Abstract
All members of the Herpesviridae family contain sequences for a highly conserved glycoprotein B (gB) gene. We investigated the phylogenetic relationships of gB sequences from eight independent rhadinovirus isolates obtained from three species: rhesus (Macaca mulatta), cynomologus (Macaca fasicularis), and pig-tailed (Macaca nemestrina) macaques. Samples were derived from monkeys housed at four separate facilities. Analysis of these eight independent gB sequences revealed five regions of heterogeneity within the 823- to 829-amino-acid polypeptides: residues 1 to 65, 120 to 185, 255 to 300, 352 to 393, and 412 to 457. The remaining regions of gB were highly conserved among the different macaque isolates. Overall divergence among these gene sequences ranged from 0.1 to 7.2% at the amino acid level. Phylogenetic trees constructed with our macaque rhadinovirus gB sequences and those derived from additional subfamilies or genera (alpha, beta, gamma-1, and gamma-2) revealed that the macaque gB sequences branched with other gamma-2 herpesvirus gB sequences and that within the gamma-2 genera, the macaque gB sequences clustered as a distinct branch. The eight macaque rhadinovirus gB sequences were all approximately equidistant from Kaposi sarcoma-associated herpesvirus (KSHV) gB sequences and had a shorter evolutionary distance to KSHV gB sequences than to any other herpesvirus, including the gamma-2 herpesvirus saimiri (HVS) of New World squirrel monkeys. The macaque gB sequences did not cluster according to the facility of origin, but did cluster according to the species of origin, displaying less intraspecies divergence (0.1 to 2.9%) than interspecies divergence (3.3 to 7.2%). These results demonstrate a close relatedness of rhadinovirus isolates from different macaque species.
The gamma-2 (Rhadinovirus) genus of herpesviruses includes Herpesvirus saimiri (HVS) (1); Human herpesvirus 8 (HHV8) (6), also known as Kaposi's sarcoma-associated herpesvirus (KSHV); Murine herpesvirus 68 (MHV68) (27); and Rhesus monkey rhadinovirus (RRV) (8). These herpesviruses have been assigned to the Rhadinovirus genus based on biological properties, similarities of genomic organization, and the relatedness of herpesvirus core genes (9). Glycoprotein B (gB) is a core herpesvirus gene that is present in all known herpesviruses (9, 23). The relatedness of gB sequences has been used to determine phylogenetic relationships among members of different herpesvirus subfamilies and between members of the same herpesvirus subfamily (3, 10, 15, 24).
We have determined gB sequences from eight rhadinovirus isolates obtained from three macaque species housed in four independent primate centers and compared these sequences with those of previously determined herpesvirus gB sequences. In this report, we describe the phylogenetic relationships of macaque rhadinovirus isolates to each other and to other members of the herpesvirus family.
Amplification and determination of gB sequences.
The macaques from which rhadinovirus isolates were obtained for this study were housed at four different primate research centers. Mm26-95 (8), Mm309-95, Mf27-97, Mf472-97, and Mf23-97 were from the New England Regional Primate Research Center (NERPRC); Mm17577 (24) and Mn19545 were from the Oregon Regional Primate Research Center (ORPRC); Mn98126 was from the University of Washington Regional Primate Research Center (WRPRC); and Mm492-98 was from the Caribbean Primate Research Center (CPRC) (Table 1). The rhadinovirus isolate and gB sequences from Mm17577 were obtained and published previously by others (24), and those from Mm26-95 were previously published by our laboratory (8). In order to grow macaque rhadinovirus isolates for this study, peripheral blood mononuclear cells from monkeys were cocultivated with rhesus fibroblast cells as previously described (8). gB sequences from three RRV isolates (RRV26-95, RRV492-98, and RRV309-95), two cynomologus monkey rhadinovirus (CRV) isolates (CRV27-97 and CRV23-97), and two pig-tailed monkey rhadinovirus (PMRV) isolates (PMRV98126 and PMRV19545) were successfully amplified directly from supernatants of infected cells in culture. Cellular DNA from CRV472-97 cocultures was isolated using a QIAMP Blood Kit (Qiagen, Valencia, Calif.) following the manufacturer's protocol; this DNA served as a template for the amplification of CRV472-97 gB sequences.
TABLE 1.
Species and origins of macaque rhadinovirus isolates
| Monkey no.a | Species | Primate center of origin |
|---|---|---|
| Mm26-95b | Macaca mulatta (rhesus macaque) | New England Regional Primate Research Center |
| Mm492-98 | Macaca mulatta | Caribbean Primate Research Center |
| Mm309-95 | Macaca mulatta | New England Regional Primate Research Center |
| Mm17577c | Macaca mulatta | Oregon Regional Primate Research Center |
| Mf27-97 | Macaca fasicularis (cynomologus macaque) | New England Regional Primate Research Center |
| Mf472-98 | Macaca fasicularis | New England Regional Primate Research Center |
| Mf23-97 | Macaca fasicularis | New England Regional Primate Research Center |
| Mn98126 | Macaca nemestrina (pig-tailed macaque) | University of Washington Regional Primate Research Center |
| Mn19545 | Macaca nemestrina | Oregon Regional Primate Research Center |
The first two letters of the monkey number refer to the species shown in column 2. Mm, Macaca mulatta (rhesus macaque); Mf, Macaca fasicularis (cynomologus macaque); Mn, Macaca nemestrina (pig-tailed macaque).
