Abstract
Of 62 Streptococcus thermophilus bacteriophages isolated from various ecological settings, half contain a lysin gene interrupted by a group IA2 intron. Phage mRNA splicing was demonstrated. Five phages possess a variant form of the intron resulting from three distinct deletion events located in the intron-harbored open reading frame (orf 253). The predicted orf 253 gene sequence showed a significantly lower GC content than the surrounding intron and lysin gene sequences, and the predicted protein shared a motif with endonucleases found in phages from both gram-positive and gram-negative bacteria. A comparison of the phage lysin genes revealed a clear division between intron-containing and intron-free alleles, leading to the establishment of a 14-bp consensus sequence associated with intron possession. The conserved intron was not found elsewhere in the phage or S. thermophilus bacterial genomes. Folding of the intron RNA revealed secondary structure elements shared with other phage introns: first, a 38-bp insertion between regions P3 and P4 that can be folded into two stem-loop structures (shared with introns from Bacillus phage SPO1 and relatives); second, a conserved P7.2 region (shared with all phage introns); third, the location of the stop codon from orf 253 in the P8 stem (shared with coliphage T4 and Bacillus phage SPO1 introns); fourth, orf 253, which has sequence similarity with the H-N-H motif of putative endonuclease genes found in introns from Lactococcus, Lactobacillus, and Bacillus phages.
Introns are regions which are transcribed and subsequently excised from the primary transcript by RNA splicing to generate the mature RNA. Group I and II introns, in which the folded structure of the intron participates directly in the splicing reaction, can be distinguished by their secondary structures and the mechanisms that they use to catalyze their own splicing. Group I introns, which are particularly well characterized, catalyze their own splicing by a series of guanosine-initiated trans-esterification reactions (for a review, see reference 20). Introns are remarkable not only in their role in encoding ribozymes but also in their ability to act as mobile genetic elements capable of efficiently inserting themselves into cognate intronless alleles. This process, known as “homing,” is mediated by different types of intron-encoded proteins which place the intron in the correct sequence context for efficient splicing (20).
The evolutionary history and biological role of introns have been the subject of much debate, and their true significance remains to be defined. The controversial debate has been nourished by the peculiar phylogenetic distribution of introns. Group I introns have been found within genes encoding mRNA, rRNA, and tRNA in diverse genetic systems, including eucaryotic (plant, fungus, and yeast) mitochondria, nuclei, and chloroplasts; bacteriophages infecting gram-positive and gram-negative bacteria; and eubacterial genomes (see references 20 and 27 for a review and a compilation, respectively). Among the eubacteria, the only group I introns described to date occur in several genera of cyanobacteria and purple proteobacteria (3, 32). Although group I introns have been described for several T-even bacteriophages of Escherichia coli (30, 36), six Bacillus subtilis bacteriophages (1, 17, 18, 22), and one bacteriophage each of Lactococcus lactis (29), Lactobacillus delbrueckii (28), and Staphylococcus aureus (21), none have been reported so far for the genomes of the respective host bacteria. Interestingly, apart from the T-even bacteriophages, all of the intron-containing phages described to date infect a group of evolutionarily related low-GC-content gram-positive bacteria. It has also been shown that the temperate phages of these bacteria are evolutionarily related (23, 24). Since Streptococcus and Lactococcus are closely related bacterial genera and since the analysis of introns from further phages of this group could constrain theories on the phylogenetic distribution and origin of introns, we investigated Streptococcus thermophilus phages (6) for the presence of introns.
Due to their economical importance for the dairy industry, large ecologically characterized S. thermophilus bacteriophage collections are available (5). Furthermore, five S. thermophilus bacteriophage genomes have been completely sequenced (25, 26, 37, 39). This fact, in addition to the fact that all S. thermophilus bacteriophages are related in terms of DNA homology (5), makes them well suited for systematic intron searches. So far, the distribution of introns within closely related bacteriophages has been studied only for T-even (31) and B. subtilis HMU phages (18).
This study describes the identification of a group I intron within the lysin gene of half of the investigated S. thermophilus bacteriophages. The relationship of this intron to other phage introns is discussed.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, and media.
S. thermophilus strains were routinely subcultured at 42°C in either LM17 (M17 supplemented with 0.5% lactose) (38) or Belliker (Elliker plus 1% beef extract) medium (Difco manual, Difco Laboratories, Detroit, Mich.). The S. thermophilus phages used in this study were obtained from the Nestlé Research Centre phage collection. The phages were propagated on the appropriate S. thermophilus strain in LM17 broth as described previously (4).
DNA techniques.
Phage purification, DNA extraction and purification, agarose gel electrophoresis, Southern blot and dot blot hybridizations, and DNA labelling were done as described previously (4).
DNA sequencing and analysis.
Phage S3b was sequenced by use of an Amersham Labstation sequencing kit based on Thermo Sequenase-labelled primer cycle sequencing with 7-deaza-dGTP. Sequencing was done on a Licor 6000L automated sequencer with fluorescence-labelled primers.
PCR products were sequenced on both strands by dideoxy chain termination with an fmol DNA sequencing system (Promega, Madison, Wis.). The sequencing primers were end labelled with [γ-33P]ATP according to the manufacturer's instructions. The thermal cycler (Perkin-Elmer) was programmed for 30 cycles at 95°C for 30 s, 50°C for 30 s, and 72°C for 1 min. The sequence obtained was analyzed as described by Lucchini et al. (23).
PCR.
