Abstract
The genetic information of retroviruses is retained within a dimeric RNA genome held together by intermolecular RNA-RNA interactions near the 5′ ends. Coencapsidation of retrovirus-derived RNA molecules allows frequent template switching of the virus-encoded reverse transcriptase during DNA synthesis in newly infected cells. We have previously shown that template shifts within the 5′ leader of murine leukemia viruses occur preferentially within the kissing stem-loop motif, a cis element crucial for in vitro RNA dimer formation. By use of a forced recombination approach based on single-cycle transfer of Akv murine leukemia virus-based vectors harboring defective primer binding site sequences, we now report that modifications of the kissing-loop structure, ranging from a deletion of the entire sequence to introduction of a single point mutation in the loop motif, significantly disturb site specificity of recombination within the highly structured 5′ leader region. In addition, we find that an intact kissing-loop sequence favors optimal RNA encapsidation and vector transduction. Our data are consistent with the kissing-loop dimerization model and suggest that a direct intermolecular RNA-RNA interaction, here mediated by palindromic loop sequences within the mature genomic RNA dimer, facilitates hotspot template switching during retroviral cDNA synthesis in vivo.
The diploid genome of retroviruses is composed of two full-length viral RNA transcripts linked by intermolecular interactions near their 5′ ends, as demonstrated by early electron microscopy (41). The primary linkage site, designated the dimer linkage structure, has been mapped within the 5′ leader sequence in a region that colocalizes with cis elements constituting the packaging signal (Ψ) required for selective RNA encapsidation into assembling virions. The conformation of this region is altered upon dimerization, suggesting that structural changes caused by dimer formation may create an overall tertiary structure or lead to the exposure of specific RNA structures that are recognized by virus-encoded proteins during the RNA packaging process (58). Close coupling between the dimerization and encapsidation processes is supported also by the finding that dimeric RNA can be extracted from virions immediately after budding from the cell surface, indicating that dimer formation occurs prior to virus assembly (21). After virus release, the dimeric genome is converted to a stable and more mature form in a process facilitated by the RNA chaperone activity of the virus-encoded nucleocapsid protein (18, 20).
The matured and structurally rearranged RNA dimer serves as substrate for double-stranded DNA synthesis by reverse transcriptase within the internalized core particle. Reverse transcription is primed by the 3′ end of a host cell-derived tRNA molecule annealed to the primer binding site (PBS) (for recent review, see reference 33). Completion of the process requires two consecutive strand transfer reactions during minus- and plus-strand synthesis. It is not known to what extent a specific secondary or tertiary dimer structure is essential for efficient initiation and completion of the process (5, 60). The presence of two genomic RNAs within each virion allows the reverse transcriptase to switch templates during RNA-dependent minus-strand DNA synthesis (26, 52) and DNA-dependent plus-strand DNA synthesis (29, 37). Reverse transcriptase-mediated recombination can occur only between RNAs copackaged into the same virion (25); at present, however, it is not known whether a physical linkage between copackaged RNAs is required for template switching to occur. In a number of cases, genetic interactions have been found among retroviral genomes harboring pronounced sequence dissimilarities within the combined RNA dimerization and packaging region (28, 62) and among copackaged heterologous RNAs of viral and cellular origin (53, 55, 63), suggesting that homology within the primary interaction site is dispensable for generation of recombinant proviral sequences. It remains uncertain, however, whether such heterologous RNAs in rare cases may be packaged as monomers or form heterodimers based on linkages at alternative sites.
Synthetic retrovirus RNA dimerizes spontaneously under appropriate salt and temperature conditions (45, 48, 58), suggesting that dimerization is facilitated by direct RNA-RNA interactions. The nucleocapsid moiety of the Gag polyprotein enhances dimer formation in vitro (13, 23, 57) and is thought to play an important role during RNA dimerization in vivo. In vitro dimer formation involves a relatively short sequence within the 5′ untranslated region (UTR), as shown by thermostability studies (48), comparison of antisense DNA oligonucleotide accessibility in monomer and dimer RNA (22, 47), chemical modification studies of monomer and dimer RNA (50, 58), and dimerization studies of truncated and internally deleted RNA fragments (22, 30, 47). Such in vitro approaches define for a number of retroviruses a narrow dimerization segment potentially forming a stem-loop structure within the highly structured 5′ leader sequence (1, 3, 58). Conformational changes of this stem-loop structure are likely to contribute to the dimerization process (22). The dimer-forming stem exposes a palindromic loop motif and therefore holds the potential to interact by Watson-Crick base pairing, or loop-loop “kissing,” with a similar loop sequence (40). Deletion of the entire stem-loop and single-base substitutions introduced into the kissing stem-loop structure abolish synthetic RNA dimerization, whereas compensatory changes introduced into the loop or stem restore dimer formation (11, 44). RNA dimerization triggered by the initial loop-loop recognition is followed, possibly, by unfolding of the stems and formation of an extended RNA-RNA duplex, representing thus a local antiparallel linkage of the two RNAs (23) and may be accompanied by intermolecular association of stem-loops elsewhere in the 5′ leader (16).
The involvement of the kissing-loop in formation of retrovirus RNA dimers remains to be directly demonstrated in vivo. The kissing-loop sequence is dispensable for virus replication, although reduced replication efficiencies and RNA encapsidation defects, possibly reflecting a defect in the level of RNA dimerization, have been reported for mutants harboring severe modifications or more fine-tuned alterations of the kissing-loop (6, 19, 30, 35, 39, 42). Dimeric RNA can be extracted from loop-mutated virions, suggesting that the kissing-loop structure is not essential for dimerization in vivo (6, 9, 24). However, viruses harboring a defective kissing-loop sequence appear to contain less dimeric RNA than wild-type viruses (24, 31). Notably, the stability of human immunodeficiency virus type 1 (HIV-1) RNA dimers remains unaffected by mutations introduced into the kissing-loop (6, 9, 24, 49). This observation possibly reflects that a kissing-loop interaction supposed to initiate dimer formation is resolved at some point after virus assembly and lends support to the idea that alternative and perhaps less stringent linkage sites exist within the RNA dimer (2, 6, 54, 56).
By utilization of a forced recombination approach based on single-cycle transfer of Akv murine leukemia virus (MLV)-derived vectors harboring defective PBS sequences, our previous work has focused on events of template switching within the 5′ leader region. Transfer of the growing minus strand was found to occur predominantly within a narrow region of the leader sequence coinciding precisely with the kissing-loop dimerization motif (36, 38), which raised the possibility that site specificity of recombination within the 5′ leader is governed by RNA secondary structures and specific intermolecular interactions at the junction site. To further assess the role played by the kissing loop in MLV replication and recombination, transduction experiments were performed with vectors harboring deletions and single-base substitutions at the recombinational hotspot kissing-loop sequence. We conclude from these investigations that an intact kissing-loop sequence is required for optimal vector transduction and RNA encapsidation and furthermore, as determined by forced recombination studies, that even minor modifications of the palindromic loop sequence may disturb site specificity and lead to nonclustered events of template switching. These results support the kissing-loop dimerization model and suggest that a restored interaction of palindromic loops within the mature dimer facilitates template switching during reverse transcription.
MATERIALS AND METHODS
Vector construction.
Alterations of the kissing-loop dimerization sequence were introduced into Akv MLV-based retroviral vectors harboring either the wild-type proline PBS (PBS-Pro) sequence or a nonfunctional mutated PBS-Umu sequence (36). These vectors, designated PBSProKLwtAkv-neo and PBSUmuKLwtAkv-neo, respectively, contain Akv MLV long terminal repeats (LTRs) as well as the neomycin resistance gene (neo) flanked upstream by the 5′ 255 bp of the MLV 5′ leader region and downstream by 480-bp Akv MLV sequences including the 3′ UTR. Kissing-loop modifications were introduced by two-step PCR-mediated site-directed mutagenesis similar to strategies previously described (32, 36). Briefly, the 5′ part of the vector was amplified, generating a 769-bp fragment carrying the desired mutations in the kissing-loop sequence. The amplified sequence was subsequently connected by overlap extension and PCR with a downstream PCR fragment harboring the 3′ part of the leader region and part of neo. The amplified fragment encompassing the 5′ LTR, wild-type or modified PBS, 5′ UTR, and part of neo was digested and cloned by standard procedures into the appropriate position of the Akv vector.
