Abstract
The All of Us Research Program’s Data and Research Center (DRC) was established to help acquire, curate, and provide access to one of the world’s largest and most diverse datasets for precision medicine research. Already, over 500,000 participants are enrolled in All of Us, 80% of whom are underrepresented in biomedical research, and data are being analyzed by a community of over 2,300 researchers. The DRC created this thriving data ecosystem by collaborating with engaged participants, innovative program partners, and empowered researchers. In this paper, we first describe how the DRC is organized to meet the needs of this broad group of stakeholders. We then outline guiding principles, common challenges, and innovative approaches used to build the All of Us data ecosystem. Finally, we share lessons learned to help others navigate important decisions and trade-offs in building a modern biomedical data platform.
Keywords: Precision medicine, data ecosystem, diversity, data privacy, data integration, electronic health records
I. The All of Us Research Program
The National Institutes of Health (NIH) All of Us Research Program is one of a growing number of big data efforts driving modern biomedical research.1–9 All of Us seeks to accelerate health research and medical breakthroughs by partnering with at least one million participants across the United States to create the most diverse biomedical data resource in history.10 A significant differentiator and core value of All of Us is its commitment to the inclusion of groups who have historically been underrepresented in biomedical research – not only racial and ethnic minorities, but also individuals with low income or low education, who live in rural areas, who identify as a gender minority, or who have a disability.11 Consequently, All of Us addresses certain limitations in prior cohorts, including small sample size,12–14 lack of diversity, and lack of granular phenotypic data.15–22 The program accomplishes this through robust community engagement and a commitment to transparency, return of value to participants, and data privacy. In addition, All of Us strives to catalyze innovative research programs and policies and make data broadly accessible to empower research.
The All of Us Research Program is enabled by a consortium of partners responsible for participant engagement, enrollment, data generation, and data sharing activities.10,23 Participant engagement and enrollment activities are led by a national network of partners who reflect the diversity of the United States. For example, enrollment partners include both regional medical centers and federally qualified health centers,24 organized into collaborative networks of Healthcare Provider Organizations (HPOs) enrolling patients in their regions. Additionally, The Participant Center was funded to build a collaborative network that enables interested individuals to sign up and donate data and biospecimens from anywhere in the United States.15,25 Most All of Us participants consent, enroll, and contribute data through an online participant portal created and managed by the Participant Technology Systems Center. Participant biospecimens, including blood, saliva, and urine, are stored by the All of Us Biobank. Blood and saliva undergo genotyping and whole genome sequencing at All of Us Genome Centers, and the return of genetic results to participants is overseen by the All of Us Genetic Counseling Resource.26,27 Finally, the organization responsible for connecting this large consortium of partners to create All of Us’ central data ecosystem and research platform is the All of Us Data and Research Center (DRC).
The DRC echoes the larger mission of the All of Us Research Program, in that it serves two important stakeholder groups - participants and researchers - to create the All of Us biomedical data resource. Building trusted relationships within underrepresented communities requires recognizing participants as partners and returning value to participants in tandem with sharing data to drive research. The DRC data ecosystem (Figure 1) must therefore support the flow of data not only across program partners and to approved researchers, but also back to participants themselves. This includes the return of hereditary disease risks resulting from genomic testing, in partnership with the All of Us Participant Technology Systems Center, Biobank, Genome Centers, and Genetic Counseling Resource (Figure 1g).26,27 In addition, All of Us’ commitment to its participants requires it to enable research that drives forward innovative approaches to individualized prevention, treatment, and care.
Figure 1.
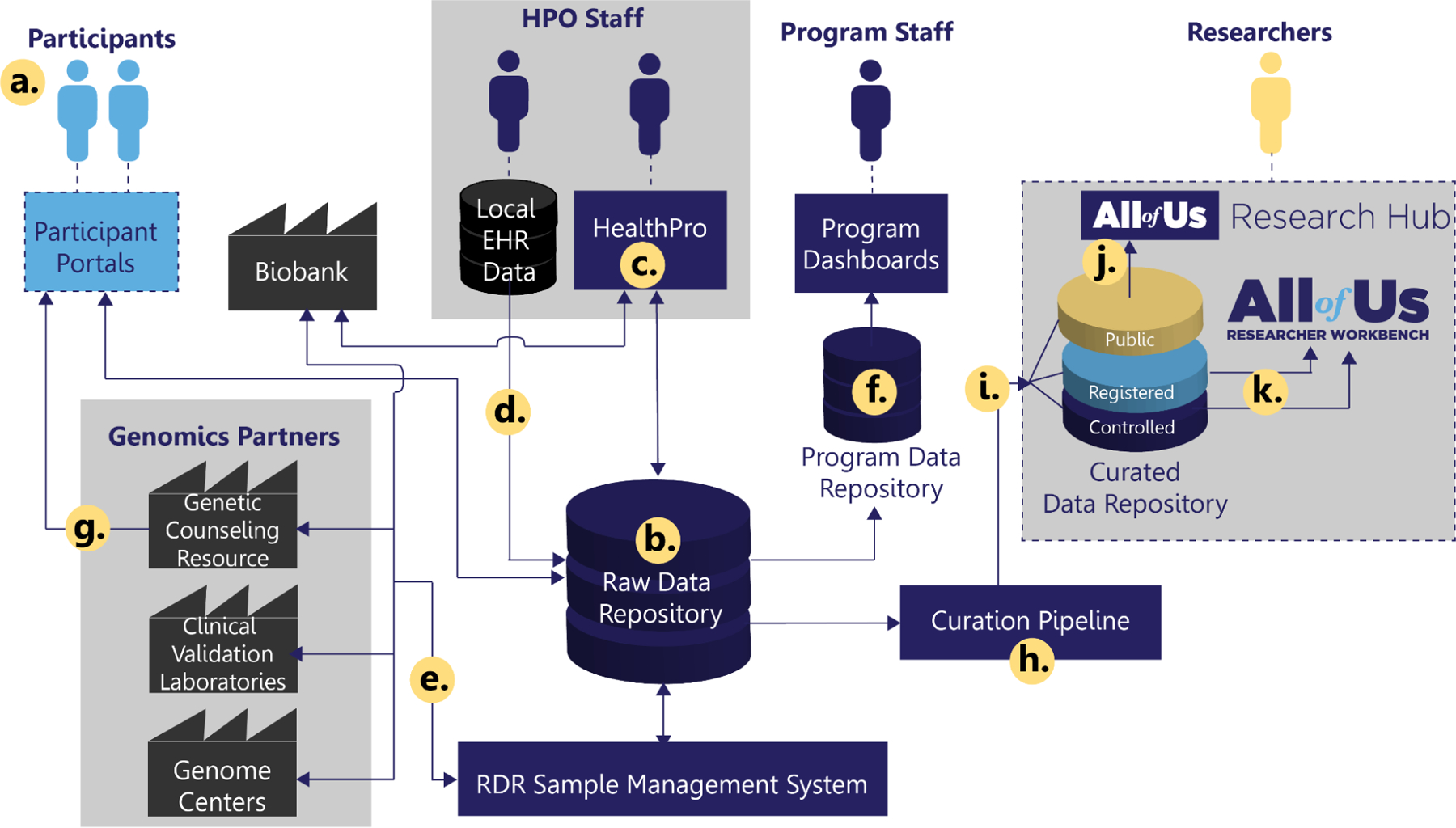
Overview of the All of Us data ecosystem. (a) Participants enroll, consent, answer questionnaires, and provide additional data through All of Us Participant Portals, managed by the Participant Technologies Systems Center and The Participant Center. (b) Data are stored in a central, secure, Raw Data Repository (RDR) at the Data and Research Center (DRC). Program staff at Healthcare Provider Organization (HPO) partners (c.) leverage DRC’s HealthPro application to complete baseline assessments, including program physical measurements and biospecimen collection, and (d.) contribute data from local clinical systems via the Electronic Health Record (EHR) data pipeline. Genomic analysis of participant biospecimen occurs via (e.) a genomic data pipeline in collaboration with the All of Us Biobank and Genomics Partners. (f.) Summary participant and additional operational data from the RDR are aggregated into a Program Data Repository to power program-staff-facing analytics and dashboards. (g.) Return of genetic results to participants is facilitated by the All of Us Genetic Counseling Resource. In parallel, participant data from the RDR is (h.) routed through a curation pipeline to create a tiered (i.) Curated Data Repository (CDR). Public CDR data are made available via the (j.) the All of Us Research Hub’s public Data Browser, while participant-level data in the Registered and Controlled Tiers are made available via the Research Hub’s secure analysis Trusted Research Environment (TRE), the (k.) Researcher Workbench.
All of Us is disease-agnostic and seeks to support research that can improve health outcomes through precision interventions and reduce health disparities to improve health equity for all populations. The program is committed to workforce diversity and training, attempting to engage and support researchers spanning disciplines, geographies, career stages, institutional settings, and demographics. The unique partnership of engaged participants, innovative program partners, and researchers empowered to conduct meaningful science represents an important new paradigm and opportunity to positively impact human health.
II. The All of Us Data and Research Center
The All of Us DRC supplies the infrastructure to acquire, organize, and provide access to one of the world’s largest and most diverse datasets for precision medicine research. Program needs include data generation, real-time data exchange between program partners, program data analytics and reporting for operational management, data ingestion from program data providers, data curation and delivery to a large and diverse research community, and outreach and training to support awareness and uptake by the larger biomedical research community.
To optimally support the All of Us Research Program vision, the DRC aligns with the data ecosystem model proposed in the NIH Strategic Plan for Data Science.28 In this manuscript, we provide an overview of the All of Us DRC, the guiding principles used to build its data ecosystem, its basic infrastructure, and key systems. We also reflect on lessons learned and highlight opportunities for the larger biomedical data community, including those creating future biomedical data platforms.