Mn26-95 was obtained from a previously published study by our laboratory (8).
Mn17577 was obtained from a previously published study (24).
In order to amplify macaque rhadinovirus gB sequences, PCR primers were designed within open reading frame 7 (ORF7) and ORF9 (DNA pol) which contain sequences flanking the gB gene. Previously determined ORF7 and ORF9 sequences from KSHV (23) and RRV26-95 (8) were aligned, and primers were made corresponding to highly conserved regions of KSHV and RRV26-95. However, these sequences were not absolutely conserved, and thus nine PCR primers in different combinations were used to amplify gB sequences from all three macaque species (Tables 2 and 3). Specific primers and PCR conditions for each macaque isolate are presented in Tables 2 and 3. Nested PCR was necessary for the amplification of gB sequences from one PMRV isolate, PMRV19545. PCR mixtures were assembled inside a PCR workstation in a laboratory that was not otherwise utilized for molecular biology or cell culture experiments. PCR of uninfected rhesus fibroblast cells did not produce gB-specific fragments, and nucleotide sequences obtained from individual samples were unique. Thus, our gB data from infected macaques did not result from contamination. For PCR, a 100-μl reaction volume was used in a 0.5-ml thin-walled PCR tube (Perkin-Elmer Cetus, Norwalk, Conn.), which included 2 U of rTth DNA polymerase XL (Perkin-Elmer Cetus), 20 mM deoxynucleoside triphosphates (dNTP), 0.1 μM each primer, and 5 μl of culture supernatant or 1 μg of cellular DNA. PCR mixtures were preheated for 1 min at 80°C in a heat block before 1 mM Mg(OAC)2, was added. The sample was then inserted into an Omnigene PCR cycler (Hybaid, Franklin, Mass.) that was preheated to 80°C, and PCR was performed. The data in this report are representative of two independent PCRs. Amplified gB gene sequences were independently digested with two restriction enzymes with 4-base recognition sites, AluI and RsaI (New England Biolabs, Beverly, Mass.), and the resulting fragments were cloned into a pUC18 SmaI/BAP kit (Amersham Pharmacia, Chicago, Ill.). The sequence of this library of random fragments was determined using universal M13 forward and reverse primers with BigDye Terminator Cycle Sequencing Ready Reaction kits (Perkin-Elmer Cetus) and an ABI377 DNA Sequencer (Perkin-Elmer Cetus). Sequences from AluI and RsaI fragments produced contiguous overlapping fragments of gB sequence (contigs). gB sequence not obtained from the AluI and RsaI fragments was determined with specific sequencing primers that annealed near the ends of individual contigs. In this manner, full-length gB sequences were obtained for five of the isolates described in this report. For two isolates, RRV492-98 and PMRV98126, gB sequences were determined directly from the PCR product, using only the specific sequencing primers that were designed to determine the intervening sequences between contigs as described above. Using these techniques, gB sequences of each macaque rhadinovirus isolate were obtained for both strands, representing two to eight determinations per base from a minimum of two independent PCRs. The macaque gB sequences were edited with Sequencer, version 3.0, software (Gene Codes Corporation, Ann Arbor, Mich.), and edited gB sequences were translated to protein sequences using MacVector, version 5.01, software (Oxford Molecular Group, Campbell, Calif.).
TABLE 2.
Primer names and sequences used for the amplification of gB sequencesa
| Primer name | Primer sequence (5′→3′) | Location |
|---|---|---|
| H1 | CCTGTACGCTCTTCTGTACCACC | ORF7 |
| H3 | GGATATTTAAAGACCTGTACGC | ORF7 |
| H4 | GAGGGCCTGCTGGAGGATGTGG | ORF9 |
| H5 | TCCTCCCAGCTGCAGTCAAACTCC | ORF9 |
| H42 | TGTACCATCACCTGCAACTG | ORF7 |
| H45 | CAGGCGGGGATGAGCCTGC | ORF9 |
| H46 | AGGTCCTCCCAGCTGCACTC | ORF9 |
| P2 | CGCGGAATTCGCAACTGTCCGACGGCCA | ORF9 |
| P3 | GCGCCTTAAGGCTGCAAGTACAGGCGGATGGGT | ORF7 |
The location of each primer is indicated by the ORF (ORF7 or ORF9 [DNA pol]) to which it anneals. All ORF7 primers are in the positive polarity, and all ORF9 primers are in the negative polarity.
TABLE 3.
