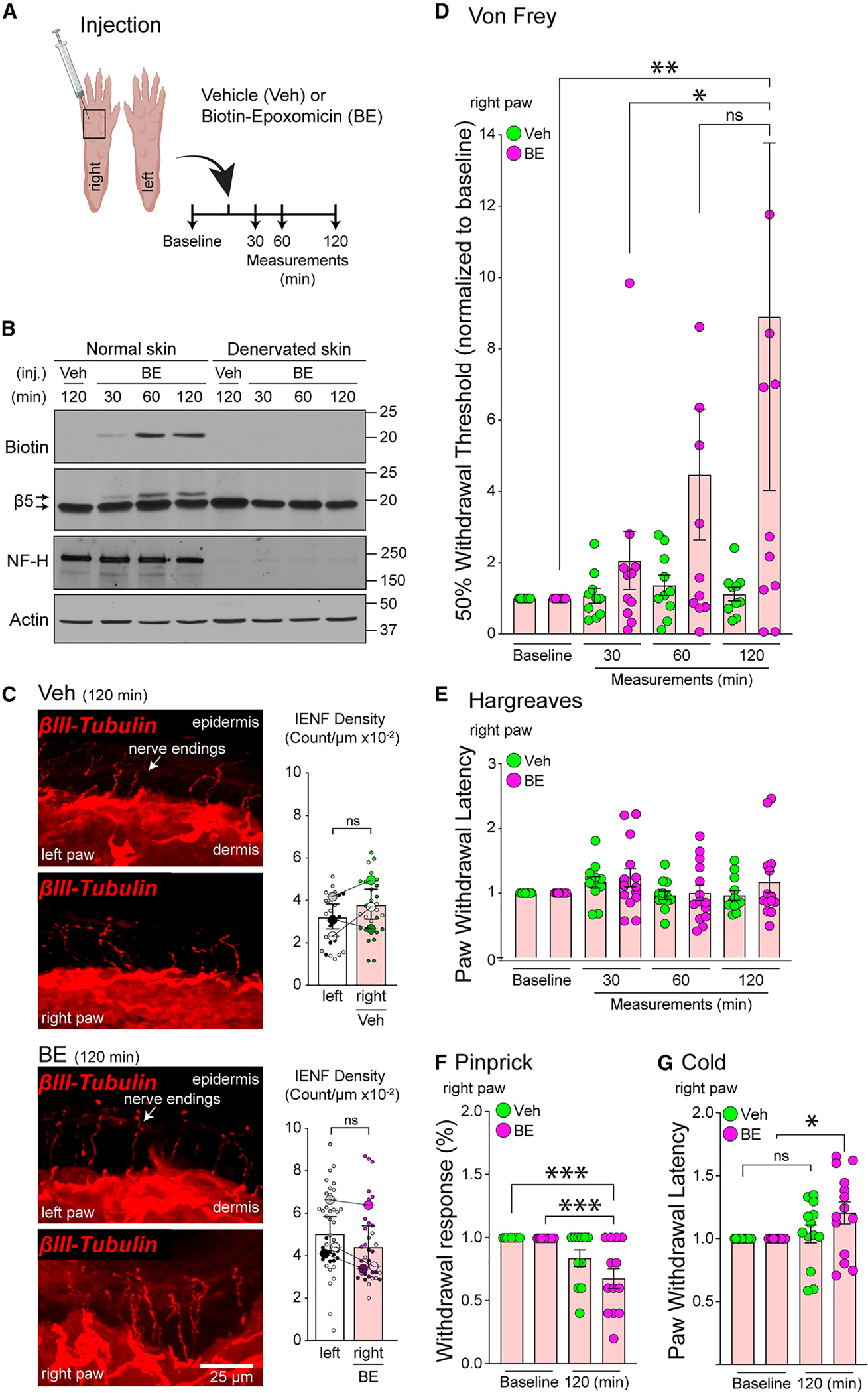
(A) Schematic of experimental design.
(B) Immunoblot of normal and denervated skin samples from (A) using indicated antibodies. β5 signal upper arrow is biotin-epoxomicin (BE)-bound.
(C) Representative images of nerve endings in paw skin sections from (A) at 120 min post treatment with vehicle (Veh) or BE. Bar graphs show intraepidermal nerve fiber density (IENF) density quantifications. The contralateral untreated (left) paw was used as an internal control (n = 3 mice per group/time point). Data are presented as super plots showing individual observations (small dots) from multiple experiments (larger dots) and mean ± SEM. No statistically significant differences were observed (unpaired t test).
(D) Von Frey up-down test quantification. Data are presented as mean ± SEM (n = 10–12 mice per treatment). *p < 0.05, **p < 0.01 (two-way ANOVA with Tukey’s post hoc test).
(E) Hargreaves test quantification. Data are presented as mean ± SEM (n = 14 mice per treatment). No statistically significant differences were observed (two-way ANOVA with Tukey’s post hoc test).
(F) Pinprick test quantification. Data are presented as mean ± SEM (n = 14 mice per treatment). ***p < 0.001 (two-way ANOVA with Tukey’s post hoc test).
(G) Cold test quantification. Data are presented as mean ± SEM (n = 14 mice per treatment). *p < 0.05 (two-way ANOVA with Tukey’s post hoc test).
All behavioral data were normalized to baseline measurements per mouse. See also Figure S3.
Cell-permeable proteasome inhibitors can promote nerve degeneration.10,14,18,19,56–60 Thus, we next investigated whether BE leads to acute nerve-ending degeneration. To do so, we injected 5 μL of 10 μM BE or vehicle (DMSO) intradermally into the right hind paw of mice and collected the skin tissue of both uninjected and injected hind paws at 30 min, 60 min, and 120 min. These tissues were prepared for intraepidermal nerve fiber density61 analysis using antibodies against β-III tubulin to label all nerve endings in the skin (Figure 3A). From these quantified data, we observed no differences in nerve-ending morphology in skin samples from the BE-treated paw compared to vehicle-treated or the uninjected (contralateral) paw at any time point (Figures 3C and S3B).
Based on our data, we were convinced that the use of BE could selectively inhibit NMPs on nerve endings in the mouse paw skin. Next, we asked whether the inhibition of the NMP in the hindlimb paw would lead to any changes in mouse sensory behavior.43,62 Toward this goal, the investigator was blinded to injection material (BE or vehicle), which was injected into the right hind paw of mice followed by Von Frey, Hargreaves, pinprick, and noxious cold behavioral tests. The Von Frey62–64 and Hargreaves62,65 tests are two methods routinely used for evaluating sensitivity to punctate mechanical stimuli and heat, respectively. The pinprick66,67 and noxious cold68,69 tests are used to assess sensitivity to painful mechanical and cold stimuli, respectively.43,62,66,67
We assessed the sensitivity to punctate mechanical stimulation by applying Von Frey filaments to the skin surface of the mouse hind paw using the up-down method62–64,70 before injection and at 30 min, 60 min, and 120 min after BE injection (Figure 3A). Our experiments showed that a 5-μL intraplantar injection of 10 μM BE significantly increased 50% withdrawal thresholds compared to vehicle-injected controls (Figure 3D). Additionally, no significant changes in 50% thresholds were observed in the left uninjected (contralateral) paw of the BE-treated mice or vehicle-treated mice (Figure S3C). Using a similar treatment protocol, we then performed the Hargreaves test. The Hargreaves test measures the sensitivity to radiant heat (<42°C) but did not reach temperatures high enough to induce pain (>42°C).62 Our results revealed no difference in sensitivity to heat stimuli in the BE-injected paw compared to the vehicle-injected paw at any of the time points tested (Figure 3E). Similarly, we saw no changes in heat sensitivity in the left (uninjected) paws of BE- or vehicle-treated mice (Figure S3D).
To investigate a potential role for NMP in modulating painful mechanical and noxious cold sensitivity, we measured the paw-withdrawal response to an insect pin being applied to the plantar skin66,67 before injection and at 120 min post injection with either BE or vehicle. Pinprick test showed that treatment significantly reduced the percent withdrawal response to painful mechanical stimulation compared to vehicle treatment (Figure 3F). Similarly, with the noxious cold sensitivity test, we observed a statistically significant increase in paw-withdrawal threshold after treatment with BE (Figure 3G). No changes in withdrawal response to pinprick or withdrawal latency to noxious cold were observed in the left uninjected paw (contralateral) of mice treated with vehicle or BE (Figures S3E and S3F).
Based on our findings, we concluded that NMPs in the PNS play an important and distinct role in mediating punctate mechanical and pain sensation. The NMP did not play a role in mediating warming temperature sensation. Given that all sensory neurons express proteasomes, it was also not immediately clear how the NMP functions to regulate a subset of behavioral responses.
The NMP is present in specific sensory neuronal subtypes
To investigate the mechanism(s) by which the NMPs modulate sensory neuron activity, we took advantage of primary DRG neuronal cultures. To ensure that the culturing process did not affect the NMP localization in DRG neurons, we performed immuno-EM proteasome labeling as well as surface protein biotinylation labeling, as described above (see Figures 1 and S1). Immuno-EM of the cultured DRG neurons (Figures 4A, S4A, and S4B) and surface protein biotinylation pull-down (Figure S4C) revealed proteasomes localized to soma and axonal membranes that were accessible from extracellular side of these neurons, similar to our findings in the spinal cord and DRG tissue that revealed an NMP. Subsequently, we wanted to ensure that the NMP in cultured neurons retained catalytic activity. Using a proteasome-specific activity-based probe (MV151), we investigated the activity of the NMP in our DRG cultures similarly to approaches described in the CNS.31,54,71–75 We observed efficient labeling of surface proteasomes with MV151 probe (Figure S4D). Based on these observations, we concluded that the culturing process retained the expression of active NMPs on DRG neurons.
Figure 4. The NMP is enriched in specific subtypes of DRG neurons.
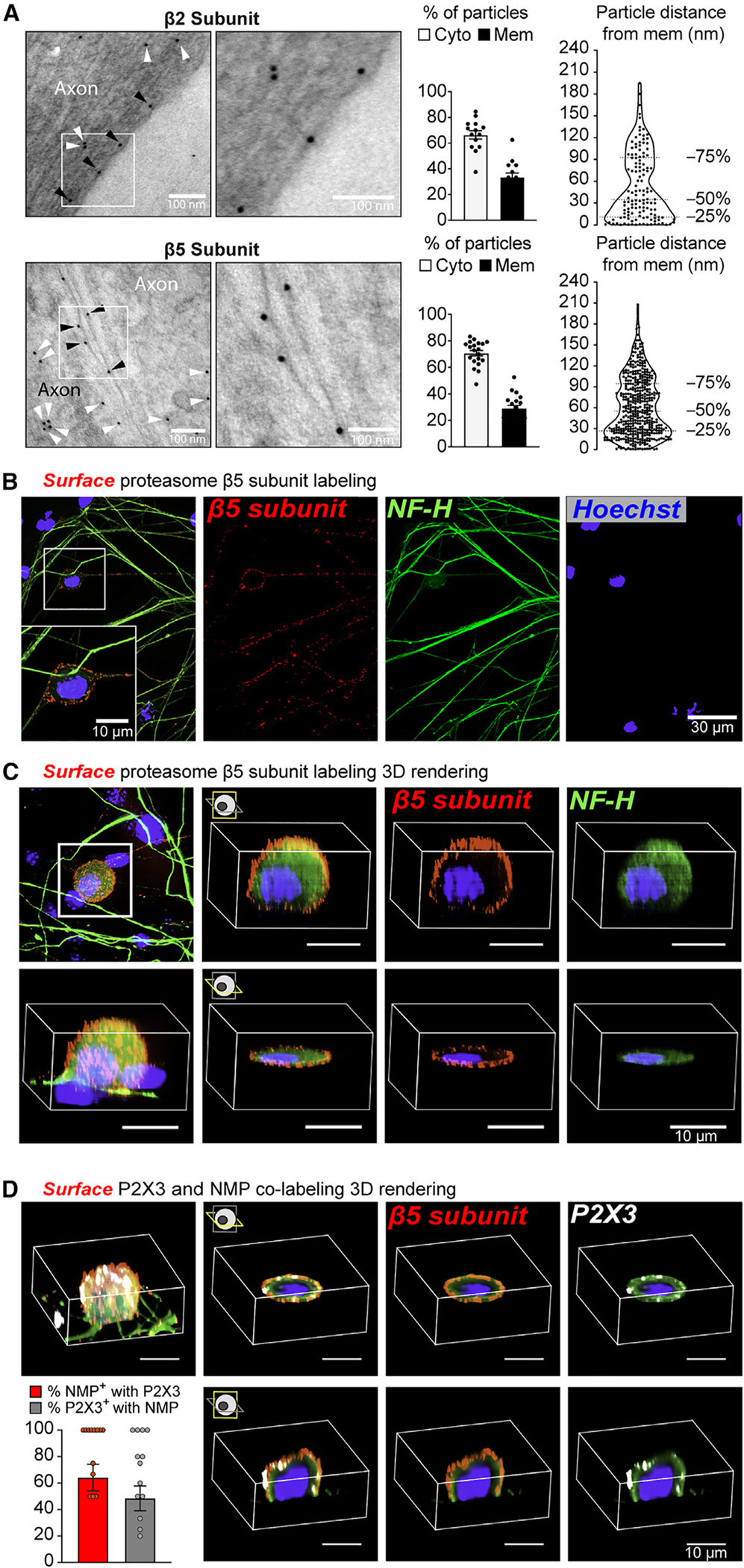
(A) Representative micrographs of immuno-EM labeling on primary DRG cultures. White arrowheads: gold particles localized on the cytoplasm. Black arrowheads: gold particles localized on the membrane. Bar graphs to the right show the proportion of gold particles localized on the cytoplasm (Cyto) and membrane (Mem). Violin plots show the distribution of gold particles within 240 nm of the neuronal membranes. Data are presented as mean ± SEM (n = 14–20 micrographs analyzed from two independent animals).
(B) Representative images of antibody feeding experiments in DRG cultures against the β5 proteasome subunit followed by cytoplasmic NF-H staining. White box: magnified section shown on bottom of composite image.
(C) Representative 3D projection images of antibody feeding experiments in DRG cultures against the β5 proteasome subunit followed by cytoplasmic NF-H staining.
(D) Representative 3D projection images of antibody feeding experiments in DRG cultures against the P2X3 and β5 proteasome subunit co-labeling. Bar graph shows quantification of surface P2X3 and β5 co-labeling under the conditions that at least one neuron in the field of view was positive for both NMP and P2X3. Data are presented as mean ± SEM (n = 13 images analyzed).
See also Figure S4.
An advantage of DRG cultures harboring active NMPs was that it allowed us to use orthogonal approaches to begin studying the mechanistic details of NMPs in the PNS. We first asked whether NMPs localize to all DRG neuronal subtypes. To address this question, we used antibody feeding approaches whereby antibodies against core proteasome subunits α2, β2, and β5 were directly applied to live DRG cultures under non-permeabilizing conditions. Using our approach, we observed specific signal that corresponded to primary proteasome antibodies localized to the surface of the soma and axons of DRG neurons (Figures 4B, S4E, S4F, and S4G). 3D-projection rendering of z stack images of the surface β5 proteasome signal further confirmed the localization of the proteasome to the surface of DRG neurons (Figure 4C). In contrast, total proteasome labeling of permeabilized cells showed more broadly localized proteasome signal that filled most of the observable neurons with 70% ± 5% of NF-H staining overlapping with proteasome staining (Figure S4H). The remaining 25% of NF-H signal that did not overlap with proteasome staining likely represents NF-H-positive areas of the neuron where proteasomes did not localize. In contrast, 30% ± 5% of total proteasome signal overlapped with NF-H staining, and the remaining 70% was likely localized to non-NF-H positive neurons. Based on these collective observations and for ease of discussion, we will refer to the surface proteasome antibody signal as the NMP.