II.1. Guiding Principles
In this section, we outline the set of principles used to guide our approach to building this important data ecosystem (Table 1). The DRC, like many other big data providers, is faced with challenges in creating systems capable of taking in Petabytes of data and maximizing its utility.29,30 Key attributes of big data have been widely characterized as the four Vs: velocity, veracity, volume, and variety.31 The four Vs represent data-based challenges and trade-offs that we evaluated and addressed in designing and implementing the All of Us DRC. In addition, the DRC addressed challenges related specifically to health-related data, and others resulting from the All of Us Research Program’s mission and core values serving researchers and participants.
Table 1.
Guiding principles used to build the DRC data ecosystem.
| Guiding Principles | Common Challenges | DRC Approaches |
|---|---|---|
| 1. Build secure, scalable, and sustainable systems | “Volume and Velocity”: Coping with rapidly growing volumes of data |
|
| Managing a complex security environment | ||
| 2. Increase utility without sacrificing integrity & richness | “Variety and Veracity”: Effectively utilizing multi-modal data |
|
| 3. Share data widely and wisely | Achieving the appropriate balance between participant privacy and broad research access and utilization |
|
| 4. Support useful, generalizable, and accessible tools and workflows | Achieving broad use by a diverse research community |
|
Healthcare data generally possess unique characteristics and complexities, such as privacy, longevity, and ownership,30 that go beyond the exigencies of the four Vs and necessitate additional infrastructure and tooling approaches. All of Us’ commitment to diversity, inclusion, and return of value also presents a set of unique and important challenges. For example, All of Us must continually balance two goals that are often in tension: protecting participant privacy and ensuring broad access and use to drive innovation and discovery. Addressing these challenges is critical to establishing a system of big data management that maintains principles of equity and privacy while endeavoring to maximize usage and value.
II.2. All of Us DRC Data Ecosystem Overview
The DRC created its data ecosystem (Figure 1) by applying the guiding principles outlined in Table 1 towards its data-centric responsibilities. These responsibilities can be categorized into the following aims: (1) Collect data, (2) Organize it, and (3) Make it useful. Here we provide a high-level overview of the DRC ecosystem, organized according to these aims.
Data Acquisition (Collect data)
The DRC provides multiple systems that facilitate the receipt and storage of data from many partners. All of Us participants enroll in the program through online portals (Figure 1a), and data regarding participant consent,32 responses to survey questionnaires, and other digital health information (including data from wearables) are passed to the DRC and stored in a raw data repository (RDR) for further use (Figure 1b). The RDR is a data store and accompanying data processing application(s), which serve as the main intake point and data repository for All of Us, the Source of Truth for both operational and research data, and the initial landing point for any data shared with All of Us by ancillary studies. The RDR adopts some principles of data lakes, such as being “append-only,” with a focus on quickly and efficiently ingesting data with varying levels of structure. The RDR serves as the DRC’s means to facilitate consistent and secure data transfer across the program. In performing its role, the system provides an array of application programming interfaces (APIs) and other integration options to both internal (DRC) and external (partner) systems.
HealthPro is one example of an internal integration (Figure 1c). HealthPro is a DRC-developed application made available to All of Us HPO partners. Its primary users are All of Us research staff, who leverage the HealthPro application to support participant baseline assessment and biospecimen collection. HealthPro facilitates the biospecimen donation workflow (in coordination with the All of Us Biobank) and captures a defined set of physical measurements, such as height, weight, and blood pressure. The HealthPro application also provides operational support to sites in the form of summary and detailed participant information displays, work queues, and dashboards.
Three examples of external system integrations required to support DRC data collection activities are those required for survey questionnaires (Figure 1a), Electronic Health Records (EHR) (Figure 1d), and genomic data (Figure 1e). To ensure concordance across the DRC, Participant Technology Systems Center,33 and Participant Center, the technology partners utilize REDCap34,35 as a joint custody framework for structural survey documentation and implementation. REDCap data dictionaries, created to define each survey’s content and organizational structure, are uploaded, and represented within each system to direct the transmission of resulting data to the DRC. The resulting data dictionary artifacts are then used as a reference for transforming the data according to the program’s data model and are disseminated broadly through the scientific community for reuse.36
EHR data are another important data collection stream that requires many external system integrations and sophisticated operational processes. EHR data are first transformed to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM)37 by HPO partners,38,39 according to a process that is supervised and facilitated by the DRC. Data stewards at these partner sites then transmit their OMOP-formatted data to the DRC via an EHR data pipeline application. This application applies OMOP conformance validation checks and returns those results to HPOs as part of a data quality feedback loop.40 The application also combines all submitted EHR data on a nightly basis into a single dataset to support downstream program operations and research activities. EHR data ingestion is supported by a robust set of operations (see Section III.1).
The genomic data pipeline refers to a collection of partners and systems, participating in a shared process coordinated by the RDR’s Sample Management Subsystem. This manifest-driven process allows for participant biospecimens to be tracked through a series of steps that begin with Biobank being notified a sample has been collected, allow samples to be sent to All of Us Genome Center partners for sequencing and analysis, and end with curated genomic data being made available to researchers. Each step has its validation and quality control activities, to help ensure data meets high-quality standards required for the return of results back to participants as well as forward to researchers.26
Data Curation (Organize it)
All incoming data are evaluated for both operational and research utility. Data needed for program operational management, such as anomaly detection, enrollment, and activity completion rates are de-identified and brought into a Program Data Repository (Figure 1f), where the information may be safely used for reporting, automated alerting, and other operational needs. The Program Data Repository is a data store and accompanying data processing application(s), including a suite of analytical dashboards maintained by the DRC for program staff.
Data with research utility is made available to a curation pipeline (Figure 1h) which applies a robust privacy methodology, a set of curation and conformance cleaning rules, as well as any data mapping necessary to ensure a consistent, appropriate data model, resulting in a curated data repository (CDR) that is made available to authorized researchers. The curation pipeline also includes a set of applications and processes for genomic data curation. These applications consume information from the RDR and genomics partners and perform additional processing, curation, and quality control to ready genomic data for research use. For example, the DRC joint-calls whole genome sequence reads from the All of Us Genome Centers to produce genetic variant information. Genomic data (including the original read data, joint-called variant data, and other auxiliary data such as QC information on genetic ancestry and relatedness) are then linked with other data from the RDR as part of the curation pipeline. The curation pipeline then applies conformance, cleaning, and privacy rules to produce the CDR - the program’s primary researcher-facing data product.
A key operational facet of the curation pipeline application is the program’s privacy methodology. The primary objective of the privacy methodology is to minimize the risk of intentional or accidental participant reidentification by researchers. Privacy methodologies fall generally into several categories, including: suppression, wherein an entire table, column, or row is suppressed based on its contents (for example, directly identifying information like names and addresses are suppressed), generalization, wherein granular information is replaced with a less specific information (substituting “More than one race” where a participant has self-reported multiple races is an example of a generalization, as is providing birth year in lieu of a participant’s full date of birth), and randomization (for example, all dates in the Registered Tier are randomly shifted by −1 to −365). Additional detail about the CDR curation processes and privacy policies are provided in Sections III.2 and III.3.
Data Dissemination (Make it useful)
Data generated from and stored in the All of Us program are valuable to both participants and researchers. As a result, the DRC facilitates data exchanges that enable the frequent return of information to participants. One of the most sophisticated examples of DRC infrastructure required to support participant-facing return of results is the return of genetic-related information (Figure 1g).
To support the return of health-related genetic results to participants, the RDR system has expanded its Sample Management System to incorporate additional non-research workflow management. This workflow includes partners involved in the return of results and ensures that appropriate steps are completed before participants receive their results. Gating events include confirmation of eligibility requirements like informed consent, quality of sequenced sample, and completion of informing loops to refresh participant understanding. If All of Us determines a participant is at risk for a hereditary disease, All of Us provides access to a Genetic Counselor and requires a participant to meet with the counselor to receive their results. This service is an outgrowth of the All of Us program’s commitment to provide value through actionable information.
In parallel, participant data in the CDR are made useful to researchers through the All of Us Research Hub (Figure 1i). The Research Hub contains a public website that connects researchers with general information about the program, research data, tools, and access process, and a secure enclave, known as the Researcher Workbench, where approved researchers access and analyze All of Us participant data.
The Research Hub public-facing website (https://researchallofus.org) includes a Data Browser allowing visitors to explore an aggregated, summary version of the CDR (Figure 1j), a Survey Explorer allowing visitors to access documentation regarding All of Us survey questionnaires, and a Research Projects Directory providing the ability to search through a library of active research projects using All of Us data. The website also provides information about how the research data are compiled, access policies, and how to request access, a robust user support hub, research spotlights, featured publications using All of Us data, and more.
All analysis of row-level participant data must be performed by authorized researchers within the Researcher Workbench (Figure 1k). The Researcher Workbench is the All of Us Research Program’s Trusted Research Environment (TRE),41,42 built using open source software running on public cloud infrastructure. Within the Workbench, researchers create a workspace for their project and share it with other researchers. They may also access a suite of custom point-and-click tools, including a Cohort Builder to select study participants based on custom criteria and a Dataset Builder for selecting a subset of data for analysis. Researchers may then conduct analyses using a growing set of analysis tools, including Jupyter notebooks, R, and Python, and bring their own analysis methods.
III. Guiding Principles in Action
In the previous sections we introduced the All of Us DRC, its guiding principles, and presented an overview of its data ecosystem. We now describe actionable approaches based on guiding principles outlined in Table 1, which enabled the DRC to successfully address major challenges to building the All of Us biomedical data resource.
III.1. First Principle: Build secure, scalable, sustainable systems
The volume of biomedical research data and the velocity at which these data are multiplying present massive opportunities for discovery.43,44 At the same time, data providers and researchers wishing to harness the power of this rapid evolution face considerable challenges. Data providers must first build secure, scalable, and sustainable systems for obtaining, storing, accessing, and analyzing this wealth of data.45,46 In this section, we describe how the DRC takes research, operations, and security requirements into consideration to build one of our major system components: the RDR.
