Abstract
Recent evidence suggests that γ‐synuclein is abnormally expressed in a high percentage of tumor tissues of diversified cancer types, but rarely expressed in tumor‐matched non‐neoplastic adjacent tissues (NNAT). The molecular mechanism of CpG island demethylation may underlie aberrant γ‐synuclein expression. To fully understand the roles of aberrant γ‐synuclein expression and demethylation in the development of colorectal cancer (CRC), we examined the expression and methylation status of γ‐synuclein in 67 CRC samples, 30 NNAT samples, and five CRC cell lines as well. By using reverse transcription–polymerase chain reaction (RT‐PCR), western blot, and immunohistochemistry analyses, γ‐synuclein expression was detected in both HT‐29 and HCT116 cells, and was much higher in CRC samples than in NNAT samples (P < 0.05). The demethylating agent, 5‐aza‐2¢‐deoxycytidine, can induce re‐expression of γ‐synuclein in COLO205, LoVo, and SW480 cells. Unmethylated γ‐synuclein alleles were detected in HT‐29, HCT116, and LoVo cells by nested methylation‐specific PCR, and the demethylated status of γ‐synuclein was much higher in CRC samples than in NNAT samples by real‐time quantitative methylation‐specific PCR (P < 0.05). The results of genomic bisulfite DNA sequencing further confirmed that the aberrant γ‐synuclein expression in CRC was primarily attributed to the demethylation of CpG island. The protein expression and demethylation status of γ‐synuclein in 67 CRC samples correlated with clinical stage, lymph node involvement, and distant metastasis. These findings suggest an involvement of aberrant γ‐synuclein expression and demethylation in progression of CRC, especially in advanced stages. (Cancer Sci 2008; 99: 1924–1932)
Colorectal cancer (CRC) is the third most common malignancy and the second leading cause of cancer‐related deaths worldwide.( 1 ) The conventional therapies involving surgery and adjuvant therapy seem to give rise to improvements in progression‐free and overall survival; nevertheless about 50% of patients die within 5 years owing to metastasis or recurrent disease. Patients with early stage CRC have an estimated 5‐year survival rate of 91%, compared to only 6% for those with later stage disease. Early detection remains the most important factor in improving long‐term survival. Furthermore, tumor invasion and regional lymph node metastasis are important factors for determining CRC prognosis.( 2 , 3 ) However, there are currently no internationally recognized molecular markers. Identification of reliable and sensitive biomarkers that indicate the development of metastasis of primary tumors will have significant clinical implications.
γ‐Synuclein is a member of the synuclein protein family comprising α‐, β‐, and γ‐synuclein, which are abundantly expressed in nervous tissues. They are characterized by 5–6 repeats of amino acid motif (KTKEGV), constituting most of the N‐terminal half of the proteins. These repeats result in the formation of conserved amphipathic A2‐helices also characteristic of apolipoproteins, which mediate reversible binding to phospholipid membranes. This property supports the role of synucleins in vesicular release at presynaptic nerve terminals.( 4 , 5 , 6 , 7 ) Although their normal cellular functions have not been clearly defined, pathologic roles in a number of human diseases have been found. α‐ and β‐synuclein participate in the pathogenesis of neurodegenerative disorders. α‐Synuclein is the major component of Lewy bodies in Parkinson's disease and has also been identified as the nonamyloid component of amyloid deposition, the hallmark of Alzheimer's disease. β‐Synuclein shares the chaperone activity of other synucleins, and is assumed to have a neuroprotective role by inhibiting α‐synuclein aggregation and toxicity.( 7 , 8 , 9 , 10 , 11 )
It is not clear whether γ‐synuclein is involved in neurodegenerative diseases. γ‐Synuclein (also referred to as breast carcinoma specific gene 1 [BCSG1]) initially was cloned from infiltrating breast carcinoma cells by using the expressed sequence tag‐based differential cDNA sequencing approach.( 12 )γ‐Synuclein expression is usually highly tissue‐specific and restricted to brain tissue and presynaptic terminals.( 12 ) However, the tissue‐specificity appears to be lost, and γ‐synuclein is abnormally expressed in a high percentage of advanced breast and ovarian cancers, but not in normal or benign tissues.( 13 ) When overexpressed, γ‐synuclein can stimulate proliferation, and induce invasion and metastasis of breast cancer cells.( 14 ) Patients with γ‐synuclein protein‐positive breast cancer have a significantly shorter disease‐free survival and overall survival period compared to patients with no γ‐synuclein expression.