The following oligonucleotides were used for introduction of modified kissing stem-loop sequences into PBS-ProKLwtAkv-neo and PBSUmuKLwtAkv-neo (kissing stem-loop sequence underlined): ON1 (Akv MLV positions 283 to 335 [59]), 5′-GAGCCGCCAGATACAGAT(Δ16)TACAGAATCAGACGCAGGC-3′, introducing a 16-bp deletion of the kissing stem-loop (KLΔ16); ON2 (Akv positions 294 to 332), 5′-CCGCCAGATACAGATCTAGTACTAGTACAGAAT-3′, introducing a 6-bp deletion of the kissing-loop (KLΔ6); ON3 (Akv positions 302 to 332), 5′-CCGCCAGATACAGATCTAGTTAGCAAACTAGT, introducing a single-base-pair substitution within the loop motif (KL-TTGCTA); ON4 and ON5 (Akv positions 294 to 332), 5′-CCGCCAGATACAGATCTAGTTAGAGCACTAGTACAGAAT-3′ and 5′-CCGCCAGATACAGATCTAGTGCTAGCACTAGTACAGAAT-3′, introducing a 3-bp substitution (KL-GCTCTA) and an alternative loop sequence (KL-GCTAGC), respectively. Mutations generating loop substitutions are indicated in boldface. ON6 (5′-TAGATCTGTATCTGGCGG-3′ [matching Akv MLV positions 315 to 332]) was used to PCR amplify downstream sequences, generating a fragment with an 18-bp overlap with fragments harboring kissing-loop modifications. Other primers used in the mutagenesis procedure have been described previously (32).
Cells, transfections, and virus infections.
Ψ2 (34) and NIH 3T3 cells were grown in Dulbecco modified Eagle medium with Glutamax-1 supplemented with 10% newborn calf serum, 100 U of penicillin per ml, and 100 μg of streptomycin per ml. Cells were incubated at 37°C in 90% relative humidity and 5.7% CO2. Ten micrograms of vector plasmid DNA was transfected by calcium phosphate treatment into Ψ2 packaging cells seeded at 5 × 103 cells per cm2 on the day before transfection. Two days after transfection, G418-containing medium (0.6 mg/ml) was added to select for stably integrated kissing-loop-modified vectors. G418-resistant colonies appearing after 12 days of selection were pooled. Transduction efficiencies were determined as follows. Producer cells were seeded at maximum density (ca. 105 cells per cm2) and allowed to attach; the medium was renewed and left on the cells for 1 day. Filtered and serially diluted virus-containing medium was transferred to NIH 3T3 cells (seeded at 5 × 103 cells per cm2 the day before infection) in the presence of Polybrene (6 μg/ml). G418-containing medium (0.6 mg/ml) was added 2 days after infection, and resistant colonies were counted and individually expanded after 10 days of selection. Obtained titers were normalized for the number of producer cells, as determined immediately after virus harvest. In transduction experiments set up for PCR-based screening for recombinant proviruses, G418-resistant NIH 3T3 colonies appearing on each plate were pooled to allow for screening among a large number of transduction events. For this purpose, virus-containing medium was diluted to obtain 10 to 30 G418-resistant colonies per plate.
Proviral DNA analysis and PCR-based screening for recombinant proviruses.
Genomic DNA from G418-resistant clones and colony pools was prepared as previously described (32). Digestion or sequence analysis of individual transduced vector sequences was performed on PCR products encompassing part of the 5′ LTR, the PBS, the 5′ UTR, and the upstream part of neo. The PCR was performed with ON7 (5′-TTCATAAGGCTTAGCCAGCTAACTGCAG-3′ [matching Akv MLV positions 7838 to 7865 [59]) and ON8 (5′-GGCGCCCCTGCGCTGACAGCCGGAACAC-3′ [matching neo positions 1656 to 1683 [4]). The resulting PCR product was digested with BstBI, which cleaves specifically within the modified PBS-Umu sequence, and potential recombinants were sequenced with ON9 (5′-TCCGAATCGTGGTCTCGCTGATCCTTGG-3′ [matching positions 69 to 96 within the Akv U5 region [59]) and ON10 (5′-CTTCCTTTAGCAGCCCTTGCGC-3′, [matching neo positions 1223 to 1244]).
PCR amplification performed on genomic DNA prepared from colony pools (obtained by pooling of all resistant colonies appearing on single plates) was used to detect among multiple transduced proviruses Akv-MLV-like endogenous virus (MLEV) recombinants that were generated during vector transduction. Screening for 5′ UTR recombination between Akv and MLEV was performed with a neo-specific primer (ON8) and ON11 (5′-GTCTTTCATTTGGAGGTCCCA-3′ [matching the MLEV-derived glutamine PBS {PBS-Gln}). The resulting PCR product (if any) was sequenced with ON10 and ON11.ON12 (5′-GCCCGGGTACCCGTATTC-3′) and ON13 (5′-GCCCGGGTACCCGTATTC-3′), specifically recognizing PBS-Umu and a genetic marker (designated IX) within the MLEV R region (37), respectively, were used to screen for Akv-MLEV recombinant proviruses that had retained PBS-Umu.
RNA preparation and dot-blot analysis.
For RNA dot blot analysis, 20-ml aliquots of virus-containing medium were collected from confluent stably transfected Ψ2 packaging cells and from NIH 3T3 cells included as a negative control. Two-milliliter aliquots of medium derived from a Ψ2 packaging cell line stably transfected with an MLV-derived retroviral vector expressing the hygromycin B resistance gene (hyg) were added to each supernatant sample. Total virion RNA was purified from 12 1.5-ml volumes of supernatant by pelleting virus particles at 4°C for 1 h at 17,000 × g in a benchtop centrifuge (Heraeus Biofuge 13) followed by guanidinium thiocyanate-phenol-chloroform extraction and subsequent precipitation (8). The number of producer cells was determined for each stably transfected cell line. Cellular RNA extraction was carried out by guanidinium thiocyanate treatment and subsequent precipitation. Virion and cellular RNA was serially diluted and blotted onto a Zetaprobe filter (Bio-Rad), using a dot blot manifold. neo and hyg, vector-specific RNA levels were determined by hybridizations with 32P-labeled probes for neo and hyg, respectively. Cellular vector RNA levels were normalized for glyceraldehyde-3-phosphate dehydrogenase (GAPDH) RNA expression determined by hybridization with a GAPDH-specific probe. Dot blots were quantified with a PhosphorImager SF (Molecular Dynamics), and the relative virion RNA contents (virion neo RNA/cell neo RNA) were normalized for hyg vector RNA level, the number of producer cells, and the cellular level of neo vector RNA relative to GAPDH RNA.
Dot blot assay measurement of NPTII activity.
Assays for neomycin phosphotransferase II (NPTII) activity were carried out essentially as described by Platt and Yang (46) and Duch et al. (17). Briefly, for each of the constructs studied, 3.75 × 105 G418-resistant Ψ2 packaging cells were centrifuged, washed twice with 200 μl of phosphate-buffered saline, and resuspended in 100 μl of 0.135 M Tris-HCl (pH 6.8)–20% glycerol–4 mM dithiothreitol. Crude cell extracts were prepared by submitting cells to four rounds of freeze-thawing and subsequently removal of cell debris by centrifugation. NPTII activities were determined by mixing 20, 10, 5, or 2.5 μl of the various cell extracts with 200 μl of 67 mM Tris-HCl (pH 7.1)–42 mM MgCl2–400 mM NH4Cl–40 μg of kanamycin SO4–2.5 to 10 μCi of [γ-32P]ATP per ml. Mixtures, left for 135 min at 27°C, were filtrated on a dot blot manifold through a filter sandwich composed of nitrocellulose (Schleicher & Schuell), which binds total protein, and phosphocellulose (Whatman), which binds positively charged kanamycin phosphate. The amount of phosphorylated kanamycin bound to the phosphocellulose membrane was quantified with a PhosphorImager SF (Molecular Dynamics). PhosphorImager scanning of the protein dots on the nitrocellulose membrane showed signals with a linear relationship with the amount of cell extract used. Hence, NPTII activities were normalized for the total protein kinase activity in the various cell extracts used as a relative measure for the total protein content.
RESULTS
Reduced replication of kissing-loop-modified vectors.