As noted in the Section II.2, the RDR is the core of the DRC’s data collection infrastructure. The RDR currently receives and facilitates transmission of a wide range of both operational and research data summarized in Table 2. Note that each stream of research data also requires operational data pipelines. EHR data serves as a good use-case to illustrate this point. Currently, All of Us collects electronic health records from more than 50 HPOs. These organizations vary in type from Federally Qualified Health Centers to major academic medical centers. Collectively, these organizations use more than 16 different EHR vendors to provide clinical services to their patient populations. Data stewards at each organization collect and share data each quarter. These data stewards match their patient records to All of Us participants and export data from their local EHR systems into a format aligned with the OMOP CDM. Data are then ingested into the RDR via an EHR Data Pipeline, conformance checks are run, and results are made available for data stewards to review. The EHR Data Pipeline is implemented and maintained by the DRC’s EHR Operations Team, which provides support and guidance to local data stewards. Furthermore, data resulting from the EHR Data Pipeline is consumed by the Program Data Repository and utilized in operational dashboards to help HPO partners and other stakeholders better understand overall data transfer performance and act on any gaps as warranted. Each quarterly EHR data submission cycle is a full data refresh to help ensure researchers have the most accurate and up-to-date participant information. Prior EHR data submissions are archived in the RDR, and only data from the most recent EHR submission is used to create a CDR. These important iterations in the EHR data upload process are the reason for the dramatic increase in the number of EHR records stored in the RDR, relative to the approximately 325,000 participants who have donated records (Table 2). Thus, in this one example of the DRC’s RDR, one can see that building a scalable, sustainable system for EHR data acquisition requires building infrastructure to support multiple teams with tightly interwoven operations and scientific components.
Table 2.
Overview of major data types currently stored in the RDR
| Data Category | Data Type | Data Source / Sharing Partner(s) | Number of Records in the RDR as of Sept 2022a |
|---|---|---|---|
| Research + Operational | Consent (Including general consent, EHR consent, and genomic return of results consent) |
Participant portalsb | 1,355,059 |
| Research + Operational | Questionnaires (18 distinct questionnaires across both portals to date) |
Participant portalsb | 2,487,109 |
| Research + Operational | Baseline physical measurements (In clinic and self-reported) |
HealthPro (in clinic) Participant portalsb (self-report) |
368,090 |
| Operational | Biospecimen orders (Blood, urine, and saliva orders) |
Biobank, Genome Centers, Clinical Validation Labs, Genetic Counseling Resources, and the DRC Genomics Curation System | 726,279 |
| Research + Operational | EHR | HPOs, Participant portalsb | 3,525,720 |
| Research + Operational | Digital Health Technology (Including Fitbit and Apple HealthKit data) |
Participant portalsb | 28,864 |
| Research + Operational | Genomic data (Array and whole genome sequencing data) |
Genome Centers, Genetic Counseling resources | 848,662 |
Record count as of September 2022 exceeds the number of enrolled participants, as participants may have multiple records, and some records are intentionally duplicated as part of operational and quality control processes. Copies are retained per the RDR’s append-only policy. Only the subset of research-grade data on currently consented participants is curated and made available for research.
All of Us participant portals are developed and managed by the Participant Technologies Systems Center and The Participant Center.
To be scalable and sustainable, it has been critical for the DRC to cultivate organizational capability to adapt quickly and engage new program partners who build relationships with and gather data in diverse communities. The DRC currently has 83 data sharing partners, including HPOs, the Participant Technologies Systems Center, the Biobank, and other program partners. Each partner may contribute many modalities of data, which have varying requirements for the timeliness of data transactions. Certain data types need to be transmitted, stored, and have receipt confirmation provided in near real-time during study visits, while other data types may be processed on other time cycles, depending on the patterns of operational activity that make those data available, and what the data will be used for. The DRC therefore supports granular configurability for asynchronous data exchanges (e.g., batch processed data), and provides synchronous data exchange methods where needed. For example, participants may not provide samples for analysis until they have completed consent in the Participant Portal, and it is synchronized to the DRC and confirmed to be present in HealthPro by clinical staff. This procedure requires the use of a highly available API with defined and managed service level targets.
In addition, as noted previously, this infrastructure must adhere to a set of stringent security requirements. Data security is fundamental to all aspects of the DRC organization. Each DRC system that touches All of Us participant data are required to achieve Federal Information Security Management Act (FISMA) Moderate+ standards47, which includes the Moderate controls as well as additional controls required to be granted the Authority to Operate (ATO) by the NIH. The DRC uses secure cloud architectures that enable elastic capacity for data acquisition, storage, and analysis. We also enlist independent, third-party reviewers to check our implementation of the FISMA Moderate security controls and test our systems on an ongoing basis to make sure we have effective security controls in place and are responsive to emerging threats. Each data sharing partner is required to have an interconnection security agreement (ISA) in place. For example, all HPOs noted above have an ISA with the DRC that covers their use of HealthPro, EHR data sharing, and access to analytical dashboards.
III.2. Second Principle: Increase data utility without sacrificing integrity and richness
Our second principle is to increase data utility without sacrificing its integrity and richness. As described in the overview above, All of Us collects and provides access to a breadth of data types from many sources. Integrating this data in a way that empowers researchers with diverse backgrounds and areas of study is a substantial challenge. It is also important to ensure that all improvements made to the data are well documented, as well as the data provenance itself.
One of the most important early decisions we made was to adopt published standards wherever possible, specifically Health Level Seven Fast Healthcare Interoperability Resources (HL7 FHIR)48 and Observational Health Data Sciences and Informatics’ (OHDSI)49–51 OMOP CDM37 standards for health data and the Global Alliance for Genomics and Health (GA4GH) standards for genomic data.52 Incorporating the experience and hard work of these international consortia into our approaches for data representation gave us a solid framework to address the challenges and connected us with existing groups in the space.
Considering OMOP, it offers two critical tools: a well-defined data model and a standardized vocabulary. We adopted OMOP in two areas: for EHR data collection from our HPO Partners and as a foundational core of our data model to share with researchers. Each of these tools provides benefits for each application.
Collecting EHR data from different sites reflects the two opportunities well suited to OMOP: data are not natively stored in the same format and may not use the same structured terminology. As noted previously, there are currently 16 EHR vendors represented; each EHR system has its own internal representation of the data. By aligning to an external standard, we have a clear target to meet that can be evaluated for accuracy and consistency.41 Many groups also had already implemented OMOP as a tool for internal researchers, and others found that institutional objectives aligned with this initiative. There are also “vendor communities” that helped many groups reach the standardization goals.
The terminology component is also critical to enabling consistent data use among EHR sites. In a healthcare setting, data are captured for multiple use cases, including billing and clinical care, which influence how they are represented. For billing, standardized vocabularies may be mandated by payors, e.g., in the United States, the Centers for Medicare and Medicaid Services require the ICD-10-CM (International Classification of Diseases, Tenth Revision, Clinical Modification) for claims reimbursement. This strongly incentivizes the capture of billing codes but does impact which codes might be recorded and why. For other aspects of clinical care, like lab measurements, aligning to standards may not have such clear benefits. In a single patient clinical care scenario, it may be evident to the treating clinician that the reported “Creatinine” measure was from a blood sample, so there may be little consequence to the lack of detail. However, in a multi-EHR setting, it is critical to ensure sufficient details are captured to properly identify the meaning of the data. By requiring OMOP conformance with standard codes, we ensure the local data stewards most familiar with the data are those ensuring compliance. This was especially important initially when there was little data context to work with; we can leverage the data at scale to assist with missing units now that we have an established dataset.
When it comes to providing data to researchers, the OMOP CDM inherently has valuable properties as a data model for analytics. First, the standardized vocabularies are designed to have a single “standard concept” for each unique “term” that are connected with “is a” parent-child relationships. These factors together mean that a user can look for “Type 2 Diabetes”, and quickly find all rows in the condition occurrence table that make assertions of Type 2 Diabetes status, no matter what source vocabulary was used. While the CDM can be intimidating, users with a technical background can also find the table structure approachable. The use of domains gives each row a “home,” and conventions provided by the OHDSI community and described in our data dictionaries help users find data. The CDR houses many different data sources within OMOP, supported in part by the inclusion of the All of Us specific vocabulary. Program survey questions and physical measurements collected have All of Us specific terms that are linked, where possible, to the standardized vocabularies within OMOP (Figure 2). This enables users to look for family history data that was self-reported or extracted from the EHR in the same way.53,54 Finally, OMOP makes is easy to expand the CDR as the program adds new health data. For example, the DRC is actively working to collect clinical documents and apply natural language processing to extract health information embedded in clinical notes,55–57 which can easily be stored and accessed in OMOP notes tables.
Figure 2.