( 15 , 16 ) Analysis of breast cancer cells did not identify any sequence variation of the γ‐synuclein gene from its original normal neuronal environment, and no gene amplification was detected either, which suggests that transcriptional activation could account for its abundant expression in breast cancer cells. The exon 1 region of the γ‐synuclein gene contains a CpG island with 15 CpG dinucleotides, and γ‐synuclein expression is activated by demethylation of the CpG island in breast and ovarian cancers.( 17 ) On the other hand, no post‐transcriptional regulation and modification of γ‐synuclein have been found so far. In the past few years, γ‐synuclein has been studied in other types of cancer. Liu et al. found that γ‐synuclein protein was abnormally expressed in a high percentage of tumor tissues of diversified cancer types, including liver, esophagus, colon, gastric, lung, prostate, cervical, and breast cancer, at a frequency of 20 cases per cancer type, but rarely expressed in tumor‐matched non‐neoplastic adjacent tissues (NNAT).( 18 ) Furthermore, reactivation of γ‐synuclein gene expression by DNA demethylation was a common critical contributing factor to malignant progression of many solid tumors.( 18 ) Some studies also suggested γ‐synuclein was a oncogene in human neoplastic diseases and was used as a new tumor marker.( 19 , 20 , 21 ) Highly punctuate γ‐synuclein expression was observed in 20% of preneoplastic lesions of the ovary, including epithelial inclusion cysts, hyperplastic epithelium, and papillary structures.( 19 )γ‐Synuclein protein was also detectable in serum samples from patients with pancreatic carcinoma and in urine samples from patients with bladder cancer.( 20 , 21 ) However, Zhou et al. had an opposite conclusion regarding esophagus cancer, for which the low expression levels of γ‐synuclein in human esophageal squamous cell carcinoma (ESCC) and the biological effects of γ‐synuclein overexpression on ESCC 9706 cells suggested that γ‐synuclein might play a role as a negative regulator in the development of human ESCC.( 22 ) Therefore, further study is needed to understand the roles of γ‐synuclein in the development of other human neoplastic diseases except breast and ovarian cancer.
To further determine whether aberrant expression and demethylation of γ‐synuclein are involved in the development of CRC, and to identify a new biomarker correlated with the disease progression, we examined γ‐synuclein expression and methylation status in 67 CRC samples, 30 NNAT samples, and five CRC cell lines, and then analyzed the relationship between the expression and demethylation status of γ‐synuclein in CRC and clinicopathological factors.
Materials and Methods
Tissue samples and cell lines. Sixty‐seven CRC samples and 30 NNAT samples were obtained from patients undergoing CRC surgery between January 2005 and October 2007 at the Department of General Surgery, Ruijin Hospital, Shanghai, China. Washed with RNase‐Free 0.9% NaCL to remove blood after surgery, one half of each sample was snap‐frozen in liquid nitrogen immediately and stored at –80°C for DNA, RNA, and protein extraction, and the other half was fixed in 10% formalin for histological assessment. For tumor samples, non‐tumor portions were trimmed off from the frozen tumor blocks and the selected areas had more than 80% tumor cells as shown by histological as assessment. Tumors were staged using the TNM and World Health Organization classification systems. The ethics committee at Ruijin Hospital approved the use of these tissues for research purposes. Five CRC cell lines, COLO205 (ATCC no. CCL‐222), HT‐29 (HTB‐38), HCT116 (CCL‐247), LoVo (CCL‐229), and SW480 (CCL‐228), with different genetic backgrounds were cultured under appropriate conditions in our laboratory. COLO205, LoVo, and SW480 cells were cultured in RPMI‐1640 medium (Gibco BRL, Life Technologies, USA) supplemented with 10% heat inactivated fetal bovine serum (FBS) (Summit Biotechnology, Fort Collins, CO, USA). HT‐29 and HCT116 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) (Gibco BRL) supplemented with 10% FBS.
5‐aza‐2¢‐deoxycytidine treatment. COLO205, LoVo, and SW480 cells were cultured for 3–6 days in medium containing 5 µM of 5‐aza‐2′‐deoxycytidine (5‐aza‐C) (Sigma, St. Louis, MO, USA), and the medium and drug were replaced everyday. After the treatment the cells were harvested for DNA and RNA extraction.
Isolation of genomic DNA, total RNA, and protein from cell lines and tissues. Cultured cells were washed two times with phosphate‐buffered saline and harvested, and tissues were ground into fine powder in liquid nitrogen before extraction of DNA, RNA, and protein. The genomic DNA was isolated using Promega's wizard DNA isolation kit (Promega, Madison, WI, USA) according to the manufacturer's instructions. Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. Extraction of total protein was carried out as described previously.( 23 ) The amount of total protein was estimated using a BCA protein assay kit (Bio‐Rad Laboratories, Hercules, CA, USA), and 50 µg of protein was mixed with SDS sample buffer and boiled for 5 min before loading.