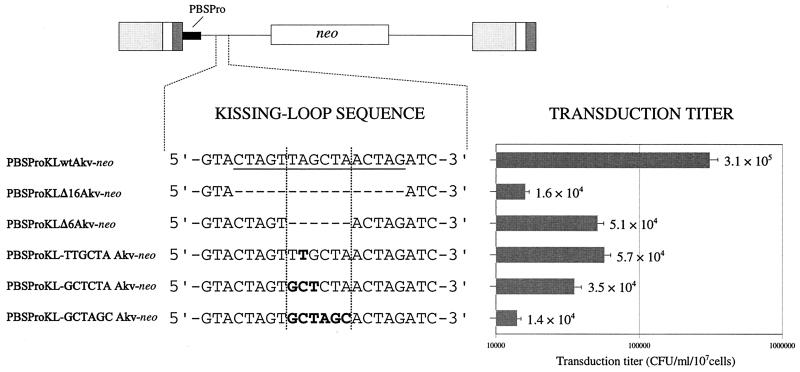
To investigate the role played by the kissing stem-loop in MLV replication and recombination, five kissing-loop-modified Akv MLV-derived vectors, carrying the neo selective marker gene, were generated. The kissing stem-loop is composed of a 5-bp RNA duplex stem and a six-nucleotide palindromic loop sequence (5′-UAGCUA-3′) is supposed to mediate a direct interaction with the kissing-loop palindrome of the copackaged genomic RNA molecule. Two vectors were designed to harbor a deletion of the entire 16-bp kissing-loop sequence (PBSProKLΔ16) and the 6-bp loop motif (PBSProKLΔ6), whereas nucleotide substitutions were introduced into the loop sequence of the remaining vector mutants (Fig. 1). In PBSProKL-TTGCTA, the second position of the loop was altered to create a loop sequence which can form only four of six potential Watson-Crick base pairs within the putative loop-loop interaction. In mutant PBSProKL-GCTCTA, the three most 5′ positions of the palindrome were modified, generating a loop motif unlikely to participate in loop-loop recognition during homodimer formation due to mismatches at all positions at the putative interaction site. In PBSProKL-GCTAGC, the entire loop sequence was replaced by an alternative palindromic sequence providing, in theory, perfect base pairing within the potential loop-loop recognition site.
FIG. 1.
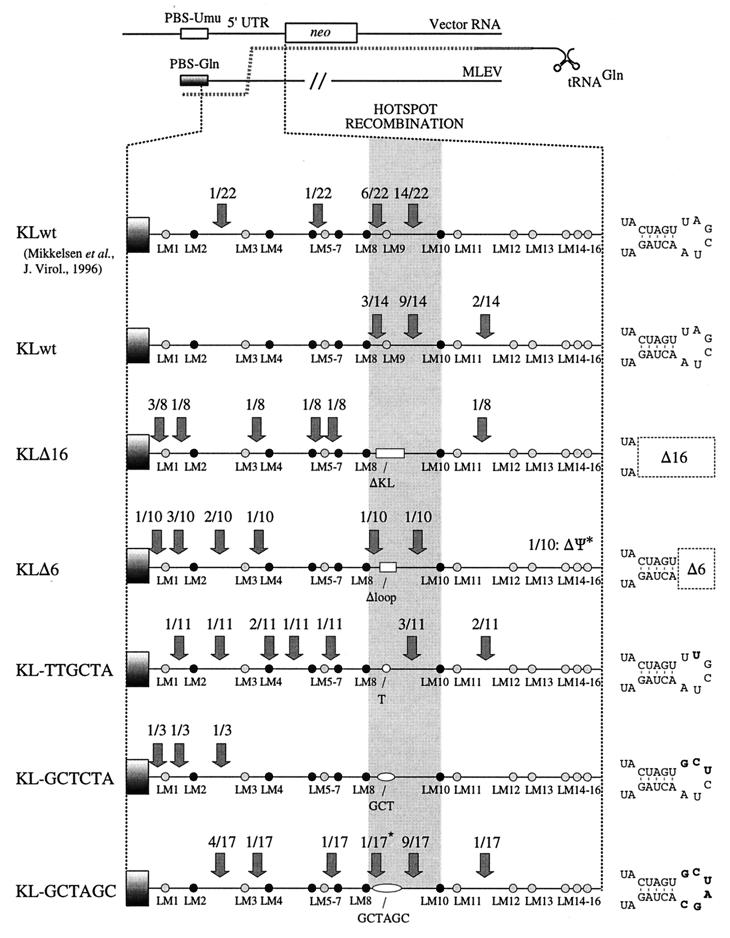
Transduction efficiencies of kissing-loop-modified vectors. The Akv kissing-loop sequence is shown at the top (underlined). Five kissing-loop-modified vector constructs, shown below the wild-type sequence, were generated. Deleted nucleotide positions are indicated by hyphens. Nucleotide substitutions within the loop motif (framed by broken lines) are indicated by boldface letters. Transduction titers were measured by counting the G418-resistant colonies appearing per milliliter of virus-containing medium transferred from stably transfected Ψ2 packaging cells to NIH 3T3 cells. Titers are based on five independent experiments performed with three independent sets of stably transfected cell lines. Standard deviations are indicated by error bars.
To analyze the effect of introducing deletions and substitutions into the kissing-loop sequence, vector transduction efficiencies were measured from Ψ2 packaging cells (34) stably transfected with the engineered constructs. Viral titers were determined by transferring the virus-containing Ψ2 supernatant to NIH 3T3 target cells followed by G418 selection. As depicted in Fig. 1, titers of kissing-loop-mutated vectors were reduced at most 22-fold compared to the unmodified vector. Hence, average titers ranged from 1.4 × 104 for PBSProKL-GCTAGC to 3.1 × 105 for the wild-type vector. Titers 19-, 9-, 6-, and 5-fold below the wild-type replication level were obtained for PBSProKLΔ16, PBSProKL-GCTCTA, PBSProKLΔ6, and PBSProKL-TTGCTA, respectively.
RNA encapsidation deficiency of kissing-loop-modified vectors.
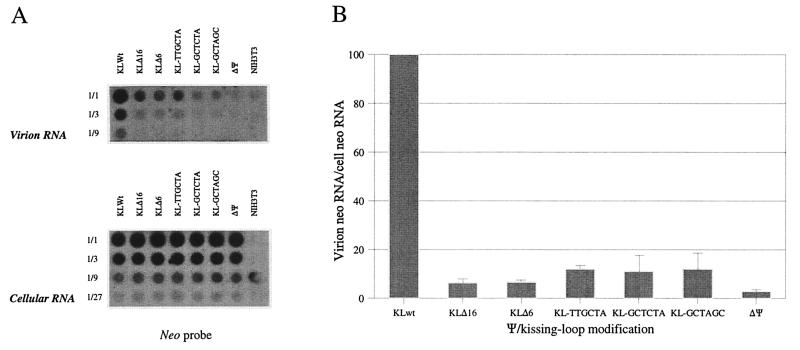
To decipher whether defects in the RNA encapsidation process account for the observed reduction in viral titers, relative quantities of genomic RNA packaged into Ψ2-derived virions were determined for the panel of kissing-loop-modified vectors. The wild-type vector and a Ψ-deficient Akv-derived vector (38a) were included in these experiments as positive and negative controls, respectively. Virus-containing supernatant was collected from confluent stably transfected Ψ2 producer cell lines and from NIH 3T3 cells included as an additional negative control. RNA extracted from virions and producer cells was serially diluted and analyzed by dot blot analysis (Fig. 2A), and the relative vector RNA content was for each construct determined as the level of virus neo RNA relative to the cellular level of neo RNA.
FIG. 2.
Reduced packaging of kissing-loop-modified RNAs into MLV particles. (A) Dot blot analysis of relative virion and cellular levels of wild-type and kissing-loop-modified vector RNAs. A Ψ-deficient vector construct, ΔΨ (Δ183 bp), stably transfected into Ψ2 cells and nontransfected NIH 3T3 cells were included as negative controls. Virion RNA was prepared from 20-ml aliquots of virus-containing supernatant collected from stably transfected Ψ2 packaging cells. Cellular RNA was prepared from transfected packaging cells following cell counting. RNA was threefold serially diluted and blotted onto a Zetaprobe filter; blots were probed with a randomly primed neo probe. As an internal control for RNA loss during viral RNA preparation, equal aliquots of virions containing hyg vector RNA were added to virions prior to RNA preparation; blots were therefore also probed with a hyg-specific probe (not shown). GAPDH probing was used as an internal control for cellular RNA levels (not shown). (B) Relative virion RNA content normalized for virion hyg vector RNA content, the number of producer cells, and the cellular level of neo vector relative to GAPDH RNA. Values are based on two independent dot blot experiments, and standard deviations are represented by error bars.