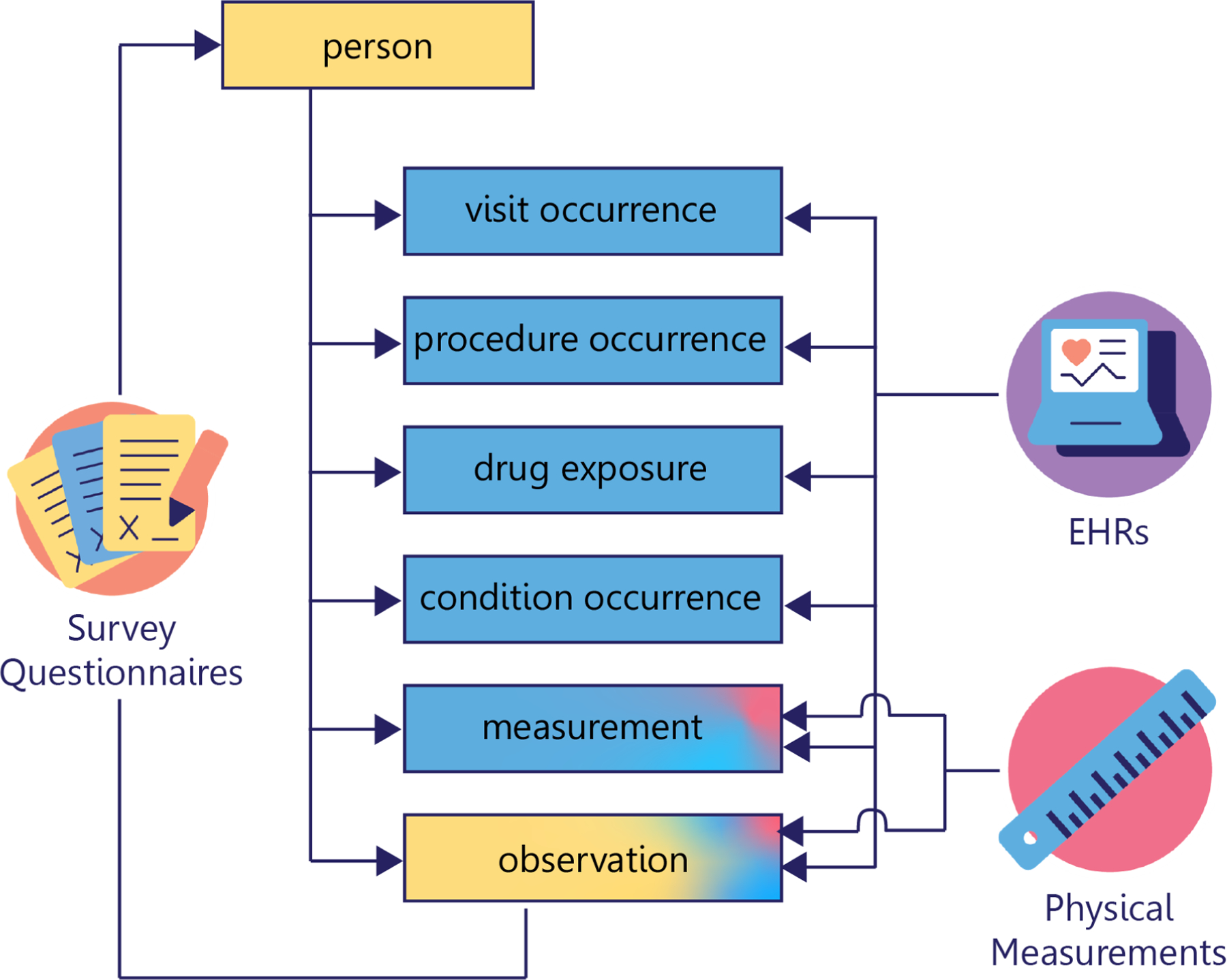
All of Us data are organized into tables according to the Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM), when possible. Self-reported demographic data from the Basics survey populates the person table. Other data obtained from surveys are found in the observation table. Program physical measurements as well as Electronic Health Record (EHR) measurements populate the measurement table. EHR data concerning visits, procedures, drugs, and conditions are arranged into their respective tables. All tables relate to the person table and the tables containing procedure, drug, condition, and measurement data relate to the visit occurrence table.
HL7 FHIR is also used in All of Us, currently as an interoperability standard. It allows us flexibility to exchange a variety of data within the Program, and even collect EHR data with exchange from individual Participants healthcare systems. Internally, we use FHIR to send data from the Participant Portals to the RDR. We also use FHIR to send data from the HealthPro application to the RDR, structuring the data exchange in a way that includes All of Us specific codes alongside the proper standardized terminologies. This dual-coding is carried into the OMOP CDM, which enables ease of use and integration with EHR data, while also allowing a detailed look at the study-specific nuances, e.g., the order of our multiple captured blood pressure measures.
The support of SMART on FHIR58 and right of access for patients also translates into exciting data sharing opportunities for research efforts like All of Us. Participants can choose to link their EHR directly with their participant portal account, allowing the All of Us Research Program to access their health data in FHIR, even if the place they receive care is not an All of Us Partner that shares EHR data directly with the program. Participants can also share data aggregated on their devices with Apple Health Records, which uses the same FHIR APIs to gather their EHR data. These tools are a crucial step towards achieving a more complete set of clinical data on participants who are likely to receive care at several different organizations. These APIs will be required to be implemented by 31 December 2022 consequent to the 21st Century Cures Act59 and Office of the National Coordinator for Health Information Technology (ONC) mandated implementation.60 Ensuring consistency, quality, and usability of this data that is not curated by local experts will be a challenge moving forward.
Due to the complexity of these many data sources and the added layers of transformation employed to support privacy and usability (discussed in more detail in Section III.3), it is also important that we maintain data provenance and provide users with details of these transformations. These goals are addressed with row-level data provenance and a breadth of support resources. The row-level data provenance describes the data source for each row, including differentiating between participant provided surveys, physical measurements collected by the program, and masked indicators of which HPO site uploaded a given element of EHR data.
We provide support and data dictionary resources61 with each release of the CDR that describe the table structures in addition to any other modifications we have made to the source data. Our researcher-facing data model includes the OMOP CDM tables as well as supplementary tables for data provenance and data like Fitbit that is not integrated into the OMOP CDM. The data dictionary clearly articulates what is available to researchers. The data dictionary also provides the details of the privacy methodology, including details about which data was suppressed or generalized, described in more detail below.
Finally, we made the decision to provide two versions of the data to researchers: a base version and a standardized version. The standardized version is more stringent with regards to data curation and provides additional transformations to support increased usability. For example, units for height and weight are standardized and measures that appear to be errors are dropped.62 This cleaner and more user friendly dataset supports a majority of research use cases. We also support users who may wish to develop their own data cleaning methods by providing the base dataset as well. This version includes more data and fewer modifications to the source data. It may be less usable to a broad audience, but is valuable to methods developers, including bioinformaticians whose work requires a rawer form of data.
III.3. Third Principle: Share data widely and wisely
All of Us is committed to honoring participants’ desire for their contributions to empower research, which requires making data broadly accessible. All of Us is also committed to honoring participants’ desire for their contributions to be used appropriately, which requires implementing privacy protections. The DRC’s approach to achieving the appropriate balance between these two goals is guided by our third principle to “Share data widely and wisely.” In this section we describe how the DRC addressed various issues affecting All of Us data accessibility, including policy and researcher support infrastructure.63
The primary set of policies governing access to All of Us data resources are described in detail on the Research Hub public website.64 Within these policies is the All of Us Data Access Framework, which explains how All of Us data are structured into different access tiers for use (Figure 3) and the steps that researchers must take to access the data.
Figure 3.
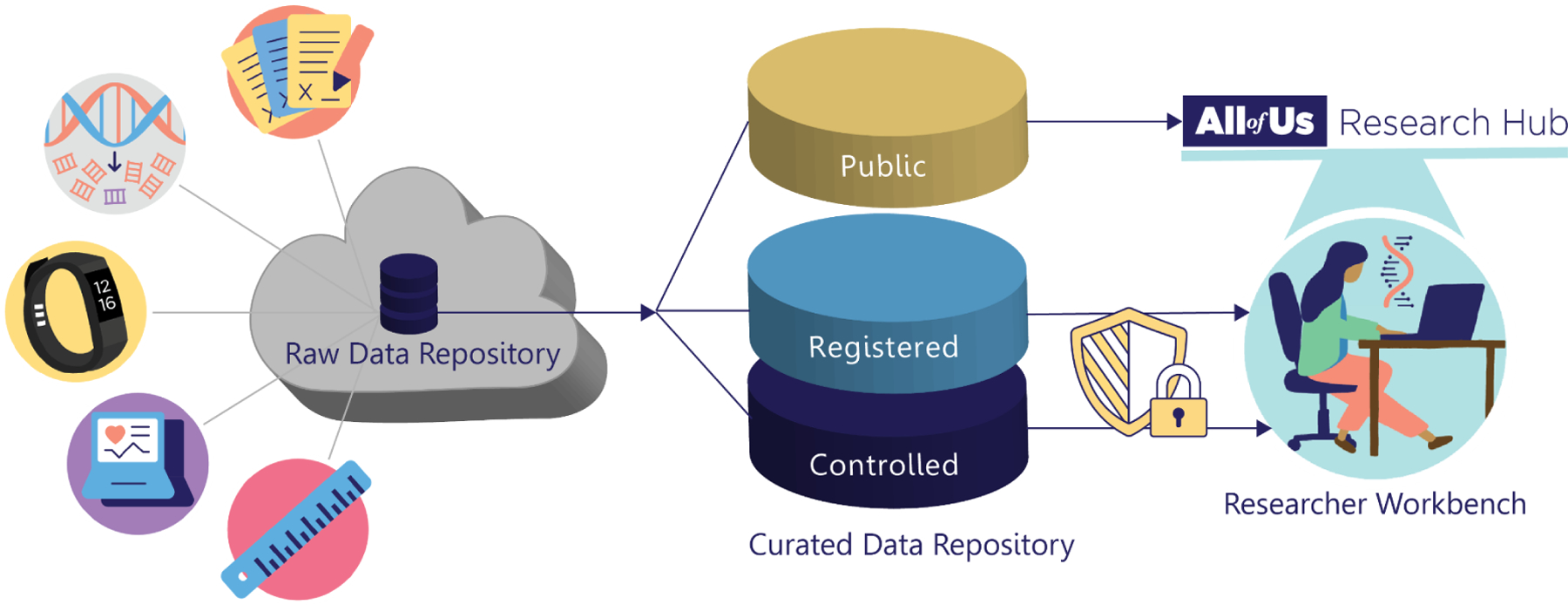
All of Us participant data are collected into the Raw Data Repository (RDR), then harmonized, organized, and further processed into a Curated Data Repository (CDR). This repository is structured into three tiers of data with corresponding tiers of access requirements: Public (no login required), Registered (login required), and Controlled (login and additional approval required).
