Abstract
A region of the bovine coronavirus (BCV) genome that functions as a packaging signal has been cloned. The 291-nucleotide clone shares 72% homology with the region of mouse hepatitis coronavirus (MHV) gene 1b that contains the packaging signal. RNA transcripts were packaged into both BCV and MHV virions when the cloned region was appended to a noncoronavirus RNA. This is the first identification of a BCV packaging signal. The data demonstrate that the BCV genome contains a sequence that is conserved at both the sequence and functional levels, thus broadening our insight into coronavirus packaging.
The coronavirus genome is a single-stranded, positive-sense, 27- to 31-kb RNA molecule, the largest among all RNA viruses. The genome is encapsidated by multiple copies of the nucleocapsid (N) protein and is packaged as a helical nucleocapsid in the mature enveloped virion (for reviews, see references 14 and 25). During infection, coronaviruses synthesize six or seven subgenomic mRNAs which share 5′ and 3′ ends that are identical to the genomic RNA (for a review, see reference 29).
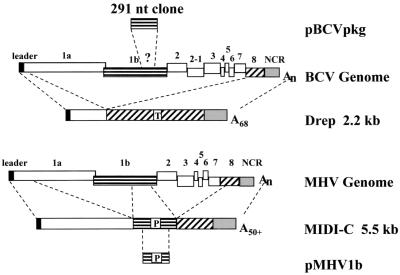
How coronaviruses recognize the viral RNA(s) to ensure specific encapsidation and packaging is only beginning to be understood. Thus far, a packaging signal has been identified only for mouse hepatitis coronavirus (MHV). By using MHV defective interfering (DI) RNAs, a 69-nucleotide (nt) packaging signal that maps approximately 20 kb from the 5′ end of the genome within the gene 1b open reading frame was identified (Fig. 1) (8, 16, 28). Inclusion of the packaging signal in a non-MHV RNA is sufficient to allow the RNA to be packaged into MHV virions (30). Mutagenesis of the predicted bulged stem-loop structure of the MHV packaging signal disrupts the ability of the sequence to function as a packaging signal (8). It was also shown that an MHV strain A59 subgenomic DI RNA is packaged into virions when it contains the DI packaging signal, but the subgenomic RNA was reported to be packaged less efficiently than its DI genomic RNA (4).
FIG. 1.
Comparison of the BCV and MHV strain A59 genomes and representative defective genomes. Drep is a cloned BCV defective RNA (6), and MIDI-C is a cloned MHV strain A59 DI RNA (7). The various parts of the genome that are included in each defective genome are indicated by dashed lines extending from the schematic of the parental genome. Drep includes a 30-nt reporter sequence (T) derived from the TGEV N gene. MIDI-C includes a known packaging signal (P). The approximate location of the packaging signal in the MHV genome (P) and the assumed position of the 291-nt cloned region of BCV (?) are indicated. Schematic representations of the 291-nt BCV RT-PCR and 312-nt MHV PCR products included in plasmids pBCVpkg and pMHV1b, respectively, are shown. NCR, noncoding region.
Sequences that are required for RNAs to be packaged by bovine coronavirus (BCV) have not been determined. BCV belongs to the same antigenic subgroup as MHV, and data based on the 3′ one-third of the genome that has been cloned and sequenced indicate that the two viruses are closely related (1, 2, 13, 15). However, multiple pieces of data suggest that BCV is different from MHV with regard to RNA packaging. The first is that BCV packages subgenomic RNAs in addition to genomic RNA, whereas MHV packages very little, if any, of its subgenomics. BCV N and M mRNAs were shown to be more abundant in virions, on a molar basis, than was genome (11). The second difference is that the BCV defective RNA Drep (Fig. 1), which is replicated and packaged by BCV, does not contain any gene 1b sequence (6).