To better understand these observations, we co-stained our surface proteasome-labeled samples with glial fibrillary acidic protein (GFAP) and NF-H and imaged areas that contained NF-H+ and NMP+ nuclei. We noted on average that for a given image ~80% of the nuclei were GFAP+, while ~6% of the nuclei were NF-H+ and the remaining ~14% of nuclei were negative for NF-H and GFAP (Figure S4I). Across these images, on average, there was ~7.5% of the total nuclei that were NMP+ and ~57% of NMP+ nuclei were NF-H+ while 0% were GFAP+ (Figure S4I). Exploring whether NMP expression was associated with populations of sensory neuronal subtypes other than NF-H+ neurons, we performed antibody feeding followed by immunostaining with the neuronal subtype biochemical markers NF-H, calcitonin gene-related protein (CGRP), or isolectin β−4 (IB4).42,43,76,77 Images were taken in areas that expressed the specific biochemical marker. We found that the NMP was also present in CGRP and IB4 sensory neuronal subtypes, albeit not in every cell for a given subtype (Figure S4J). From these data, NMPs were clearly expressed in DRG neurons, and various DRG neurons within subtypes can exist with and without NMP.
NF-H, CGRP, and IB4 sensory neurons can be further classified into more specific cell types depending on function, area of innervation, and response properties, which have various soma sizes relevant to their somatosensory function.42,78 For example, NF-H+ neurons can be distinguished into large-size proprioceptors, medium-size Aβ and Aδ low-threshold mechanoreceptors (LTMRs), and some small-size C-type sensory neurons.42,43,76,77,79–82 Thus, we quantified NMP soma signal intensity compared to NF-H+ DRG soma size and found that the NMP predominantly localized to small- to medium-size (<600 μm2) neurons, which constitute the majority of C-type, Aδ, and Aβ nociceptors and a small portion of Aδ and Aβ LTMRs (Figure S4K).79–82 To further validate our NMP antibody labeling of nociceptive and LTMR neurons, we performed similar experiments measuring surface localization and that of the P2X3 receptor. P2X3, a purinoreceptor, is a non-selective ion channel specifically expressed in peripheral nociceptive neurons including those which are NF-H+.83–85 We performed dual antibody feeding experiments to label NMP and P2X3 receptor using a validated antibody that recognizes the P2X3 extracellular domain.86,87 Images were taken in areas that contain at least one P2X3+/NMP+ neuron (Figures 4D and S4L). Note that, despite significant P2X3+/NMP+ overlapping cells, we did observe multiple examples of images that had NMP signal that did not overlap with P2X3 signal.
These results revealed that NMPs localize to a subset of somatosensory DRG neurons and were most enriched, but not exclusively located, in a portion of small- to medium-size nociceptors and LTMRs. Given that we observed an active NMP in DRG neuronal cultures and that in vivo we observed inhibition of NMP activity and reduced sensitivity of the neurons to pain or mechanical touch, we hypothesized that the active NMP might function to alter intrinsic responsiveness of these neuron populations to stimulation.42,43,88 However, based on these findings, it was not immediately clear to us how the NMP may be functioning. There were three possibilities: (1) the NMP was working to cell-autonomously regulate the function of NMP-expressing neurons; (2) the NMP was working to non-cell-autonomously regulate the function of neighboring non-NMP-expressing neurons; or (3) a combination of the two. To begin to address this line of questioning, we tested whether inhibition of the NMP changes DRG neuronal response to stimulation.
The NMP modulates DRG neuron sensitivity to depolarization- and P2X3-mediated stimulation
Previous descriptions in the CNS indicate that the NMP localizes to the plasma membrane in a manner that allows for the regulation of calcium-dependent neuronal signaling.29,30 To investigate NMPs in the PNS, we used calcium imaging to measure cellular responsiveness under conditions where the NMP was selectively inhibited using BE. DRG neuronal responsiveness was assayed using the following paradigm: three initial sequential stimulations with 15 mM KCl with 2-min washout in between, followed by a 10-min treatment with vehicle or 10 μM BE, then four sequential 15-mM KCl and one final 25-mM KCl stimulation (Figure 5A). For each neuron, the change in fluorescence intensity from the baseline to the peak of the response curve was used to calculate the overall response. The response to 15 mM KCl in stimulations 4, 5, and 6 were similar across stimulations; therefore, for ease of presentation and understanding we focused on the last 15-mM KCl and the 25-mM KCl stimulation results. These experiments revealed that inhibition of the NMP under basal conditions results in a reduction in peak response to depolarization (Figures 5B and 5C). We then addressed whether the attenuation in neuronal response to stimulation by inhibition of the NMP was differential among DRG neuronal subtypes. To do so, we classified the neuronal responses according to soma size as a surrogate for sensory neuronal subtype.42,43,76,77,79–82 Classifying the overall peak response to 15 mM KCl and 25 mM KCl revealed that neurons in the small-size (soma area <300 μm2) and medium-size (soma area 300–600 μm2) groups, which are enriched for nociceptors and purinoceptors, were the most affected, as the peak responses to stimulation in these groups were significantly reduced after NMP inhibition (Figure 5D).
Figure 5. Inhibition of PNS NMP function reduces depolarization and P2X3-signaling-evoked neuronal responses.
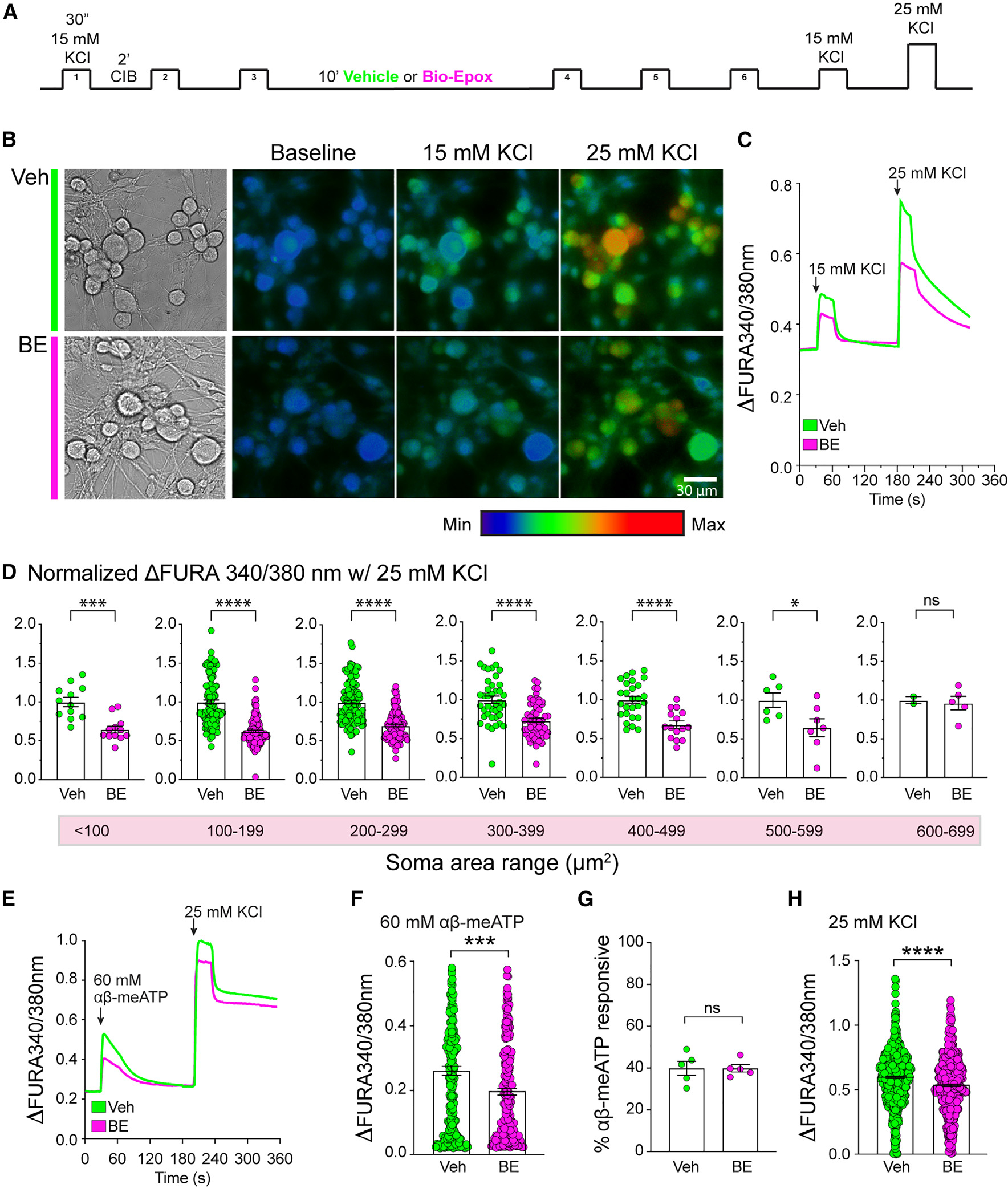
(A) Schematic representation of experimental paradigm.
(B) Representative images of DRG neuron calcium imaging.
(C) Representative calcium response traces of DRGs in culture showing KCl stimulations in the presence of vehicle (Veh) or 10 μM BE.
(D) Normalized peak response to 25 mM KCl in different-sized neuron groups. Data are presented as mean ± SEM (n = 4). *p < 0.05, ***p < 0.001, ****p < 0.0001 (Welch’s t test).
(E) Representative calcium traces of DRGs in culture showing response to αβ-meATP followed by 25 mM KCl stimulation in the presence of Veh or 10 μM BE. (F) Quantification of DRG peak calcium response to αβ-meATP in the presence of Veh or 10 μM BE. Data are presented as mean ± SEM (n = 5). ***p < 0.001 (Welch’s t test).
(G) Quantification of the proportion of αβ-meATP-responsive DRG neurons. Data are presented as mean ± SEM (n = 5). Statistically significant differences between samples were not observed (Welch’s t test).
(H) Quantification of DRG peak calcium response to 25 mM KCl following αβ-meATP treatment. Data are presented as mean ± SEM (n = 5). ****p < 0.0001 (Welch’s t test).
See also Figure S5.
The P2X3 purinoreceptor is specifically expressed in peripheral nociceptive neurons83–85 that are activated by α,β-methylene-ATP (αβ-meATP) and can modulate pain and innocuous warmth sensation.89–92 Recent studies have found a link between the use of proteasome inhibitors and the attenuation of P2X3 signaling in PNS sensory neurons.11 However, the mechanisms of this attenuation are not understood. We therefore used calcium imaging to explore the effects of inhibiting the NMP on P2X3 signaling in DRG neurons. In these experiments, we incubated the neurons for 10 min with vehicle or 10 μM BE and then stimulated them with 60 μM αβ-meATP, a highly selective P2X3 receptor agonist, for 30 s followed by a 2-min washout.91 The cells were then stimulated with 25 mM KCl for 30 s. Calcium imaging response curves and quantification revealed that inhibition of the NMP results in a reduction in P2X3 receptor signaling peak response (Figures 5E and 5F). We observed no difference in the proportion of αβ-meATP-responsive cells (~40%) between vehicle and BE treatment (Figure 5G). All cells, whether αβ-meATP-responsive or non-responsive, showed a reduction in peak response to 25 mM KCl (Figures 5H and S5A). Based on these data, NMPs in the PNS appeared to regulate DRG nociceptor responsiveness to both depolarization and αβ-meATP-induced stimulation.
The PNS NMP modulates sensory neuron activity in a non-cell-autonomous manner
Unlike neurons in the CNS, the PNS DRG sensory neurons do not form synapses to communicate with one another,43,93 so interactions between neurons could be mediated via release of diffusible molecules and/or interactions with other cells. This postulates that proximity between neurons in the PNS is important for signaling molecule-dependent paracrine communication. In the hindlimb paw, evidence supports that there are ~500 nerve endings per mm2.56,58 In our densely populated neuronal cultures, we have a density of ~250 neurons/mm2 (Figure S5B). Consistent with NMP function in a non-cell-autonomous manner, we found that reducing this density to ~50 neurons/mm2 (Figure S5B) and, thus, limiting possible interactions between neighboring neurons results in a reduction in stimulation-induced calcium signaling that was not further reduced upon inhibition of NMP (Figure S5C). We noted that BE addition to densely populated neuronal cultures have reduced activity, which was similar to the activity of sparsely populated neurons without BE (Figure S5C).
Based on these data and our antibody feeding experiments (Figures 4 and S4), we hypothesized that the NMP was expressed in a subset of DRG neurons and modulates the response to stimulation of neighboring non-NMP-expressing DRG neurons in a non-cell-autonomous manner. To test this hypothesis, we sought to identify the pool of NMP-expressing somatosensory DRG neurons.
Single-cell RNA sequencing reveals the NMP-expressing DRG neuronal populations
Morphological, physiological, and molecular characterization of somatosensory neurons have allowed their classification into several distinct types of neurons that are highly specific and sensitive to selective environmental inputs.94–98 Our functional and cellular characterization of PNS NMP-expressing neurons indicated that this complex was only found on a subset of DRG neurons and that it appeared to have a specific function related to mechanical and pain sensation. To acquire a deeper understanding of the mechanisms that govern the function of the PNS NMP, we aimed to identify the NMP-expressing somatosensory neurons. For this, we dissected DRGs from all axial levels from wild-type mice. Once the cells were dissociated into single-cell suspension, we fluorescently labeled the NMP by antibody feeding approaches. This allowed the identification of NMP-positive and NMP-negative cells using fluorescence-activated cell sorting (Figures S6A and S6B). scRNA-seq analysis was then performed on these isolated cell populations. Clustering and visualization of the transcriptomes by uniform manifold approximation and projection (UMAP) analysis revealed 20 transcriptionally distinct clusters (Figure 6A). Interestingly, cells in the NMP-positive group clustered only to 3 of the 20 clusters (Figure 6A). We then used previously identified marker gene sets from the most current published scRNA-seq analyses to identify the transcriptionally distinct neuronal subtypes.94,95 Our analysis identified 13 known somatosensory neuronal subtypes mapping to 13 distinct clusters (Figures 6B and 6C). Differential gene expression analysis identified the top ten genes that were selectively expressed in each cluster, which are presented in a heatmap (Figure 6D and Table S1). Using known gene markers of different sensory neuron types,94–97 we found that the NMP-positive clusters were selectively identified by MrgprA3 and Cysltr2, which corresponded to CGRP-θ and SST neuronal subtypes, respectively (Figures 6A–6C). We were not able to detect distinct gene sets to label the third NMP-positive cluster as a specific neuronal subtype, and it was thus left as unassigned.
Figure 6. Transcriptional characterization of NMP-positive DRG neurons.
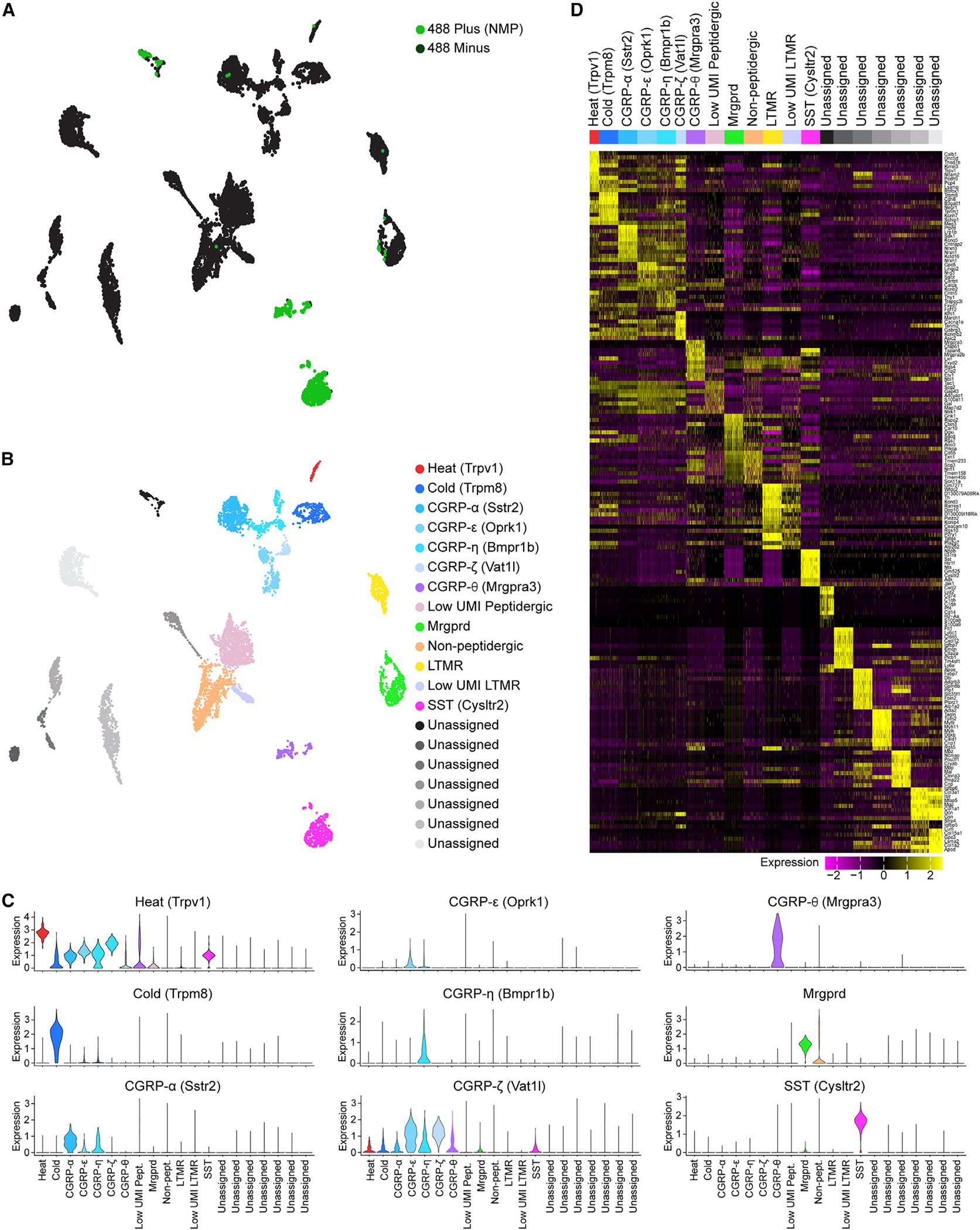
(A) UMAP visualization of scRNA-seq analysis of NMP-positive (green) and NMP-negative (black) neurons showing cell clusters of putative different DRG neuronal subtypes.