As evident from the normalized values depicted in Fig. 2B, RNA encapsidation was reduced for all kissing-loop-modified vectors. Packaging efficiencies of kissing-loop mutants thus ranged from 8- to 15-fold below the wild-type level, consistent with the notion that the kissing loop, although perhaps indirectly, is part of the MLV packaging signal. Notably, the decrease in encapsidation of RNA measured for all mutants was found to reasonably mimic the observed replication deficiency, indicating that the effect of introducing mutations into the kissing loop was exerted at the level of RNA packaging in particular. However, it cannot be excluded that packaging defects may only partly explain differences in viral titers and that alternative stages of the replication cycle were also affected by the kissing-loop alterations. Therefore, as an attempt to evaluate whether the introduced kissing-loop deletions and substitutions influenced the level of neo production at a posttranscriptional stage, we used a sensitive dot blot assay to measure the activity of the neo-encoded NPTII in cell extracts prepared from G418-selected packaging cells (17). NPTII activities, normalized for the total protein content, were found to be reduced twofold or less in all Ψ2 cell lines harboring kissing-loop-modified proviruses compared to the wild-type level (data not shown). Since the cellular level of vector-derived RNA was higher (approximately twofold) for all mutant-containing cell lines (Fig. 2A, lower panel), the relative NPTII protein/neo RNA ratios were reduced between three- and fourfold in comparison with the wild type. These results likely reflect that a higher level of kissing-loop-mutated neo RNA is required to obtain G418 resistance, reflecting that the introduced mutations might affect also posttranscriptional events of MLV gene expression.
Forced recombination in PBS- and kissing-loop-modified vectors.
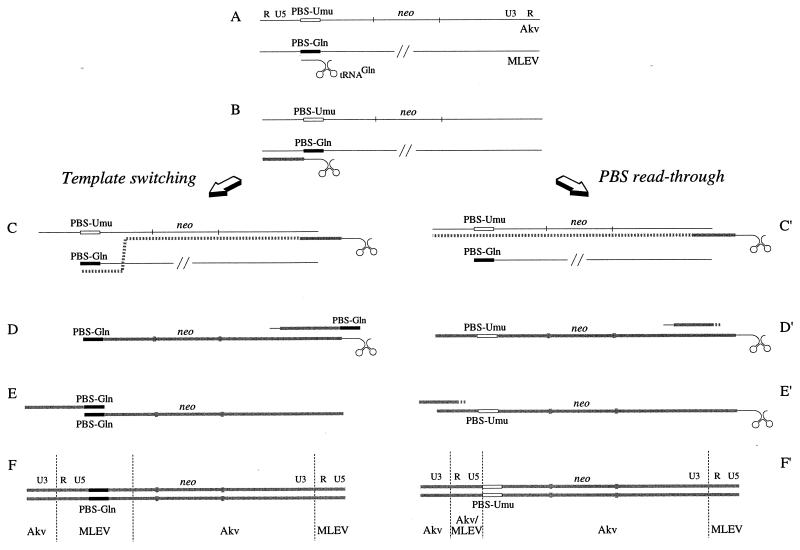
The replication of MLV-derived vectors harboring nonsense PBS sequences is strongly restricted (36), and such impaired vectors may thus rely on copackaging with viral sequences harboring functional PBS sequences in order to be reverse transcribed in the early stage of infection. At present, two recombination-based rescue pathways have been identified (Fig. 3); both are based on initiation of minus-strand synthesis from the functional PBS-Gln of MLEV (36, 37). Recombinant proviruses harboring the repaired or the originally mutated PBS sequence are results of 5′ UTR template switching and PBS read-through during minus-strand DNA synthesis, respectively (Fig. 3F and F′).
FIG. 3.
Models for forced recombination of PBS-modified retroviral vectors. (A) Copackaging of vector with MLEV sequence harboring a functional PBS-Gln sequence bound by a host cell-derived tRNAGln. (B) Initiation of minus-strand synthesis on MLEV leads to the generation of a MLEV-derived minus strand strong-stop DNA. (C and C′). Following minus-strand transfer, minus-strand synthesis is continued through the neo gene. Template switching may occur between neo and the mutated PBS (36), or the nascent reverse transcriptase may read through the mutated PBS and continue until reaching the 5′ terminus of the vector RNA (37). (D and D′) Generation of full-length and incomplete plus-strand strong-stop DNAs, respectively. (E and E′) Second-strand transfer facilitated by complementary PBS-Gln sequences or by R-U5 sequences. (F and F′) Akv-MLEV recombinant proviruses harboring repaired and originally mutated PBS, respectively.
In case of recombination within the 5′ UTR, template switching of nascent minus-strand DNA occurs site preferentially within the kissing-loop dimerization sequence (36, 38). Therefore, to further dissect the role played by the kissing-loop structure in recombination, we wanted to test whether mutations of the kissing-loop affect the crossover site specificity. For use in recombination studies, the PBS-Pro sequence of all kissing-loop-modified vectors was replaced by a defective PBS-Umu sequence, which does not match the 3′ end of any known tRNA. In Akv-MLEV heterodimer formation involving the wild-type vector, perfect base pairing between the distinct kissing loops of Akv (5′-UAGCUA-3′) and MLEV (5′-UGGCUA-3′) is restored by U (Akv):G (MLEV) base pairing at the genetic marker position LM9 (Fig. 4). In contrast, vector RNAs harboring mutated kissing-loop sequences may possibly differ in their capabilities to form Akv-Akv homodimers and Akv-MLEV heterodimers, respectively. Hence, for the substitutions mutants KL-TTGCTA and KL-GCTCTA, only one and three mismatches were introduced into a possible RNA duplex formed with the MLEV kissing loop in Akv-MLEV heterodimers, whereas twice as many mismatches were expected to affect Akv-Akv homodimer formation of these mutants. For the remaining mutants, Akv-MLEV heterodimers involving putative kissing-loop interactions were not likely to form more efficiently than Akv-Akv homodimers.
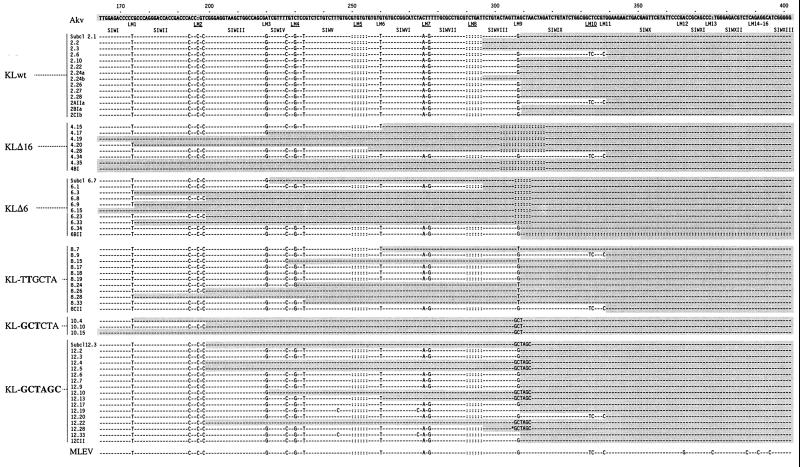
FIG. 4.
5′ UTR sequences of PBS-Gln-harboring transduced proviruses resulting from transfer of wild-type and kissing-loop-modified Akv vectors. The sequences of individual transduced proviruses were determined by sequence analysis of PCR fragments encompassing the Akv-MLEV chimeric 5′ UTR. Transduced viral sequences are compared with homologous regions of Akv (top) and MLEV (bottom). Subclones 2.1, 6.7, and 12.3 were derived from individual G418-resistant clones; the remaining sequences (except 2.24-a and -b, derived from the same plate) originate from PCR-based screenings of colony pools (Table 1) obtained from separate plates and subsequent sequence analysis of resulting PCR products. Nucleotides similar to positions in Akv are indicated by hyphens; deleted nucleotides compared to Akv are indicated by colons (:); insertions are indicated by introduction of colons in the Akv sequence. Hatched areas indicate Akv-derived sequences. The asterisk in KL-GCTAGC 12.28 indicates the finding of the two most 5′ nucleotides of wild-type loop motif, demonstrating that template switching was inaccurate in recombinational repair of this mutant. The transduced proviruses 12.19 and 12.33 were generated by recombination not with MLEV but with a related sequence similar to the previously identified clone 621 (12).