All of Us chose to create three tiers of data access: Public (no login required), Registered (login required), and Controlled (additional approval required). The Public Tier provides aggregate participant data (e.g. number of participants with a Type 2 Diabetes diagnoses) for broad use via a public Data Browser.65 In contrast, access to participant-level records in either the Registered or Controlled Tiers requires users to complete a multi-step access process (Figure 4). To support efforts at broadening the range of users who can analyze participant-level All of Us data, the program intends Registered Tier to be easier to access compared to the Controlled Tier. Therefore, the DRC applies various transformations to lower participant re-identification risk for the Registered Tier. Registered Tier re-identification risk mitigation policy includes suppression, generalization, and randomization (including date-shifting and removal of all direct identifiers), consistent with the Health Insurance Portability and Accountability Act (HIPAA) Safe Harbor Standards for de-identification.66 The Controlled Tier contains the most granular and potentially sensitive data. For example, the Controlled Tier contains participant whole genome sequences and unshifted dates of events. Consequently, access requirements for the Controlled Tier build on those for the Registered Tier to further establish user trustworthiness. Researchers are therefore able to apply for the level of access required for their specific research purpose, and the requirements for accessing data are appropriate to the level of participant re-identification risk in each tier.
Figure 4.
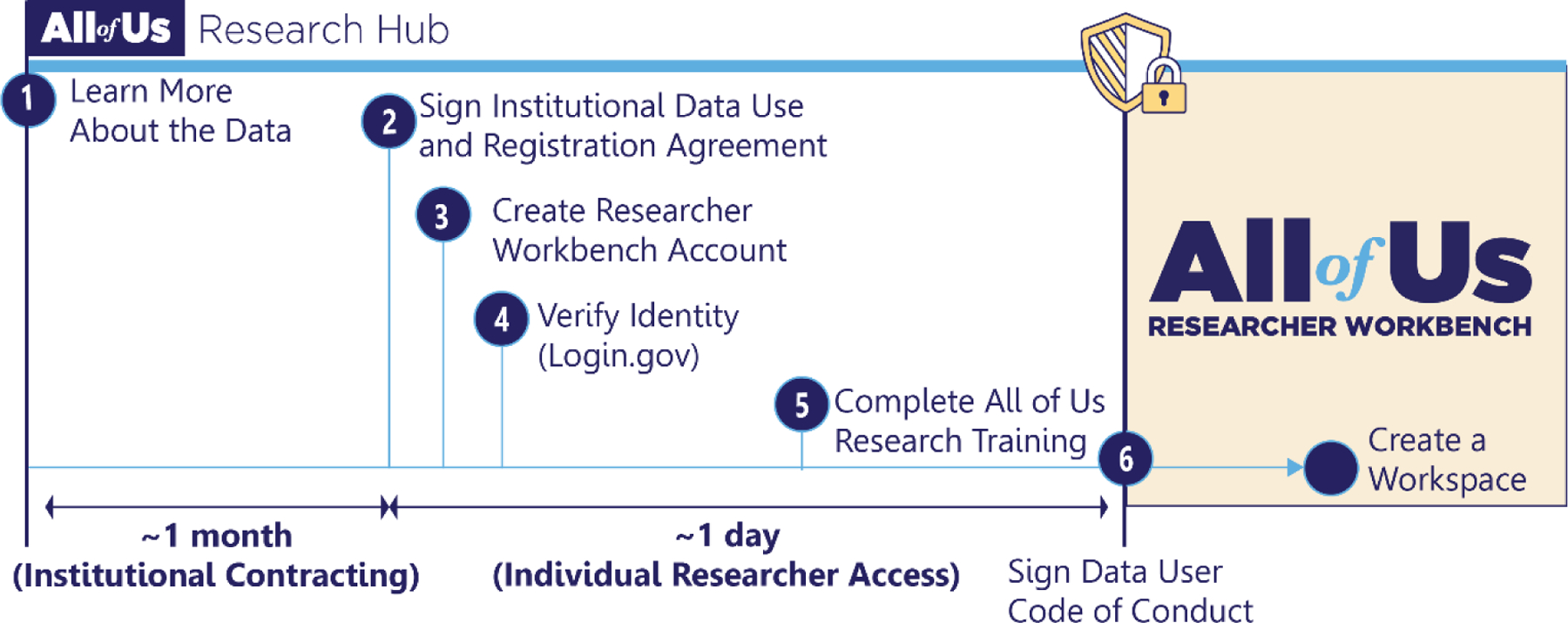
Researchers wishing to analyze participant-level data must follow a 6-step access process: (1) explore data and policies on the Research Hub’s public website, (2) check that their institution has signed a Data Use and Registration Agreement. The indicated period of institutional contracting is only required if a researcher’s institution does not have an agreement in place. Researchers may then (3) create a Researcher Workbench account, (4) verify their identity using Login.gov, (5) complete required responsible and ethical research training, and (6) sign an individual data user code of conduct. Upon approval, researchers are granted a “data passport” enabling them to create a workspace to access and analyze All of Us participant data within the Researcher Workbench.
Authorization for access to both Registered and Controlled Tier data in All of Us is user based, rather than project based. Researchers apply for access via the Research Hub, following the process outlined in Figure 4, and receive a “data passport” indicating which tier of data they are authorized to use. Currently, this process requires researchers to be affiliated with a U.S.-based academic, nonprofit, or health care institution to enter into our Data Use and Registration Agreement. The program is, however, actively working to broaden access to international researchers, industry researchers, and community scientists conducting research outside of academic medical centers. Approved researchers must complete analyses within the Researcher Workbench, by creating workspaces for use by them and their collaborators. Upon workspace creation, users are required to submit a description of their project. These descriptions are made available publicly on the Research Hub’s Research Projects Directory,67 furthering our commitment to partnership and transparency with participants,68 and in compliance with the 21st Century Cures Act (Pub.L. 114–255). Together, these measures are aimed at removing unnecessary barriers while protecting participant privacy and helping to ensure appropriate data use.
As of September 30, 2022, 2,367 researchers from 409 different institutions across the United States have completed this process to become authorized users of All of Us data. Together, they have created more than 2,553 workspaces to analyze All of Us data. Over 66% of these researchers are authorized to access Controlled Tier data, and these researchers created over 890 workspaces in the seven months since the initial launch of the Controlled Tier within the Researcher Workbench. The breadth of this research activity is evidenced by the diseases that researchers are exploring. To identify the top diseases that researchers are currently investigating using the All of Us data, we used Clinical Language Annotation, Modeling, and Processing (CLAMP),57 a natural language extraction tool, to extract medical terms from the projects’ titles. As Figure 5 depicts, All of Us researchers are studying a wide range of diseases. Currently, hypertensive disorders, malignant neoplasm, diabetes, and mental depression are the four diseases most frequently investigated using the All of Us data. Additional analysis of workspace descriptions reveals that beyond disease-focused research, the All of Us research community is leveraging this resource for educational purposes and methods / tools development.
Figure 5.
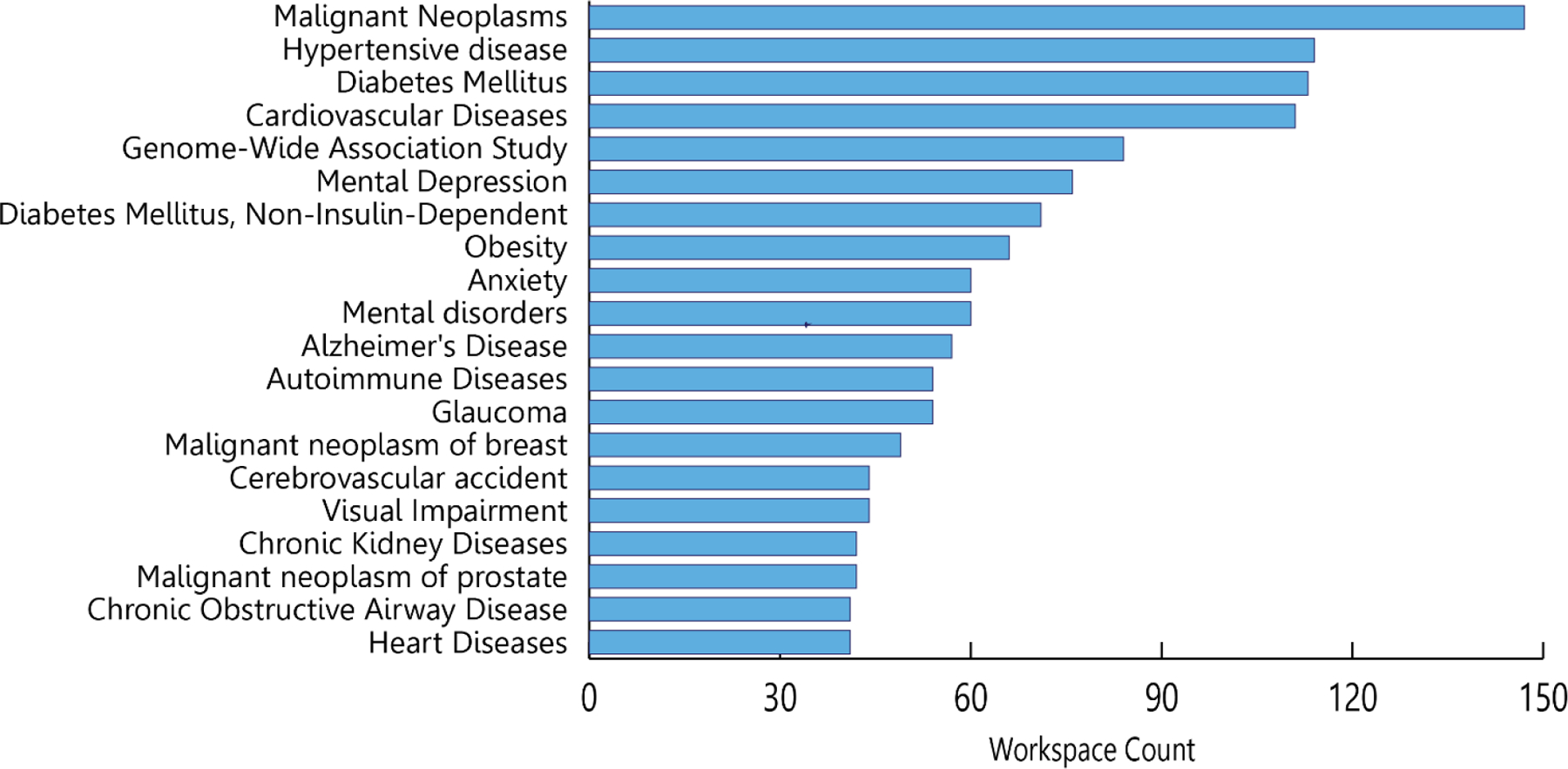
The 20 most common areas of researcher investigation according to medical terms in the projects’ self-reported descriptive summaries.