(B) UMAP showing identification of 13 cell clusters as distinct sensory neuronal subtypes and 7 unassigned clusters using previously defined gene sets.94,95 UMAP legend shows the marker gene that can be used to identify the different sensory neuronal subtype clusters.
(C) Violin plots showing expression profile of the marker genes that identify the different neuronal subtypes.
(D) Heatmap showing up to ten differentially expressed genes that specifically map to each cluster and can be used to identify the different sensory neuronal subtypes. Differential expression was performed by comparing each cluster to all other cells in the dataset using a Mann-Whitney U test. Genes were selected by fold change. Expression scale is shown below the heatmap. A random selection of up to 200 cells was selected in clusters with more than 200 cells to allow for visualization of gene expression patterns in smaller clusters.
In addition, we performed differential gene expression analysis to identify genes that were unique to the peptidergic MrgprA3+ and Cysltr2+ neurons versus other somatosensory neurons that were NMP negative (Figure S6C; Tables S2 and S3). We then compared the two lists and identified genes that were unique to both types of NMP-positive neurons (Figure S6D and Table S4). While this analysis identified several genes unique to the NMP-expressing cells, further studies are needed to tease apart the foundation of why MrgprA3+ and Cysltr2+ sensory neurons selectively express the NMP.
Pruritogen stimulation mediates NMP-dependent inhibition of non-pruritogen-responsive neurons
MrgprA3+ and Cysltr2+ subtypes have been found to innervate the glabrous skin of the paw.99–101 Classically, activation of MrgprA3+ and Cysltr2+ neurons has been shown to mediate itch sensation through a variety of pruritogens.100,102–107 In addition, these populations of DRG somatosensory neurons have low mechanosensitivity compared to other somatosensory neuronal subtypes.94 Here, we found that inhibition of NMP in the paw was sufficient to cause a decrease in mechanosensation and pain (Figure 3). Taken together, these data indicated that NMP expressed on MrgprA3+ and Cysltr2+ DRGs may act to promote enhanced sensitivity to mechanical and pain sensation through potentially more sensitive neighboring neurons. For this to function in this manner, inhibition of the NMP on pruritogen-responsive neurons (such as MrgprA3+ and Cysltr2+) would likely be able to modulate the activity of neighboring cells that are not responsive to pruritogens. This was a challenging hypothesis to test, as we did not know how to reduce NMP function in just these MrgprA3+ and Cysltr2+ neuronal subtypes. However, we considered that the selective activation of these cells using the pruritogen histamine in the presence and absence of NMP inhibitor may shed some light on this line of thinking. To test this idea, we incubated the neurons for 10 min with vehicle or 10 μM BE and then stimulated them with 8 mM histamine for 30 s followed by a 2-min washout and a subsequent 25-mM KCl stimulation for 30 s with a 2-min washout. Consistent with previous reports, we found that histamine activates a subpopulation of cells (Figure 7A). After the histamine stimulation, both histamine-responsive and cells non-responsive to histamine were activated by KCl (Figure 7B). We found that the addition of NMP inhibitor led to no change in the histamine responsiveness in the dish (Figure 7C). When combined with histamine stimulation, however, the inhibition of NMP led to a significant increase in subsequent KCl depolarization in both histamine-responsive and -non-responsive cells.
Figure 7. Pruritogen stimulation mediates NMP-dependent inhibition of pruritogen-responsive and pruritogen-non-responsive neurons.
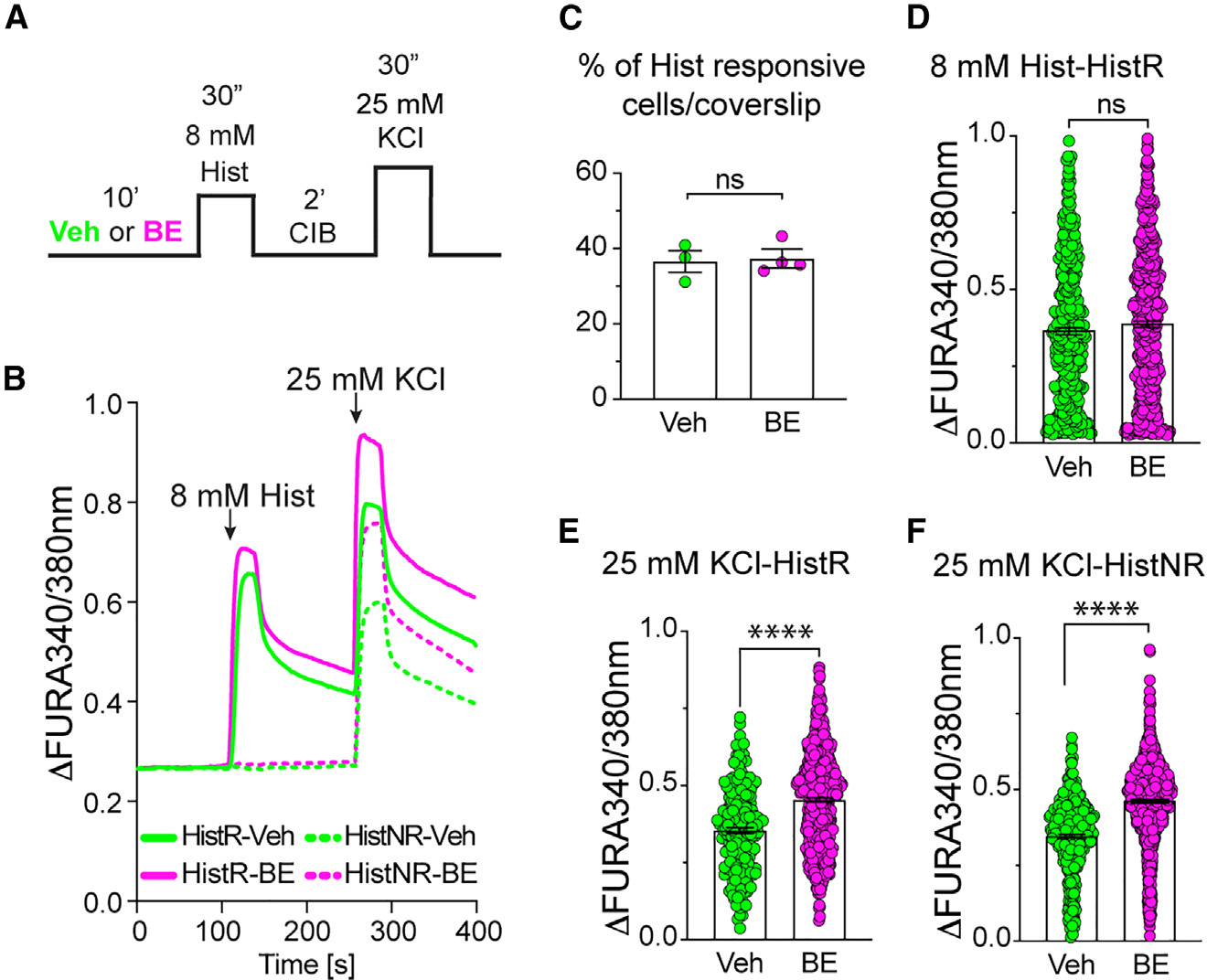
(A) Schematic representation of experimental paradigm. (B) Representative calcium response traces of DRGs in culture showing response to histamine (Hist) followed by KCl stimulation in the presence of vehicle (Veh) or 10 μM BE. Traces show both histamine-responsive neurons (HistR, solid line) and histamine-non-responsive neurons (HisNR, dashed line).
(C) Quantification of the percentage of histamine-responsive (HisR) neurons in the presence of Veh or 10 μM BE. Data are presented as mean ± SEM (n = 4). No statistically significant differences were observed (Welch’s t test).
(D) Quantification of DRG peak calcium response to histamine for HisR neurons in the presence of Veh or 10 μM BE. Data are presented as mean ± SEM (n = 4). No statistically significant differences were observed (Welch’s t test).
(E and F) Quantification of DRG peak calcium response to KCl stimulation for both HisR (E) and HisNR (F) neurons in the presence of Veh or 10 μM BE. Data are presented as mean ± SEM (n = 4). ****p < 0.0001 (Welch’s t test).
Based on these observations, we concluded that the NMP can function to suppress neuronal response to stimulation across different types of sensory neurons (responsive and non-responsive to histamine) following pruritogen-induced stimulation. These findings were distinct from our observations with KCl and αβ-meATP and suggest a context-dependent role for NMP in promoting or inhibiting DRG neuron response to stimulation. Moreover, these data support our hypothesis that NMP-expressing somatosensory neurons can modulate NMP non-expressing neurons and the idea that the NMP mediates neuronal “crosstalk” important for PNS function.
DISCUSSION
In this study, we revealed the presence and function of NMPs in the PNS. Anatomical and transcriptomic analysis showed that the NMP was not found on all DRG neurons, but rather was enriched almost exclusively (>95%) in two transcriptionally distinct populations of DRG neurons; MrgprA3+ and Cysltr2+. MrgprA3+ and Cysltr2+ neurons are C-type nociceptors that overlap with IB4, CGRP, and NF-H,94 are sensitive to mechanical and heat stimulation,94 respond to pruritogens such as histamine,103,104,106,108,109 and have been found to innervate the paw skin.99–101 Interestingly, we found that histamine pre-treatment of DRG neurons resulted in NMP-mediated suppression of response to KCl stimulation in all neurons (i.e., histamine-responsive and histamine-non-responsive). These data support the idea that the PNS NMP functions to mediate cell-autonomous and non-cell-autonomous somatosensory neuron “crosstalk” that modulates context-dependent response to stimulation (Figure S7). We believe that the PNS may have evolved to use the NMP to discriminate between distinct sensory modalities and that this is necessary for an animal’s behavioral response to diverse environmental sensory stimuli.
Decades of studies aimed at understanding the mechanisms that control peripheral nociceptor activity have revealed that PNS sensory neurons are able to communicate with one another110–115 and that this “crosstalk,” to some extent, mediates a heightened sensitivity to stimuli resulting in neuropathic pain.110,112,114 For example, in one of these studies, the authors demonstrated neuronal electrical coupling following nerve injury in neurons that are in close proximity within a DRG.114 This electrical coupling of DRG neurons results in mechanical pain hypersensitivity and is mediated, in part, through the upregulation of gap junctions between satellite glial cells surrounding DRG neuronal soma.114 Other studies have shown that the depolarization of axons from a group of neurons leads to depolarization coupling of nearby neurons in the same DRG. A prevailing hypothesis is that this “crosstalk” is mediated through the release and detection of diffusible molecules from the somata or axons of sensory neurons.113,115 While these studies were focused on the DRG in the spinal cord, whether similar mechanisms exist in the skin where the fibers are more distant from one another remains to be determined. However, our in vivo data using BE injection in the hindlimb paw indicate this possibility.
Diffusible molecules that mediate communication between somatosensory neurons could include PNS NMP-derived signaling peptides. Indeed, previous studies have shown that CNS neurons express the NMP, which generates extracellular bioactive peptides capable of signaling to nearby neurons.30,31 PNS NMP-derived signaling peptides could mediate cell-autonomous and non-cell-autonomous stimulation-dependent regulation through interaction with somatosensory neuron receptors. For example, the NMP-derived peptides could directly or indirectly regulate P2X3 sensitivity to its agonist αβ-meATP via binding to the channel’s extracellular domain or its modulators. The P2X3 receptor is a large transmembrane complex that has most of its structure exposed to the extracellular space and contains three ATP-binding sites and several putative binding sites for molecules other than ATP.116–118 In addition, while we did not investigate this in our study, NMP-derived peptides could regulate the activity of non-neuronal cells. Several studies have demonstrated that peripheral nociceptors communicate with other cells, such as immune cells, to regulate pain sensitivity.119,120 Future work is needed to delineate the exact pathway mediating NMP-dependent “crosstalk” between the diverse populations of somatosensory neurons in the PNS.
In the PNS, a diverse array of somatosensory neurons innervate peripheral organs, such as the paw skin, and carry sensory information to the neuronal somata located in the DRG and the spinal cord. The use of single-cell transcriptomics, expression atlases, and fluorescently labeled mouse lines has led to the identification and characterization of many distinct somatosensory neuronal types.97,98,121–123 For example, unbiased scRNA-seq analyses identified 18–26 different neuron types in mice.94,95,97,98 Adding to the complexity of sensory neuron diversity, many neuronal markers vary in expression in neurons within and across groups. For instance, in mice transient receptor potential vanilloid 1 (TRPV1), a non-selective ion channel that is activated by capsaicin and heat, is expressed differentially in non-peptidergic neuron type 2 (NP2), NP3, and peptidergic neuron type 1.98,121,122 Similarly, the purinoreceptor P2X3, a non-selective ion channel activated by ATP that is involved in mediating pain sensation, is highly expressed in NP1 but has low expression in NP2 and NP3 neurons.98,122 Such differential molecular profiles among sensory neurons are correlated with different patterns of physiological responsiveness and function. Consistent with this notion, we observed that the NMP was also differentially localized among sensory neuronal subtypes (MrgprA3+ and Cysltr2+). Interestingly, while the NMP mainly localized to MrgprA3+ and Cysltr2+ neurons, its function appeared to be necessary to modulate the sensitivity of all neuronal subtypes (NMP-expressing and non-NMP-expressing). For example, in our data we observed that inhibition of the NMP, which localized to MrgprA3+ and Cysltr2+ neurons that have been shown to be sensitive to punctate mechanical forces between 20 and 40 mN, led to attenuation in light punctate mechanical sensitivity, which is known to be mediated by Mrgprd+ (non-NMP-expressing, this study) neurons that exhibit force thresholds between 5 and 20 mN.94 Thus, we believe that NMP-expressing neurons have the capacity to regulate neighboring neurons in a manner that affects their response to incoming stimuli. Based on this idea, we believe, that, in the case of punctate mechanical stimulation, neurons with low-threshold activation can be regulated in a positive manner by these higher-threshold mechanically sensitive somatosensory neuron subtypes through the NMP in the periphery near nerve bundles (in dermis) or at nerve terminals (in epidermis). This “crosstalk” between neurons associated with distinct sensory modalities likely plays a role important for PNS function. Moreover, depending on the context, NMP-mediated regulation of neuronal response to stimulation is different. For example, under basal conditions, the NMP functions to enhance the sensitivity of neurons to depolarization, while pre-treatment with histamine switches the function of the NMP to be restrictive to subsequent depolarization stimulations. These results could be explained by the fact that different somatosensory neuronal subtypes are transcriptionally distinct and express a unique proteome depending on the type of incoming stimuli and the neuron’s relative state of stimulation. Given that the proteome of a cell is the source of the NMP-derived peptides, there is a direct relationship between incoming diverse stimuli and NMP-dependent control of peripheral neurons’ response that is relevant to animal sensory behaviors.