Primers, annealing temperatures, PCR template, and methods of sequence determination for the gB of each macaque rhadinovirus isolatea
| Macaque rhadinovirus | Primers | Primer annealing temp (°C) | PCR template and method of sequence determination |
|---|---|---|---|
| RRV492-98 | H3 and H4 | 58 | PCR fragment, directly sequenced with specific primers |
| RRV309-95 | H1 and H45 | 58 | Cloned AluI and RsaI restriction fragments, sequenced with M13 forward and reverse primers, gaps filled with specific primers |
| CRV27-97 | H1 and H46 | 50 | Cloned AluI and RsaI restriction fragments, sequenced with M13 forward and reverse primers |
| CRV472-97 | H1 and H5 | 50 | Cloned AluI and RsaI restriction fragments, sequenced with M13 forward and reverse primers |
| CRV23-97 | H1 and H5 | 50 | Cloned AluI and RsaI restriction fragments, sequenced with M13 forward and reverse primers |
| PMRV98126 | P2 and P3 | 58 | PCR fragment, directly sequenced with specific primers |
| PMRV19545b | H1 and H5, H42 and H45 | 50 | Cloned AluI and RsaI restriction fragments, sequenced with M13 forward and reverse primers, gaps filled with specific primers |
The sequence of each primer is shown in Table 2.
Amplification of PMRV19545 gB sequences required nested PCR with the four primers shown.
Alignment of gB sequences.
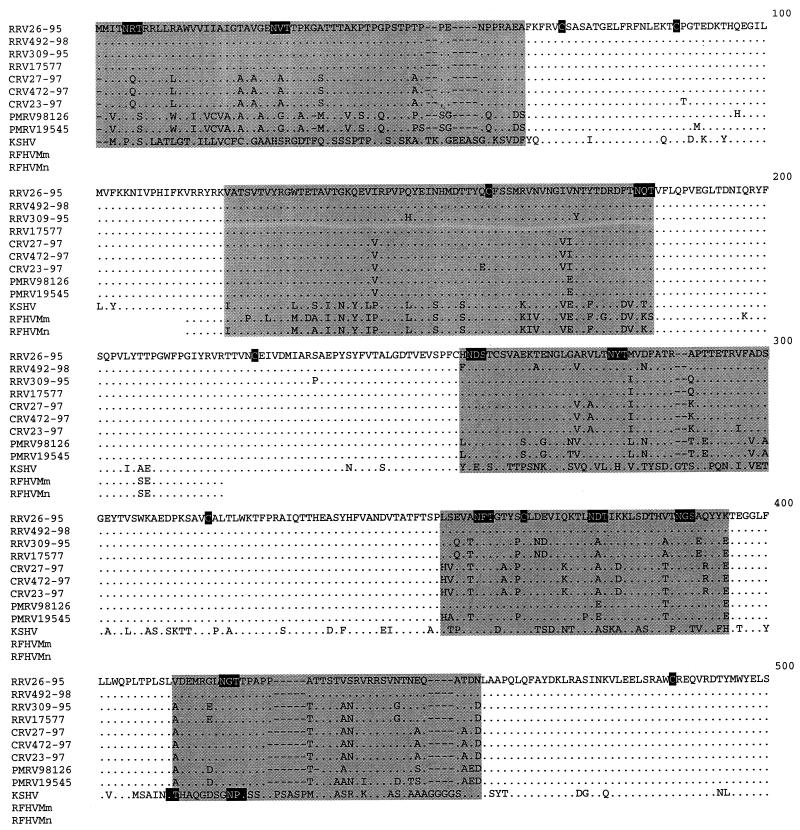
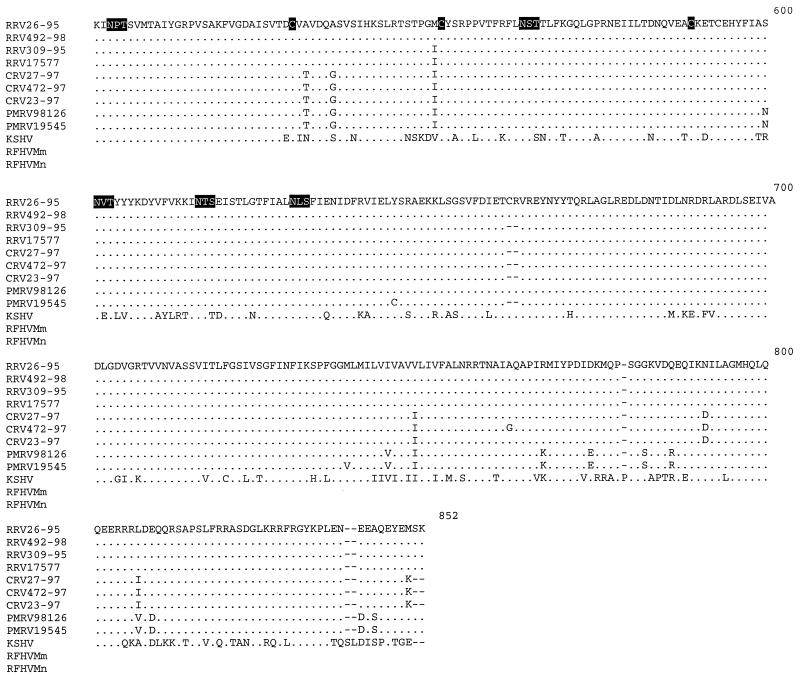
An alignment of gB amino acid sequences from macaque rhadinoviruses, including those previously documented for RRV26-95 (8), and RRV17577 (24), KSHV (23), and partial sequences from retroperitoneal fibromatosis tissues from Macaca mulatta, and Macaca nemestrina (RFHVMm and RFHVMn, respectively) (3), was constructed with ClustalW multiple alignment software which was manually adjusted (EMBL, Heidelberg, Germany) (Fig. 1). Amino acid sequence analysis revealed that 10 cysteine residues were conserved at equivalent positions among the nine macaque isolates (Fig. 1). These cysteines are conserved among several gB sequences of the documented alpha, beta, and gamma herpesviruses, including KSHV (13, 16). Furthermore, 14 potential N-linked glycosylation sites (N-X-S or N-X-T) were conserved among all macaque rhadinovirus isolates presented in this study. Twelve of the 14 potential N-linked glycosylation sites were also conserved in the KSHV gB sequence. The KSHV gB sequence contained two additional potential N-linked glycosylation sites that were not present in the macaque rhadinovirus gB sequences, and these two sites were in very close proximity to each other at residues 409 and 420 (residue numbering based on the alignment in Fig. 1). Our analysis also revealed that the full-length macaque gB sequences contained regions of conserved sequence blocks interspersed with five regions of variability: amino acids 1 to 65, 120 to 185, 255 to 300, 352 to 393, and 412 to 457 (Fig. 1). Although amino acids 1 to 65 were variable between macaque species, this region was well conserved within a single species. In fact, among the four RRV isolates, residues 1 to 65 of gB were completely conserved (Fig. 1). Conversely, these 65 amino acids were highly divergent when individual rhadinovirus isolates from different species were compared with each other or with KSHV (Fig. 1). Nineteen amino acid differences have been previously described between the gB sequences of RRV26-95 and RRV17577 between residues 279 and 551, while the remaining amino acids were identical between the two isolates (24). Of these 19 differences, 16 were located between residues 354 and 457, which include two of the gB variable regions (residues 352 to 393 and 412 to 457 [Fig. 1]). Our analysis revealed that RRV17577 and RRV309-95 have identical amino acid sequences between residues 279 and 551, whereas, RRV26-95 and RRV492-98 differ by only one amino acid in this region and are thus divergent from RRV17577 and RRV309-95 sequences (Fig. 1). Despite the observed heterogeneity, five potential N-linked glycosylation sites and four cysteine residues are conserved within this region among these RRV isolates.
FIG. 1.
Alignment of gB amino acid sequences. An alignment was constructed with the full-length gB amino acid sequences from nine macaque rhadinovirus isolates, including RRV (Macaca mulatta), CRV (Macaca fasicularis), and PMRV (Macaca nemestrina); one KSHV isolate (accession no. AAC57085); and two partial sequences from retroperitoneal fibromatosis tissues from Macaca mulatta (RFHMm, accession no. AAC72187) and Macaca nemestrina (RFHMn, accession no. AAC72188). RRV26-95 and RRV17577 gB sequences were obtained from GenBank (accession no. AAC58686 and AAD21335, respectively). ClustalW software was used to determine alignments of the gB sequences. The gB sequences were compared to the RRV26-95 sequence (8) (shown on the top line). Deletion polymorphisms between sequences are indicated by dashes (−). Conserved cysteine residues and conserved potential N-linked glycosylation sites (N-X-S or N-X-T) are highlighted in black. The five variable regions (residues 1 to 65, 120 to 185, 255 to 300, 352 to 393, and 412 to 457) are highlighted in gray.
We have included the partial gB sequences from the previously sequenced RFHVMm and RFHVMn in the gB amino acid alignment (Fig. 1) (3, 21). The 106-amino-acid fragment of documented RFHV gB sequences corresponds to residues 114 to 219 in the numbering of our alignments in Fig. 1. We determined that 13 of the 106 amino acids of RFHV gB sequence which are identical to KSHV sequences are divergent in macaque rhadinovirus sequences (Fig. 1). Seven of the 106 amino acids are unique to the two RFHV gB sequences, and 5 residues are unique to one of the RFHV gB sequences (Fig. 1). The remaining amino acids are identical to the macaque rhadinovirus gB sequences (Fig. 1). There are 15 amino acid differences between KSHV and RFHVMm, 8 differences between KSHV and RFHVMn, and 20 to 22 differences between KSHV and the nine macaque isolates in our analysis (Table 4). Despite the observed heterogeneity, one conserved cysteine and one potential N-linked glycosylation site are maintained in all of the macaque rhadinovirus, KSHV, and RFHV gB sequences (Fig. 1).