As expected, the replication efficiency of vectors harboring modifications within both the PBS and the kissing-loop region was strongly reduced compared to unmodified vectors (Fig. 1 and Table 1). To test whether rarely transduced proviruses were generated through recombinational patch repair of the PBS, the PBS composition of a total of 84 individually transduced proviruses conferring G418 resistance on NIH 3T3 target cells was analyzed. Fragments encompassing the transduced PBS were PCR amplified and subsequently digested with BstBI, which cleaves specifically within the PBS-Umu sequence (data not shown). The majority of the transduced proviruses contained the PBS-Umu sequence, revealing a high background of transduction pathways in which the PBS had not been repaired. In 3 of 19 wild-type proviruses and in 3 of 65 kissing-loop-modified vector transfers, the PBS modification had been replaced by the MLEV-derived PBS-Gln (Table 1). Alternative recombination mechanisms based on PBS read-through (Fig. 3) or hitherto unknown transduction pathways may account for these background transfer events. Notably, this result also suggests that the kissing-loop structure is not essential for recombination with copackaged endogenous viral RNA.
TABLE 1.
Replication efficiencies of kissing-loop-modified vectors and detection of recombination with MLEV by analysis of single colonies and PCR-based screening of colony pools
| Vector construct | Single-colony analysisa
|
Colony pool analysisd
|
||||
|---|---|---|---|---|---|---|
| Transduction titerb (101 CFU/ml/107 cells) | Transduced PBSc (PBS-Gln/total) | No. of colony pools | Avg. no. of colonies/ pool | Transduction pathway (no. of Akv-MLEV recombinants/total no. of colony pools)
|
||
| 5′ UTR minus-strand recombinatione | R-U5-mediated second-strand transfer recombinationf | |||||
| PBSUmuKLwtAkv-neo | 0.2 | 3/19 | 31 | 25 | 13/31 | 5/31 |
| PBSUmuKLΔ16Akv-neo | 0.03 | 1/18 | 27 | 9 | 8/27 | 6/27 |
| PBSUmuKLΔ6Akv-neo | 0.07 | 1/17 | 24 | 9 | 9/24 | 6/24 |
| PBSUmuKL-TTGCTA Akv-neo | 0.8 | 0/17 | 32 | 36 | 11/32 | 6/32 |
| PBSUmuKL-GCTCTA Akv-neo | 0.02 | 0/3 | 18 | 4 | 3/18 | 6/18 |
| PBSUmuKL-GCTAGC Akv-neo | 0.06 | 1/10 | 27 | 13 | 16/27 | 4/27 |
G418-resistant colonies were isolated, individually expanded, and analyzed by PCR amplification, restriction enzyme digestion, and sequencing.
Based on two independent experiments.
Detection of MLEV-derived PBS-Gln given as number of proviruses harboring PBS-Gln relative to the total number of transduced proviruses analyzed.
PCR-based screening of colony pools obtained by pooling of all G418-resistant colonies obtained on single plates.
Transduced PBS-Gln originates from recombination with MLEV through template switching within the 5′ UTR (Fig. 3A). Transduced proviral sequences are given in Fig. 4.
Transduction mediated through initial priming on copackaged MLEV RNA, read-through of PBS-Umu during minus-strand synthesis, and subsequent second-strand transfer facilitated by R-U5 complementarity of plus-strand and extended minus-strand DNA (Fig. 3B).
Sequence analysis of transduced recombinant 5′ UTRs detected by PCR screening of colony pools.
To investigate the effect on site specificity of template switching within kissing-loop-modified 5′ UTR sequences, we set out to screen a large number of transduced proviral sequences for Akv-MLEV recombinants harboring PBS-Gln. For the various kissing-loop-modified constructs, a total of 128 colony pools were generated by pooling of G418-resistant NIH 3T3 colonies obtained on 128 separate plates, each containing on average 4 to 36 colonies, depending on the mutant studied (Table 1). Colony pools were screened with primers specifically matching PBS-Gln and sequences within the neo gene, and amplicons were reproducibly obtained in 47 of the 128 colony pools for the kissing-loop-modified vectors and in 13 of 31 wild-type colony pools. These findings indicated that each colony pool contained one or less 5′ UTR Akv-MLEV recombinant and that recombinant sequences did not originate from PCR recombination between distinct recombinants within a pool or between transduced vectors and viral sequences endogenous to the target cells. All PCR products were sequenced with primers matching the PBS and the upstream part of neo to elucidate the pattern of genetic markers in each recombinant 5′ UTR sequence (Fig. 4) and thereby map the site of template shifting in individual repair events.
The 5′ UTRs of Akv and MLEV differ at scattered nucleotide leader marker (LM) positions dispersed throughout the region. This genetic marker-based division of the recombinational target sequence defines a panel of sequence identity windows (SIWI to SIWXIII), ranging in size from 6 to 27 nucleotides (Fig. 4). For the wild-type vector (referred to as PBSUmuKLwt), 12 of 14 template shifts were mapped within the 12- and 24-nucleotide Akv-MLEV SIWVIII and SIWIX intervening markers LM8 and LM10 (Fig. 5), providing independent confirmation that recombination within the 5′ UTR occurs site preferentially (36). In PBSUmuKLΔ16, a significant region of the recombinogenic site was removed and the lengths of SIWVIII and SIWIX were reduced to 6 and 15 nucleotides, respectively (Fig. 4). For this vector, all eight registered junction sites were mapped outside the hotspot region, perhaps as a result of reduced lengths of the SIWs flanking the deletion. For PBSUmuKLΔ6, 7 of 10 recombinant proviruses had been generated through template switching events in SIWs close to the PBS. One event was registered within each of the hotspot SIWs SIWVIII and SIWIX. In addition, one provirus was found to harbor a deletion of the downstream part of the 5′ UTR, indicating that template shifting in this case was not accurate. PBSUmuKL-TTGCTA harbors a single-nucleotide substitution exactly at the Akv-MLEV marker position LM9. Hence, this alteration may interfere with functions of the kissing-loop but does not change lengths of the flanking SIWs. Interestingly, recombination between this vector and MLEV occurred randomly within the target sequence; only 3 of 11 crossover sites were identified within the hotspot region, whereas the remaining junction sites were mapped within six distinct SIWs upstream and downstream of the kissing-loop domain (Fig. 5). For PBSUmuKL-GCTCTA, none of three identified template shifts had occurred within the kissing-loop region, indicating that function of the kissing-loop was disturbed by the nucleotide alterations introduced into the loop motif. PBSUmuKL-GCTAGC harbored an alternative palindromic loop sequence, which did not facilitate optimal kissing-loop function, as determined by virus titer experiments (Fig. 1). The kissing-loop sequence in this vector was therefore not expected to confer site specificity of recombinational PBS repair. However unexpectedly, more than half of the registered template switching events (10 of 17) had occurred within the hotspot region and predominantly within SIWIX.
FIG. 5.
Sites of 5′ UTR template switching from kissing-loop-modified vector RNAs to copackaged MLEV-derived RNA. Arrows point to sites of recombination; ratios indicate number of proviruses with specific Akv-MLEV junction site/total number of proviruses analyzed. The hatched box indicates the hotspot region for recombination of wild-type vectors (between markers LM8 and LM10). Twenty-two previously mapped recombination junction sites identified for the wild-type vector (36) are included for comparison. Hatched dots indicate single nucleotide differences, whereas clusters of differences are indicated by black dots. White boxes (KLΔ16 and KLΔ6) and white ellipses (KL-TTGCTA, KL-GCTCTA, and KL-GCTAGC) indicate kissing-loop deletions and nucleotide substitutions, respectively. Graphic representations of the modified kissing stem-loops are shown at the right.