Pain and itch are distinct unpleasant sensations that share common anatomical activation pathways.100,124,125 Both pain and itch are initiated by activation of primary afferent neurons in the periphery by direct thermal, mechanical, or chemical stimuli and influence one another.100,126–128 Pain is typically initiated through the activation of specific receptors on nociceptive neurons, either via direct mechanical or thermal stimulation or following exposure to exogenous or endogenously released chemical signals from nearby tissues in the setting of damaging or potentially damaging conditions.129 Itch is typically initiated through a response to mechanical or chemical irritants.126,130 At the level of the spinal cord, pain can inhibit itch through inhibitory interneuron activation.102 In contrast, it has been reported that the primary afferent neurons, such as MrgprA3+, can generate a painful or itch response by discriminating between the incoming stimuli at the nerve terminals.100 While this multi-modality feature of C fibers can be driven by specific receptor activation on these cells, the resulting behavioral sensory response might be non-cell autonomous. If so, one possible mechanism for the modulation could involve the NMP-regulated pathways described in this study.
Proteasomes have been shown to be associated with pain.7,14–17 The use of proteasome inhibitors in human patients has been shown to be highly effective for the treatment of various human cancers.14,131–134 A key feature of the currently available proteasome inhibitors used for the treatment of cancers is their ease in crossing the plasma membrane and reaching all proteasomes in the cell.12,135 However, despite success in treating cancer, a dose-limiting side effect in many patients treated with cell-permeable pan-proteasome inhibitors is the development of PNS neuropathology that leads to neuropathic pain and degeneration of sensory nerve endings.7,14–17 Interestingly, other studies using pan-proteasome inhibitors, such as MG132 and epoxomicin, have reported that administration of low doses of these agents suppress mechanically and thermally evoked pain in animal models without inducing cellular damage or degeneration.11,18–21 While the mechanisms behind these observations remain unclear, they imply alternative, non-pathological roles for proteasomes in regulating pain. The differential neurological effects of high versus low concentrations of proteasome inhibitors lead us to hypothesize that low concentrations of proteasome inhibitors might predominantly target membrane-localized proteasomes as opposed to intracellular proteasomes, thereby producing differential analgesic versus neuropathic outcomes. Consistent with this idea, our findings reveal that local subcutaneous administration of BE to selectively inhibit NMPs results in reduced mechanical sensitivity without inducing acute nerve-ending degeneration. Thus, our results provide evidence for the presence of a PNS NMP with a tonic function in modulating sensory neuron activity relevant to pain. This may present the PNS NMP as a therapeutic target for pain control.
It will be important in future efforts to further understand the cell-type-specific function of the NMP, the nature and sequence of NMP-derived bioactive peptides, and the critical NMP functions related to modulating the multi-modality of somatosensory neuronal subtypes and their relevance to animals’ sensory behavioral response to diverse stimuli.
Limitations of the study
In our study, we present data demonstrating the presence of the NMP in the PNS and the role it plays in modulating somatosensory neuron activity. A technological limitation that prevents cell-specific analysis of the NMP function is the lack of tools to knock out or knock down the NMP. Instead we rely on the use of the selective NMP inhibitor BE. The need to exogenously administer NMP inhibitors leaves questions about the in vivo function of the NMP. NMP-deletion mouse models will allow for cell-specific electrophysiology and in vivo imaging. In addition, we turned to traditional reflexive behavioral analysis to elucidate the effects of the NMP on somatosensory behaviors. Future studies will require the use of established pain mouse models to investigate the in vivo role of the NMP on pain modulation. Moreover, translation potential of the NMP was not examined, thus leaving the question of whether the NMP is a molecular target for controlling pain.
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Seth S. Margolis (smargol7@jhmi.edu).
Materials availability
All materials generated in this study are available from the lead contact upon request.
Data and code availability
The scRNA-seq dataset generated in this study has been deposited to the Gene Expression Omnibus (GEO: GSE261727). Other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
All animal procedures were performed according to protocols approved by the IACUC of The Johns Hopkins University School of Medicine. For all experiments, we used wild-type male C57/Bl6 mice (Stock number 027; RRID: IMSR_CRL:27). All mice were raised in standard light/dark cycle with free access to food and water.
Cell culture models
Primary DRG cultures
For primary DRG cultures, 21 days-old male mice were obtained from Charles River Laboratories. On the day of culture, mice were euthanized with carbon-dioxide-induced anoxia and decapitated as a secondary method of euthanasia. The entire spinal column was dissected and cleaned out from any muscle and fat tissue on the dorsal side. The spinal column was then transferred to a dissecting dish containing ice-cold Hanks Balanced Salt Solution (HBSS). HBSS consisted of 5 mM HEPES, and 1.8 mg/mL glucose. Under a dissecting scope, all dorsal root ganglion (DRG) neurons were dissected from 2 to 4 mice, combined, and placed in HBSS solution on ice. Once all dissections were finished, the DRGs were collected and transferred to Complete Saline Solution (CSS) (137 mM NaCl, 5.3 mM KCl, 1 mM MgCl2-6H2O, 25 mM Sorbitol, 10 mM HEPES, and 3 mM CaCl2-2H2O, pH 7.2). The DRGs were digested for 20 min at 37°C with 0.35 U/ml of Liberase TM followed by 15 min at 37°C in 0.25 U/ml Liberase TM and 30 U/ml papain in CSS containing 0.5 mM EDTA. The DRGs were then triturated using a fire-polished Pasteur pipette and resuspended in DMEM containing 1 mg/mL trypsin inhibitor and 1 mg/mL bovine serum albumin. Dissociated cells were plated on plates or coverslips coated with poly-L-lysine/laminin and incubated in DMEM/F12 with 10% fetal calf serum, 1% penicillin/streptomycin, and 0.1 μg/mL nerve growth factor and B27 supplement. Cells were maintained in a humidified incubator at 37°C/5% CO2. 50% media changes were performed every 3 days. On days in vitro (DIV) 2 the media was supplemented with 10 μM 5-Fluoro-2′-deoxyuridine to prevent glial cell proliferation. DRG cultures were used for experiments on DIV 6.
METHOD DETAILS
Antibodies
The following antibodies were used according to manufacturer’s and/or published suggestions: anti-β2 (1:200, Santa Cruz), anti-β5 (1:200, Santa Cruz), anti-α2 (1:200, Santa Cruz), anti-β2 (1:500, Novus), anti-β5 (1:500, Enzo), anti-α1–7 (1:1000, Enzo), anti-β-Actin (1:2000, Abcam), anti-NF-H (1:10000, Millipore Sigma), anti-βIII-tubulin (1:5000, Millipore Sigma), IB4-biotin (1:500, Millipore Sigma), anti-CGRP (1:500 Immunostar), anti Na+/K+-ATPase (1:1000, Millipore Sigma), anti-Transferrin receptor, (1:1000, Thermo Fisher Scientific), anti-PGP9.5 (1:50 Thermo Fisher Scientific), anti-GFAP (1:500, Thermo Fisher Scientific), anti-Biotin (1:1000, Cell Signaling), anti-rabbit IgG-HRP (1:4000, Cell Signaling), anti-mouse IgG-HRP (1:5000, Cell Signaling), anti-goat IgG-HRP (1:2000, Promega), anti-rabbit-IgG-HRP (1:4000, Cell Signaling), anti-chicken IgY-HRP (1:5000, Thermo Fisher).
Immunoblot analysis
Fresh tissue or cultured cells were lysed in cold RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% Sodium Deoxycholate, 0.1% SDS, 5 mM EDTA, complete protease inhibitor cocktail tablet (Roche), 1 mM sodium orthovanadate, 1 mM b-glycerophosphate). Samples were spun down and Laemmli SDS sample buffer was added. Samples were boiled for 5 min and resolved on 12% Tris/Glycine SDS-PAGE gels made fresh. Proteins were transferred to nitrocellulose membranes at 60 mA for 12 h in 20% methanol tris/glycine-based transfer buffer. The membranes were then blocked for 1 h at room temperature with 5% bovine serum albumin (BSA) in 1x tris-buffered saline (TBS) plus 0.1% Tween 20 (TBST). All antibodies were diluted in 5% BSA, 0.05% TBST buffer made fresh. Membranes were incubated in primary antibody solution overnight at 4°C and washed 3x (5 min/wash) with 0.05% TBST. membranes were incubated in corresponding HRP-conjugated secondary antibodies diluted in 5% BSA, 0.05% TBST at room temp for 1–2 h and washed at least 5X with 0.05% TBST at room temp for 5 min each wash. Western blots were then incubated with ECL and exposed to film in a dark room. The films were then scanned to obtain a digital image and analyzed using ImageJ.
Immunoelectron microscopy
Processing
For immunoelectron microscopy, 1-month-old C57/Bl6 male mice were euthanized with CO2. Animals were perfused with freshly made EM grade 4% paraformaldehyde (PFA) 1% glutaraldehyde 50 mM phosphate buffer (Sorenson’s), 50 mM sodium cacodylate, 5 mM MgCl2 pH 7.4 at 4°C. Cell culture plates were fixed with the same fixative minus the paraformaldehyde. Animals were perfused at a rate of 2 mL/min for a total of 30 min. The lumbar 3–5 region of the spinal cord containing the corresponding DRGs was dissected out. Then, the mouse tissues and culture plates were placed in pH 8.5 perfusate overnight with slow rocking.37 All subsequent steps were done at 4°C until 70% ethanol dehydration. Spinal cord regions and DRGs were carefully dissected in fixative measuring no more than 2 mm3. All samples were rinsed in buffer containing 75 mM PO4 buffer, 75 mM cacodylate, 3% sucrose, 5 mM MgCl2 pH 7.4 (428 mOsmols, 3 × 15 min). Only tissue was then microwaved 2x (fume hood vented, Pelco 3400 laboratory microwave) in secondary fixative. This was performed in 1.5% potassium ferrocyanide reduced 0.75% osmium tetroxide in rinsing buffer without sucrose, containing 5 mM MgCl2. To ensure adequate osmication, cell cultures were osmicated at 4°C without microwave assistance. Samples were placed in opened 5 mL vials containing 1.5 mL of reduced osmium in an ice tray. The ice level was matched to the fixative level in the vial. Two 500 mL beakers with D-H2O were placed on each side of the ice tray to serve as heat traps. Samples were pulsed for 10 s at 50% power, rested for 20 s, and then pulsed again for 10 s. Vials were then held with fixative for 5 min, and then the microwave procedure was repeated. After the second 5-min pause, samples were then osmicated for 2 h at 4°C. They were then rinsed in 100 mM maleate buffer (3 × 5 min) containing 3% sucrose, then en-bloc stained with 2% filtered uranyl acetate in the same buffer for 1 h. Samples were dehydrated at 4°C up to 70% ethanol, then they were brought to room temp and further dehydrated to 100% ethanol. Spinal cords were embedded with Eponate 12 after a brief propylene oxide transition and finally cured in a 60°C oven for two days. Cell plates were dehydrated and infiltrated similarly except the propylene oxide step was omitted. Plates were cured for 3 days at 37°C then overnight at 60°C for final curing.
Labeling
70–80 nm ultra-thin sections were picked up on formvar-coated 200 mesh nickel grids. Sections were floated on all subsequent steps. All solutions were filtered except for antibodies which were centrifuged at 13K for 5 min. Grids were placed on (3%) sodium meta periodate (aq) for 30 min.136 After a 3 × 5 min rinse in D-H2O, grids were floated on 10 mM Citrate buffer pH 6.2 for 20 min at 95°C. Grids were floated on buffer in a porcelain staining dish, covered with a glass chamber, and placed on a pre-set hot plate. After 20 min cool down, grids were rinsed in tris-buffered saline (TBS) 3 × 1 min, then placed on 100 mM glycine in TBS for 10 min, followed by 30 min block in 1% NGS +1% BSA in TBS blocking solution. Other blocking solutions used were 1% NGS for goat secondary antibodies. Antibody incubation was done at appropriate dilutions (10x more than fluorescent dilution) in the same block. No primary antibody, blocking solution only, served as negative controls. Incubations were carried out at 4°C overnight. After 1 h to equilibrate to room temperature, grids were placed on blocking solution for 10 min, followed by a 1 min rinse in TBS. Gold conjugated secondary antibodies were diluted 1:40 in TBS and sections were incubated for 2 h at room temperature in a humidity chamber (i.e., for a mouse-derived primary antibody, a 6 nm gold conjugated goat anti-mouse antibody was used, for a rabbit-derived primary, a 6 nm gold conjugated goat anti-rabbit was used). After a 10-min TBS incubation followed by a quick D-H2O rinse, grids were hard fixed in 2% glutaraldehyde in 100 mM sodium cacodylate buffer pH 7.4 for 5 min. After a brief D-H2O rinse, grids were stained with 2% uranyl acetate (aq.) for 20 min, rinsed again with D-H2O, aspirated, and then blot-dried and placed in grid boxes overnight before viewing.137 Immuno-EM grids were imaged using a Hitachi 7600 transmission electron microscope and imaged using an XR80 high-resolution high-speed (8 Mpixel, 16-bit) camera at 100000X to 150000× magnifications.
Surface biotin-labeling, neutravidin pulldown and western blotting
Surface biotinylation and neutravidin pulldown experiments were done as previously described.30 All steps were done on ice. Briefly, all tissue samples were collected and kept on ice (plates with cultured cells were placed on ice). Mouse tissues (100–300 mg) were chopped using a blade, placed in an Eppendorf tube and 250 μL of Dulbecco’s Phosphate-Buffered Saline with magnesium and calcium (DPBSMC) pH 7.4 was added. For culture plates, the media was removed and replaced with 1 mL of DPBSMC. The samples were then treated by adding equal volumes (250 μL or 1 mL) of 1.5 mg/mL Sulfo-NHS-SS-biotin dissolved in DPBSCM and incubated on light shaker for 12 min. Then the liquid was removed and replaced with 250 μL or 1 mL of DPBSMC, and 250 μL or 1 mL of 100 mM glycine was added to quench the reaction and allowed to incubate for 10 min with light shaking. Tissues were then collected by spinning down the tubes for 3 min at 4000 rpm. Cells were scraped off the plates and placed in Eppendorf tubes, spun down, and the supernatant was removed. The tissues were then lysed using 500 μL of RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 5 mM EDTA, complete protease inhibitor cocktail tablet (Roche), 1 mM β-glycerophosphate) and sheered tissue by pipetting up and down. The samples were then clarified by spinning the tubes at 14000 rpm for 5 min, and then the supernatant was transferred to a new tube. The pellet was discarded. 25 μL from each sample was placed in a new tube and Laemmli SDS sample buffer was added. To the rest of the sample 50 μL of neutravidin-agarose beads were added and allowed to incubate overnight with light agitation. The next day the beads were collected by spinning the tubes at 5000 rpm for 3 min. The supernatant was removed and saved. The beads were then rinsed 5x using RIPA buffer, shaking for 5 min and spinning down at 5000 rpm for 3 min in between washes. The washing eluate was discarded. Washed beads were treated with Laemmli SDS sample buffer, boiled at 65°C for 5 min and loaded on 12% SDS denaturing gels and immunoblot analysis.