TABLE 4.
Amino acid differences between gB sequencesa
| gB sequence | No. of amino acid differences vs. sequence of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RRV26-95 | RRV492-98 | RRV309-95 | RRV17577 | CRV27-97 | CRV472-97 | CRV23-97 | PMRV98126 | PMRV19545 | RFHVMm | RFHVMn | KSHV | |
| RRV26-95 | ||||||||||||
| RRV492-98 | 0 | |||||||||||
| RRV309-95 | 2 | 2 | ||||||||||
| RRV17577 | 0 | 0 | 2 | |||||||||
| CRV27-97 | 3 | 3 | 5 | 3 | ||||||||
| CRV472-97 | 3 | 3 | 5 | 3 | 0 | |||||||
| CRV23-97 | 4 | 4 | 6 | 4 | 1 | 1 | ||||||
| PMRV98126 | 2 | 2 | 4 | 2 | 2 | 2 | 3 | |||||
| PMRV19545 | 2 | 2 | 4 | 2 | 2 | 2 | 3 | 0 | ||||
| RFHVMm | 27 | 27 | 28 | 27 | 26 | 26 | 27 | 26 | 26 | |||
| RFHVMn | 22 | 22 | 23 | 22 | 21 | 21 | 27 | 21 | 21 | 7 | ||
| KSHV | 21 | 21 | 22 | 21 | 20 | 20 | 21 | 20 | 20 | 15 | 8 | |
Pairwise comparisons by Gap analysis (GCG software) were made between partial (106 amino acids) gB sequences from the macaque isolates, KSHV, and two RFHV fragments in the region of gB (residues 114 to 219, based on numbering in Fig. 1) for which RFHV sequences have been determined (3). The numbers indicate the number of amino acid differences between the gB sequences in this region. KSHV, RRV26-95, RRV17577, and the two RFHV sequences were obtained from GenBank.
Amino acid divergence between gB sequences.
Degrees of amino acid divergence between herpesvirus gB sequences were calculated by using Gap analysis software (Genetics Computer Group [GCG], version 9.1; Madison, Wis.). Default gap penalties and gap extension parameters were used to determine sequence relatedness. Although there are five areas of heterogeneity, the overall divergence between the macaque rhadinovirus gB sequences ranged from only 0.1 to 7.2% at the amino acid level (Table 5). The 0.1 to 7.2% amino acid divergence corresponds to 0.1 to 11% divergence at the nucleotide level. The degree of synonymous changes (i.e., changes that do not change an amino acid) is high between different macaque rhadinovirus gB sequences. Approximately 74% of the nucleotide substitutions between RRV26-95 and PMRV19545 are synonymous. However, in the defined variable regions, only 25% of the nucleotide substitutions were synonymous. Thus, the patterns of sequence variation suggest pressure to conserve amino acids in the conserved regions and selective advantage for amino acid changes in the variable regions. The levels of divergence between the CRV isolates were the smallest among the three macaque species (0.1 to 0.2%), corresponding to one to three amino acid differences among the three CRV isolates in this study (Table 5). Although the gB sequences of the three CRV isolates were very closely related at the amino acid level, six nucleotide differences were observed in the CRV472-97 versus CRV23-97 comparison and in the CRV23-97 versus CRV27-97 comparison. Our analysis also revealed that the RRV gB sequences displayed the most intraspecies diversity. Within the four RRV gB sequences presented here, amino acid divergence ranged between 0.4 and 2.9% (Table 5). gB sequences from all macaque rhadinovirus isolates were approximately equidistant to KSHV, ranging from 25.3 to 27.4% in amino acid divergence (Table 5).
TABLE 5.
Amino acid divergence in gB sequences in macaque rhadinovirus isolates and KSHVa
| gB sequence | No. of amino acid difference vs. sequence of:
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| RRV26-95 | RRV492-98 | RRV309-95 | RRV17577 | CRV27-97 | CRV472-97 | CRV23-97 | PMRV98126 | PMRV19545 | KSHV | |
| RRV26-95 | ||||||||||
| RRV492-98 | 0.5 | |||||||||
| RRV309-95 | 2.4 | 2.9 | ||||||||
| RRV17577 | 2.1 | 2.5 | 0.4 | |||||||
| CRV27-97 | 4.3 | 4.4 | 3.6 | 3.3 | ||||||
| CRV472-97 | 4.3 | 4.5 | 3.8 | 3.4 | 0.1 | |||||
| CRV23-97 | 4.2 | 4.5 | 3.8 | 3.4 | 0.2 | 0.1 | ||||
| PMRV98126 | 5.6 | 5.5 | 6.1 | 5.7 | 6.0 | 6.1 | 6.0 | |||
| PMRV19545 | 7.2 | 7.0 | 6.7 | 6.3 | 6.3 | 6.5 | 6.4 | 1.8 | ||
| KSHV | 25.3 | 25.2 | 26.0 | 25.7 | 27.2 | 27.4 | 27.4 | 26.6 | 27.4 | |
Pairwise comparisons were made by using full-length gB amino acid sequences (823 to 829 amino acids). Divergence between sequences was calculated by Gap analysis (GCG software). The numbers indicate the percentage of amino acid difference between the macaque rhadinovirus and KSHV gB sequences. KSHV, RRV26-95, and RRV17577 gB sequences were obtained from GenBank.