DNA samples were amplified in a Perkin-Elmer thermal cycler programmed for 30 cycles each consisting of 94°C for 30 s, 55°C for 30 s, and 72°C for 1.5 min. Synthetic primers were designed according to established S. thermophilus phage DNA sequences and used together with the relevant DNA template and Taq polymerase (Fermentas). PCR products were gel purified with Ultrafree-MC Centrifugal Filter Units (Millipore) by following the manufacturer's instructions. The sequences of relevant primers used for PCR and reverse transcription (RT)-PCR are as follows: A (5′ TGT CCC ACA ATC TCT TGT 3′), B (5′ TTG TGA CTA CTC AAC TCA AGG AGC 3′), C (5′ GAA GCC AAT GAA GTC AAA TAC G 3′), D1 (5′ GGT CTG CTC CAT CTG GAA GGT CGT T 3′), D2 (5′ GGT CAG CTC CGT CTG GAA GGT CGT T 3′), E (5′ GGT CTG CTC CAT CTG GAA 3′), and F (5′ GTG GTC TAT TGG TAG TAG TTT ACC 3′). Oligonucleotides G1 and G2, H1 and H2 and I1 and I2 anneal to positions 29400 and 31476, 31159 and 33515, and 33367 and 35460, respectively, of the published phage Sfi21 sequence (GenBank accession no. AF115103). They are all 18 nucleotides long. Oligonucleotides J (5′ GGC AAT ACC GTG CCA AGT C 3′) and K (5′ CCC AAC TTG GAT TCT AGC 3′) were used to generate the S3b intron probe for dot blot hybridization.
RT-PCR.
Fifty milliliters of Belliker medium was inoculated with the relevant S. thermophilus strain and grown to an optical density at 600 nm of 0.2. CaCl2 (final concentration, 10 mM) was added together with the phage (105 to 106 PFU) to be tested. Following 15 min of incubation at 42°C, the cultures were harvested by centrifugation. The pellets obtained were washed with diethyl pyrocarbonate-treated water and resuspended in 200 μl of RNase-free 10 mM Tris–1 mM EDTA solution (pH 7.5). The cells were ruptured by agitation for 5 min with 150 μl of RNase-free glass beads (Sigma; 106 μm) in a bead beater at 4°C. The lysate was rapidly centrifuged at 4°C, and the supernatant was recovered.
RNA was isolated from the supernatant with an RNeasy kit (Qiagen) by following the manufacturer's instructions. cDNA was synthesized and amplified with an Access RT-PCR system (Promega) and the following thermal cycling profile: 48°C for 45 min; 94°C for 2 min; 40 cycles at 94°C for 30 s, 55°C for 1 min, and 68°C for 2 min; 68°C for 7 min; and a 4°C soak.
Nucleotide sequence accession numbers.
The sequence data for phages S3b, ST3, J, S92, Sfi16A, and ST64 have been submitted to the GenBank database under accession no. AF148561, AF148565, AF148566, AF148563, AF148564, and AF148562, respectively.
RESULTS
Identification of an interrupted lysin gene in phage S3b.
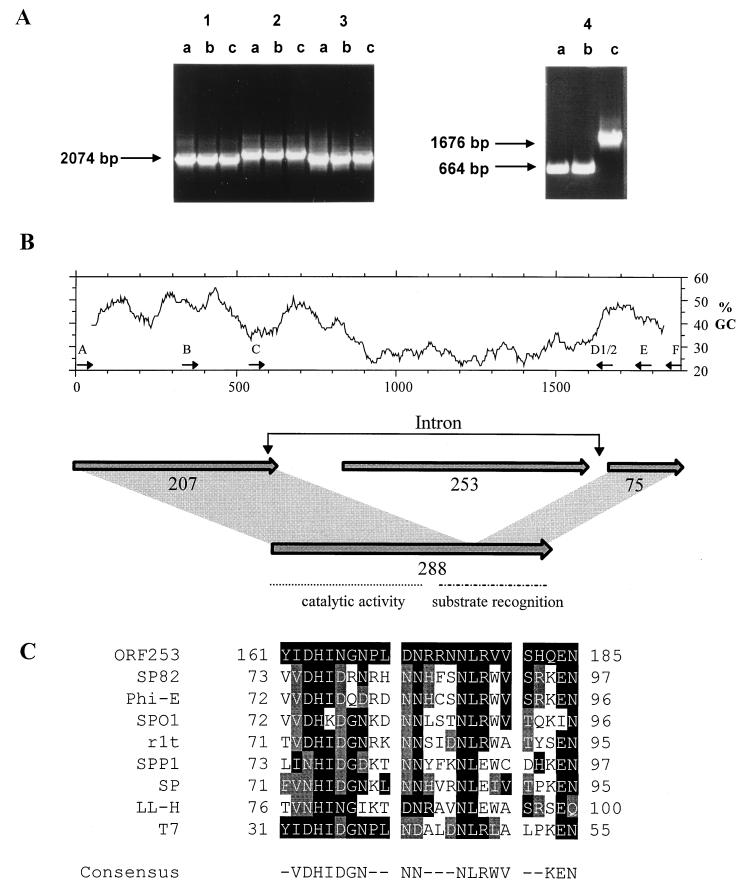
Several S. thermophilus phages have been entirely sequenced (25, 26, 37, 39). For one of them, phage Sfi21, deletion derivatives and variants have been reported (7). The availability of the Sfi21 genome sequence allowed a rapid characterization of these phages by use of PCR and primers selected such that the entire genome was amplified as overlapping fragments of approximately 2 kb (data not shown). The use of primers A and E, which are located within the Sfi21 putative lysin gene (orf 288) (Fig. 1B), resulted in an amplified DNA product of approximately 1.6 kb for variant phage S3b, in contrast to the expected 664-bp product for Sfi21 and S3 (Fig. 1A). This region of S3b was sequenced; analysis led to the prediction of three similarly oriented open reading frames with coding potential for 207, 253, and 75 amino acids (aa) (Fig. 1B). The proteins predicted for both orf 207 and orf 75 demonstrated high sequence similarity to the putative enzymatic and substrate recognition domains, respectively, of bacteriophage lysins (Fig. 1B).
FIG. 1.