To gain further insight into the requirements for coronavirus packaging, we asked whether the BCV genome contains sequence information equivalent to the MHV packaging signal. Reverse transcription followed by PCR (RT-PCR) was carried out by using BCV virion RNA as the initial template. RT-PCR products were generated with primers 5′ EcoRI (5′-GCAGAATTCAATGGCGTAGTGG-3′) and 3′ BamHI (5′-GGCCTCGGATCCATACTTCTGAAT-3′). Primers were designed based on published sequence information for MHV (5) and unpublished sequence information from D. Yoo, University of Guelph, Guelph, Ontario, Canada. EcoRI and BamHI restriction sites (underlined) were included for subcloning. Multiple RT-PCRs were performed, and the products were pooled prior to isolation of the 291-nt DNA product, digestion with EcoRI and BamHI, and ligation into EcoRI/BamHI-cut pGEM3Zf(+). Four clones (designated pBCVpkg1 to pBCVpkg4) were sequenced by using Sequenase (USB). All four clones had the same sequence. The sequence revealed that the BCV genome contains a region that is homologous to the part of MHV gene 1b where the packaging signal maps (Fig. 2). Alignment of the BCV and MHV sequences, using the Gap program in the Genetics Computer Group software package (27), indicated that the regions have 72% homology overall. The 69 nt of the BCV cDNA that aligned with the MHV packaging signal is 74% conserved (underlined region in Fig. 2).
FIG. 2.
Sequence of the cloned BCV region and alignment with the homologous region from the MHV strain A59 genome. The region of the BCV clone that is homologous to the MHV packaging signal is underlined. Only nonidentical nucleotides are shown for MHV.
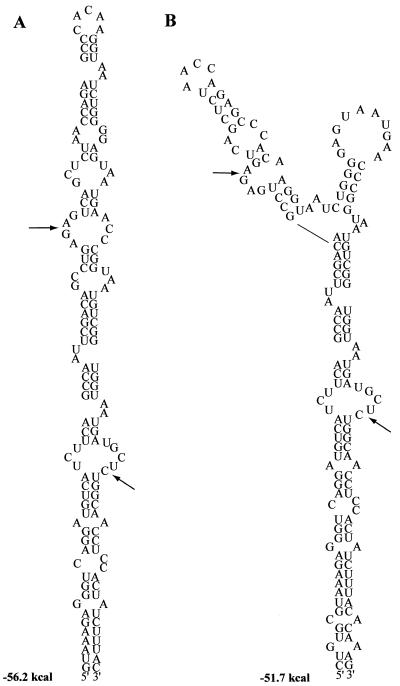
After determining that a sequence exists in the BCV genome that shares homology with the MHV packaging signal, we analyzed the RNA sequence to determine its predicted secondary structure. We were interested in determining the predicted structure of the putative BCV packaging signal, since it was already demonstrated that the structure of the MHV packaging signal is important for its function (8). We used mFOLD 3.0, the most current version, with parameters set at 37°C, 1 M NaCl, and no divalent cations for our analyses (17, 32). Analysis was initially done in the context of a 190-nt region (nt 42 to 232 in Fig. 2) that corresponds to the region of MHV that was previously examined (8). Two structures with overall free energies of −56.2 and −51.7 kcal/mol were predicted for the BCV 190-nt RNA (Fig. 3). We also examined the 69-nt region in the context of the entire 290-nt cloned region. Identical or similar structures were predicted for the sequence in the context of the larger RNA. Fifteen of the 23 structures predicted for BCV were identical or similar to the one shown for BCV in Fig. 3A.
FIG. 3.
Predicted secondary RNA structures of the cloned BCV region. Predictions were made in the context of both the 190-nt (nt 42 to 232 in Fig. 2) and 291-nt (nt 1 to 291) RNAs. Only nt 59 to 200 (Fig. 2) are shown for simplicity. Arrows indicate the position of the 69-nt region that shares homology with the MHV packaging signal.
Parallel analysis of the 190-nt homologous region from MHV predicted only one structure (data not shown) that was similar to the predicted BCV bulged stem-loop with the lower free energy (Fig. 3A). However, the predicted structure was different from the structure that was reported previously for the MHV packaging signal (8). This was surprising, since the reported structure was at least partially confirmed by mutations in base-paired regions that either disrupted or restored the packaging signal function (8). What accounts for this difference is not clear. It is possible that changes in parameters used to predict RNA secondary structures account for the difference between the previously reported structure and our observations. It should be stressed that the secondary structures shown here are only speculative in the absence of experimental data. Further experimental analysis is required to determine the validity of the predicted BCV structure(s). Nonetheless, the conservation of the BCV and MHV structures predicted by our analysis justified further experiments to determine if the newly cloned BCV sequence contains a functional packaging signal. The results also suggested that the putative BCV packaging signal has the potential to fold into different structures.