As Figure 5 illustrates, researchers from a range of disciplines currently intend to use All of Us data. With such a diverse research community, data access alone is insufficient to ensure usage. For these researchers to maximize the innate potential of the All of Us dataset requires multidisciplinary knowledge in: 1) health literacy, defined as the ability to extract, process, and comprehend health information, and 2) data literacy, defined as the ability to examine a phenomena by extracting, analyzing, and comprehend the data.69,70 Providing resources and venues to address their questions, comments, and feedback is crucial to ensure the use of these data.
The DRC, therefore, made the decision to integrate extensive research support infrastructure within the Research Hub. This includes a curated repository of onboarding, training, and support materials known as the User Support Hub. It also includes a help desk through which researchers can get expert support from the DRC’s Research Support Team. The Research Support Team also holds monthly orientation and weekly office hours sessions, available to all authorized researchers, to introduce researchers to the data, answer questions, and introduce researchers to recently published projects. Moreover, this team works with subject matter experts across the All of Us consortium to create a Featured Workspace Library. This library contains detailed tutorials and end-to-end example analysis projects. Collectively, these resources can help researchers to understand All of Us data and identify the best methods to extract and analyze data for their research projects.
III.4. Fourth Principle: Support useful, generalizable, and accessible tools and workflows
Our fourth principle is to support useful, generalizable, and accessible tools and workflows. This goal aligns with the earlier principle to share data widely, and stems from the program’s charter to advance diversity in precision medicine, which applies to both participants and researchers. We aim to make this rich dataset usable for researchers across the globe, spanning disciplines, career stages, resource levels, and analytical approaches. Whereas the previous section focused on All of Us data access policies and research support infrastructure, in this section we dive more deeply into our approach to building the technical infrastructure behind the Researcher Workbench.
The broad intended research audience for All of Us makes it challenging to define and achieve success. Additionally, this vision of broad use must be accomplished within the constraints set by the All of Us Research Program policy framework ‒ individual-level participant data must remain on the cloud-based Researcher Workbench Trusted Research Environment, and researchers must login and perform all their work on this platform. To overcome these challenges, the DRC chose to take a user-centered, iterative approach to develop the Researcher Workbench. Put simply, we chose to “launch small, and iterate” based on real-world user feedback from the All of Us researcher community.
The first step the DRC took to building the Researcher Workbench was to conduct user research to better understand researcher needs. Initial reports from this work recommended that the Researcher Workbench begin by ensuring success for computationally sophisticated, traditional researchers, then grow data and tool depth and capability over time. They also underscored the importance of collaboration and team-based approaches to realize the full potential inherent in big data. As a result, we made the decision to build the Researcher Workbench on the established Terra platform.71 Terra is a secure, scalable, open-source platform for biomedical researchers to access data, run analysis tools, and collaborate. Terra extends cloud-native tools and capabilities with researcher-focused features to address today’s research challenges. The Terra platform provides the infrastructure to enable reproducible and collaborative research, in accordance with FAIR principles.72
We also made the choice to store All of Us data in BigQuery, a multicloud data warehouse for hosting relational databases like the OMOP-formatted CDR (curated data repository) data described in the previous section. As a result, extracting data directly from this BigQuery database requires a combination of health and data literacy to identify clinical concepts and write the queries to retrieve the data. This presented a significant barrier to extracting data for those researchers without this expertise. The DRC therefore developed point-and-click graphical user interface tools built on top of BigQuery that allow researchers to extract the data more easily: the Cohort Builder and Dataset Builder.
Researchers use the Cohort Builder to select participants for analysis based on custom inclusion and exclusion criteria. This tool is powered by flexible search methods that utilize hierarchical relationships between concepts to extract possible concepts from the underlying data. Researchers may then use the Dataset Builder to create an analysis-ready dataset by selecting specific health concepts of relevance to their study. Importantly, neither of these tools require researchers to have extensive knowledge in programming, BigQuery, or OMOP.
Another important decision we made was to select the initial analysis tool for the Researcher Workbench minimum viable product. Jupyter Notebooks,73 SAS,74 SPSS,75 RStudio,76 and Stata77 were identified as potential candidates for an initial data analysis tool within the Researcher Workbench. Subsequent evaluation of these tools rated candidates according to their ability to provide support for big data analysis and visualization, versatility, collaboration support, reproducibility, extensibility, cloud integration, and cost. From this evaluation, Jupyter Notebooks with support for SQL, Python, and R programming languages were selected as our initial data analysis tool. The DRC then initiated extensive user testing to collect feedback on the Workbench, first conducting alpha testing with a small group of <100 users from within the All of Us Research Program consortium and then launching a beta version publicly in May of 2020.41
Just as the study collects data longitudinally, adapting to new opportunities in enrollment and data collection, our offerings to researchers must also evolve over time. Since its initial launch in May of 2020, the DRC has released 5 increasingly rich versions of the CDR to a growing community of researchers. As the CDR increases in volume and new data types are added, we make corresponding improvements in the Workbench software to facilitate analysis. This continual evolution is made possible by our Workbench engineering team, who employ Agile-like software development practices,78 releasing updated versions of the Workbench Software on an approximately bi-weekly cadence. For example, as part of the launch of genomics data to All of Us researchers in March 2022, the DRC incorporated new tools, including Hail79 and Plink,80 to allow access and analysis of genomic data, while reducing the cost and computational burden on researchers. We also upgraded the computational environment to support custom Dataproc clusters suited to large-scale genomic analysis. Due to the popularity of deep learning methods, we added Graphics Processing Unit support for researchers who want to train deep learning models using the All of Us dataset. To accommodate the large scale of genomic data available, we have also added command line support for popular batch genomic workflow engines: Nextflow81 and Cromwell.82 Finally, users are often able to install their own tools to support analyses. For example, pip is available for users to install new python libraries developed by the community. These tools, together with other tools available in the Workbench, enable researchers to utilize linked phenotypic and genotypic data to perform a broad spectrum of research on any disease of interest.
III. Lessons Learned from All of Us
The All of Us Research Program is helping to shape the future of biomedical research, and the practice of implementing the All of Us data ecosystem has surfaced learnings relevant to the broader biomedical research community.
Fortify trust in the community to encourage participation
Participants and the data they contribute are at the core of the research program. The better informed they are of the intended use of data, the more likely they are to contribute. Finding new methods to engage with the community transparently, including openly sharing the benefits and risks of health research, and research progress and impact, will help maintain trust, mitigate concerns, and encourage more active participation.11 Moreover, frequent security assessments to ensure compliance with the evolving regulatory and security landscape will strengthen public trust and safety.
Strengthen data infrastructure and capacity to support big data for health
New modalities and increasing volumes of healthcare data will require increasingly flexible compute power and storage. Building a solution to maintain data integrity and quality while facilitating secure and compliant data linkage and harmonization is fundamental to generating longitudinal patient insights.
Operate as a production grade software engineering organization
Scalable and sustainable systems require production grade engineering and operational processes. This is imperative to the secure and effective management of complex systems of data generators and consumers.
Standardize data management and support data quality across modalities
Data heterogeneity introduces complexities even before analysis begins. Building a platform to ingest, integrate, and activate different modalities of healthcare data without sacrificing clinically rich information is critical to generating representative insights. Standardization requires multi-disciplinary collaborations to create uniform clinical definitions, data structure, and format. Furthermore, data quality and provenance are key factors to power clinical-grade research. Providing transparency into these measures will enhance research insights and findings.
Advance an extensible governance framework for collaboration
Collaborations across disciplines and industry, whereby researchers share data and methods, are increasingly becoming the standard for research.83,84 As regulatory and security requirements evolve, biomedical research platforms must continue to adjust to stakeholder and system requirements to protect personal health information and create an auditable trail to ensure research is reproducible and safeguards the integrity of the science. Creating safe passages to access valuable health data will enable innovations to flourish.
Simplify and support the researcher experience to accelerate science
The abundance of healthcare data poses increasing challenges, calling for the need of modern artificial intelligence/machine learning methods and algorithms to facilitate discoveries. However, the heterogeneity of user types creates opportunities for platforms to build more interactive tooling to power different research studies. Creating interactive tools encourages more research participation and activity. In addition, although recent training programs in biomedical, health, and clinical informatics have been established to train and equip the next generation of scientists with the necessary knowledge of clinical terminologies and taxonomies, ethical usage of information technologies, and skills to extract and process information,85,86 many researchers do not have all necessary health literacy or data literacy knowledge to use these massive health datasets efficiently. It is therefore imperative to provide researchers with robust onboarding, training, and support resources in parallel to data and tools.