Phylogenetic analysis.
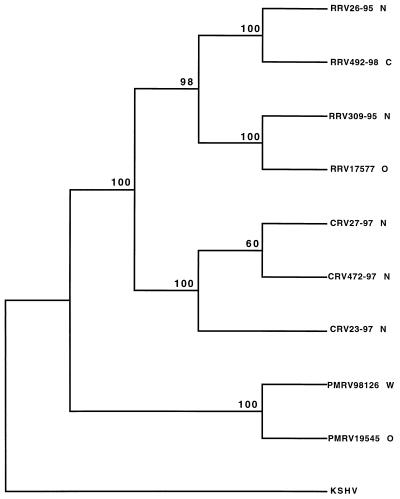
A phylogenetic tree was built with previously determined gB amino acid sequences from KSHV (23), RRV26-95 (8), and RRV17577 (24), along with sequences from the two additional RRV isolates, three CRV isolates, and two PMRV isolates by using PAUP* software (26). These data were manually adjusted, and a neighbor-joining tree was constructed with KSHV serving as the outgroup (Fig. 2). Parsimony and nucleotide sequence analyses produced trees with identical topologies (not shown). There is strong support (bootstrap values of 98% and higher) for the branches separating the three species of macaque rhadinovirus gB sequences in this analysis (Fig. 2). Within the RRV grouping, RRV26-95 clustered with RRV492-98 and RRV309-95 clustered with RRV17577. RRV26-95 and RRV309-95 originated at the NERPRC, whereas RRV492-98 and RRV17577 originated at the CPRC and WRPRC, respectively. Thus, gB sequences within the RRV isolates clustered in an origin-independent manner (Fig. 2).
FIG. 2.
Phylogenetic analysis of gB genes from nine macaque rhadinovirus isolates and KSHV. N, NEPRC; C, CPRC; O, ORPRC; W, WRPRC. ClustalW software was used to align gB amino acid sequences. The neighbor-joining method was used to generate this phylogeny by using PAUP* software (26), with KSHV sequences serving as the outgroup. Bootstrap values from 1,000 replications (repeated three times) are shown for each branch point.
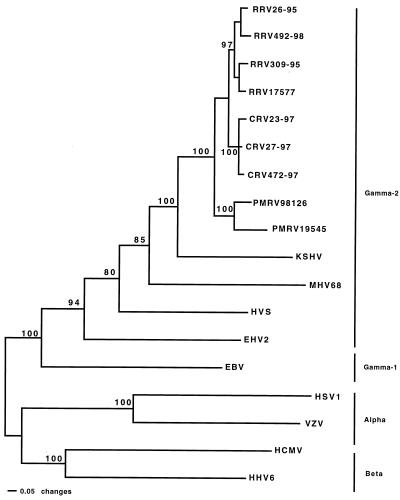
To determine the relationships of the macaque rhadinovirus gB sequences with other members of the herpesvirus family, phylogenetic analysis was performed with full-length aligned gB sequences from representative members of the alpha, beta, and gamma subfamilies and the nine macaque rhadinovirus isolates (Fig. 3). This analysis agreed with the known phylogeny of the herpesvirus subfamily divisions (15). Epstein-Barr virus (EBV), designated as a gamma-1 (Lymphocryptovirus) herpesvirus grouped within the gamma subfamily, was separate from the gamma-2 (Rhadinovirus) subgroup (Fig. 3). In this analysis, the macaque rhadinovirus isolates grouped identically to the analysis displayed in Fig. 2. There is strong support (bootstrap values of 80% and higher) for the branches separating the three major subfamilies of herpesviruses in this analysis (Fig. 3). The macaque rhadinovirus gB sequences had a shorter evolutionary distance to the KSHV gB sequence than to any other herpesvirus (Fig. 3). Parsimony and unweighted pair group method by arithmetic averaging analyses were also performed and produced the same tree topology (not shown), except that the positions of HSV and MHV68 were exchanged. Our data reveal that the macaque rhadinovirus isolates in this report are most closely related to KSHV among all the herpesvirus isolates for which significant sequence information has been determined (Fig. 3).
FIG. 3.