Activity-based probe and in-gel visualization of surface proteasome activity
For proteasome activity assays, live DIV 6 mouse dorsal root ganglion neurons were treated with 2.5 μM BODIPY(TMR)-Ahx3-L3-VS (MV151) in calcium imaging buffer for 30 min at 37°C. Cells were rinsed with PBSCM (pH 8.0, with 1 mM CaCl2 and 2 mM MgCl2; Gibco) and all proceeding steps were performed on ice. Cells were treated with 1.0 mg/mL Sulfo-NHS-SS-biotin dissolved in PBSCM and incubated at 4°C for 30 min, gently rocking every 5 min. Following treatment, cells were rinsed once with PBSCM, followed by 3 5-min washes with 100 mM glycine to quench the unreacted biotin. After quenching the reaction, cells were rinsed 3 times with PBSCM and then lysed in 500 μL of RIPA buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 5 mM EDTA, complete protease inhibitor cocktail tablet (Roche), 1 mM β-glycerophosphate). A cell scraper was used to fully detach cells, which were transferred to Eppendorf tubes. The cells where further lysed by aspirating the suspension through a 25 g needle 10 times. Samples were clarified by spinning down at 14,000 rpm for 20 min. The pellet was discarded and 50 μL of the clarified sample was saved as “input,” and was diluted in 6X SDS sample buffer. The rest of the homogenate was mixed with 40 μL of neutravidin-agarose beads and incubated at 4°C overnight with light agitation. The next day, beads were collected by spinning the tubes at 5000 rpm for 1 min. The supernatant was removed and saved. The beads were washed with RIPA buffer by incubating them for 5 min with light agitation, followed by spinning down at 5000 rpm for 1 min in between washes. The washing cycle was repeated 5 times. The eluate was discarded after each wash. The washed beads were then treated with 80 μL 2x SDS sample buffer supplemented with 50 μM DTT and incubated at room temperature for 5 min with light agitation. The samples were then boiled at 95°C for 5 min and spun at 5000 rpm for 1 min to pellet the beads. The supernatant was then removed and loaded on 12% SDS denaturing gels for in-gel fluorescent imaging and immunoblot analysis. Gels were first imaged with the Cy3/Tamra setting (λex: 532, λem: 560) on the Typhoon Variable Mode Imager to detect MV151 signal, and then transferred to nitrocellulose membranes for immunoblot analysis.
Antibody feeding
DIV 6 DRG cultures that had been grown in 12 mm round microscope cover glasses were placed in the tissue culture hood, and 50% of the media was removed and used to dilute primary antibodies against proteasome subunits (goat anti-α2, goat anti-β2, and goat anti-β5, all diluted at 1:50). All incubations and treatments were done at room temperature in the tissue culture hood. The left-over media on the cells was removed and replaced with the media mixed with antibodies and incubated for 30 min. The cells were then rinsed with DPBSMC 3x for 3 min. Secondary fluorophore-conjugated antibodies were diluted in fresh culture media, added to the cells, and incubated for 40 min. The cells were then rinsed with DPBSMC 3x for 3 min. The cells were then fixed with 4% PFA, 4% sucrose, 1X phosphate buffered saline (PBS) for 10 min followed by rinsing with PBSMC 3x for 3 min. Then, cells were permeabilized with 0.1% Triton X-100 in 1X PBS for 5 min and rinsed with PBSMC 3x. This was followed by blocking with 5% donkey serum, 5% BSA in 1x PBS for 30 min. Then the cells were incubated with primary antibodies against cytosolic cell markers (anti-NF-H (1:10000), anti-anti-CGRP (1:500), IB4, anti-GFAP (1:500)) diluted in blocking solution for 45 min and rinsed with PBSMC 3x for 3 min. Then corresponding fluorescent-labeled secondary antibodies were added diluted in blocking solution and allowed to incubate for 45 min. The cells were then rinsed 3x for 3 min and incubated with HOECHST (1:10000) in diH2O for 10–20 s and immediately rinsed by dipping into beaker with diH2O. The coverslips were then mounted on microscope slides with Fluoromount-G and allowed to dry overnight at room temp. The coverslips were then sealed with clear nail polish and stored at 4°C or used immediately for imaging.
Calcium imaging
DRG neuron cultures were prepared by combining DRGs dissected from 4 mice. Coverslips were seeded with 600–800 neurons per coverslip. On the day of calcium imaging experiments, the coverslips were taken from the incubator, and the media was removed and washed once with room temperature calcium imaging buffer (CIB) (130 mM NaCl, 3 mM KCl, 2.5 mM CaCl2, 10 mM HEPES, 1.2 mM NaHCO3, 10 mM glucose, pH 7.5, 290 mOsm adjusted with mannitol). The coverslips were then incubated for 30 min at room temperature in the dark in a solution of 2 μM Fura-2-acetoxymethyl ester diluted in CIB. The coverslips were rinsed 2x with CIB for at least 10 min. Then the coverslips were mounted on a perfusion calcium imaging chamber on an inverted fluorescent microscope (TE200, Nikon). Continuous images were captured at a rate of 1 image every 2 s with a cMOS camera (NEO, Andor) using an excitation filter wheel (Ludl) equipped with 340 and 380 nm filters. The data was acquired using NIS Elements imaging software (Nikon). Fluorescence changes are represented as the ratio of fluorescence emission upon stimulation at 340 nm to that upon stimulation at 380 nm (F340/F380). The experiments were recorded continuously while imaging at 2-s intervals under continuous CIB perfusion. which the cells.
KCL depolarization
The cells were perfused with normal CIB for 1 min, and then stimulated with three sequential stimuli of 15 mM KCl in CIB for 30 s with a recovery period of 2 min in CIB buffer between stimulations. Then the cells were incubated with vehicle (DMSO, 0.1%) or biotin-epoxomicin (BE, 10 μM) diluted in CIB buffer for 10 min. During this period, the perfusion was paused. Once the 10-min incubation was over, the perfusion was resumed. At this time, the cells were stimulated with four consecutive pulses of 15 mM KCl and a final 25 mM KCl pulse for 30 s with a recovery period of 2 min in CIB buffer between pulses. The response curve and soma area of each neuron was exported and analyzed.
αβ-meATP stimulation
Under continuous perfusion with normal CIB the cells were treated with vehicle or 10 μM BE for 10 min and then stimulated with 60 μM αβ-meATP for 30 s followed by a 2-min washout. Then, the cells were stimulated with 25 mM KCl for 30 s followed by a 2-min washout. The response curve for αβ-meATP-responsive and αβ-meATP-non-responsive neurons were exported and analyzed.
Histamine stimulation
Under continuous prefusion with normal CIB buffer the cells were treated with vehicle or 10 μM BE for 10 min and then stimulated with 8 mM histamine for 30 s followed by a 2-min washout. The cells were then stimulated with a 25 mM KCl pulse for 30 s followed by a 2-min washout. The response curves of histamine-responsive and histamine-non-responsive neurons were exported and analyzed.
Mouse hind paw injections
Local injection into the paw skin was performed using hypodermic needles and a Hamilton syringe. 1-month-old C57Bl/6 naive male mice were placed under isoflurane anesthesia (10% in oxygen in an induction chamber and then maintained with a nose cone), and the needle was inserted from the heel toward the middle of the paw just under the skin. 5 μL of vehicle (0.1% DMSO), 10 μM BE or 10 μM epoxomicin was injected always under the right foot pads. The mice were allowed to recover, and behavioral tests were performed at the following time points after injection; 30 min, 1 h, and 2 h. 12–16 mice were used per treatment group, and separate groups were used for each behavioral test. For non-recovery treatments, the mice were euthanized at each time point indicated above, and tissues was collected, including paw skin, sciatic nerves, and spinal cord.
Plantar skin antibody feeding and labeling of nerve endings
Antibody feeding
Plantar skin was harvested fresh and placed on ice immediately. The tissues were then placed in primary antibody against proteasome subunit β5 (goat anti-β5 (1:50)) diluted in blocking solution containing 5% horse serum, 5% BSA, in PBS and incubated on a shaker for 30 min at room temperature followed by 3 rinses of 5 min with PBS. Then the tissues were incubated in secondary antibody solution containing donkey anti-goat-555 antibody (1:250), 5% horse serum, 5% BSA, in PBS on a shaker for 30 min at room temperature followed by 3 rinses of 5 min with PBS. The tissues were then immersion fixed in 4% PFA, 4% sucrose in 1X PBS, pH 7.4 for 15 min at room temperature and processed for labeling of nerve endings.
Labeling of nerve endings
Paw skin from untreated, DMSO, or BE-treated animals was collected immediately after treatment and immersion fixed in 4% PFA, 4% sucrose in 1X PBS, pH 7.4 overnight at 4°C. The tissue was cryoprotected in 30% sucrose overnight at 4°C. The tissues were then embedded in Tissue-Tek O.C.T. Compound and cryosection at a thickness of 15 μm. The sections were used for free-floating immunostaining. All steps were done at room temperature on a shaker. The sections were permeabilized with 0.1% Tritton X-100 in 1X PBS, pH 7.4 while free floating for 10 min. The sections were then rinsed 3X with PBS for 5 min/wash and blocked with 5% horse serum, 5% BSA, in PBS for 1 h. Then, incubated in primary antibody solution (anti-βIII-tubulin at 1:1000, anti-NF-H at 1:1000, or anti-PGP9.5 at 1:50) diluted in blocking solution for 2 h followed by 3 rinses of 5 min with PBS. Corresponding fluorophore-conjugated secondary antibody diluted in blocking solution was added and incubated for 2 h. Finally, the sections were rinsed 3x with PBS and mounted on microscope slides using Fluoromount-G. The slides were allowed to dry overnight before imaging. Images were collected using a spinning disc confocal microscope (Leica Microsystems, Inc). Images were analyzed using ImageJ.138
Von Frey test
Mechanical allodynia was tested using Von Frey filaments by the Up-Down method.62,63,70,139,140 Twelve 1 month-old C57Bl/6 naive male mice were used per test group. The mice were placed under ventilated plexiglass boxes over a stainless-steel wire mesh and habituated for 2 h for two consecutive days before the test. On the test day, the mice were habituated for 2 h, and then a series of Von Frey filaments were applied perpendicular to the glabrous sural area of the plantar surface of the hind paws to the point of bending following the Up-Down method. The first set of measurements (pre-injection) were recorded as baseline measurements. The animals were then given a treatment injection on the right hind paw, and the Von Frey filaments were applied again at 30 min, 1 h and 2 h post-injection. The forces of each Von Frey filament used are as follows in grams: 0.04, 0.07, 0.16, 0.4, 0.6, 1, and 1.4, 2, and 4. Positive responses were defined as paw withdrawal, shaking, or flinching. The 50% withdrawal thresholds were calculated using a script provided by Christensen et al. (https://github.com/MikkelAStorm/Up.Down.Method).139 In simple terms, the higher the 50% withdrawal threshold, the higher the force necessary to elicit a response and the less sensitive the animals are. The left hind paw served as internal, untreated control to compare the responses of the treated paw (right).
Hargreaves test
To test for thermal hyperalgesia, we used the Hargreaves test. Twelve to sixteen 1-month-old C57Bl/6 naive male mice were used for this experiment per treatment. Mice were placed on a glass platform under a ventilated plexiglass box and allowed to habituate for 2 h for two consecutive days before the test. On test day, the animals were allowed to habituate for 2 h, and then a radiant heat stimulus was focused on the plantar surface of the hind paws. Next, the paw withdrawal latency was recorded. These measurements were set as baseline withdrawal thresholds. Then animals received a treatment injection, as described previously, on the right hind paw, and withdrawal latencies were measured at 30 min, 1 h, and 2 h post-injection. The intensity of the heat source (IITC Life Science, Model 336) was adjusted so that the baseline latency was 10 s across population. A cutoff time to the heat source was set at 15 s to prevent tissue damage. Each paw was tested three times with a 10-min rest period in between stimulations, and the average of the three was used for calculations. Stimulations on both hind paws were recorded. The left hind paw served as internal, untreated control to compare the responses of the treated paw (right).
Pinprick test
To test sensitivity to fast acute mechanical pain we used the pinprick test. Twelve to sixteen 1 month-old C57Bl/6 naive male mice were placed on a wire mesh, similar to that used for the Von Frey test, under a ventilated plexiglass box and allowed to habituate for 2 h for two consecutive days before testing. An Austerlitz pin was gently applied to the glabrous skin in the paw without breaking the skin. Each paw was stimulated 10 times with a 3-min rest in between stimulation. The percentage of paw withdrawal to the stimulation was calculated from the total number of stimulations. The responses of each individual mouse were normalized to itself to baseline and presented as an average per treatment group. The left hind paw served as internal, untreated control to compare the responses of the treated paw (right).
Noxious cold test
To test for sensitivity to cold painful stimuli we used the noxious cold test as previously described.68,69 For this a cohort of 12 1 month-old male C57Bl/6 mice were placed on a glass platform under a ventilated plexiglass box and allowed to habituate for 2 h on two consecutive days before the test. On the day of the test, dry ice was crushed down to a fine powder and used to pack the bore of a 3 mL syringe that had the tip cut off to be used as a cold probe. The open side of the syringe was held against the glass surface underneath the mouse paw while pressure was applied to the plunger to keep constant contact between the dry ice and the glass plate. Withdrawal was defined as movement of the paw in any direction away from the cold glass. The paw withdrawal response latency was recorded in seconds and each paw was tested 3 times with at least 5 min of rest in between stimulation. The 3 measurements were averaged to get a number for each paw per mouse. The averages per paw were normalized to themselves at baseline and then the averages per treatment group were presented. The left hind paw served as internal, untreated control to compare the responses of the treated paw (right).
FACS, single-cell RNA seq, and analysis
For single-cell RNA sequencing (scRNA-Seq) experiments, we harvested all the DRGs from six 1 month-old male mice and dissociated them as explained in the DRG culture protocols. Immediately after dissociation of DRG neurons, the NMP was labeled using a modified antibody feeding protocol. For this we incubated the dissociated cells with antibodies against the β5 proteasome subunit (goat anti-β5) diluted in DRG neuron media (1:50) for 20 min with gentle agitation at room temperature. The cells were then spun down at 800 rpm for 1 min and the media was removed. The cells were resuspended in 1x PBS and lightly agitated for 3 min. This process was repeated 3X for washing. Then, cells were incubated in secondary anti-goat-488 secondary antibody diluted in DRG neuron culture media (1:250) for 30 min at room temperature. The cells were rinsed 3X as previously described. Washed cells were resuspended in flow cytometry buffer (1X PBS, 1.0% fetal bovine serum (FBS), 25mM HEPES, and 1% penicillin/streptomycin. Fluorescence-activated cell sorting (FACS) was used to sort cells using a 100 μm nozzle gating for 488-plus (NMP positive) and 488-minus (NMP negative) neurons. Cell sorting was performed at the Flow Cytometry Core Facility using an Aria IIu Cell sorter. The two populations of cells were then immediately used for 10X Genomics single-cell RNA sequencing using a NovaSeq 6000 platform. Cells in the 488-negative group were read at a depth of 36,000 reads per cell and those in the 488-positive group at 88,000 reads per cell translating to about 2104 and 7848 genes per cell, respectively. Sequencing data was then outsourced for detailed analysis (https://www.cmcherryconsulting.com). Automatically called cells were further filtered to ensure usage of high-quality droplets with captured cells. RNA barcodes were then filtered on total UMI count (>500 UMIs), feature counts (>250 features), and percentage of mitochondrial genes (<25%). Seurat v4.3.0141 was used for handling of normalization, identification of variable genes, scaling, principal component analysis, UMAP dimensional reduction, and SNN generation followed by Leiden clustering. Batch effect correction was performed using Harmony.142 Clusters were identified using a combination of marker genes and differential expression comparing each cluster to all other cells in the dataset. Differential expression analyses were performed using Mann-Whitney U test. Fgsea v1.24.0143 was used to run gene set enrichment analysis (GSEA) with gene sets obtained from the Molecular Signatures Database.143 Features were ranked by −log(p value) * sign (foldchange). Raw data and additional information required for analysis is available from the lead contact upon request.
QUANTIFICATION AND STATISTICAL ANALYSIS
All DRG culture experiments represent at least three independent culture experiments where the cultures were obtained by dissecting and combining DRGs from four animals per culture. For immunofluorescent and calcium imaging experiments, when comparing two conditions, a two-tailed Welch t test was used. For immunofluorescent experiments when comparing two treatment groups across multiple time points a two-way ANOVA was used. For behavioral experiments when comparing treatments effects on the same paw over multiple time points, a two-way repeated measures ANOVA was used. ANOVA tests were followed by Bonferroni post hoc tests for multiple comparisons with corrections for multiple time points (where applicable). All data were expressed as mean ± SEM, and the criterion for statistical significance was a p value of <0.05. The n values represent biological repeats from independent experiments as specified in each figure legend. The specific statistical test used for each experiment is indicated in the Figure legend. All statistical analyses were carried out using GraphPad Prism 9 (Version 9.3.0 (345), GraphPad, San Diego, CA, USA).