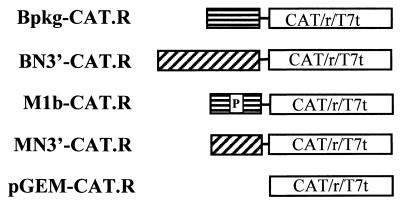
To determine directly whether the region we cloned from BCV could function as a packaging signal, Bpkg-CAT.R was constructed by appending the chloramphenicol acetyltransferase (CAT) gene at the 3′ end of this sequence (Fig. 4). To accomplish this, the hepatitis delta virus ribozyme and T7 terminator from plasmid v2.0 (21) were subcloned downstream of the CAT gene in pcDNA3.1/Zeo/CAT (Invitrogen) to yield pCAT.R. The CAT-ribozyme-terminator cassette was then released from pCAT.R by cutting with XhoI and SphI and subcloned into plasmid pBCVpkg. The CAT cassette was also cloned downstream of the MHV packaging signal in pMHV1b to generate M1b-CAT.R (Fig. 4). Plasmid pMHV1b contains a 312-nt fragment that was PCR amplified from MHV DI MIDI-C (7) and subcloned into EcoRI and BamHI sites of pGEM3Zf(+). This construct was used as a control for packaging in MHV-infected cells and to determine if the BCV and MHV sequences are functionally equivalent. Two additional constructs, BN3′-CAT.R and MN3′-CAT.R, which had been made previously for other reasons were also used as controls in the experiments described below. These included an 830-nt region and a 385-nt region from the 3′ ends of the BCV and MHV N genes, respectively, upstream of the CAT cassette (Fig. 4).
FIG. 4.
Schematic representation of constructs used to test the ability of the BCV clone to function as a packaging signal. All coronavirus sequences were placed under the control of the T7 promoter. A cassette containing the CAT gene, the hepatitis delta virus ribozyme (r), and the T7 terminator (T7t) was cloned at the 3′ end of each coronavirus sequence.
To determine if the addition of the cloned BCV sequence was sufficient for packaging of the CAT gene, we performed the following experiments. BHK cells for the BCV experiments or 17Cl1 cells for the MHV experiments were infected with vTF7-3 (9), the vaccinia virus recombinant that expresses T7 RNA polymerase. After infection, cells were transfected with the different plasmids essentially as described previously (19) (Fig. 4). Plasmid pMHV4S, which contains the MHV4 spike (S) gene (10), was included in transfections in the BCV experiments since fusion of the monolayer by the expressed S protein helped increase the number of dually infected and transfected cells. Four hours after transfection, cells were either mock infected or infected with BCV or MHV. At 24 h after infection with BCV or 12 h after infection with MHV, both intracellular RNA and the media were harvested. The media were clarified and treated with both DNase and RNase prior to being either pelleted through a 30% sucrose cushion or sedimented on a 20 to 60% sucrose gradient. After gradient fractionation, the fractions with densities ranging from 1.15 to 1.20 g/cm3 were pooled and virions were pelleted. RNAs were extracted from pelleted virions. One-tenth of the total intracellular RNA (Fig. 5A and B, lanes 1 to 10) and all of the RNA from pelleted virions (Fig. 5A and B, lanes 11 to 20) were analyzed by Northern blotting using a 32P-labeled CAT-specific riboprobe. Densitometry was used to quantify bands after autoradiography of Northern blots. After autoradiography, the membranes were reprobed with N gene-specific probes to confirm successful coronavirus infection and the absence of coronavirus infection in the controls (data not shown).
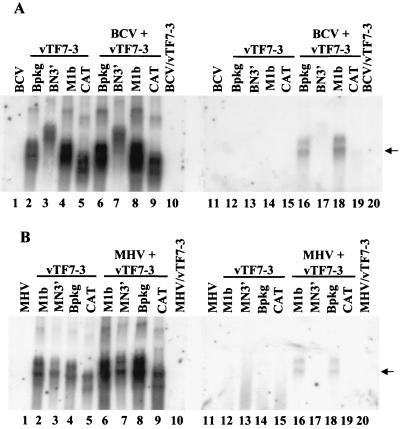
FIG. 5.