Phylogenetic analysis of gB gene sequences from macaque rhadinoviruses and representative alpha, beta, gamma-1, and gamma-2 herpesviruses. The alignment was created by using ClustalW software. The neighbor-joining method was used to generate this phylogeny by using PAUP* software (26) with alpha and beta herpesvirus gB sequences serving as the outgroup. Bootstrap values from 1,000 replications (repeated three times) are shown for each branch point. Branch lengths are drawn in proportion to distances. Sequences not determined in this study were acquired from GenBank. Accession numbers are as follows: KSHV, AAC57085; MHV68, AAB06229; HVS, P24905; Equine herpesvirus 2, (EHV2); AAC13795; EBV, P02188; Human simplex virus 1 (HSV1), P10211; Varicella-zoster virus (VZV), P09257; Human cytomegalovirus (HCMV), AAA45928; and Human herpesvirus 6 (HHV6), P36319.
gB sequences have been utilized for estimating phylogenetic relationships between closely related herpesvirus isolates (10, 11). Furthermore, gB sequences have proven predictive of the relatedness of complete herpesvirus genome sequences (15). Our data reveal that macaque rhadinovirus gB sequences are very closely related (Table 5 and Fig. 2 and 3) and suggest that the overall structure and sequence of complete rhadinovirus genomes are likely to also be very closely related. Our analysis also indicates that the original RRV isolate, RRV26-95 (8), contains gB sequences very similar to those contained in other macaque rhadinoviruses and thus is not unusual in this respect.
We have also included the 106-amino-acid gB sequence fragments from RFHVMm and RFHVMn in our analysis (3). In this region, there is less divergence between RFHV isolates and KSHV than between macaque rhadinovirus isolates and KSHV (Table 4). Based on previous analysis of the RFHVMm, RFHVMn, KSHV, and RRV26-95 partial DNA pol and gB sequences, it has been suggested that RFHV and macaque rhadinoviruses constitute independent phylogenetic branches and that RFHV is more similar to KSHV than are macaque rhadinoviruses (3). Our results extending these comparisons to eight additional macaque rhadinovirus isolates are consistent with this interpretation. However, additional RFHV data will be needed to confirm the relatedness of RFHV to macaque rhadinovirus isolates and KSHV.
While 90% or more of adult macaques are seropositive for RRV (8), newborn macaques can be raised free of RRV (17). It has been recently documented that cross-species infection with macaque rhadinoviruses does readily occur following experimental inoculation (17). The NERPRC houses monkeys according to species, including infants that are housed with their natural mothers. The species specificity of gB sequences presented here is consistent with other findings suggesting transmission from infected adults or juveniles to uninfected juveniles or infants through close contact (17). Our data suggest that cross-species infection in a primate center setting is not common and that macaque rhadinovirus sequences have evolved with their specific hosts.
The herpesvirus gB gene is a highly conserved gene that is essential for infectivity (5, 7, 12, 18). The data presented here reveal that macaque rhadinovirus gB sequences cluster in a species-specific manner. Heterogeneity among different macaque rhadinovirus gB sequences from the three species in this study is confined to five distinct regions in the N-terminal half of the gB protein (Fig. 1). The first and third of these regions (amino acids 1 to 65 and 255 to 300, respectively) correspond to sequences in herpes simplex virus (HSV) gB that express strain-specific neutralizing epitopes (18). Furthermore, envelope functions including fusion, cell-to-cell spread, and syncytium formation have been mapped to HSV gB sequences that correspond to the second, third, fourth, and fifth regions of heterogeneity (amino acids 120 to 185, 255 to 300, 352 to 393, and 412 to 457) (18). It seems likely that these putative functional domains of macaque rhadinovirus gB have evolved in a species-specific manner. Also contained within the heterogeneous regions are 8 of the 14 potential N-linked glycosylation sites that are conserved among all the macaque rhadinovirus gB sequences. It has been previously documented that gB is a highly immunogenic protein (4, 14, 22). Selected N-linked glycosylation sites in a variable region of simian immunodeficiency virus (SIV) envelope (gp120 surface protein) (20) and of other viruses (2, 25) act to shield the virus from antibody recognition (20). It is possible that some of the potential N-linked sites contained in variable regions of macaque rhadinovirus gB sequences are playing an analogous role in limiting immune recognition. It is also likely that, as seen with SIV (19), some of these N-linked sites in gB contribute to gB structure and function.
Acknowledgments
We thank Daniel Silva, Dong Ling Xia, and Allan McPhee for technical assistance and Joanne Newton for manuscript preparation. We also thank Deborah Glanister and Patrick Delio from the WRPRC, Lisa Hoeber and Steven Kelly from the ORPRC, and Prabhat Sehgal from the NERPRC for supplying macaque samples. We thank Jae Jung for comments on the manuscript.
This study was supported by PHS grants AI38131 and RR00168.