(A) PCR screening of S. thermophilus bacteriophage genomes for insertion or deletion events. PCR amplification products obtained with primer pairs G1-G2 (block 1), H1-H2 (block 2), I1-I2 (block 3), and A-E (block 4) and phage DNAs from Sfi21 (lanes a), S3 (lanes b), and S3b (lanes c) are shown. Molecular sizes of the PCR products are provided for blocks 1 and 4. (B) (Top) Percent GC content distribution of the investigated 1.9-kb region of phage S3b encompassing the lysin gene (window length, 100 bp). Primers referred to throughout the text are indicated by thin arrows above the ruler providing the nucleotide scale in base pairs. (Bottom) Prediction of open reading frames in the region of the phage S3b lysin gene. The open reading frames are indicated by shaded arrows, with their lengths given in amino acids. orf 288, which results from an orf 207-orf 75 fusion, is shown together with the domain structure deduced from knowledge of pneumococcal phage Dp-1 (33). (C) Multiple alignment of a 25-aa block from the S3b orf 253 gene product and the gene product of coliphage T7 gene 3.8 (V01146), the gene product of Bacillus phage SPP1 orf 36.1 (X67865), and the intron-encoded gene products of Bacillus phages SP82 (U04812), phi-E (U04813), SPO1 (P34081), and SP (L31962), L. lactis phage r1t (U38906), and Lactobacillus phage LL-H (L37351). Amino acid positions identical to those of S3b are shaded in black, while conserved amino acids are shaded in grey. The consensus sequence indicates identical amino acids present in five or more sequences.
Database searches yielded no significant matches with the noncoding regions, while the predicted orf 253 gene product showed similarity over the N- and C-terminal halves to an unattributed 54-aa protein of S. thermophilus phage DT1 and to the predicted protein for gene 3.8 of coliphage T7, respectively. Closer scrutiny revealed a 25-aa region corresponding to the recently described H-N-H motif found in group I intron endonucleases of phages infecting gram-positive bacteria (34) (Fig. 1C). These data suggest that there is an intron in the lysin genes of S. thermophilus phages S3b and DT1.
Identification of the presence of a group I intron.
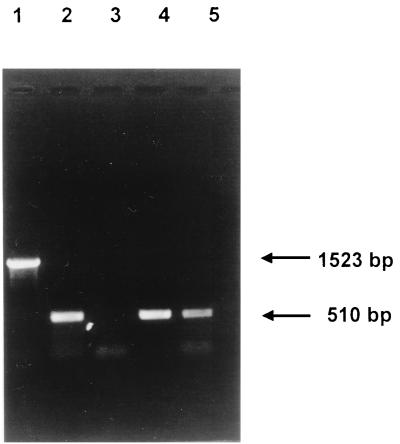
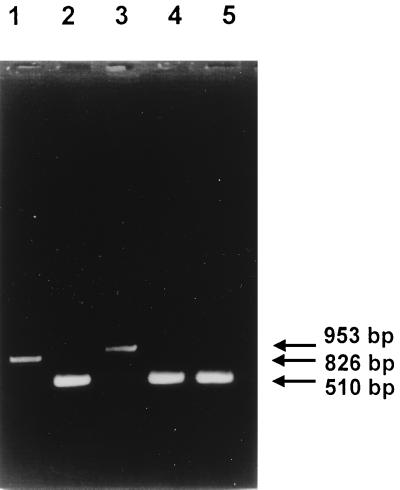
In order to test for in vivo splicing of RNA transcripts, reverse transcription (RT)-PCR was performed with oligonucleotides which specifically anneal to orf 207 and orf 75 of S3b. RNA was isolated from S. thermophilus Sfi1 15 min following infection by S3b and used for the synthesis of cDNA and subsequent PCR amplification with primers B and F (Fig. 1B). Phage Sfi21-infected S. thermophilus Sfi1 was also included in this assay for comparison. As a control, Sfi21 and S3b phage DNAs were PCR amplified with the same primers. A 510-bp RT-PCR product was obtained for S3b-infected Sfi1, in contrast to the 1,523-bp fragment obtained for S3b phage DNA (Fig. 2A). This result indicates that RNA splicing has occurred, resulting in the excision of 1,013 nucleotides at the level of the RNA precursor. Furthermore, the S3b RT-PCR product is identical in size to that obtained from Sfi21-infected Sfi1.
FIG. 2.
In vivo splicing of S3b intron RNA. Lanes 1 and 4, RT-PCR products obtained with phage S3b and Sfi21 DNAs, respectively, as templates; lanes 2 and 5, RT-PCR products obtained with RNA isolated from S3b- and Sfi21-infected cells, respectively, 15 min after infection; lane 3, PCR product (no reverse transcriptase) obtained with RNA isolated from S3b-infected cells 15 min after infection. Primers B and F (Fig. 1B) were used. The sizes of the products obtained are indicated in base pairs.
In order to determine the exact intron boundaries, the RT-PCR product was gel purified and sequenced. Sequence analysis revealed that splicing has occurred, as is typical of group I introns, following uridine and guanosine residues at positions 617 and 1630, respectively, resulting in the excision of a 1,013-bp intron (Fig. 3A). This event results in the generation of an open reading frame with a coding potential for a 288-aa protein. Although a new codon (GUA rather than GUU) is created at the splice junction, the coding potential for valine is conserved. The predicted orf 288 gene product demonstrates >85% identity with the putative lysins of S. thermophilus phages (Sfi21, Sfi19, Sfi18, Sfi11, DT1, and O1205) present in the database.