In summary, these data support the idea that site-specific transfer of nascent minus-strand DNA within the 5′ UTR is closely related to the structural and functional properties of the kissing loop and to a lesser extent depends on sequence identity within the recombination-prone region of the 5′ UTR.
Rescue of modified vectors through R-U5-mediated second-strand transfer.
Deletion of the kissing stem-loop has previously been claimed to impair second strand-transfer during HIV-1 reverse transcription (42). We therefore examined whether all PBS-modified kissing-loop mutants could be rescued through a transduction pathway based on mutated PBS read-through and an unconventional R-U5-mediated second-strand transfer (Fig. 3B). Proviruses that originate from this rescue mechanism are characterized by harboring a mutated PBS (PBS-Umu) flanked upstream by MLEV-derived R and U5 sequences (37). Hence, to detect recombination with MLEV, a primer recognizing specifically an MLEV genetic marker in the R region (in the 5′ LTR) was used together with a PBS-Umu primer in a PCR-based screening of all colony pools. R-U5 recombinants were found with comparable incidences for all vector constructs analyzed (Table 1). Notably, exact ratios defined as recombinants per screened provirus do not directly reflect the recombination rates, as the background level of nonrecombination-based transduction may vary significantly among the kissing-loop mutants. Nevertheless, these observations show that mutations of the kissing loop do not prevent Akv vectors from interacting genetically with endogenous viral RNAs outside the 5′ leader.
DISCUSSION
The 5′ leader region of retroviruses consists of a panel of stem-loop structures, some of which are essential for optimal RNA dimerization and packaging, reverse transcription, and translation. Predicted RNA secondary structures have been shown to arrest reverse transcriptase during viral cDNA synthesis in vitro (15). Pausing of the polymerase due to secondary structures or reduction of deoxynucleoside triphosphate concentrations may promote its dissociation and enhance chances of template switching (7, 14, 61). Recombination within the kissing-loop dimerization sequence occurs at both sides of the loop motif (references 36 and 38 and this study); therefore, the hotspot distribution of transfer sites within the 5′ UTR is not compatible with a model in which the reverse transcriptase is arrested and forced to switch template before traversing a stable intramolecular RNA structure. This view is supported also by the finding that other stem-loops, some longer than the kissing stem-loop, do not promote template switching within the highly structured leader region. To study in more detail the functional properties of the kissing loop, in this study we introduced various mutations into the dimerization sequence and examined the recombination preference of the resulting vectors.
Sixty-three individual strand transfer events were analyzed in the present study. For all kissing-loop-modified PBS-impaired vectors except the vector harboring the GCTAGC palindromic sequence, the sites of recombinational patch repair were mapped predominantly upstream from kissing-loop sequence and found distributed without preference for any specific SIW throughout the 5′ UTR. Since most Akv-MLEV chimeric proviruses, generated by template switching in sequence identity windows near the PBS, harbored the mutated vector-derived kissing-loop sequence, it remained a formal possibility that recombinant proviruses identified after G418 selection were results of a selective bias exerted at the level of marker gene expression. However, since reduced NPTII-to-RNA ratios were measured in all cell lines stably transfected with kissing-loop-modified vector constructs, it seems unlikely that a selective advantage for the recombinant provirus could be obtained by read-through of the altered kissing-loop sequence during minus-strand synthesis. Rather, the nonclustered site distribution pattern described above may reflect a functional defect in kissing-loop function or for some vectors modified lengths of the SIWs encompassing the kissing-loop sequence, rendering these identity stretches less prone for switching between homologous donor and acceptor templates.
In KL-TTGCTA, one position coinciding with the Akv-MLEV marker LM9 was altered. Hence, in this vector the lengths of the flanking SIWVIII and SIWIX were not modified. The pattern of template switching within the 5′ UTR was markedly altered by this single-base mutation, since 8 of 11 template shifts were mapped outside the hotspot region. Therefore, a specific constitution of vector-MLEV sequence identity within the recombination-prone region is not the primary cis-acting determinant for frequent template shifting within the kissing-loop sequence. Rather, we suggest that a single-nucleotide substitution within the six-nucleotide loop motif disrupts the functional properties of the entire kissing stem-loop and thereby strongly influences strand transfer at the dimerization site. Consistent with this view, the single-base mutation was found to diminish viral replication fivefold and RNA encapsidation about eightfold. Removal of the entire stem-loop (39) or insertion of an 18-bp linker sequence within the kissing stem-loop (19) has been found to reduce encapsidation of genomic RNA six- or threefold, respectively, and to inhibit overall Moloney MLV replication. Detailed mutation studies of the loop motif of the kissing stem-loop have not been carried out in an MLV-based system. Interestingly, however, single-nucleotide changes within the HIV-1 loop-loop kissing motif have been found to hamper RNA dimerization of synthetic RNAs, suggesting that two mismatches at the putative 6-bp recognition site are disruptive for dimer formation (11, 24, 44). Provided that a changed recombination pattern reflects irregularities in Akv-MLEV RNA dimerization, our observations lend credence to the notion that a perfect 6-bp loop-loop interaction is essential for optimal function of the kissing-loop in vivo. Results obtained with deletion mutants (KLΔ16 and KLΔ6) and the three-nucleotide substitution mutant (KL-GCTCTA) confirm that an intact loop sequence is a prerequisite for efficient RNA encapsidation and hotspot recombinational repair within the 5′ UTR, since recombination junction sites for these three mutants were mapped in nonclustered patterns predominantly within SIWs upstream from the introduced mutation. Notably, genetic interactions between MLEV and Akv were not blocked by introduced kissing-loop deletions or nucleotide substitutions, supporting the recently reported notion that rates of recombination are unaffected by kissing-loop nonhomology between interacting HIV-1-derived RNAs (51).
Preferred template switching within the kissing-loop dimerization site was partly restored in the KL-GCTAGC vector harboring the alternative autocomplementary sequence. Template shifts during patch repair occurred predominantly within SIWIX, which was four nucleotides shorter than the corresponding wild-type Akv-MLEV window (Fig. 4). This site preference clearly differs from the lack of specificity observed during repair of the KLΔ6 vector, which harbors identical sequence similarity windows within the modified kissing-loop region. Hence, reduced lengths of the SIWs flanking LM9 do not solely explain the altered properties of recombination observed for the KLΔ6 vector. The kissing stem-loop in the KL-GCTAGC vector appears to be nonfunctional, as indicated by a 22-fold titer reduction relative to the wild-type level. A similar reduction was found for the KLΔ16 vector, possibly indicating that loop-loop complementarity is not sufficient for optimal dimer formation and that a specific loop composition is required for its function. This notion finds support in HIV-1 studies, demonstrating that within the context of a retroviral genome a specific loop sequence is required to facilitate optimal RNA dimerization (11). However, an alternative possibility is that introduction of the alternative palindrome interferes with leader functions other than the intermolecular kissing-loop interaction and that the altered loop sequence may in fact support homodimer formation of vector RNA. Regardless of the reasons for the titer reduction of this mutant, it seems unlikely that the modified sequence (5′-GCUAGC-3′) would facilitate traditional loop kissing with the loop sequence of MLEV (5′-UGGCUA-3) during formation of Akv-MLEV heterodimers, leading to the observed hotspot pattern of template switching. As an alternative explanation, introduction of the palindrome might in the KL-GCTAGC vector lead to the generation of artificial RNA secondary structures within the 5′ leader RNA; e.g., as one possibility we note that five of six inserted bases may putatively form base pairs with nucleotides further downstream, perhaps leading to an alternative folding of the RNA. Notably, purely speculative models for such changes suggest that an alternative loop with an eight-nucleotide palindromic sequence could be exposed. Such possible structural alterations may disturb normal functions of the region in the KL-GCTAGC vector and could be hypothesized to facilitate an intermolecular interaction with MLEV.
In the present study we have found that an intact kissing-loop sequence is essential for both optimal virus replication and hotspot recombination within the 5′ leader region, suggesting that the panel of introduced mutations interferes with functional properties of the kissing loop and perhaps flanking structures. Retroviruses are highly sensitive to base changes introduced into the multifunctional and dynamic 5′ leader region that may possess different conformations of importance for functions at various stages of retrovirus replication. Structural rearrangements, perhaps involving both intramolecular and intermolecular interactions, are likely to be crucial for efficient reverse transcription in newly infected cells and may also affect RNA binding of Gag and nucleocapsid proteins during genomic RNA encapsidation and early steps of infection.