Supplementary Material
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER (Cat#) |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse anti-β-Actin (8226) | Abcam | ab8226; RRID: AB_306371 |
| Rabbit anti-Biotin (D5A7) | Cell Signaling | 5597S; RRID: AB_10828011 |
| Goat anti-Rabbit IgG, (H + L), HRP | Cell Signaling | 7074; RRID: AB_2099233 |
| Horse anti-Mouse IgG, (H + L), HRP | Cell Signaling | 7076; RRID: AB_330924 |
| Mouse anti-α1–7 proteasome subunit | Enzo | BML-PW8195; RRID: AB_2052368 |
| Rabbit anti-β5 proteasome subunit | Enzo | BML-PW8895; RRID: AB_10540901 |
| Rabbit anti-CGRP | Immunostar | 24112; RRID: AB_572217 |
| Chicken anti-NF-H | Millipore Sigma | AB5539; RRID: AB_11212161 |
| Mouse anti-Na+/K+-ATPase | Millipore Sigma | 05–369; RRID: AB_309699 |
| Mouse anti-βIII-tubulin | Millipore Sigma | T8578; RRID: AB_1841228 |
| IB4, biotin conjugate | Millipore Sigma | L2140; RRID: AB_2313663 |
| Rabbit anti-β2 | Novus | NBP1–92294; RRID: AB_1104294 |
| Donkey anti-Goat IgG, (H + L), HRP | Promega | V8051; RRID: AB_430838 |
| Goat anti-β2 | Santa Cruz | sc-54676; RRID:AB_2172024 |
| Goat anti-β5 | Santa Cruz | sc-55009; RRID: AB_2237739 |
| Goat anti-α2 | Santa Cruz | sc-54715; RRID: AB_2171410 |
| Rabbit anti-Chicken IgY (H + L), HRP | Thermo Fisher Sci. | 31401; RRID: AB_228385 |
| Mouse anti-Transferrin receptor (H68.4) | Thermo Fisher Sci. | 136800; RRID: AB_2533029 |
| Anti-P2X3 | Thermo Fisher Sci. | PA5–77682; RRID: AB_2736292 |
| Anti-goat-488 | Thermo Fisher Sci. | A32814TR; RRID: AB_2866497 |
| Rabbit anti-PGP9.5 | Thermo Fisher Sci. | PA5–16825; RRID: AB_10986012 |
| Rabbit anti-GFAP | Thermo Fisher Sci. | PA-110019; RRID: AB_1074611 |
|
| ||
| Biological samples | ||
|
| ||
| Mouse spinal cord, DRG, and nerve | Charles River | RRID: IMSR_CRL:27 |
| Mouse skin tissue | Charles River | RRID: IMSR_CRL:27 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Precision Plus Standards All Blue | Bio-Rad | 1610393 |
| TEMED | Bio-Rad | 1610800 |
| 40% Acrylamide/Bis Solution, 37.5:1 | Bio-Rad | 1610148 |
| 30% Acrylamide/Bis Solution, 29:1 | Bio-Rad | 1610156 |
| Phosphate Buffered Saline (PBS) | Corning | 46–013-CM |
| Paraformaldehyde | EMS | 15710 |
| Horse serum | GE Lifesciences | SH30074.03 |
| Fura-2-acetoxymethyl ester | Invitrogen | F1201 |
| Tris | Millipore Sigma | 77–86-1 |
| Sodium deoxycholate | Millipore Sigma | 264103 |
| DMSO | Millipore Sigma | D8418 |
| β-methylene ATP | Millipore Sigma | 5054190001 |
| 5-Fluoro-2’ -deoxyuridine | Millipore Sigma | F0503 |
| Fetal calf serum (FCS) | Millipore Sigma | SH30071 |
| Fetal bovine serum (FBS) | Millipore Sigma | A9647 |
| Nerve growth factor (NGF) | Millipore Sigma | N0513 |
| Donkey serum | Millipore Sigma | 566460 |
| Potassium Chloride (KCI) | Millipore Sigma | P5405 |
| Magnesium Chloride (MgCI2) | Millipore Sigma | M2393 |
| Calcium chloride (CaCI2) | Millipore Sigma | C7902 |
| Sorbitol | Millipore Sigma | 56755-M |
| β-glycerophosphate | Millipore Sigma | G5422 |
| NaHCO3 | Millipore Sigma | S5761 |
| Glucose | Millipore Sigma | G7021 |
| Liberase TM | Millipore Sigma | 5401119001 |
| Liberase TL | Millipore Sigma | 5401020001 |
| Glycine | Millipore Sigma | 4840 |
| Epoxomicin | Millipore Sigma | E3652 |
| cOmplete Protease inhibitor | Millipore Sigma | 11836170001 |
| ECL Cytiva, RPN2106 | Millipore Sigma | Ref# GERPN2106 |
| Poly-L-Lysine | Millipore Sigma | P4707 |
| Adenosine triphosphate (ATP) | Millipore Sigma | A26209; CAS:34369–07-8 |
| Penicillin/streptomycin | Millipore Sigma | 15140122 |
| Trypsin inhibitor | Millipore Sigma | T9253; CAS: 9035–81-8 |
| L-cysteine | Millipore Sigma | 168149; CAS: 52–90-4 |
| Bovine Serum Albumin (BSA) | Millipore Sigma | A9647; CAS: 9048–46-8 |
| Glycerol | Millipore Sigma | G6279; CAS: 56–81-5 |
| Xylene cyanol FF | Millipore Sigma | 335940; CAS: 2650–17-1 |
| Sodium dodecyl sulfate (SDS) | Millipore Sigma | L3771; CAS: 151–21-3 |
| Ammonium persulfate (APS) | Millipore Sigma | A3678; CAS: 7727–54-0 |
| Sodium chloride (NaCl) | Millipore Sigma | S9888; CAS: 7647–14-5 |
| Tween 20 | Millipore Sigma | P2287 |
| EDTA | Millipore Sigma | E5134; CAS: 6381–92-6 |
| Sodium orthovanadate | Millipore Sigma | 567540 |
| HEPES buffer (1M), pH 7.3 | Quality Biological | 118–089-721 |
| DPBSMC | Quality Biological | 114–059-101 |
| Fluoromount-G | Southern Biotech | 0100–01 |
| Tissue-Tek O.C.T. | Sakura Finetek | 4583 |
| DMEM/F12 | Thermo Fisher Sci. | 11320033 |
| Triton X-100 | Thermo Fisher Sci. | X100 |
| B27 | Thermo Fisher Sci. | 17504044 |
| HOECHST | Thermo Fisher Sci. | H21492 |
| Hanks Balanced Salt Solution (HBSS) | Thermo Fisher Sci. | H9394; CAS: 14170112 |
| L-Glutamine | Thermo Fisher Sci. | 25030081 |
| Dithiothreitol (DTT) | Thermo Fisher Sci. | R0861 |
| DMSO | Thermo Fisher Sci. | 25–950-CQC |
| Sulfo-NHS-SS-biotin | Thermo Fisher Sci. | 21335 |
| Neutravidin agarose | Thermo Fisher Sci. | 29202 |
| DMEM | Thermo Fisher Sci. | 11995065 |
| Papain | Worthington | Cat#:9001–73-4 |
| Biotin-epoxomicin | Synthesized | In-house |
| Bodipy TMR-Ahx3L3VS (MV151) | Lab of Dr. Alexei F. Kisselev | https://doi.org/10.1016/j.chembiol.2006.09.013 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Quick Start Bradford Protein Assay | Bio-Rad | 5000201 |
|
| ||
| Deposited Data | ||
|
| ||
| scRNA-Seq | This paper | GEO: GSE261727 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| C57BL/6 | Charles River | RRID: IMSR_CRL:27 |
|
| ||
| Software and algorithms | ||
|
| ||
| ImageJ | Schindelin et al.138 | https://imagej.nih.gov/ij/ |
| GraphPad Prism | GraphPad | http://www.graphpad.com/ |
| Von Frey Up Down Method calculator | Christensen et al.139 | https://github.com/MikkelAStorm/Up.Down.Method |
| NIS Elements imaging software | Nikon | https://www.microscope.healthcare.nikon.com/ |
| Seurat | Hao et al.141 | https://github.com/satijalab/seurat |
| Harmony | Korsunsky et al.142 | https://github.com/immunogenomics/harmony |
| Fgsea v1.24.0 | Korotkevich et al.143 | https://github.com/ctlab/fgsea |
|
| ||
| Other | ||
|
| ||
| Gel Doc EZ System | Bio-Rad | 1708270EDU |
| GE Typhoon FLA 9500 | Cytiva | N/A |
| Austerlitz pin | Fine Science Tools | 000 |
| Von Frey filaments | North Coast Medical | NC12775–02 to NC12775–09 |
Highlights.
The neuronal membrane proteasome (NMP) is found in peripheral somatosensory neurons
The NMP selectively localizes to specific types of somatosensory neurons
The NMP mediates crosstalk between sensory neurons to modulate sensitivity to stimulation
Inhibition of the NMP reduces sensitivity to mechanical and painful stimuli
ACKNOWLEDGMENTS
We thank the members of the Margolis laboratory for critical reading and feedback. We thank Dr. Sang-Min Jeon for training on Von Frey assays and intraplantar paw injections. We thank Dean Zhang for his assistance with immune-EM analysis. T.R.C. was supported by NIH-NINDS NRSA F31NS134239. E.V.L. was supported by NIH-NIGMS NRSA F32NS119202 and Merkin Peripheral Neuropathy and Nerve Regeneration Center grant 22DF-C1/232. This work was funded by institutional funding and NIH grant R01 NS110754 (to S.S.M.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2024.114058.
REFERENCES
- 1.Thibaudeau TA, and Smith DM (2019). A Practical Review of Proteasome Pharmacology. Pharmacol. Rev. 71, 170–197. 10.1124/pr.117.015370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortun J, Li J, Go J, Fenstermaker A, Fletcher BS, and Notterpek L (2005). Impaired proteasome activity and accumulation of ubiquitinated substrates in a hereditary neuropathy model. J. Neurochem. 92, 1531–1541. 10.1111/j.1471-4159.2004.02987.x. [DOI] [PubMed] [Google Scholar]
- 3.VerPlank JJS, Lokireddy S, Feltri ML, Goldberg AL, and Wrabetz L (2018). Impairment of protein degradation and proteasome function in hereditary neuropathies. Glia 66, 379–395. 10.1002/glia.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cartelli D, Cavaletti G, Lauria G, and Meregalli C (2022). Ubiquitin Proteasome System and Microtubules Are Master Regulators of Central and Peripheral Nervous System Axon Degeneration. Cells 11, 1358. 10.3390/cells11081358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavaletti G, Gilardini A, Canta A, Rigamonti L, Rodriguez-Menendez V, Ceresa C, Marmiroli P, Bossi M, Oggioni N, D’Incalci M, and De Coster R (2007). Bortezomib-induced peripheral neurotoxicity: a neurophysiological and pathological study in the rat. Exp. Neurol. 204, 317–325. 10.1016/j.expneurol.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Palanca A, Casafont I, Berciano MT, and Lafarga M (2014). Reactive nucleolar and Cajal body responses to proteasome inhibition in sensory ganglion neurons. Biochim. Biophys. Acta 1842, 848–859. 10.1016/j.bbadis.2013.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Staff NP, Grisold A, Grisold W, and Windebank AJ (2017). Chemotherapy-induced peripheral neuropathy: A current review. Ann. Neurol. 81, 772–781. 10.1002/ana.24951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staff NP, Podratz JL, Grassner L, Bader M, Paz J, Knight AM, Loprinzi CL, Trushina E, and Windebank AJ (2013). Bortezomib alters microtubule polymerization and axonal transport in rat dorsal root ganglion neurons. Neurotoxicology 39, 124–131. 10.1016/j.neuro.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meregalli C, Canta A, Carozzi VA, Chiorazzi A, Oggioni N, Gilardini A, Ceresa C, Avezza F, Crippa L, Marmiroli P, and Cavaletti G (2010). Bortezomib-induced painful neuropathy in rats: a behavioral, neurophysiological and pathological study in rats. Eur. J. Pain 14, 343–350. 10.1016/j.ejpain.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 10.Carozzi VA, Canta A, Oggioni N, Sala B, Chiorazzi A, Meregalli C, Bossi M, Marmiroli P, and Cavaletti G (2010). Neurophysiological and neuropathological characterization of new murine models of chemotherapy-induced chronic peripheral neuropathies. Exp. Neurol. 226, 301–309. 10.1016/j.expneurol.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Holzer AK, Suciu I, Karreman C, Goj T, and Leist M (2022). Specific Attenuation of Purinergic Signaling during Bortezomib-Induced Peripheral Neuropathy In Vitro. Int. J. Mol. Sci. 23, 3734. 10.3390/ijms23073734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adams J, and Kauffman M (2004). Development of the proteasome inhibitor Velcade (Bortezomib). Cancer Invest. 22, 304–311. 10.1081/cnv-120030218. [DOI] [PubMed] [Google Scholar]
- 13.Sun CY, Li JY, Chu ZB, Zhang L, Chen L, and Hu Y (2017). Efficacy and safety of bortezomib maintenance in patients with newly diagnosed multiple myeloma: a meta-analysis. Biosci. Rep. 37. 10.1042/BSR20170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Velasco R, Alberti P, Bruna J, Psimaras D, and Argyriou AA (2019). Bortezomib and other proteosome inhibitors-induced peripheral neurotoxicity: From pathogenesis to treatment. J. Peripher. Nerv. Syst. 24 (Suppl 2), S52–S62. 10.1111/jns.12338. [DOI] [PubMed] [Google Scholar]
- 15.Meregalli C (2015). An Overview of Bortezomib-Induced Neurotoxicity. Toxics 3, 294–303. 10.3390/toxics3030294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cata JP, Weng HR, Burton AW, Villareal H, Giralt S, and Dougherty PM (2007). Quantitative sensory findings in patients with bortezomib-induced pain. J. Pain 8, 296–306. 10.1016/j.jpain.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 17.Meng L, Huang J, Qiu F, Shan X, Chen L, Sun S, Wang Y, and Yang J (2022). Peripheral Neuropathy During Concomitant Administration of Proteasome Inhibitors and Factor Xa Inhibitors: Identifying the Likelihood of Drug-Drug Interactions. Front. Pharmacol. 13, 757415. 10.3389/fphar.2022.757415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed AS, Ahmed M, Li J, Gu HF, Bakalkin G, Stark A, and Harris HE (2017). Proteasome inhibitor MG132 modulates inflammatory pain by central mechanisms in adjuvant arthritis. Int. J. Rheum. Dis. 20, 25–32. 10.1111/1756-185X.12353. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed AS, Li J, Erlandsson-Harris H, Stark A, Bakalkin G, and Ahmed M (2012). Suppression of pain and joint destruction by inhibition of the proteasome system in experimental osteoarthritis. Pain 153, 18–26. 10.1016/j.pain.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Ossipov MH, Bazov I, Gardell LR, Kowal J, Yakovleva T, Usynin I, Ekströ m TJ, Porreca F, and Bakalkin G (2007). Control of chronic pain by the ubiquitin proteasome system in the spinal cord. J. Neurosci. 27, 8226–8237. 10.1523/JNEUROSCI.5126-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moss A, Blackburn-Munro G, Garry EM, Blakemore JA, Dickinson T, Rosie R, Mitchell R, and Fleetwood-Walker SM (2002). A role of the ubiquitin-proteasome system in neuropathic pain. J. Neurosci. 22, 1363–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coux O, Tanaka K, and Goldberg AL (1996). Structure and functions of the 20S and 26S proteasomes. Annu. Rev. Biochem. 65, 801–847. 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 23.Ciechanover A (2010). The ubiquitin system: historical perspective. Proc. Am. Thorac. Soc. 7, 11–12. 10.1513/pats.200908-095JS. [DOI] [PubMed] [Google Scholar]
- 24.Ciechanover A (1998). The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17, 7151–7160. 10.1093/emboj/17.24.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciechanover A, and Schwartz AL (1998). The ubiquitin-proteasome pathway: the complexity and myriad functions of proteins death. Proc. Natl. Acad. Sci. USA 95, 2727–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ben-Nissan G, and Sharon M (2014). Regulating the 20S proteasome ubiquitin-independent degradation pathway. Biomolecules 4, 862–884. 10.3390/biom4030862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Türker F, Cook EK, and Margolis SS (2021). The proteasome and its role in the nervous system. Cell Chem. Biol. 28, 903–917. 10.1016/j.chembiol.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pines J, and Lindon C (2005). Proteolysis: anytime, any place, anywhere? Nat. Cell Biol. 7, 731–735. 10.1038/ncb0805-731. [DOI] [PubMed] [Google Scholar]
- 29.Ramachandran KV, Fu JM, Schaffer TB, Na CH, Delannoy M, and Margolis SS (2018). Activity-Dependent Degradation of the Nascentome by the Neuronal Membrane Proteasome. Mol. Cell 71, 169–177.e6. 10.1016/j.molcel.2018.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramachandran KV, and Margolis SS (2017). A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nat. Struct. Mol. Biol. 24, 419–430. 10.1038/nsmb.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Türker F, Brennan A, and Margolis SS (2024). Neuronal membrane proteasome-derived peptides modulate NMDAR-dependent neuronal signaling to promote changes in gene expression. Mol. Biol. Cell 35, ar6. 10.1091/mbc.E23-06-0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fleck RA, and Humbel BM (2019). Biological Field Emission Scanning Electron Microscopy (Wiley; )). [Google Scholar]