Northern blot analysis of expressed RNA transcripts packaged by BCV (A) and MHV strain A59 (B). Plasmid DNAs were transfected into vTF7-3-infected cells (lanes 2 to 9 and 12 to 19). Both intracellular RNAs (lanes 1 to 10) and extracellular virion RNAs (lanes 11 to 20) were analyzed by Northern blotting after cells were mock infected (lanes 2 to 5 and 12 to 15) or infected (lanes 6 to 9 and 16 to 19) with BCV (A) or MHV strain A59 (B). Extracellular media were treated with both DNase and RNase prior to isolation of virions. RNAs were analyzed by using a CAT-specific riboprobe. Arrows indicate the position of the packaged Bpkg-CAT and M1b-CAT RNAs.
Northern blotting clearly demonstrated that RNA transcribed from our cloned BCV sequence was sufficient to target CAT for packaging into extracellular BCV virions (Fig. 5A, lane 16). The two bands detected by the CAT-specific probe likely resulted from incomplete ribozyme activity. Interestingly, the data also showed that the BCV RNA is functionally homologous to the MHV RNA since it was also sufficient for packaging of a non-MHV RNA into MHV virions (Fig. 5B, lane 18). Consistent with previously published data, the MHV packaging signal was functional in MHV-infected cells (Fig. 5B, lane 16) (4, 30). In addition, the results provided new data indicating that the MHV signal is also functional in BCV-infected cells (Fig. 5A, lane 18). In the case of extracellular BCV virions, small amounts of both the BN3′-CAT and CAT transcripts were detected in virions in some experiments (Fig. 5B, lanes 17 and 19). Only very small amounts of CAT and MN3′-CAT were detected in extracellular MHV virions in some experiments (Fig. 5B, lanes 17 and 19).
The ratio of each packaged RNA relative to the amount of its intracellular RNA was determined by densitometric scanning of RNA bands on Northern blots from multiple experiments like those shown in Fig. 5. The average of multiple experiments indicated that the Bpkg-CAT and M1b-CAT RNAs were packaged into BCV virions about five- and sixfold, respectively, better than the CAT or BN3′-CAT transcripts. Both RNAs were packaged about sixfold better in MHV strain A59-infected cells.
Taken together, the results of this study demonstrated for the first time that the BCV genome contains a sequence that functions as a packaging signal, at least when appended to a noncoronavirus RNA. The BCV signal exhibits 74% homology with the MHV packaging signal. This conservation is sufficient to allow the two sequences to be reciprocally functional. It remains to be determined whether the sequence we have identified, like that of MHV, is the functional packaging signal in the context of the wild-type BCV genome. For both viruses, this must await the construction of an infectious clone.
The MHV packaging signal is necessary for packaging of several DI RNAs (8, 28). Since the BCV defective genome Drep does not contain any of the genomic sequence we have identified here, it would be informative to determine if the BCV signal increases the packaging efficiency of Drep when incorporated into the defective RNA. Thus far, our attempts to assay the effects of incorporation of the sequence into Drep have not been successful. We have introduced the sequence in frame into multiple sites within the defective genome, and in all cases replication of the defective genome was completely compromised (R. Cologna and B. G. Hogue, unpublished data).
Since BCV and MHV contain highly homologous packaging signals, why does BCV not discriminate among the different viral RNAs whereas MHV is more selective in packaging? Since BCV packages subgenomics and the Drep defective genome, none of which contain the identified BCV packaging signal, other factors or sequences may function as alternative signals for these RNAs. Alternatively, subtle differences between the nucleocapsid proteins of the two viruses may account for the packaging difference. In the case of alphaviruses, the ability to package genomic RNA over the subgenomic RNAs was lost when a small region of the capsid protein was deleted (20). The nucleocapsid interacts with genomic and subgenomic RNAs in both MHV-infected cells (3, 26) and BCV-infected cells (R. Cologna and B. G. Hogue, unpublished data). The BCV N-subgenomic RNA interactions may be more stable than the comparable MHV complexes, thus allowing BCV subgenomics to be readily incorporated into assembling virions. Another possibility is that replication and assembly complexes are compartmentalized in MHV-infected cells, whereas these complexes may not be separated in BCV-infected cells. This might allow the abundant subgenomic RNAs to be nonselectively included during assembly. Our data suggest that BCV is less selective than MHV in packaging since a small amount of the CAT RNA was consistently detected in BCV virions. Finally, differences in the translation efficiencies of subgenomic RNAs from the two viruses might account for which RNAs are packaged. For example, if MHV mRNAs are translated more efficiently than BCV mRNAs, more of the subgenomics in MHV-infected cells may be associated with polyribosomes, resulting in their being unavailable for packaging.