Impairment of RNA packaging and changes in patterns of template switching may reflect that mutations of the kissing-loop influence (i) intramolecular, (ii) intermolecular RNA-RNA, and/or (iii) RNA-protein interactions, possibly leading to RNA structural and functional defects. The conformation of the dimeric RNA genome is essential for minus-strand DNA transfer in reverse transcription, since a heat-denatured template does not support generation of extended cDNAs (5). In addition, the viral environment provided by nucleocapsid core architecture appears to have importance for the strand transfer reaction (60), consistent with the notion that exact structural properties of the reverse-transcribed dimer are important for efficient reverse transcription to occur. The kissing-loop sequence may thus serve an essential role in maintaining a correct structure of the dimer, as indicated by the finding that plus-strand DNA transfer is impaired upon deletion of the entire stem-loop (42). Multiple lines of biochemical evidence support that a direct intermolecular interaction at the kissing loop is crucial for correct dimer formation (22, 30, 40, 43). It remains unclear whether loop-loop kissing is followed by local structural rearrangements, leading to the formation of an extended RNA duplex at the interaction site (22, 43). However, we propose that the proximity of donor and acceptor templates facilitated by loop-loop recognition increases the chance of intermolecular template switching at the dimerization site, in accordance with the finding that intramolecular template shifts are more efficient than intermolecular template switches and that the reverse transcriptase may therefore tend to reassociate with the template from which it dissociates during minus-strand DNA synthesis (27). Since the HIV-1 kissing stem-loop may serve as an independent low-affinity Gag binding site in vitro (10), we cannot at this stage exclude that kissing-loop alterations may affect not only direct RNA-RNA interactions but also protein binding onto monomeric or dimeric RNA.
In conclusion, we propose that alterations of the kissing loop, ranging from a deletion of the entire sequence to a single point mutation within the loop motif, primarily interfere with intermolecular recognition at the modified site. This would imply that the kissing-loop interaction is maintained within the mature dimer and suggests that the kissing-loop sequence may be one of several genomic interaction points that facilitate RNA dimerization and subsequent encapsidation and which may therefore contribute to an optimal structure of the genome required for early events of retrovirus infection. Our data may hence serve as a first demonstration that direct intermolecular interactions facilitate template switching during retroviral cDNA synthesis.
ACKNOWLEDGMENTS
This work was supported by the Danish Biotechnology Programme, the Danish Cancer Society, the Danish Natural Sciences and Medical Research Councils, the Karen Elise Jensen Foundation, and contracts Biotech CT95-0100 and Biomed2 CT95-0675 from the European Commission.
REFERENCES
- 1.Alford R L, Honda S, Lawrence C B, Belmont J W. RNA secondary structure analysis of the packaging signal for Moloney murine leukemia virus. Virology. 1991;183:611–619. doi: 10.1016/0042-6822(91)90990-s. [DOI] [PubMed] [Google Scholar]
- 2.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 3.Baudin F, Marquet R, Isel C, Darlix J-L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 4.Beck E, Ludwig G, Auerswald E A, Reiss B, Schaller H. Nucleotide sequence and exact localization of the neomycin phosphotransferase gene from transposon Tn5. Gene. 1982;19:327–336. doi: 10.1016/0378-1119(82)90023-3. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout B, Das A T, van Wamel J L B. The native structure of the human immunodeficiency virus type 1 RNA genome is required for the first strand transfer of reverse transcription. Virology. 1998;249:211–218. doi: 10.1006/viro.1998.9321. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buiser R G, Bambara R A, Fay P J. Pausing by retroviral DNA polymerases promotes strand transfer from internal regions of RNA donor templates to homopolymeric acceptor templates. Biochim Biophys Acta. 1993;1216:20–30. doi: 10.1016/0167-4781(93)90033-a. [DOI] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization and encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clever J L, Wong M L, Parslow T G. Requirements for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Colicelli J, Goff S P. Identification of endogenous retroviral sequences as potential donors for recombinational repair of mutant retroviruses: positions of crossover points. Virology. 1987;160:518–522. doi: 10.1016/0042-6822(87)90030-4. [DOI] [PubMed] [Google Scholar]
- 13.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski M C, Roques B, Darlix J-L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc Natl Acad Sci USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeStefano J J, Buiser R G, Mallaber L M, Fay P J, Bambara R A. Parameters that influence synthesis and site-specific termination by human immunodeficiency virus reverse transcriptase on RNA and DNA templates. Biochim Biophys Acta. 1992;1131:270–280. doi: 10.1016/0167-4781(92)90025-u. [DOI] [PubMed] [Google Scholar]
- 15.DeStefano J J, Mallaber L M, Rodriguez-Rodriguez L, Fay P J, Bambara R A. Requirements for strand transfer between internal regions of heteropolymer templates by human immunodeficiency virus reverse transcriptase. J Virol. 1992;66:6370–6378. doi: 10.1128/jvi.66.11.6370-6378.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Tapia M, Metzler V, Mougel M, Ehresmann B, Ehresmann C. Dimerization of MoMuLV genomic RNA: redefinition of the role of the palindromic stem-loop H1 (278-303) and new roles for stem-loops H2 (310-352) and H3 (355-374) Biochemistry. 1998;37:6077–6085. doi: 10.1021/bi9800303. [DOI] [PubMed] [Google Scholar]
- 17.Duch M, Paludan K, Pedersen L, Jørgensen P, Kjeldgaard N O, Pedersen F S. Determination of transient or stable neo expression levels in mammalian cells. Gene. 1990;95:285–288. doi: 10.1016/0378-1119(90)90373-y. [DOI] [PubMed] [Google Scholar]
- 18.Feng Y-X, Copeland T D, Henderson L E, Gorelick R J, Bosche W J, Levin J G, Rein A. HIV-1 nucleocapsid protein induces “maturation” of dimeric retroviral RNA in vitro. Proc Natl Acad Sci USA. 1996;93:7577–7581. doi: 10.1073/pnas.93.15.7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fisher J, Goff S P. Mutational analysis of stem-loops in the RNA packaging signal of the Moloney murine leukemia virus. Virology. 1998;244:133–145. doi: 10.1006/viro.1998.9090. [DOI] [PubMed] [Google Scholar]
- 20.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wildtype and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu W, Rein A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J Virol. 1993;67:5443–5449. doi: 10.1128/jvi.67.9.5443-5449.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Girard P-M, Bonnet-Mathonière B, Muriaux D, Paoletti J. A short autocomplementary sequence in the 5′ leader region is responsible for dimerization of MoMuLV genomic RNA. Biochemistry. 1995;34:9785–9794. doi: 10.1021/bi00030a016. [DOI] [PubMed] [Google Scholar]
- 23.Girard P-M, de Rocquigny H, Roques B-P, Paoletti J. A model of PSI dimerization: destabilization of the C278-G303 stem-loop by the nucleocapsid protein (NCp10) of MoMuLV. Biochemistry. 1996;35:8705–8714. doi: 10.1021/bi952454s. [DOI] [PubMed] [Google Scholar]
- 24.Haddrick M, Lear A L, Cann A J, Heaphy S. Evidence that a kissing loop structure facilitates genomic RNA dimerisation in HIV-1. J Mol Biol. 1996;259:58–68. doi: 10.1006/jmbi.1996.0301. [DOI] [PubMed] [Google Scholar]
- 25.Hu W-S, Temin H M. Genetic consequences of packaging two RNA genomes in one retroviral particle: pseudoploidy and high rate of genetic recombination. Proc Natl Acad Sci USA. 1990;87:1556–1560. doi: 10.1073/pnas.87.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu W-S, Temin H M. Retroviral recombination and reverse transcription. Science. 1990;250:1227–1233. doi: 10.1126/science.1700865. [DOI] [PubMed] [Google Scholar]