- 33.Rigaud M, Gemes G, Barabas ME, Chernoff DI, Abram SE, Stucky CL, and Hogan QH (2008). Species and strain differences in rodent sciatic nerve anatomy: implications for studies of neuropathic pain. Pain 136, 188–201. 10.1016/j.pain.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Catala M, and Kubis N (2013). Gross anatomy and development of the peripheral nervous system. Handb. Clin. Neurol. 115, 29–41. 10.1016/B978-0-444-52902-2.00003-5. [DOI] [PubMed] [Google Scholar]
- 35.Palazzo E, Marabese I, Gargano F, Guida F, Belardo C, and Maione S (2021). Methods for Evaluating Sensory, Affective and Cognitive Disorders in Neuropathic Rodents. Curr. Neuropharmacol. 19, 736–746. 10.2174/1570159X18666200831153117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le Pichon CE, and Chesler AT (2014). The functional and anatomical dissection of somatosensory subpopulations using mouse genetics. Front. Neuroanat. 8, 21. 10.3389/fnana.2014.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamashita S (2010). The post-embedding method for immunoelectron microscopy of mammalian tissues: a standardized procedure based on heat-induced antigen retrieval. Methods Mol. Biol. 657, 237–248. 10.1007/978-1-60761-783-9_19. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka K (2009). The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 12–36. 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caccamo D, Katsetos CD, Herman MM, Frankfurter A, Collins VP, and Rubinstein LJ (1989). Immunohistochemistry of a spontaneous murine ovarian teratoma with neuroepithelial differentiation. Neuron-associated beta-tubulin as a marker for primitive neuroepithelium. Lab. Invest. 60, 390–398. [PubMed] [Google Scholar]
- 40.Duly AMP, Kao FCL, Teo WS, and Kavallaris M (2022). betaIII-Tubulin Gene Regulation in Health and Disease. Front. Cell Dev. Biol. 10, 851542. 10.3389/fcell.2022.851542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sell GL, Schaffer TB, and Margolis SS (2017). Reducing expression of synapse-restricting protein Ephexin5 ameliorates Alzheimer’s-like impairment in mice. J. Clin. Invest. 127, 1646–1650. 10.1172/JCI85504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emery EC, and Ernfors P (2020). Dorsal Root Ganglion Neuron Types and Their Functional Specialization. In The Oxford Handbook of the Neurobiology of Pain, Wood JN, ed. (Oxford University Press; ), pp. 128–155. 10.1093/oxfordhb/9780190860509.013.4. [DOI] [Google Scholar]
- 43.Crawford LK, and Caterina MJ (2020). Functional Anatomy of the Sensory Nervous System: Updates From the Neuroscience Bench. Toxicol. Pathol. 48, 174–189. 10.1177/0192623319869011. [DOI] [PubMed] [Google Scholar]
- 44.Hanley JG, Khatri L, Hanson PI, and Ziff EB (2002). NSF ATPase and alpha-/beta-SNAPs disassemble the AMPA receptor-PICK1 complex. Neuron 34, 53–67. 10.1016/s0896-6273(02)00638-4. [DOI] [PubMed] [Google Scholar]
- 45.Kim MJ, Dunah AW, Wang YT, and Sheng M (2005). Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron 46, 745–760. 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- 46.Lauria G, Borgna M, Morbin M, Lombardi R, Mazzoleni G, Sghirlanzoni A, and Pareyson D (2004). Tubule and neurofilament immunoreactivity in human hairy skin: markers for intraepidermal nerve fibers. Muscle Nerve 30, 310–316. 10.1002/mus.20098. [DOI] [PubMed] [Google Scholar]
- 47.Johansson O, Wang L, Hilliges M, and Liang Y (1999). Intraepidermal nerves in human skin: PGP 9.5 immunohistochemistry with special reference to the nerve density in skin from different body regions. J. Peripher. Nerv. Syst. 4, 43–52. [PubMed] [Google Scholar]
- 48.Oaklander AL, and Brown JM (2004). Unilateral nerve injury produces bilateral loss of distal innervation. Ann. Neurol. 55, 639–644. 10.1002/ana.20048. [DOI] [PubMed] [Google Scholar]
- 49.Van Acker N, Ragé M, Sluydts E, Knaapen MWM, De Bie M, Timmers M, Fransen E, Duymelinck C, De Schepper S, Anand P, et al. (2016). Automated PGP9.5 immunofluorescence staining: a valuable tool in the assessment of small fiber neuropathy? BMC Res. Notes 9, 280. 10.1186/s13104-016-2085-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gaudet AD, Popovich PG, and Ramer MS (2011). Wallerian degeneration: gaining perspective on inflammatory events after peripheral nerve injury. J. Neuroinflammation 8, 110. 10.1186/1742-2094-8-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoll G, and Müller HW (1999). Nerve injury, axonal degeneration and neural regeneration: basic insights. Brain Pathol. 9, 313–325. 10.1111/j.1750-3639.1999.tb00229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li N, Kuo CL, Paniagua G, van den Elst H, Verdoes M, Willems LI, van der Linden WA, Ruben M, van Genderen E, Gubbens J, et al. (2013). Relative quantification of proteasome activity by activity-based protein profiling and LC-MS/MS. Nat. Protoc. 8, 1155–1168. 10.1038/nprot.2013.065. [DOI] [PubMed] [Google Scholar]
- 53.He HY, Ahsan A, Bera R, McLain N, Faulkner R, Ramachandran KV, Margolis SS, and Cline HT (2023). Neuronal membrane proteasomes regulate neuronal circuit activity in vivo and are required for learning-induced behavioral plasticity. Proc. Natl. Acad. Sci. USA 120, e2216537120. 10.1073/pnas.2216537120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Türker F, Bharadwaj RA, Kleinman JE, Weinberger DR, Hyde TM, White CJ, Williams DW, and Margolis SS (2023). Orthogonal approaches required to measure proteasome composition and activity in mammalian brain tissue. J. Biol. Chem. 299, 104811. 10.1016/j.jbc.2023.104811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, and Crews CM (1999). Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc. Natl. Acad. Sci. USA 96, 10403–10408. 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geisler S, Doan RA, Cheng GC, Cetinkaya-Fisgin A, Huang SX, Höke A, Milbrandt J, and DiAntonio A (2019). Vincristine and bortezomib use distinct upstream mechanisms to activate a common SARM1-dependent axon degeneration program. JCI Insight 4, e129920. 10.1172/jci.insight.129920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Richardson PG, Xie W, Mitsiades C, Chanan-Khan AA, Lonial S, Hassoun H, Avigan DE, Oaklander AL, Kuter DJ, Wen PY, et al. (2009). Single-agent bortezomib in previously untreated multiple myeloma: efficacy, characterization of peripheral neuropathy, and molecular correlations with response and neuropathy. J. Clin. Oncol. 27, 3518–3525. 10.1200/JCO.2008.18.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bloomingdale P, Meregalli C, Pollard K, Canta A, Chiorazzi A, Fumagalli G, Monza L, Pozzi E, Alberti P, Ballarini E, et al. (2021). Systems Pharmacology Modeling Identifies a Novel Treatment Strategy for Bortezomib-Induced Neuropathic Pain. Front. Pharmacol. 12, 817236. 10.3389/fphar.2021.817236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zheng H, Xiao WH, and Bennett GJ (2012). Mitotoxicity and bortezomib-induced chronic painful peripheral neuropathy. Exp. Neurol. 238, 225–234. 10.1016/j.expneurol.2012.08.023. [DOI] [PubMed] [Google Scholar]
- 60.Poruchynsky MS, Sackett DL, Robey RW, Ward Y, Annunziata C, and Fojo T (2008). Proteasome inhibitors increase tubulin polymerization and stabilization in tissue culture cells: a possible mechanism contributing to peripheral neuropathy and cellular toxicity following proteasome inhibition. Cell Cycle 7, 940–949. 10.4161/cc.7.7.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsieh CH, Jeng SF, Lu TH, Yang JCS, Hsieh MW, Chen YC, and Rau CS (2009). Correlation between skin biopsy with quantification of intraepidermal nerve fiber and the severity of sciatic nerve traction injury in rats. J. Trauma 66, 737–742. 10.1097/TA.0b013e3181623332. [DOI] [PubMed] [Google Scholar]
- 62.Deuis JR, Dvorakova LS, and Vetter I (2017). Methods Used to Evaluate Pain Behaviors in Rodents. Front. Mol. Neurosci. 10, 284. 10.3389/fnmol.2017.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chaplan SR, Bach FW, Pogrel JW, Chung JM, and Yaksh TL (1994). Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53, 55–63. 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 64.Dixon WJ (1980). Efficient analysis of experimental observations. Annu. Rev. Pharmacol. Toxicol. 20, 441–462. 10.1146/annurev.pa.20.040180.002301. [DOI] [PubMed] [Google Scholar]
- 65.Hargreaves K, Dubner R, Brown F, Flores C, and Joris J (1988). A new and sensitive method for measuring thermal nociception in cutaneous hyperalgesia. Pain 32, 77–88. [DOI] [PubMed] [Google Scholar]
- 66.Latremoliere A, Cheng L, DeLisle M, Wu C, Chew S, Hutchinson EB, Sheridan A, Alexandre C, Latremoliere F, Sheu SH, et al. (2018). Neuronal-Specific TUBB3 Is Not Required for Normal Neuronal Function but Is Essential for Timely Axon Regeneration. Cell Rep. 24, 1865–1879.e9. 10.1016/j.celrep.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hill RZ, Loud MC, Dubin AE, Peet B, and Patapoutian A (2022). PIEZO1 transduces mechanical itch in mice. Nature 607, 104–110. 10.1038/s41586-022-04860-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brenner DS, Golden JP, and Gereau RW 4th. (2012). A novel behavioral assay for measuring cold sensation in mice. PLoS One 7, e39765. 10.1371/journal.pone.0039765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brenner DS, Golden JP, Vogt SK, and Gereau RW 4th. (2015). A simple and inexpensive method for determining cold sensitivity and adaptation in mice. J. Vis. Exp. 97, 52640. 10.3791/52640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonzalez-Cano R, Boivin B, Bullock D, Cornelissen L, Andrews N, and Costigan M (2018). Up-Down Reader: An Open Source Program for Efficiently Processing 50% von Frey Thresholds. Front. Pharmacol. 9, 433. 10.3389/fphar.2018.00433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Carmony KC, and Kim KB (2013). Activity-based imaging probes of the proteasome. Cell Biochem. Biophys. 67, 91–101. 10.1007/s12013-013-9626-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gan J, Leestemaker Y, Sapmaz A, and Ovaa H (2019). Highlighting the Proteasome: Using Fluorescence to Visualize Proteasome Activity and Distribution. Front. Mol. Biosci. 6, 14. 10.3389/fmolb.2019.00014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Verdoes M, Berkers CR, Florea BI, van Swieten PF, Overkleeft HS, and Ovaa H (2006). Chemical proteomics profiling of proteasome activity. Methods Mol. Biol. 328, 51–69. 10.1385/1-59745-026-X:51. [DOI] [PubMed] [Google Scholar]
- 74.Verdoes M, Florea BI, Menendez-Benito V, Maynard CJ, Witte MD, van der Linden WA, van den Nieuwendijk AMCH, Hofmann T, Berkers CR, van Leeuwen FWB, et al. (2006). A fluorescent broad-spectrum proteasome inhibitor for labeling proteasomes in vitro and in vivo. Chem. Biol. 13, 1217–1226. 10.1016/j.chembiol.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 75.Verdoes M, Willems LI, van der Linden WA, Duivenvoorden BA, van der Marel GA, Florea BI, Kisselev AF, and Overkleeft HS (2010). A panel of subunit-selective activity-based proteasome probes. Org. Biomol. Chem. 8, 2719–2727. 10.1039/c001036g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rice FL, and Albrecht PJ (2020). 4.04 - Cutaneous Mechanisms of Tactile Perception: Morphological and Chemical Organization of the Innervation to the Skin. In The Senses: A Comprehensive Reference, Second Edition, Fritzsch B, ed. (Elsevier; ), pp. 35–60. 10.1016/B978-0-12-805408-6.00340-7. [DOI] [Google Scholar]
- 77.Fornaro M, Lee JM, Raimondo S, Nicolino S, Geuna S, and Giacobini-Robecchi M (2008). Neuronal intermediate filament expression in rat dorsal root ganglia sensory neurons: an in vivo and in vitro study. Neuroscience 153, 1153–1163. 10.1016/j.neuroscience.2008.02.080. [DOI] [PubMed] [Google Scholar]
- 78.Ruscheweyh R, Forsthuber L, Schoffnegger D, and Sandkühler J (2007). Modification of classical neurochemical markers in identified primary afferent neurons with Abeta-Adelta-and C-fibers after chronic constriction injury in mice. J. Comp. Neurol. 502, 325–336. 10.1002/cne.21311. [DOI] [PubMed] [Google Scholar]
- 79.Fang X, Djouhri L, McMullan S, Berry C, Waxman SG, Okuse K, and Lawson SN (2006). Intense isolectin-B4 binding in rat dorsal root ganglion neurons distinguishes C-fiber nociceptors with broad action potentials and high Nav1.9 expression. J. Neurosci. 26, 7281–7292. 10.1523/JNEUROSCI.1072-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Harper AA, and Lawson SN (1985). Conduction velocity is related to morphological cell type in rat dorsal root ganglion neurones. J. Physiol. 359, 31–46. 10.1113/jphysiol.1985.sp015573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lawson SN, Fang X, and Djouhri L (2019). Nociceptor subtypes and their incidence in rat lumbar dorsal root ganglia (DRGs): focussing on C-polymodal nociceptors, Abeta-nociceptors, moderate pressure receptors and their receptive field depths. Curr. Opin. Physiol. 11, 125–146. 10.1016/j.cophys.2019.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lawson SN, and Waddell PJ (1991). Soma neurofilament immunoreactivity is related to cell size and fibre conduction velocity in rat primary sensory neurons. J. Physiol. 435, 41–63. 10.1113/jphysiol.1991.sp018497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen CC, Akopian AN, Sivilotti L, Colquhoun D, Burnstock G, and Wood JN (1995). A P2X purinoceptor expressed by a subset of sensory neurons. Nature 377, 428–431. 10.1038/377428a0. [DOI] [PubMed] [Google Scholar]
- 84.Cook SP, Vulchanova L, Hargreaves KM, Elde R, and McCleskey EW (1997). Distinct ATP receptors on pain-sensing and stretch-sensing neurons. Nature 387, 505–508. 10.1038/387505a0. [DOI] [PubMed] [Google Scholar]
- 85.Bradbury EJ, Burnstock G, and McMahon SB (1998). The expression of P2X3 purinoreceptors in sensory neurons: effects of axotomy and glial-derived neurotrophic factor. Mol. Cell. Neurosci. 12, 256–268. 10.1006/mcne.1998.0719. [DOI] [PubMed] [Google Scholar]
- 86.Gu Y, Wang C, Li G, and Huang LYM (2016). EXPRESS: F-actin links Epac-PKC signaling to purinergic P2X3 receptors sensitization in dorsal root ganglia following inflammation. Mol. Pain 12, 1744806916660557. 10.1177/1744806916660557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sheng X, Dan Y, Dai B, Guo J, Ji H, Zhang X, Xie Z, Song S, Pan Q, Wang J, et al. (2018). Knockdown the P2X3 receptor in the stellate ganglia of rats relieved the diabetic cardiac autonomic neuropathy. Neurochem. Int. 120, 206–212. 10.1016/j.neuint.2018.09.002. [DOI] [PubMed] [Google Scholar]