Other coronavirus DI RNAs are currently being studied, but no packaging signal for them has been definitively identified. Defective RNAs have been examined for the avian infectious bronchitis virus, an antigenic group III virus. The data indicate that information at the 5′ end of the 1b gene may be involved in the packaging of these defective RNAs (22, 23). DI RNAs of transmissible gastroenteritis coronavirus (TGEV), a member of antigenic group I, have also been isolated and studied (12, 18). Recent analysis of one of the DI RNAs suggests that two regions play a role in packaging (12). These regions include about 1 kb from the 5′ end of the genome which consists of sequences from both genes 1a and 1b in the DI RNA and about 4.1 kb at the 3′ end of gene 1b. The exact role of the regions in packaging remains to be shown. Both TGEV and infectious bronchitis virus package subgenomic RNAs (24, 31). Clearly, further studies are necessary to comprehensively understand the requirements for coronavirus packaging. Our data provide a step in that direction.
Nucleotide sequence accession number.
The packaging signal sequence was deposited in GenBank under accession no. AF159126.
Acknowledgments
We thank Dongwan Yoo for discussions and sharing unpublished sequence information with us. We thank Willy Spaan for pMIDI-C, Andy Ball for the v2.0 vector, Julian Leibowitz for the 17Cl1 cells and MHV A59, Tom Gallagher and Michael Buchmeier for the MHV4S clone, and Bernard Moss for the recombinant vaccinia virus vTF7.3. We thank Phuc Nguyen and Jeannie Spagnolo for many discussions.
This work was supported by Public Health Service grant AI33500 to B.G.H. from the National Institute of Allergy and Infectious Diseases and in part by National Institutes of Health predoctoral traineeship AI07471 to R.C.
REFERENCES
- 1.Abraham S, Kienzle T E, Lapps W, Brian D A. Deduced sequence of the bovine coronavirus spike protein and identification of the internal proteolytic cleavage site. Virology. 1990;176:296–300. doi: 10.1016/0042-6822(90)90257-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abraham S, Kienzle T E, Lapps W E, Brian D A. Sequence and expression analysis of potential nonstructural proteins of 4.9, 4.8, 12.7, and 9.5 kDa encoded between the spike and membrane protein genes of the bovine coronavirus. Virology. 1990;177:488–495. doi: 10.1016/0042-6822(90)90513-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric R S, Nelson G W, Fleming J O, Deans R J, Keck J G, Casteel N, Stohlman S A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bos E C W, Dobbe J C, Luytjes W, Spaan W J M. A subgenomic mRNA transcript of the coronavirus mouse hepatitis virus strain A59 defective interfering (DI) RNA is packaged when it contains the DI packaging signal. J Virol. 1997;71:5684–5687. doi: 10.1128/jvi.71.7.5684-5687.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bredenbeek B J, Pachuk C J, Notern A F H, Charite J, Luytjes W, Weiss S R, Spaan W J M. The primary structure and expression of the 2 open reading frame of the polymerase gene of the coronavirus MHV-A59—a highly conserved polymerase is expressed by an efficient ribosomal frame-shifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang R-Y, Hofmann M A, Sethna P B, Brian D A. A cis-acting function for the coronavirus leader in defective interfering RNA replication. J Virol. 1994;68:8223–8231. doi: 10.1128/jvi.68.12.8223-8231.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Groot R J, van der Most R G, Spaan W J M. The fitness of defective interfering murine coronavirus DI-a and its derivatives is decreased by nonsense and frameshift mutations. J Virol. 1992;66:5898–5905. doi: 10.1128/jvi.66.10.5898-5905.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fosmire J A, Hwang K, Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallagher T M, Escarmis C, Buchmeier M J. Alteration of the pH dependence of coronavirus-induced cell fusion: effect of mutations in the spike glycoprotein. J Virol. 1991;65:1916–1928. doi: 10.1128/jvi.65.4.1916-1928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann M A, Sethna P B, Brian D A. Bovine coronavirus mRNA replication continues throughout persistent infection in cell culture. J Virol. 1990;64:4108–4114. doi: 10.1128/jvi.64.9.4108-4114.