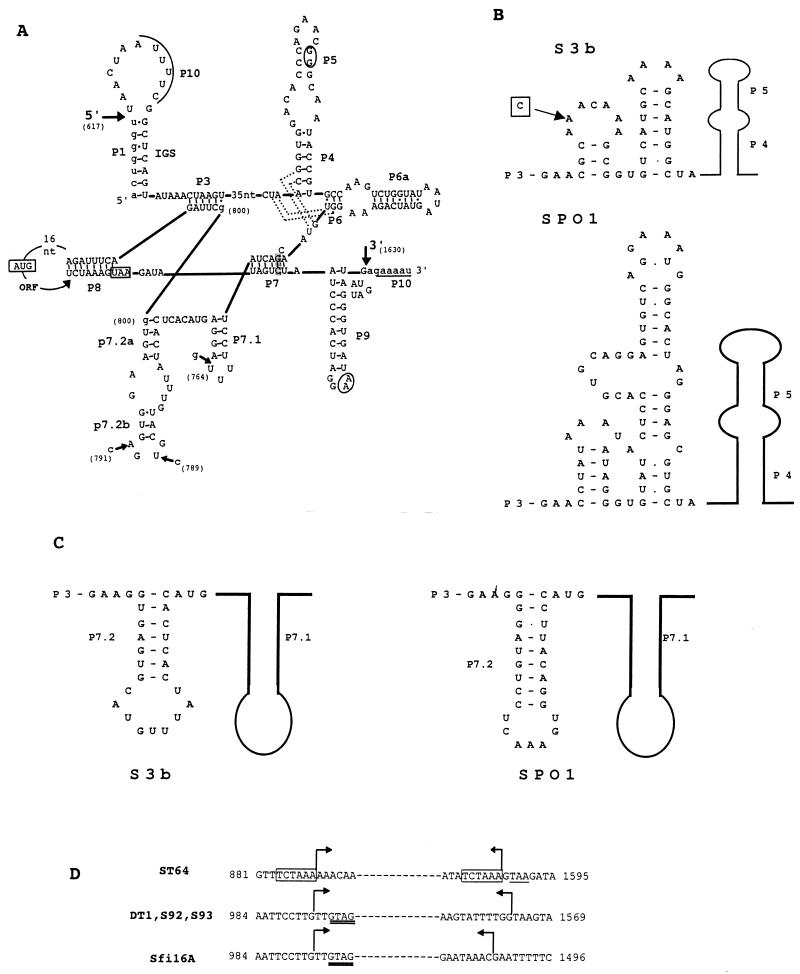
FIG. 3.
(A) Secondary structure prediction for the S3b intron represented according to the structural convention of Burke et al. (8). Large arrows indicate the 5′ and 3′ splice sites. The exon and intron sequences are denoted by lower- and uppercase letters, respectively. Underlined sequences denoted with P10 indicate the regions which can anneal to form P10. Predicted tertiary interactions are indicated by dotted lines (P4/J6-7 and J3-4/P6) and by circled nucleotides (P5/P9). The shaded nucleotides in P7 represent the putative guanosine binding site. The start and stop codons of orf 253 are boxed. Sequence differences within the structural regions of phage Sfi16A (nucleotide [nt] positions 764 and 789), S92 (nt positions 764 and 791), ST64 (nt positions 764 and 789), and DT1 (nt position 791) introns are indicated by small arrows. The two possible locations for the G residue (nt position 800) are indicated. Nucleotide positions are numbered according to the S3b sequence (accession no. AF148561). The numbering begins with the start codon of orf 207. (B) Folding of the joining region between P3 and P4 (J3-4) of the S3b intron and the intron of B. subtilis phage SPO1. The sequence difference between both the phage ST64 and Sfi16A introns and the S3b intron is boxed. A belongs to phage S3b. (C) Alternative folding of a P7.2 stem for the phage S3b intron in comparison with the P7.2 stem for the phage SPO1 intron. (D) Comparison of the deletion points of the variant introns of phages ST64, DT1, S92, S93, and Sfi16A. The sequence given is that of S3b. Bent arrows indicate the region which is deleted in the respective phages. The 6-bp direct repeat flanking the deletion in the phage ST64 intron is boxed, and the stop codon of orf 253 is underlined. The GTAG sequence matching the intron insertion site (see Fig. 6) is doubly underlined in variant introns of phages DT1 and Sfi16A.
Secondary structure analysis of the phage S3b intron.
By exploiting the secondary structure predictions based on a comparative sequence analysis (27) and the recently established three-dimensional structure of a group I intron (16), it was possible to predict a secondary structure for the S. thermophilus phage intron (Fig. 3A) (T. R. Cech, personal communication). The S. thermophilus phage intron possesses a conserved core of seven base-paired stems (P3 to P9), the integrity of which is essential for the self-splicing activity of introns (9, 10). The additional stem-loop structures P7.1, P7.2a, and P7.2b (located between P7 and P3) indicate the presence of a subgroup IA2 intron (27). An internal guide sequence (Fig. 3A) which could bring P1 and P10 sequences in proximity to facilitate the splicing process was identified (14). Tertiary interactions were also predicted (P5/P9; joining segments J3-4/P6 and J6-7/P4) (Fig. 3A) (T. R. Cech, personal communication).
Several interesting features were noted for this intron. The S. thermophilus phage intron lacks the P2 hairpin, a property shared with phage T4 sunY (41). The intron possesses a small catalytic core of about 230 nucleotides starting from the internal guide sequence. A similarly small core has recently been reported for the group I intron of S. aureus phage Twort (21). The orf 253 termination codon from phage S3b is located within the intron core, in the 3′ portion of the P8 stem, which is critical for catalysis (Fig. 3A). Similar observations have been made for the three T4 introns and the SPO1 intron (17, 35). Due to the location of the stop codon, a link between translation and the splicing and expression of genes containing these introns has been suggested (35).
Two further regions of the S3b intron showed a relationship to the intron in the DNA polymerase gene of B. subtilis phage SPO1 and its derivatives (18). First, the region between P3 and P4, which is characteristically only a few nucleotides long, has a long extension in both SPO1 and S3b. This structure has two stems flanked by short single-stranded regions. As shown in Fig. 3B, the size and sequence of these regions are identical between SPO1 and S3b, indicating a possible relationship between the respective introns. Second, an alternative structure that resembles the structure suggested for the phage SPO1 intron can be proposed for P7.2 (Fig. 3C).
The GC content of 41% for the noncoding intron sequences did not differ from the value of 43% for the surrounding phage sequences. In contrast, orf 253, located in the intron, had a remarkably low GC content of 28%, clearly indicating different origins for the intron coding and noncoding segments.
Distribution of introns within lysin genes of S. thermophilus phages.