- 88.Zhang J, Cavanaugh DJ, Nemenov MI, and Basbaum AI (2013). The modality-specific contribution of peptidergic and non-peptidergic nociceptors is manifest at the level of dorsal horn nociresponsive neurons. J. Physiol. 591, 1097–1110. 10.1113/jphysiol.2012.242115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bleehen T, and Keele CA (1977). Observations on the algogenic actions of adenosine compounds on the human blister base preparation. Pain 3, 367–377. 10.1016/0304-3959(77)90066-5. [DOI] [PubMed] [Google Scholar]
- 90.Honore P, Kage K, Mikusa J, Watt AT, Johnston JF, Wyatt JR, Faltynek CR, Jarvis MF, and Lynch K (2002). Analgesic profile of intrathecal P2X(3) antisense oligonucleotide treatment in chronic inflammatory and neuropathic pain states in rats. Pain 99, 11–19. 10.1016/s0304-3959(02)00032-5. [DOI] [PubMed] [Google Scholar]
- 91.Jarvis MF, Burgard EC, McGaraughty S, Honore P, Lynch K, Brennan TJ, Subieta A, Van Biesen T, Cartmell J, Bianchi B, et al. (2002). A-317491, a novel potent and selective non-nucleotide antagonist of P2X3 and P2X2/3 receptors, reduces chronic inflammatory and neuropathic pain in the rat. Proc. Natl. Acad. Sci. USA 99, 17179–17184. 10.1073/pnas.252537299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Souslova V, Cesare P, Ding Y, Akopian AN, Stanfa L, Suzuki R, Carpenter K, Dickenson A, Boyce S, Hill R, et al. (2000). Warm-coding deficits and aberrant inflammatory pain in mice lacking P2X3 receptors. Nature 407, 1015–1017. 10.1038/35039526. [DOI] [PubMed] [Google Scholar]
- 93.Krames ES (2014). The role of the dorsal root ganglion in the development of neuropathic pain. Pain Med. 15, 1669–1685. 10.1111/pme.12413. [DOI] [PubMed] [Google Scholar]
- 94.Qi L, Iskols M, Shi D, Reddy P, Walker C, Lezgiyeva K, Voisin T, Pawlak M, Kuchroo VK, Chiu IM, et al. (2024). A mouse DRG genetic toolkit reveals morphological and physiological diversity of somatosensory neuron subtypes. Cell 187, 1508–1526.e16. 10.1016/j.cell.2024.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zheng Y, Liu P, Bai L, Trimmer JS, Bean BP, and Ginty DD (2019). Deep Sequencing of Somatosensory Neurons Reveals Molecular Determinants of Intrinsic Physiological Properties. Neuron 103, 598–616.e7. 10.1016/j.neuron.2019.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sharma N, Flaherty K, Lezgiyeva K, Wagner DE, Klein AM, and Ginty DD (2020). The emergence of transcriptional identity in somatosensory neurons. Nature 577, 392–398. 10.1038/s41586-019-1900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zeisel A, Hochgerner H, Lönnerberg P, Johnsson A, Memic F, van der Zwan J, Häring M, Braun E, Borm LE, La Manno G, et al. (2018). Molecular Architecture of the Mouse Nervous System. Cell 174, 999–1014.e22. 10.1016/j.cell.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Usoskin D, Furlan A, Islam S, Abdo H, Lönnerberg P, Lou D, Hjerling-Leffler J, Haeggström J, Kharchenko O, Kharchenko PV, et al. (2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153. 10.1038/nn.3881. [DOI] [PubMed] [Google Scholar]
- 99.Steele HR, Xing Y, Zhu Y, Hilley HB, Lawson K, Nho Y, Niehoff T, and Han L (2021). MrgprC11(+) sensory neurons mediate glabrous skin itch. Proc. Natl. Acad. Sci. USA 118, e2022874118. 10.1073/pnas.2022874118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Sharif B, Ase AR, Ribeiro-da-Silva A, and Séguéla P (2020). Differential Coding of Itch and Pain by a Subpopulation of Primary Afferent Neurons. Neuron 106, 940–951.e4. 10.1016/j.neuron.2020.03.021. [DOI] [PubMed] [Google Scholar]
- 101.Olson W, and Luo W (2018). Somatotopic organization of central arbors from nociceptive afferents develops independently of their intact peripheral target innervation. J. Comp. Neurol. 526, 3058–3065. 10.1002/cne.24533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang J, Polgár E, Solinski HJ, Mishra SK, Tseng PY, Iwagaki N, Boyle KA, Dickie AC, Kriegbaum MC, Wildner H, et al. (2018). Circuit dissection of the role of somatostatin in itch and pain. Nat. Neurosci. 21, 707–716. 10.1038/s41593-018-0119-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Voisin T, Perner C, Messou MA, Shiers S, Ualiyeva S, Kanaoka Y, Price TJ, Sokol CL, Bankova LG, Austen KF, and Chiu IM (2021). The CysLT(2)R receptor mediates leukotriene C(4)-driven acute and chronic itch. Proc. Natl. Acad. Sci. USA 118, e2022087118. 10.1073/pnas.2022087118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Stantcheva KK, Iovino L, Dhandapani R, Martinez C, Castaldi L, Nocchi L, Perlas E, Portulano C, Pesaresi M, Shirlekar KS, et al. (2016). A subpopulation of itch-sensing neurons marked by Ret and somatostatin expression. EMBO Rep. 17, 585–600. 10.15252/embr.201540983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Han L, Ma C, Liu Q, Weng HJ, Cui Y, Tang Z, Kim Y, Nie H, Qu L, Patel KN, et al. (2013). A subpopulation of nociceptors specifically linked to itch. Nat. Neurosci. 16, 174–182. 10.1038/nn.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu P, Zhao Y, Xie Z, Zhou H, Dong X, Wu GF, Kim BS, Feng J, and Hu H (2023). MrgprA3-expressing pruriceptors drive pruritogen-induced alloknesis through mechanosensitive Piezo2 channel. Cell Rep. 42, 112283. 10.1016/j.celrep.2023.112283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mishra SK, and Hoon MA (2013). The cells and circuitry for itch responses in mice. Science 340, 968–971. 10.1126/science.1233765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Roberson DP, Gudes S, Sprague JM, Patoski HAW, Robson VK, Blasl F, Duan B, Oh SB, Bean BP, Ma Q, et al. (2013). Activity-dependent silencing reveals functionally distinct itch-generating sensory neurons. Nat. Neurosci. 16, 910–918. 10.1038/nn.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Solinski HJ, Kriegbaum MC, Tseng PY, Earnest TW, Gu X, Barik A, Chesler AT, and Hoon MA (2019). Nppb Neurons Are Sensors of Mast Cell-Induced Itch. Cell Rep. 26, 3561–3573.e4. 10.1016/j.celrep.2019.02.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hanani M (2012). Intercellular communication in sensory ganglia by purinergic receptors and gap junctions: implications for chronic pain. Brain Res. 1487, 183–191. 10.1016/j.brainres.2012.03.070. [DOI] [PubMed] [Google Scholar]
- 111.Huang TY, Belzer V, and Hanani M (2010). Gap junctions in dorsal root ganglia: possible contribution to visceral pain. Eur. J. Pain 14, 49. e1–11. 10.1016/j.ejpain.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 112.Devor M, and Wall PD (1990). Cross-excitation in dorsal root ganglia of nerve-injured and intact rats. J. Neurophysiol. 64, 1733–1746. 10.1152/jn.1990.64.6.1733. [DOI] [PubMed] [Google Scholar]
- 113.Amir R, and Devor M (1996). Chemically mediated cross-excitation in rat dorsal root ganglia. J. Neurosci. 16, 4733–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kim YS, Anderson M, Park K, Zheng Q, Agarwal A, Gong C, Saijilafu Young, L., He S, LaVinka PC, et al. (2016). Coupled Activation of Primary Sensory Neurons Contributes to Chronic Pain. Neuron 91, 1085–1096. 10.1016/j.neuron.2016.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Utzschneider D, Kocsis J, and Devor M (1992). Mutual excitation among dorsal root ganglion neurons in the rat. Neurosci. Lett. 146, 53–56. 10.1016/0304-3940(92)90170-c. [DOI] [PubMed] [Google Scholar]
- 116.Illes P, Müller CE, Jacobson KA, Grutter T, Nicke A, Fountain SJ, Kennedy C, Schmalzing G, Jarvis MF, Stojilkovic SS, et al. (2021). Update of P2X receptor properties and their pharmacology: IUPHAR Review 30. Br. J. Pharmacol. 178, 489–514. 10.1111/bph.15299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.North RA (2002). Molecular physiology of P2X receptors. Physiol. Rev. 82, 1013–1067. 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- 118.Bae J, Kang KM, and Kim YC (2022). Discovery of 5-methyl-1H-benzo[d]imidazole derivatives as novel P2X3 Receptor antagonists. Bio-org. Med. Chem. Lett. 72, 128820. 10.1016/j.bmcl.2022.128820. [DOI] [PubMed] [Google Scholar]
- 119.Pinho-Ribeiro FA, Verri WA Jr., and Chiu IM (2017). Nociceptor Sensory Neuron-Immune Interactions in Pain and Inflammation. Trends Immunol. 38, 5–19. 10.1016/j.it.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Klein Wolterink RGJ, Wu GS, Chiu IM, and Veiga-Fernandes H (2022). Neuroimmune Interactions in Peripheral Organs. Annu. Rev. Neurosci. 45, 339–360. 10.1146/annurev-neuro-111020-105359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Michael GJ, and Priestley JV (1999). Differential expression of the mRNA for the vanilloid receptor subtype 1 in cells of the adult rat dorsal root and nodose ganglia and its downregulation by axotomy. J. Neurosci. 19, 1844–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tominaga M, Caterina MJ, Malmberg AB, Rosen TA, Gilbert H, Skinner K, Raumann BE, Basbaum AI, and Julius D (1998). The cloned capsaicin receptor integrates multiple pain-producing stimuli. Neuron 21, 531–543. 10.1016/s0896-6273(00)80564-4. [DOI] [PubMed] [Google Scholar]
- 123.Luo W, Enomoto H, Rice FL, Milbrandt J, and Ginty DD (2009). Molecular identification of rapidly adapting mechanoreceptors and their developmental dependence on ret signaling. Neuron 64, 841–856. 10.1016/j.neuron.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Davidson S, and Giesler GJ (2010). The multiple pathways for itch and their interactions with pain. Trends Neurosci. 33, 550–558. 10.1016/j.tins.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Klein A, Carstens MI, and Carstens E (2011). Facial injections of pruritogens or algogens elicit distinct behavior responses in rats and excite overlapping populations of primary sensory and trigeminal subnucleus caudalis neurons. J. Neurophysiol. 106, 1078–1088. 10.1152/jn.00302.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Simone DA, Zhang X, Li J, Zhang JM, Honda CN, LaMotte RH, and Giesler GJ Jr. (2004). Comparison of responses of primate spinothalamic tract neurons to pruritic and algogenic stimuli. J. Neurophysiol. 91, 213–222. 10.1152/jn.00527.2003. [DOI] [PubMed] [Google Scholar]
- 127.Nilsson HJ, Levinsson A, and Schouenborg J (1997). Cutaneous field stimulation (CFS): a new powerful method to combat itch. Pain 71, 49–55. 10.1016/s0304-3959(97)03339-3. [DOI] [PubMed] [Google Scholar]
- 128.Brull SJ, Atanassoff PG, Silverman DG, Zhang J, and Lamotte RH (1999). Attenuation of experimental pruritus and mechanically evoked dysesthesiae in an area of cutaneous allodynia. Somatosens. Mot. Res. 16, 299–303. 10.1080/08990229970366. [DOI] [PubMed] [Google Scholar]
- 129.Institute of Medicine Committee on Pain, D.; Chronic Illness B (1987). In Pain and Disability: Clinical, Behavioral, and Public Policy Perspectives, Osterweis M, Kleinman A, and Mechanic D, eds. (National Academies Press (US) Copyright © 1987 by the National Academy of Sciences; ). 10.17226/991. [DOI] [PubMed] [Google Scholar]
- 130.Sikand P, Shimada SG, Green BG, and LaMotte RH (2009). Similar itch and nociceptive sensations evoked by punctate cutaneous application of capsaicin, histamine and cowhage. Pain 144, 66–75. 10.1016/j.pain.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kaplan GS, Torcun CC, Grune T, Ozer NK, and Karademir B (2017). Proteasome inhibitors in cancer therapy: Treatment regimen and peripheral neuropathy as a side effect. Free Radic. Biol. Med. 103, 1–13. 10.1016/j.freeradbiomed.2016.12.007. [DOI] [PubMed] [Google Scholar]
- 132.Alsina M, Trudel S, Furman RR, Rosen PJ, O’Connor OA, Comenzo RL, Wong A, Kunkel LA, Molineaux CJ, and Goy A (2012). A phase I single-agent study of twice-weekly consecutive-day dosing of the proteasome inhibitor carfilzomib in patients with relapsed or refractory multiple myeloma or lymphoma. Clin. Cancer Res. 18, 4830–4840. 10.1158/1078-0432.CCR-11-3007. [DOI] [PubMed] [Google Scholar]
- 133.O’Connor OA, Stewart AK, Vallone M, Molineaux CJ, Kunkel LA, Gerecitano JF, and Orlowski RZ (2009). A phase 1 dose escalation study of the safety and pharmacokinetics of the novel proteasome inhibitor carfilzomib (PR-171) in patients with hematologic malignancies. Clin. Cancer Res. 15, 7085–7091. 10.1158/1078-0432.CCR-09-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kisselev AF, van der Linden WA, and Overkleeft HS (2012). Proteasome inhibitors: an expanding army attacking a unique target. Chem. Biol. 19, 99–115. 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kortuem KM, and Stewart AK (2013). Carfilzomib. Blood 121, 893–897. 10.1182/blood-2012-10-459883. [DOI] [PubMed] [Google Scholar]
- 136.Morris RE, and Ciraolo GM (1997). A universal post-embedding protocol for immunogold labelling of osmium-fixed, epoxy resin-embedded tissue. J. Electron. Microsc. 46, 315–319. 10.1093/oxfordjournals.jmicro.a023525. [DOI] [PubMed] [Google Scholar]
- 137.Stirling JW, and Graff PS (1995). Antigen unmasking for immunoelectron microscopy: labeling is improved by treating with sodium ethoxide or sodium metaperiodate, then heating on retrieval medium. J. Histochem. Cytochem. 43, 115–123. 10.1177/43.2.7529784. [DOI] [PubMed] [Google Scholar]
- 138.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Christensen SL, Hansen RB, Storm MA, Olesen J, Hansen TF, Ossipov M, Izarzugaza JMG, Porreca F, and Kristensen DM (2020). Von Frey testing revisited: Provision of an online algorithm for improved accuracy of 50% thresholds. Eur. J. Pain 24, 783–790. 10.1002/ejp.1528. [DOI] [PubMed] [Google Scholar]
- 140.Mills C, Leblond D, Joshi S, Zhu C, Hsieh G, Jacobson P, Meyer M, and Decker M (2012). Estimating efficacy and drug ED50’s using von Frey thresholds: impact of Weber’s law and log transformation. J. Pain 13, 519–523. 10.1016/j.jpain.2012.02.009. [DOI] [PubMed] [Google Scholar]
- 141.Hao Y, Hao S, Andersen-Nissen E, Mauck WM 3rd, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, Baglaenko Y, Brenner M, Loh PR, and Raychaudhuri S (2019). Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 16, 1289–1296. 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, and Sergushichev A (2021). Fast gene set enrichment analysis. Preprint at bioRxiv, 060012. 10.1101/060012. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq dataset generated in this study has been deposited to the Gene Expression Omnibus (GEO: GSE261727). Other data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