- 27.Hu W-S, Bowman E H, Delviks K A, Pathak V K. Homologous recombination occurs in a distinct retroviral subpopulation and exhibits high negative interference. J Virol. 1997;71:6028–6036. doi: 10.1128/jvi.71.8.6028-6036.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Itin A, Keshet E. Apparent recombinants between virus-like (VL30) and murine leukemia virus-related sequences in mouse DNA. J Virol. 1983;47:178–184. doi: 10.1128/jvi.47.1.178-184.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Junghans R P, Boone L R, Skalka A M. Retroviral DNA H structures: displacement-assimilation model for recombination. Cell. 1982;30:53–62. doi: 10.1016/0092-8674(82)90011-3. [DOI] [PubMed] [Google Scholar]
- 30.Laughrea M, Jetté L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms and all sequences upstream and downstream of hairpin 248-271 are dispensable for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 31.Laughrea M, Jetté L, Mak J, Kleiman L, Liang C, Wainberg M A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J Virol. 1997;71:3397–3406. doi: 10.1128/jvi.71.5.3397-3406.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lund A H, Duch M, Lovmand J, Jørgensen P, Pedersen F S. Mutated primer binding sites interacting with different tRNAs allow efficient murine leukemia virus replication. J Virol. 1993;67:7125–7130. doi: 10.1128/jvi.67.12.7125-7130.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mak J, Kleiman L. Primer tRNAs for reverse transcription. J Virol. 1997;71:8087–8095. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mann R, Mulligan R C, Baltimore D. Construction of a retrovirus packaging mutant and its use to produce helper-free defective retrovirus. Cell. 1983;33:153–159. doi: 10.1016/0092-8674(83)90344-6. [DOI] [PubMed] [Google Scholar]
- 35.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikkelsen J G, Lund A H, Kristensen K D, Duch M, Sørensen M S, Jørgensen P, Pedersen F S. A preferred region for recombinational patch repair in the 5′ untranslated region of primer binding site-impaired murine leukemia virus vectors. J Virol. 1996;70:1439–1447. doi: 10.1128/jvi.70.3.1439-1447.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikkelsen J G, Lund A H, Dybkær K, Duch M, Pedersen F S. Extended minus-strand DNA as template for R-U5-mediated second-strand transfer in recombinational rescue of primer binding site-modified retroviral vectors. J Virol. 1998;72:2519–2525. doi: 10.1128/jvi.72.3.2519-2525.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikkelsen J G, Lund A H, Duch M, Pedersen F S. Recombination in the 5′ leader of murine leukemia virus is accurate and influenced by sequence identity with a strong bias toward the kissing-loop dimerization domain. J Virol. 1998;72:6967–6978. doi: 10.1128/jvi.72.9.6967-6978.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38a.Mikkelsen J G, Lund A H, Duch M, Pedersen F S. Forced recombination of ψ-modified murine leukaemia virus-based vectors with murine leukaemia virus-like and VL30 murine endogenous retroviruses. J Gen Virol. 1999;80:2957–2967. doi: 10.1099/0022-1317-80-11-2957. [DOI] [PubMed] [Google Scholar]
- 39.Mougel M, Zhang Y, Barklis E. cis-active structural motifs involved in specific encapsidation of Moloney murine leukemia virus RNA. J Virol. 1996;70:5043–5050. doi: 10.1128/jvi.70.8.5043-5050.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mujeeb A, Clever J L, Billeci T M, James T L, Parslow T G. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat Struct Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 41.Murti K G, Bondurant M, Tereba A. Secondary structural features in the 70S RNAs of Moloney murine leukemia virus and Rous sarcoma viruses as observed by electron microscopy. J Virol. 1981;37:411–419. doi: 10.1128/jvi.37.1.411-419.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paillart J-C, Berthoux L, Ottman M, Darlix J-L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paillart J-C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Paillart J-C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 45.Paoletti J, Mougel M, Tounekti N, Girard P M, Ehresmann C, Ehresmann B. Spontaneous dimerization of retroviral MoMuLV RNA. Biochemie. 1993;75:681–686. doi: 10.1016/0300-9084(93)90099-e. [DOI] [PubMed] [Google Scholar]
- 46.Platt S G, Yang N-S. Dot assay for neomycin phosphotransferase activity in crude cell extracts. Anal Biochem. 1987;162:529–535. doi: 10.1016/0003-2697(87)90429-5. [DOI] [PubMed] [Google Scholar]
- 47.Prats A-C, Roy C, Wang P A, Erard M, Housset V, Gabus C, Paoletti C, Darlix J-L. cis elements and trans-acting factors involved in dimer formation of murine leukemia virus RNA. J Virol. 1990;64:774–783. doi: 10.1128/jvi.64.2.774-783.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy C, Tounekti N, Mougel M, Darlix J-L, Paoletti C, Ehresmann C, Ehresmann B, Paoletti J. An analytical study of the dimerization of in vitro generated RNA of Moloney murine leukemia virus MoMuLV. Nucleic Acids Res. 1990;18:7287–7292. doi: 10.1093/nar/18.24.7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sakuragi J-I, Panganiban A T. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J Virol. 1997;71:3250–3254. doi: 10.1128/jvi.71.4.3250-3254.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skripkin E, Paillart J-C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.St. Louis D C, Gotte D, Sanders-Buell E, Ritchey D W, Salminen M O, Carr J K, McCutchan F E. Infectious molecular clones with the nonhomologous dimer initiation sequences found in different subtypes of human immunodeficiency virus type 1 can recombine and initiate a spreading infection in vitro. J Virol. 1998;72:3991–3998. doi: 10.1128/jvi.72.5.3991-3998.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stuhlmann H, Berg P. Homologous recombination of copackaged retrovirus RNAs during reverse transcription. J Virol. 1992;66:2378–2388. doi: 10.1128/jvi.66.4.2378-2388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stuhlmann H, Dieckmann M, Berg P. Transduction of cellular neo mRNA by retrovirus-mediated recombination. J Virol. 1990;64:5783–5796. doi: 10.1128/jvi.64.12.5783-5796.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sundquist W I, Heaphy S. Evidence for intrastrand quadruplex formation in the dimerization of human immunodeficiency virus 1 genomic RNA. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Swain A, Coffin J M. Mechanism of transduction by retroviruses. Science. 1992;255:841–845. doi: 10.1126/science.1371365. [DOI] [PubMed] [Google Scholar]
- 56.Tchénio T, Heidmann T. The dimerization/packaging sequence is dispensable for both the formation of high-molecular-weight RNA complexes within retroviral particles and the synthesis of proviruses of normal structure. J Virol. 1995;69:1079–1084. doi: 10.1128/jvi.69.2.1079-1084.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Torrent C, Bordet T, Darlix J-L. Analytical study of rat retrotransposon VL30 RNA dimerization in vitro and packaging in murine leukemia virus. J Mol Biol. 1994;240:434–444. doi: 10.1006/jmbi.1994.1459. [DOI] [PubMed] [Google Scholar]
- 58.Tounekti N, Mougel M, Roy C, Marquet R, Darlix J-L, Paoletti J, Ehresmann B, Ehresmann C. Effect of dimerization on the conformation of the encapsidation psi domain of Moloney murine leukemia virus RNA. J Mol Biol. 1992;223:205–220. doi: 10.1016/0022-2836(92)90726-z. [DOI] [PubMed] [Google Scholar]
- 59.Van Beveren C, Coffin J M, Hughes S. Nucleotide sequences complemented with functional and structural analysis. In: Weiss R, Teich N, Varmus H, Coffin J, editors. RNA tumor viruses. Vol. 2. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1985. pp. 790–805. [Google Scholar]
- 60.van Wamel J L B, Berkhout B. The first strand transfer during HIV-1 reverse transcription can occur either intramolecularly or intermolecularly. Virology. 1998;244:245–251. doi: 10.1006/viro.1998.9096. [DOI] [PubMed] [Google Scholar]
- 61.Wu W, Blumberg B M, Fay P J, Bambara R A. Strand transfer mediated by human immunodeficiency virus reverse transcriptase in vitro is promoted by pausing and results in misincorporation. J Biol Chem. 1995;270:325–332. doi: 10.1074/jbc.270.1.325. [DOI] [PubMed] [Google Scholar]
- 62.Yin P D, Hu W-S. RNAs from genetically distinct retroviruses can copackage and exchange genetic information in vivo. J Virol. 1997;71:6237–6242. doi: 10.1128/jvi.71.8.6237-6242.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang J, Temin H M. 3′ junctions of oncogene-virus sequences and the mechanisms for formation of highly oncogenic retroviruses. J Virol. 1993;67:1747–1751. doi: 10.1128/jvi.67.4.1747-1751.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]