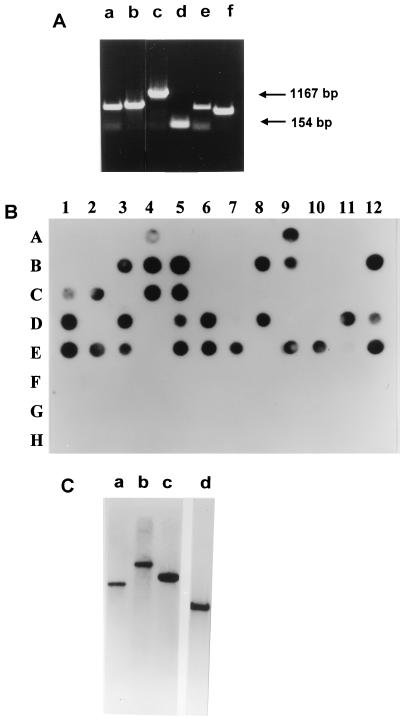
Our S. thermophilus phage collection, which comprises isolates from different ecological settings in France, Italy, Germany, Switzerland, and Austria, was screened for the presence of an interrupted lysin gene. Of 61 phages tested, 31 yielded a PCR product larger than expected for a noninterrupted lysin gene. Twenty-seven of these phages yielded a fragment similar in size to that obtained for S3b, while smaller PCR amplification products were obtained for phages Sfi16A, S92, S93, and ST64 (Fig. 4A). It should be noted that the presence or absence of the lysin intron could not be correlated with the ecological context of the streptococcal phages (data not shown).
FIG. 4.
Screening of S. thermophilus bacteriophages and starter cultures for the presence of the S3b-type intron. (A) PCR amplification with primer pair C-D1 (or, alternatively, C-D2 in the case of nonannealing of D1) and the following phage DNAs as templates: S93, Sfi16A, S3b, Sfi21, S92, and ST64 (lanes a to f, respectively). The sizes of the DNA products obtained for S3b and Sfi21 are shown in base pairs. (B) Dot blot of S. thermophilus bacteria and bacteriophage DNA with S3b intron DNA as a probe. (A) Lanes 1 to 12, Sfi phages 21, 11, 13, 16A, 18, 19, and 121 and HN phages 1, 4, 6, 7, and 9. (B) Lanes 1 to 12, ST phages 1, 2, 3, 9, 12, 13, 20, 30, 40, 41, 42, and 44. (C) Lanes 1 to 5, ST phages 64, 72, 84, 128, and J1; lanes 7 to 12, TN starter cultures 2, 17, 36, 44, 21, and 62. (D) Lanes 1 to 12, S phages 3b, 1, 2, 3, 5, 6, 7, 8, 9, 10, 11, and 12. (E) Lanes 1 to 10 and 12, phages 13, 14, 15, 17, 18, 19, 56, 66, 77, 89, and 94. (F) Lanes 1 and 2, S phages 96 and 97; lanes 6 to 12, TN starter cultures 49, 70, 39, 41, 42, 45, and 58. (G) Lanes 1 to 12, Sfi starter cultures 1, 2, 3, 6, 8, 10, 11, 13, 16, 18, 19, and 20. (H) Lanes 1 to 5, Sfi starter cultures 25, 26, 39, 41, and IL; lanes 7 to 9, S starter cultures 3, 17, and 97. (C) Southern hybridization with the phage S3 intron DNA as a probe. Lanes a to c, PvuII-digested genomic DNAs from phages S18, S89, and S94; lane d, HindIII-digested DNA from phage S3b. Molecular sizes for the signals in lanes a to d were approximately 8.5, 12, 7, and 3.5 kb, respectively.
Phages which yielded a PCR result indicative of an intron sequence were all found positive by dot blot hybridization when primers J and K were used to generate S3b intron DNA as a probe (Fig. 4B). Conversely, all phages which yielded PCR products that comigrated with the phage Sfi21 signal were negative with this probe in dot blot hybridization experiments. Southern hybridizations were performed with a selected number of phages possessing the S3b-type intron with S3b intron DNA as a probe. When a restriction enzyme which does not cut within the intron sequence was used, a single hybridization signal which corresponded to the lysin-located intron was obtained (Fig. 4C). These experiments indicated that the S3b-type intron is not located in areas of the S. thermophilus bacteriophage genome other than the lysin gene.
The genomic DNAs of 33 S. thermophilus strains (including raw milk isolates and yogurt and mozzarella cheese starter cultures) were examined for evidence of the presence of an S3b-type intron by dot blot analysis. No hybridization signals were obtained when a probe consisting of S3b intron DNA was used (Fig. 4B), indicating the absence of this group I intron in these strains.
Total RNA was isolated from S. thermophilus cultures infected with phages S92 and ST64. RT-PCR analysis yielded products which were smaller than those obtained with the respective phage DNAs as templates and which comigrated with the RT-PCR product obtained from Sfi21-infected cells (Fig. 5). This result confirms the presence of an intron in both phages.
FIG. 5.
In vivo splicing of phage ST64 and S92 intron RNAs. Lanes 2, 4, and 5, RT-PCR products obtained with RNA isolated from ST64-, S92-, and Sfi21-infected cells 15 min after infection; lanes 1 and 3, RT-PCR products obtained with phage ST64 and S92 DNAs as templates. The oligonucleotide pair B-E (Fig. 1B) was used. The sizes of the products obtained are indicated in base pairs.
The variant introns have deletions in the intron open reading frame.
The variant introns detected in phages Sfi16A, S92, and ST64 were sequenced. Phages ST3 and J, which have a lysin gene interruption similar in size to that of phage S3b, were also chosen randomly for sequence comparison. Analysis revealed that the introns of phages J and ST3 are identical, while those of ST3 and S3b differ by one nucleotide substitution. Sequence alignment indicated that phages Sfi16A, S92, and ST64 possess introns of 519, 443, and 316 bp, respectively. The phage DT1 intron differs from that of phage S92 by one base-pair substitution. The size differences between the variant introns were caused by deletion events within orf 253 (Fig. 3D). Since RNA splicing was observed for the variant introns (Fig. 5), the orf 253 gene product is dispensable for this process.