IV. Conclusions and Outlook
Building a modern biomedical data ecosystem is a balancing act. Builders must consider the needs of participants, program staff, and researchers. Data must be kept private and secure and be shared widely for use. Data must be organized and transformed into more usable forms without losing accuracy or provenance. Researcher onboarding must be made easy, but with enough controls deployed to reduce risk of bad actors. Researcher-facing tools must be both powerful and easy to use. Ultimately, the guiding principles and lessons learned while building the All of Us Data and Research Center are available for others to use to navigate these important decisions and trade-offs.
Acknowledgements:
The authors would like to note that the Researcher Workbench is made possible by the Terra platform, which is co-developed by The Broad Institute of MIT and Harvard, Microsoft, and Verily. The All of Us Research Program is supported by the National Institutes of Health, Office of the Director: Regional Medical Centers: 1 OT2 OD026549; 1 OT2 OD026554; 1 OT2 OD026557; 1 OT2 OD026556; 1 OT2 OD026550; 1 OT2 OD 026552; 1 OT2 OD026553; 1 OT2 OD026548; 1 OT2 OD026551; 1 OT2 OD026555; IAA #: AOD21037, AOD22003, AOD16037; Federally Qualified Health Centers: HHSN 263201600085U; Data and Research Center: 5 U2C OD023196; Biobank: 1 U24 OD023121; The Participant Center: U24 OD023176; Participant Technology Systems Center: 1 U24 OD023163; Communications and Engagement: 3 OT2 OD023205; 3 OT2 OD023206; and Community Partners: 1 OT2 OD025277; 3 OT2 OD025315; 1 OT2 OD025337; 1 OT2 OD025276. In addition, the All of Us Research Program would not be possible without the partnership of its participants.
LITERATURE CITED
- 1.The Cancer Genome Atlas Program - NCI. 2018. URL: https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga (Accessed 30 September 2022).
- 2.ENCODE. n.d. URL: https://www.encodeproject.org/ (Accessed 5 October 2022).
- 3.Human Genome Diversity Project. n.d. URL: https://hagsc.org/hgdp/ (Accessed 5 October 2022).
- 4.Reuter MS, Walker S, Thiruvahindrapuram B, Whitney J, Cohn I, Sondheimer N, et al. The Personal Genome Project Canada: findings from whole genome sequences of the inaugural 56 participants. CMAJ 2018;190:E126–36. 10.1503/cmaj.171151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.CHARGE Consortium. n.d. URL: https://www.chargeconsortium.com/ (Accessed 5 October 2022).
- 6.UK Biobank. n.d. URL: https://www.nature.com/collections/bpthhnywqk (Accessed 5 October 2022). [Google Scholar]
- 7.Chen Z, Group on behalf of the CKB (CKB) collaborative, Chen J, Group on behalf of the CKB (CKB) collaborative, Collins R, Group on behalf of the CKB (CKB) collaborative, et al. China Kadoorie Biobank of 0.5 million people: survey methods, baseline characteristics and long-term follow-up. Int J Epidemiol 2011;40:1652–66. 10.1093/ije/dyr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schatz MC, Philippakis AA, Afgan E, Banks E, Carey VJ, Carroll RJ, et al. Inverting the model of genomics data sharing with the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-space. Cell Genom 2022;2:100085. 10.1016/j.xgen.2021.100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Bajari R, Andric D, Gerthoffert F, Lepsa A, Nahal-Bose H, et al. The International Cancer Genome Consortium Data Portal. Nat Biotechnol 2019;37:367–9. 10.1038/s41587-019-0055-9. [DOI] [PubMed] [Google Scholar]
- 10.The “All of Us” Research Program | NEJM. n.d. URL: https://www.nejm.org/doi/full/10.1056/NEJMsr1809937 (Accessed 6 October 2022).
- 11.Mapes BM, Foster CS, Kusnoor SV, Epelbaum MI, AuYoung M, Jenkins G, et al. Diversity and inclusion for the All of Us research program: A scoping review. PLOS ONE 2020;15:e0234962. 10.1371/journal.pone.0234962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ginsburg GS, Phillips KA. Precision Medicine: From Science To Value. Health Aff (Millwood) 2018;37:694–701. 10.1377/hlthaff.2017.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robinson PN. Deep phenotyping for precision medicine. Hum Mutat 2012;33:777–80. 10.1002/humu.22080. [DOI] [PubMed] [Google Scholar]
- 14.Torous J, Kiang MV, Lorme J, Onnela J-P. New Tools for New Research in Psychiatry: A Scalable and Customizable Platform to Empower Data Driven Smartphone Research. JMIR Ment Health 2016;3:e16. 10.2196/mental.5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khodyakov D, Bromley E, Evans SK, Sieck K. Best Practices for Participant and Stakeholder Engagement in the All of Us Research Program. RAND Corporation; 2018. [Google Scholar]
- 16.Fighting Unfairness in Genetic Medicine - Scientific American. n.d. URL: https://www.scientificamerican.com/article/fighting-unfairness-in-genetic-medicine/ (Accessed 21 October 2022).
- 17.Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature 2016;538:161–4. 10.1038/538161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baxter SL, Saseendrakumar BR, Paul P, Kim J, Bonomi L, Kuo T-T, et al. Predictive Analytics for Glaucoma Using Data From the All of Us Research Program. Am J Ophthalmol 2021;227:74–86. 10.1016/j.ajo.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lyles CR, Lunn MR, Obedin-Maliver J, Bibbins-Domingo K. The new era of precision population health: insights for the All of Us Research Program and beyond. Journal of Translational Medicine 2018;16:211. 10.1186/s12967-018-1585-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bohnert K Thematic analysis of sexual and gender minority enrollment in the all of us Pennsylvania project: implications for public health research 2019:151. [Google Scholar]
- 21.Tabak LA, Collins FS. Weaving a Richer Tapestry in Biomedical Science. Science 2011;333:940–1. 10.1126/science.1211704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oh SS, Galanter J, Thakur N, Pino-Yanes M, Barcelo NE, White MJ, et al. Diversity in Clinical and Biomedical Research: A Promise Yet to Be Fulfilled. PLOS Medicine 2015;12:e1001918. 10.1371/journal.pmed.1001918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Advisory Committee to the Director, NIH. ThePrecision Medicine Initiative Cohort Program–Building a Research Foundation for 21st Century Medicine. 2015.
- 24.Doerr M, Moore S, Barone V, Sutherland S, Bot BM, Suver C, et al. Assessment of the All of Us research program’s informed consent process. AJOB Empirical Bioethics 2021;12:72–83. 10.1080/23294515.2020.1847214. [DOI] [PubMed] [Google Scholar]
- 25.Aschebrook-Kilfoy B, Zakin P, Craver A, Shah S, Kibriya MG, Stepniak E, et al. An Overview of Cancer in the First 315,000 All of Us Participants. PLOS ONE 2022;17:e0272522. 10.1371/journal.pone.0272522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harrison SM, Austin-Tse CA, Kim S, Lebo M, Leon A, Murdock D, et al. Harmonizing variant classification for return of results in the All of Us Research Program. Human Mutation 2022;43:1114–21. 10.1002/humu.24317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Venner E, Muzny D, Smith JD, Walker K, Neben CL, Lockwood CM, et al. Whole-genome sequencing as an investigational device for return of hereditary disease risk and pharmacogenomic results as part of the All of Us Research Program. Genome Med 2022;14:1–13. 10.1186/s13073-022-01031-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.NIH Strategic Plan for Data Science | Data Science at NIH. n.d. URL: https://datascience.nih.gov/nih-strategic-plan-data-science (Accessed 4 October 2022). [Google Scholar]
- 29.Rossi RL, Grifantini RM. Big Data: Challenge and Opportunity for Translational and Industrial Research in Healthcare. Frontiers in Digital Humanities 2018;5:. [Google Scholar]
- 30.Hong L, Luo M, Wang R, Lu P, Lu W, Lu L. Big Data in Health Care: Applications and Challenges. Data and Information Management 2018;2:175–97. 10.2478/dim-2018-0014. [DOI] [Google Scholar]
- 31.Discover the 4 V’s of Big Data. OpenSistemas 2020. URL: https://opensistemas.com/en/the-four-vs-of-big-data/ (Accessed 4 October 2022).
- 32.Doerr M, Grayson S, Moore S, Suver C, Wilbanks J, Wagner J. Implementing a universal informed consent process for the All of Us Research Program. Pac Symp Biocomput 2019;24:427–38. [PMC free article] [PubMed] [Google Scholar]
- 33.Participant Technology Systems Center. All of Us Research Program | NIH. 2020. URL: https://allofus.nih.gov/funding-and-program-partners/participant-technology-systems-center (Accessed 22 November 2022). [Google Scholar]
- 34.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. Journal of Biomedical Informatics 2009;42:377–81. 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris PA, Taylor R, Minor BL, Elliott V, Fernandez M, O’Neal L, et al. The REDCap consortium: Building an international community of software platform partners. Journal of Biomedical Informatics 2019;95:103208. 10.1016/j.jbi.2019.103208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cronin RM, Jerome RN, Mapes B, Andrade R, Johnston R, Ayala J, et al. Development of the Initial Surveys for the All of Us Research Program. Epidemiology 2019;30:597–608. 10.1097/EDE.0000000000001028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Overhage JM, Ryan PB, Reich CG, Hartzema AG, Stang PE. Validation of a common data model for active safety surveillance research. J Am Med Inform Assoc 2012;19:54–60. 10.1136/amiajnl-2011-000376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner SP, Pompea ST, Williams KL, Kraemer DA, Sholle ET, Chen C, et al. Implementation of Informatics to Support the NIH All of Us Research Program in a Healthcare Provider Organization. AMIA Jt Summits Transl Sci Proc 2019;2019:602–9. [PMC free article] [PubMed] [Google Scholar]
- 39.Klann JG, Joss MAH, Embree K, Murphy SN. Data model harmonization for the All Of Us Research Program: Transforming i2b2 data into the OMOP common data model. PLOS ONE 2019;14:e0212463. 10.1371/journal.pone.0212463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engel N, Wang H, Jiang X, Lau CY, Patterson J, Acharya N, et al. EHR Data Quality Assessment Tools and Issue Reporting Workflows for the ‘All of Us’ Research Program Clinical Data Research Network. AMIA Annu Symp Proc 2022;2022:186–95. [PMC free article] [PubMed] [Google Scholar]
- 41.Ramirez AH, Sulieman L, Schlueter DJ, Halvorson A, Qian J, Ratsimbazafy F, et al. The All of Us Research Program: Data quality, utility, and diversity. Patterns 2022;3:100570. 10.1016/j.patter.2022.100570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhou W, Kanai M, Wu K-HH, Humaira R, Tsuo K, Hirbo JB, et al. Global Biobank Meta-analysis Initiative: powering genetic discovery across human diseases 2021:2021.11.19.21266436. 10.1101/2021.11.19.21266436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fan J, Han F, Liu H. Challenges of Big Data analysis. National Science Review 2014;1:293–314. 10.1093/nsr/nwt032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Healthcare Big Data and the Promise of Value-Based Care. https://catalyst.nejm.org/doi/full/10.1056/CAT.18.0290. NEJM Catalyst 2018.