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Izeta A, Smerdou C, Alonso S, Penzes Z, Mendez A, Plana-Duran J, Enjuanes L. Replication and packaging of transmissible gastroenteritis coronavirus-derived synthetic minigenomes. J Virol. 1999;73:1535–1545. doi: 10.1128/jvi.73.2.1535-1545.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kienzle R T E, Abraham S, Hogue B G, Brian D A. Structure and orientation of expressed bovine coronavirus hemagglutinin esterase protein. J Virol. 1990;64:1834–1838. doi: 10.1128/jvi.64.4.1834-1838.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–77. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lapps W, Hogue B G, Brian D A. Sequence analysis of the bovine coronavirus nucleocapsid and matrix protein genes. Virology. 1987;157:47–57. doi: 10.1016/0042-6822(87)90312-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makino S, Yokomori K, Lai M M C. Analysis of efficiently packaged defective interfering RNAs of murine coronavirus: localization of a possible RNA-packaging signal. J Virol. 1990;64:6045–6053. doi: 10.1128/jvi.64.12.6045-6053.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mathews D H, Sabina J, Zuker M, Turne D H. Expanded sequence dependence of thermodynamic parameters provides robust prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 18.Mendez A, Smerdou C, Izeta A, Gebauer F, Enjuanes L. Molecular characterization of transmissible gastroenteritis coronavirus defective interfering genomes: packaging and heterogeneity. Virology. 1996;217:495–507. doi: 10.1006/viro.1996.0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen V-P, Hogue B G. Protein interactions during coronavirus assembly. J Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Owen K E, Kuhn R J. Identification of a region in the Sindbis virus nucleocapsid protein that is involved in specificity of RNA encapsidation. J Virol. 1996;70:2757–2763. doi: 10.1128/jvi.70.5.2757-2763.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pattnaik A K, Ball L A, LeGrone A W, Wertz G W. Infectious defective interfering particles of VSV from transcripts of a cDNA clone. Cell. 1992;69:1011–1020. doi: 10.1016/0092-8674(92)90619-n. [DOI] [PubMed] [Google Scholar]
- 22.Penzes Z, Tibbles K, Shaw K, Britton P, Brown T D K, Cavanagh D. Characterization of a replicating and packaged defective RNA of avian coronavirus infectious bronchitis virus. Virology. 1994;203:286–293. doi: 10.1006/viro.1994.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Penzes Z, Wroe C, Brown T D K, Britton P, Cavanagh D. Replication and packaging of coronavirus infectious bronchitis virus defective RNAs lacking a long open reading frame. J Virol. 1996;70:8660–8668. doi: 10.1128/jvi.70.12.8660-8668.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sethna P B, Hung S-H, Brian D A. Coronavirus subgenomic minus-strand RNAs and the potential for mRNA replicons. Proc Natl Acad Sci USA. 1989;86:5626–5630. doi: 10.1073/pnas.86.14.5626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siddell S G. The Coronaviridae: an introduction. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 1–10. [Google Scholar]
- 26.Stohlman S A, Baric R S, Nelson G N, Soe L H, Welter L M, Deans R J. Specific interactions between coronavirus leader RNA and nucleocapsid protein. J Virol. 1988;62:4288–4295. doi: 10.1128/jvi.62.11.4288-4295.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.University of Wisconsin Genetics Computer Group. Wisconsin package, version 10.0. Madison: University of Wisconsin Genetics Computer Group; 1999. [Google Scholar]
- 28.van der Most R G, Bredenbeek P J, Spaan W J. A domain at the 3′ end of the polymerase gene is essential for encapsidation of coronavirus defective interfering RNAs. J Virol. 1991;65:3219–3226. doi: 10.1128/jvi.65.6.3219-3226.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Most R G, Spaan W J M. Coronavirus replication, transcription, and RNA recombination. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 11–31. [Google Scholar]
- 30.Woo K, Joo M, Narayanan K, Kim K H, Makino S. Murine coronavirus packaging signal confers packaging to nonviral RNA. J Virol. 1997;71:824–827. doi: 10.1128/jvi.71.1.824-827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhao S, Shaw K, Cavanagh D. Presence of subgenomic mRNAs in virions of coronavirus IBV. Virology. 1993;196:172–178. doi: 10.1006/viro.1993.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction. NATO ASI (Adv Sci Inst) Ser A Life Sci. 1999;1999:11–43. [Google Scholar]