An alignment of the ST64 and S3b introns revealed that each extremity of the S3b intron region, absent in ST64, was preceded by an identical 6-bp motif (Fig. 3D). Since only one of the repeats was retained in ST64, the deletion event probably resulted from slippage of the DNA polymerase. A similar observation was made by Eddy and Gold (15) for the nonmobile nrdB intron of bacteriophage T4. No such repeats flanked the deletion sites in the phage S93, S92, DT1, and Sfi16A introns. However, all of these phages share the same deletion start site, located in codon 55 of orf 253, and S93, S92, and DT1 share the same deletion stop site (Fig. 3D). Furthermore, the four phages are not ecologically or genetically related in that they possess nonoverlapping host ranges and distinct restriction patterns and are of diverse geographical origins. Phages S92, S93, and DT1 are mozzarella isolates from Italy, Italy, and Canada, respectively, while Sfi16A originates from a French yogurt.
Apart from the deletion event, alignment of the variant introns with that of S3b revealed the presence of nearly identical intron sequences. The highest diversification was seen between S3b and Sfi16A, which differed in 15 nucleotide positions. The secondary structures of the variant introns do not differ greatly from that of S3b, since the majority of the differences are located in the looped-out region of P8. In contrast to the high degree of sequence conservation within the intron sequences, substantial sequence diversity was detected over the adjacent lysin-encoding sequences (data not shown). For example, alignment of the 1-kb intron sequences revealed differences of 3 and 1 nucleotides for the S3b-DT1 and S3b-ST3 comparisons, respectively, while alignment of the 0.25-kb region 3′ of the intron revealed 24 and 54 nucleotide differences for the S3b-DT1 and S3b-ST3 comparisons, respectively.
Comparison of sequences surrounding the splice site.
The DNA sequence of a 40-bp region surrounding the intron insertion site of 10 phages was compared with the equivalent region of 10 intron-free S. thermophilus lysin genes (Fig. 6). The sequence alignment demonstrated that phages possessing the intron and those lacking it differed clearly in a 14-bp segment centered around the intron insertion point. A screening of the five complete phage genome sequences available indicated that no unoccupied 14-bp sequence was present. The nearest matches were two segments with 12-bp identity (nucleotide substitutions at the −2 G and +1 A positions), possibly reflecting the critical nature of these nucleotide positions for intron homing and/or splicing (data not shown).
FIG. 6.
Nucleotide sequence alignment of the region surrounding the splice site of the intron-containing lysin genes (in bold) with the equivalent region from intron-free lysin genes. Sequence differences relative to the phage S3b sequence are indicated. The intron insertion site is indicated by a vertical arrow. The horizontal arrows represent a 6-bp inverted repeat. The nucleotide positions indicated are based on the numbering of the S3b sequence (accession no. AF148561).
DISCUSSION
This study describes a group I intron present within the lysin gene of over half of the investigated S. thermophilus bacteriophages. No phenotypic or ecological differences were observed between S. thermophilus phages possessing an intron-containing lysin gene or an intron-free lysin gene. The possession of the intron is thus either without selective advantage or is used in subtle regulatory circuits which do not detectably affect the laboratory growth or environmental distribution of S. thermophilus phages. A comparison of the intron insertion site for intron-containing and intron-free lysin genes led to the identification of a 14-bp consensus sequence associated with intron possession. Since no unoccupied consensus sequence was identified, one might deduce that intron invasion has reached a saturation point in S. thermophilus phages, thereby suggesting the presence of a very invasive molecular parasite. However, the consensus sequence could also be a consequence of the homing event if it causes gene conversion of the flanking exon sequences.
The intron itself was apparently molecularly parasitized by orf 253, which has sequence similarity to H-N-H-type endonuclease genes commonly found in introns from bacteriophages infecting gram-positive bacteria (34). This gene conferred intron mobility in L. lactis phage r1t (29). The invasive character of orf 253 was deduced from its distinct GC content. On the basis of theoretical consideration, one would expect pressure to remove the intron (13), and indeed five phages possess large deletions within orf 253. Examination of the DNA sequence indicated the presence of a deletion hot spot, i.e., codon position 55 in orf 253, in three of the five introns. The partial removal of the intron-bracketed orf 253, but maintenance of the intron in the phage genome is reminiscent of a similar process in coliphage T4 (15) and may be explained by the difficulty in removing the intron precisely without compromising lysin gene expression.
The initial discovery of self-splicing introns in coliphage T4 (11, 12) has attracted much interest among biologists interested in the evolutionary origin of introns (2, 19, 32, 40). The discovery of a group I intron in B. subtilis phage SPO1 was taken as evidence favoring the antiquity of introns in phages (17). To the discovery of additional type I introns in phages infecting the bacterial genera Bacillus, Lactococcus, Lactobacillus, and Staphylococcus can now be added the discovery of a type I intron in phages infecting the genus Streptococcus. All of these bacteriophages infect bacteria belonging to the low-GC-content branch of gram-positive bacteria. Comparative sequence analysis has demonstrated that temperate Siphoviridae from this branch of gram-positive bacteria are also evolutionarily related (23, 24).
The intron of S. thermophilus phages did not share significant overall sequence similarity with the introns from other phages. It is, however, premature to exclude a common origin for phage introns from that observation. In fact, the S. thermophilus phage intron looks like a typical subgroup IA2 intron, like many other phage introns. In addition, the secondary structure prediction provided strong evidence for a relationship to the introns in other phage systems. First, the intron of S. thermophilus phages possesses a 38-bp insert between the 5′ portions of P3 and P4 which can be folded into two stable stem-loop structures (Fig. 3C). In most group I introns, this region comprises a few nucleotides (typically 3 or 4). The only group IA introns which contain extra nucleotides in this region are those of Bacillus phages SPO1, SP82, and E, which possess 69 nucleotides in this interval folded into two thermodynamically stable stem-loop structures (18). Second, the location of the stop codon for orf 253 within the P8 stem is shared not only with the B. subtilis phage SPO1 intron but also with the three introns of E. coli bacteriophage T4 (17, 35). Third, the P7.2 region is highly similar in all phage introns described so far. It is always a perfect base-paired stem that is separated from P7.1 by the sequence GUA. Fourth, the intron-encoded proteins from Lactococcus, Streptococcus, Lactobacillus, and Bacillus phages have sequence similarity over the H-N-H motif. Fifth, the S3b intron lacks the P2 hairpin, a property shared with the phage T4 sunY intron.