- 45.Navale V, von Kaeppler D, McAuliffe M. An overview of biomedical platforms for managing research data. J of Data, Inf and Manag 2021;3:21–7. 10.1007/s42488-020-00040-0. [DOI] [Google Scholar]
- 46.Yin Z, Lan H, Tan G, Lu M, Vasilakos AV, Liu W. Computing Platforms for Big Biological Data Analytics: Perspectives and Challenges. Comput Struct Biotechnol J 2017;15:403–11. 10.1016/j.csbj.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stine KM, Kissel RL, Barker WC, Lee A, Fahlsing J, Gulick J. SP 800–60 Rev. 1. Volume I: Guide for Mapping Types of Information and Information Systems to Security Categories; Volume II: Appendices to Guide for Mapping Types of Information and Information Systems to Security Categories. [Google Scholar]
- 48.HL7 Fast Healthcare Interoperability Resources - FHIR v4.3.0. n.d. URL: https://hl7.org/fhir/ (Accessed 4 October 2022).
- 49.OHDSI – Observational Health Data Sciences and Informatics n.d. URL: https://www.ohdsi.org/ (Accessed 5 October 2022). [Google Scholar]
- 50.Hripcsak G, Schuemie MJ, Madigan D, Ryan PB, Suchard MA. Drawing Reproducible Conclusions from Observational Clinical Data with OHDSI. Yearb Med Inform 2021;30:283–9. 10.1055/s-0041-1726481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Stud Health Technol Inform 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 52.THE GLOBAL ALLIANCE FOR GENOMICS AND HEALTH. A federated ecosystem for sharing genomic, clinical data. Science 2016;352:1278–80. 10.1126/science.aaf6162. [DOI] [PubMed] [Google Scholar]
- 53.Cronin RM, Halvorson AE, Springer C, Feng X, Sulieman L, Loperena-Cortes R, et al. Comparison of family health history in surveys vs electronic health record data mapped to the observational medical outcomes partnership data model in the All of Us Research Program. Journal of the American Medical Informatics Association 2021;28:695–703. 10.1093/jamia/ocaa315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sulieman L, Cronin RM, Carroll RJ, Natarajan K, Marginean K, Mapes B, et al. Comparing medical history data derived from electronic health records and survey answers in the All of Us Research Program. Journal of the American Medical Informatics Association 2022;29:1131–41. 10.1093/jamia/ocac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. Journal of the American Medical Informatics Association 2010;17:19–24. 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Y, Denny JC, Trent Rosenbloom S, Miller RA, Giuse DA, Wang L, et al. A long journey to short abbreviations: developing an open-source framework for clinical abbreviation recognition and disambiguation (CARD). Journal of the American Medical Informatics Association 2017;24:e79–86. 10.1093/jamia/ocw109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soysal E, Wang J, Jiang M, Wu Y, Pakhomov S, Liu H, et al. CLAMP – a toolkit for efficiently building customized clinical natural language processing pipelines. Journal of the American Medical Informatics Association 2018;25:331–6. 10.1093/jamia/ocx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.SMART Health IT. SMART Health IT. n.d. URL: https://smarthealthit.org/ (Accessed 4 October 2022). [Google Scholar]
- 59.21st Century Cures Act. FDA. FDA; 2020. URL: https://www.fda.gov/regulatory-information/selected-amendments-fdc-act/21st-century-cures-act (Accessed 4 October 2022).
- 60.Information Blocking and the ONC Health IT Certification Program: Extension of Compliance Dates and Timeframes in Response to the COVID-19 Public Health Emergency. Federal Register. 2020. URL: https://www.federalregister.gov/documents/2020/11/04/2020-24376/information-blocking-and-the-onc-health-it-certification-program-extension-of-compliance-dates-and (Accessed 4 October 2022). [Google Scholar]
- 61.Data Methods – All of Us Research Hub n.d. URL: https://www.researchallofus.org/data-tools/methods/ (Accessed 10 October 2022).
- 62.Khan MS, Carroll RJ. Inference-based correction of multi-site height and weight measurement data in the All of Us research program. Journal of the American Medical Informatics Association 2022;29:626–30. 10.1093/jamia/ocab251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cimino JJ, Ayres EJ. The Clinical Research Data Repository of the US National Institutes of Health. Stud Health Technol Inform 2010;160:1299–303. [PMC free article] [PubMed] [Google Scholar]
- 64.Data Access Tiers – All of Us Research Hub n.d. URL: https://www.researchallofus.org/data-tools/data-access/ (Accessed 7 October 2022).
- 65.Data Browser | All of Us Public Data Browser. n.d. URL: https://databrowser.researchallofus.org/ (Accessed 21 October 2022).
- 66.Guidance Regarding Methods for De-identification of Protected Health Information in Accordance with the Health Insurance Portability and Accountability Act (HIPAA) Privacy Rule. HHS.Gov. 2012. URL: https://www.hhs.gov/hipaa/for-professionals/privacy/special-topics/de-identification/index.html (Accessed 6 October 2022).
- 67.Research Projects Directory – All of Us Research Hub n.d. URL: https://www.researchallofus.org/research-projects-directory/ (Accessed 10 October 2022).
- 68.Precision Medicine Initiative: Privacy and Trust Principles. All of Us Research Program | NIH. 2020. URL: https://allofus.nih.gov/protecting-data-and-privacy/precision-medicine-initiative-privacy-and-trust-principles (Accessed 6 October 2022). [Google Scholar]
- 69.Diwadkar AR, Yoon S, Shim J, Gonzalez M, Urbanowicz R, Himes BE. Integrating Biomedical Informatics Training into Existing High School Curricula. AMIA Jt Summits Transl Sci Proc 2021;2021:190–9. [PMC free article] [PubMed] [Google Scholar]
- 70.Wolff A, Gooch D, Montaner JJC, Rashid U, Kortuem G. Creating an Understanding of Data Literacy for a Data-driven Society. The Journal of Community Informatics 2016;12:. 10.15353/joci.v12i3.3275. [DOI] [Google Scholar]
- 71.Terra.Bio. 2020. URL: https://terra.bio/ (Accessed 6 October 2022).
- 72.Wilkinson MD, Dumontier M, Aalbersberg IjJ, Appleton G, Axton M, Baak A, et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci Data 2016;3:1–9. 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Project Jupyter. n.d. URL: https://jupyter.org (Accessed 5 October 2022).
- 74.Statistical Analysis Software, SAS/STAT | SAS. n.d. URL: https://www.sas.com/en_us/software/stat.html (Accessed 5 October 2022).
- 75.SPSS Software. 2022. URL: https://www.ibm.com/spss (Accessed 5 October 2022).
- 76.RStudio | Open source & professional software for data science teams. n.d. URL: https://www.rstudio.com/ (Accessed 5 October 2022).
- 77.Statistical software for data science | Stata. n.d. URL: https://www.stata.com/ (Accessed 5 October 2022).
- 78.Agile Essentials. Agile Alliance | 2019. URL: https://www.agilealliance.org/agile-essentials/ (Accessed 5 October 2022).
- 79.Hail Team HT. Hail 2022. [Google Scholar]
- 80.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. The American Journal of Human Genetics 2007;81:559–75. 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nextflow | A DSL for parallel and scalable computational pipelines. n.d. URL: https://www.nextflow.io/ (Accessed 7 October 2022).
- 82.Cromwell. n.d. URL: https://cromwell.readthedocs.io/en/stable/ (Accessed 7 October 2022).
- 83.Cirillo D, Valencia A. Big data analytics for personalized medicine. Current Opinion in Biotechnology 2019;58:161–7. 10.1016/j.copbio.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 84.Hulsen T, Jamuar SS, Moody AR, Karnes JH, Varga O, Hedensted S, et al. From Big Data to Precision Medicine. Frontiers in Medicine 2019;6:. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Florance V Training for Informatics Research Careers: History of Extramural Informatics Training at the National Library of Medicine. In: Berner ES, editor. Informatics Education in Healthcare: Lessons Learned. London: Springer London; 2014. p. 27–42. [Google Scholar]
- 86.Staggers N, Gassert CA, Skiba DJ. Health Professionals’ Views of Informatics Education: Findings from the AMIA 1999 Spring Conference. Journal of the American Medical Informatics Association 2000;7:550–8. 10.1136/jamia.2000.0070550. [DOI] [PMC free article] [PubMed] [Google Scholar]