REFERENCES
- 1.Albrecht J-C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, Honess R W. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander S, Elder J H. Carbohydrate dramatically influences immune reactivity of antisera to viral glycoprotein antigens. Science. 1984;226:1328–1330. doi: 10.1126/science.6505693. [DOI] [PubMed] [Google Scholar]
- 3.Bosch M L, Strand K B, Rose T M. Gammaherpesvirus sequence comparisons. J Virol. 1998;72:8458–8459. doi: 10.1128/jvi.72.10.8458-8459.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyle K A, Pietropaolo R L, Compton T. Engagement of the cellular receptor for glycoprotein B of human cytomegalovirus activates the interferon-responsive pathway. Mol Cell Biol. 1999;19:3607–3613. doi: 10.1128/mcb.19.5.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai W, Gu B, Person S. Role of glycoprotein B of herpes simplex virus type 1 in viral entry and cell fusion. J Virol. 1988;62:2596–2604. doi: 10.1128/jvi.62.8.2596-2604.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 7.DeLuca N, Bzik D J, Bond V C, Person S, Snipes W. Nucleotide sequences of herpes simplex virus type 1 (HSV-1) affecting virus entry, cell fusion, and production of glycoprotein gB (VP7) Virology. 1982;122:411–423. doi: 10.1016/0042-6822(82)90240-9. [DOI] [PubMed] [Google Scholar]
- 8.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fields B N, Knipe T P, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2221–2230. [Google Scholar]
- 10.Goltz M, Broll H, Mankertz A, Weigelt W, Ludwig H, Buhk H J, Brochers K. Glycoprotein B of bovine herpesvirus type 4: its phylogenetic relationship to gB equivalents of the herpesviruses. Virus Genes. 1994;9:53–59. doi: 10.1007/BF01703435. [DOI] [PubMed] [Google Scholar]
- 11.Griffin A M. The nucleotide sequence of the glycoprotein gB gene of infectious laryngotracheitis virus: analysis and evolutionary relationship to the homologous gene from other herpesviruses. J Gen Virol. 1991;72:393–398. doi: 10.1099/0022-1317-72-2-393. [DOI] [PubMed] [Google Scholar]
- 12.Herrold R E, Marchini A, Fruehling S, Longnecker R. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J Virol. 1996;70:2049–2054. doi: 10.1128/jvi.70.3.2049-2054.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holloway S A, Studdert M J, Drummer H E. Characterization of glycoprotein B of the gammaherpesvirus equine herpesvirus-2. J Gen Virol. 1998;79:1619–1629. doi: 10.1099/0022-1317-79-7-1619. [DOI] [PubMed] [Google Scholar]
- 14.Hwang E S, Kwon K B, Park J W, Kim D J, Park C G, Cha C Y. Induction of neutralizing antibody against human cytomegalovirus (HCMV) with DNA-mediated immunization of HCMV glycoprotein B in mice. Microbiol Immunol. 1999;43:307–310. doi: 10.1111/j.1348-0421.1999.tb02409.x. [DOI] [PubMed] [Google Scholar]
- 15.Karlin S, Mocarski E S, Schachtel G A. Molecular evolution of herpesviruses: genomic and protein sequence comparisons. J Virol. 1994;68:1886–1902. doi: 10.1128/jvi.68.3.1886-1902.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomonte P, Filee P, Lyaku J R, Bublot M, Pastoret P-P, Thiry E. Glycoprotein B of bovine herpesvirus 4 is a major component of the virion, unlike that of two other gammaherpesviruses, Epstein-Barr virus and murine gammaherpesvirus 68. J Virol. 1997;71:3332–3335. doi: 10.1128/jvi.71.4.3332-3335.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mansfield K G, Westmoreland S V, DeBakker C D, Czajak S, Lackner A A, Desrosiers R C. Experimental infection of rhesus and pig-tailed macaques with macaque rhadinoviruses. J Virol. 1999;73:10320–10328. doi: 10.1128/jvi.73.12.10320-10328.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pereira L. Function of glycoprotein B homologues of the family herpesviridae. Infect Agents Dis. 1994;3:9–28. [PubMed] [Google Scholar]
- 19.Reitter J N, Desrosiers R C. Identification of replication-competent strains of simian immunodeficiency virus lacking multiple attachment sites for N-linked carbohydrates in variable regions 1 and 2 of the surface envelope protein. J Virol. 1998;72:5399–5407. doi: 10.1128/jvi.72.7.5399-5407.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 21.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Jr, Thouless M E, Tsai C-C, Bosch M L. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy D M, Grundy J E, Emery V C. Sequence variation within neutralizing epitopes of the envelope glycoprotein B of human cytomegalovirus: comparison of isolates from renal transplant recipients and AIDS patients. J Gen Virol. 1993;74:2499–2505. doi: 10.1099/0022-1317-74-11-2499. [DOI] [PubMed] [Google Scholar]
- 23.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skehel J J, Stevens D J, Daniels R S, Douglas A R, Knossow M, Wilson I A, Wiley D C. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci USA. 1984;81:1779–1783. doi: 10.1073/pnas.81.6.1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swofford D L. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods), version 4 ed. Sunderland, Mass: Sinauer Associates; 1998. [Google Scholar]
- 27.Virgin H W, IV, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]