Taken together, these observations suggest a certain conservation of structural features in phage introns. Whether this conservation reflects common ancestry or functional constraints of phage introns cannot currently be decided. We suspect that introns are much more widely distributed in phages than initially anticipated. The identification of phage introns from further genera of gram-positive bacteria could shed light on the evolutionary origin of phage introns.
Another observation is worth noting. While all group I introns of bacteria and eucaryotic organelles have been localized to the anticodon position of tRNA genes, phage introns have been detected in a number of distinct genes (DNA polymerase, thymidylate synthase, ribonucleotide reductase, structural proteins, large-subunit terminase, and now lysin), demonstrating flexibility in the intron homing or invasion process between phages.
ACKNOWLEDGMENTS
We express our gratitude to Thomas Cech for providing the secondary structure analysis of the phage S3b intron.
We acknowledge the Swiss National Science Foundation for financial support of Sophie Foley in the framework of its Biotechnology Module (grant 5002-044545/1).
REFERENCES
- 1.Bechhofer D H, Hue K K, Shub D A. An intron in the thymidylate synthase gene of Bacillus bacteriophage β22: evidence for independent evolution of a gene, its group I intron, and the intron open reading frame. Proc Natl Acad Sci USA. 1994;91:11669–11673. doi: 10.1073/pnas.91.24.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belfort M. Self-splicing introns in prokaryotes: migrant fossils? Cell. 1991;64:9–11. doi: 10.1016/0092-8674(91)90201-9. [DOI] [PubMed] [Google Scholar]
- 3.Biniszkiewicz D, Cesnaviciene E, Shub D A. Self-splicing group I intron in cyanobacterial initiator methionine tRNA: evidence for lateral transfer of introns in bacteria. EMBO J. 1994;13:4629–4635. doi: 10.1002/j.1460-2075.1994.tb06785.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brüssow H, Bruttin A. Characterization of a temperate Streptococcus thermophilus bacteriophage and its genetic relationship with lytic phages. Virology. 1995;212:632–640. doi: 10.1006/viro.1995.1521. [DOI] [PubMed] [Google Scholar]
- 5.Brüssow H, Bruttin A, Desiere F, Lucchini S, Foley S. Molecular ecology and evolution of Streptococcus thermophilus bacteriophages—a review. Virus Genes. 1998;16:95–109. doi: 10.1023/a:1007957911848. [DOI] [PubMed] [Google Scholar]
- 6.Brüssow H. Phages of Streptococcus thermophilus. In: Webster R, Granoff A, editors. Encyclopedia of virology. Vol. 2. London, United Kingdom: Academic Press Ltd.; 1999. pp. 1253–1262. [Google Scholar]
- 7.Bruttin A, Brüssow H. Site-specific spontaneous deletions in three genome regions of a temperate Streptococcus thermophilus phage. Virology. 1996;219:96–104. doi: 10.1006/viro.1996.0226. [DOI] [PubMed] [Google Scholar]
- 8.Burke J M, Belfort M, Cech T R, Davies R W, Schweyen R J, Shub D A, Szostak J W, Tabak H F. Structural conventions for group I introns. Nucleic Acids Res. 1987;15:7217–7221. doi: 10.1093/nar/15.18.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cech T R. Conserved sequences and structures of group I introns: building an active site for RNA catalysis—a review. Gene. 1988;73:259–271. doi: 10.1016/0378-1119(88)90492-1. [DOI] [PubMed] [Google Scholar]
- 10.Cech T R. Self-splicing of group I introns. Annu Rev Biochem. 1990;59:543–568. doi: 10.1146/annurev.bi.59.070190.002551. [DOI] [PubMed] [Google Scholar]
- 11.Chu F K, Maley G F, Maley F, Belfort M. Intervening sequence in the thymidylate synthase gene of bacteriophage T4. Proc Natl Acad Sci USA. 1984;81:3049–3053. doi: 10.1073/pnas.81.10.3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu F K, Maley G F, West D K, Belfort M, Maley F. Characterization of the intron in the phage T4 thymidylate synthase gene and evidence for its self-excision from the primary transcript. Cell. 1986;45:157–166. doi: 10.1016/0092-8674(86)90379-x. [DOI] [PubMed] [Google Scholar]
- 13.Darnell J E, Doolittle W F. Speculations on the early course of evolution. Proc Natl Acad Sci USA. 1986;83:1271–1275. doi: 10.1073/pnas.83.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Davies R W, Waring R B, Ray J A, Brown T A, Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982;300:719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- 15.Eddy S R, Gold L. The phage T4 nrdB intron: a deletion mutant of a version found in the wild. Genes Dev. 1991;5:1032–1041. doi: 10.1101/gad.5.6.1032. [DOI] [PubMed] [Google Scholar]
- 16.Golden B L, Gooding A R, Podell E R, Cech T R. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 17.Goodrich-Blair H, Scarlato V, Gott J M, Xu M-Q, Shub D A. A self-splicing group I intron in the DNA polymerase gene of Bacillus subtilis bacteriophage SPO1. Cell. 1990;63:417–424. doi: 10.1016/0092-8674(90)90174-d. [DOI] [PubMed] [Google Scholar]
- 18.Goodrich-Blair H, Shub D A. The DNA polymerase genes of several HMU-bacteriophages have similar group I introns with highly divergent open reading frames. Nucleic Acids Res. 1994;22:3715–3721. doi: 10.1093/nar/22.18.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhsel M G, Strickland R, Palmer J D. An ancient group I intron shared by eubacteria and chloroplasts. Science. 1990;250:1570–1573. doi: 10.1126/science.2125748. [DOI] [PubMed] [Google Scholar]
- 20.Lambowitz A M, Belfort M. Introns as mobile genetic elements. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 21.Landthaler M, Shub D A. Unexpected abundance of self-splicing introns in the genome of bacteriophage Twort: introns in multiple genes, a single gene with three introns, and exon skipping by group I ribozymes. Proc Natl Acad Sci USA. 1999;96:7005–7010. doi: 10.1073/pnas.96.12.7005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazarevic V, Soldo B, Dusterhoft A, Hilbert H, Mauel C, Karamata D. Introns and intein coding sequences in the ribonucleotide reductase genes of Bacillus subtilis temperate bacteriophage SPβ. Proc Natl Acad Sci USA. 1998;95:1692–1697. doi: 10.1073/pnas.95.4.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lucchini S, Desiere F, Brüssow H. The structural gene module in Streptococcus thermophilus bacteriophage φSfi11 shows a hierarchy of relatedness to Siphoviridae from a wide range of bacterial hosts. Virology. 1998;246:63–73. doi: 10.1006/viro.1998.9190. [DOI] [PubMed] [Google Scholar]
- 24.Lucchini S, Desiere F, Brüssow H. Similarly organized lysogeny modules in temperate Siphoviridae from low GC content Gram-positive bacteria. Virology. 1999;263:427–435. doi: 10.1006/viro.1999.9959. [DOI] [PubMed] [Google Scholar]
- 25.Lucchini S, Desiere F, Brüssow H. The genetic relationship between virulent and temperate Streptococcus thermophilus bacteriophages: whole genome sequence comparisons of cos-site phages Sfi19 and Sfi21. Virology. 1999;260:232–243. doi: 10.1006/viro.1999.9814. [DOI] [PubMed] [Google Scholar]
- 26.Lucchini S, Desiere F, Brüssow H. Comparative genomics of Streptococcus thermophilus phage species supports a modular evolution theory. J Virol. 1999;73:8647–8656. doi: 10.1128/jvi.73.10.8647-8656.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Michel F, Westhof E. Modelling of the three-dimensional architecture of group I catalytic introns based on comparative sequence analysis. J Mol Biol. 1990;216:585–610. doi: 10.1016/0022-2836(90)90386-Z. [DOI] [PubMed] [Google Scholar]
- 28.Mikkonen M, Alatossava T. A group I intron in the terminase gene of Lactobacillus delbrueckii subsp. lactis phage LL-H. Microbiology. 1995;141:2183–2190. doi: 10.1099/13500872-141-9-2183. [DOI] [PubMed] [Google Scholar]
- 29.Nauta A. Molecular characterization and exploitation of the temperate Lactococcus lactis bacteriophage r1t. Ph.D. thesis. Groningen, The Netherlands: University of Groningen; 1997. [Google Scholar]
- 30.Pedersen-Lane J, Belfort M. Variable occurrence of the nrdB intron in the T-even phages suggests intron mobility. Science. 1987;237:182–183. doi: 10.1126/science.3037701. [DOI] [PubMed] [Google Scholar]
- 31.Quirk S M, Bell-Pedersen D, Tomaschewski J, Rüger W, Belfort M. The inconsistent distribution of introns in the T-even phages indicates recent genetic exchanges. Nucleic Acids Res. 1989;17:301–315. doi: 10.1093/nar/17.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reinhold-Hurek B, Shub D A. Self-splicing introns in tRNA genes of widely divergent bacteria. Nature. 1992;357:173–176. doi: 10.1038/357173a0. [DOI] [PubMed] [Google Scholar]
- 33.Sheehan M M, García J L, López R, García P. The lytic enzyme of the pneumococcal phage Dp-1: a chimeric lysin of intergenic origin. Mol Microbiol. 1997;24:717–725. doi: 10.1046/j.1365-2958.1997.5101880.x. [DOI] [PubMed] [Google Scholar]
- 34.Shub D A, Goodrich-Blair H, Eddy S R. Amino acid sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 35.Shub D A, Gott J M, Xu M-Q, Lang B F, Michel F, Tomaschewski J, Pedersen-Lane J, Belfort M. Structural conservation among three homologous introns of bacteriophage T4 and the group I introns of eukaryotes. Proc Natl Acad Sci USA. 1988;85:1151–1155. doi: 10.1073/pnas.85.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shub D A, Xu M-Q, Gott J M, Zeeh A, Wilson L D. A family of autocatalytic group I introns in bacteriophage T4. Cold Spring Harbor Symp Quant Biol. 1987;52:193–200. doi: 10.1101/sqb.1987.052.01.024. [DOI] [PubMed] [Google Scholar]
- 37.Stanley E, Fitzgerald G F, Le Marrec C, Fayard B, van Sinderen D. Sequence analysis and characterization of φO1205, a temperate bacteriophage infecting Streptococcus thermophilus CNRZ1205. Microbiology. 1997;143:3417–3429. doi: 10.1099/00221287-143-11-3417. [DOI] [PubMed] [Google Scholar]
- 38.Terzaghi B E, Sandine W E. Improved media for lactic streptococci and their bacteriophages. Appl Microbiol. 1975;29:807–813. doi: 10.1128/am.29.6.807-813.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tremblay D M, Moineau S. Complete genomic sequence of the lytic bacteriophage DT1 of Streptococcus thermophilus. Virology. 1999;255:63–76. doi: 10.1006/viro.1998.9525. [DOI] [PubMed] [Google Scholar]
- 40.Xu M-Q, Kathe S D, Goodrich-Blair H, Nierzwicki-Bauer S A, Shub D A. Bacterial origin of a chloroplast intron: conserved self-splicing group I introns in cyanobacteria. Science. 1990;250:1566–1570. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- 41.Xu M-Q, Shub D A. The catalytic core of the sunY intron of bacteriophage T4. Gene. 1989;82:77–82. doi: 10.1016/0378-1119(89)90032-2. [DOI] [PubMed] [Google Scholar]