Reverse transcription–polymerase chain reaction (RT‐PCR) analysis of γ‐synuclein mRNA. cDNA was synthesized from 2 µg of total RNA as described previously,( 23 ) and stored at –20°C until use. PCR reactions were done in a volume of 25 µL containing 1 × PCR buffer, 2.5 mM MgCl2, 200 µM of dNTP mix, 100 pmol of each primer, 0.625 U of HotstarTaq DNA Polymerase (Qiagen, Hilden, Germany), and 1 µL cDNA template. Amplification conditions were a hold of 95°C for 15 min to activate the hotstart enzyme, followed by 34 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s, and finally 72°C for 5 min. The expression of GAPDH mRNA was taken as an internal loading control. H2O was used as a negative control. The PCR products were separated on 1.5% agarose gels, stained with ethidium bromide. In addition, cDNA was used as a template for real‐time quantitative RT‐PCR (Q‐RT‐PCR). Q‐RT‐PCR was carried out in 96‐well polypropylene microplates on an ABI Prism 7000 (Applied Biosystems, Foster City, CA, USA) using SYBR Green Realtime PCR Master Mix (TOYOBO, Tokyo, Japan) according to the manufacturer's instructions. Amplification were carried out with the following profile: 1 cycle at 95°C for 1 min, and 40 cycles each at 95°C for 15 s, 59°C for 15 s, and 72°C for 45 s. All PCR reactions were run in triplicate wells. Specificity of resulting PCR products was confirmed by melting curves. H2O was used as a negative control. Data were analyzed by using the comparative Ct (ΔCt) method; the amount of γ‐synuclein relative to the GAPDH was expressed as 10 000 × 2−(ΔCt). Table 1 provides the sequences of the primers used in these studies.
Table 1.
Sequences of γ‐synuclein gene‐specific primers
| Primer | Sequence (5’‐3’) | Product size (bp) | |
|---|---|---|---|
| RT‐PCR primers 17 | |||
| γ‐synuclein‐RT‐5′ | CAAGAAGGGCTTCTCCATCGCCAAGG | 338 (60 to 397) | |
| γ‐synuclein‐RT‐3′ | CCTCTTTCTCTTTGGATGCCACACCC | ||
| GAPDH‐RT‐5′ | GAAGGTGAAGGTCGGAGTC | 226 (108 to 333) | |
| GAPDH‐RT‐3′ | GAAGATGGTGATGGGATTTC | ||
| Real‐time RT‐PCR primers | |||
| γ‐synuclein‐Real‐RT‐5′ | GGAGGACTTGAGGCCATCTG | 73 (339 to 411) | |
| γ‐synuclein‐Real‐RT‐3′ | CTCCTCTGCCACTTCCTCTTTC | ||
| GAPDH‐Real‐RT‐5′ | GGACCTGACCTGCCGTCTAG | 100 (831 to 930) | |
| GAPDH‐Real‐RT‐3′ | GTAGCCCAGGATGCCCTTGA | ||
| Bisulfite sequencing PCR primers 18 | |||
| γ‐synuclein‐SF | GGTTGAGTTAGTAGGAGTTTA | 415 (−275 to +140) | |
| γ‐synuclein‐SR | CCTACCATACCCCACTTACCC | ||
| Nested MSP primers 18 | |||
| Methylated | γ‐synuclein‐MF | TCGTATTAATATTTTATCGGCGT | 194 (−139 to +55) |
| γ‐synuclein‐MR | CCGCACCCACCACGCCCTCCTTAACGA | ||
| Unmethylated | γ‐synuclein‐UF | TTGGTGTTAATAGGAGGTATTGGGGATAGTTGTTGTG | 177 (−123 to +54) |
| γ‐synuclein‐UR | CACACCCACCACACCCTCCTTAACAAT | ||
| Real‐time Q‐MSP primers | |||
| Methylated | γ‐synuclein‐Real‐MF | TTCGTATTAATATTTTATCGGCGT | 104 (−140 to −37) |
| γ‐synuclein‐Real‐MR | GAAACTAAATCTCCCTACGAACTACG | ||
| Unmethylated | γ‐synuclein‐Real‐UF | GGTTTTTGTATTAATATTTTATTGGTG | 110 (−144 to −35) |
| γ‐synuclein‐Real‐UR | ACAAAACTAAATCTCCCTACAAACTACAA | ||
| WT primers 18 | |||
| γ‐synuclein‐WF | ACGCAGGGCTGGCTGGGCTCCA | 242 (−169 to +73) | |
| γ‐synuclein‐WR | CCTGCTTGGTCTTTTCCACC | ||
The CpG sites within the primer sequences are underlined
Genomic bisulfite DNA sequencing, nested methylation‐specific PCR (NMSP) and real‐time quantitative methylation‐specific PCR (real‐time Q‐MSP). Two µg of genomic DNA from each sample was denatured by 3 M NaOH for 30 min at 42°C, followed by the treatment of 3.6 M sodium bisulfite (Sigma) and 10 mM hydroquinone (Sigma) at 50°C for 16 h. The modified DNA was purified using DNA cleanup kit (Promega) in a total volume of 20 µL, and 2 µL was used for genomic sequencing PCR. For genomic sequencing PCR, the modified DNA that includes the entire CpG island with 15 CpG sites was amplified. Amplification conditions were a hold of 95°C for 15 min, and 34 cycles of 94°C for 30 s, 58°C for 30 s, 72°C for 30 s, and finally 72°C for 5 min. The 415‐bp PCR product was gel purified and ligated into pGEM‐T Easy Vector (Promega). After transformation, plasmid DNAs were isolated from individual colonies and subjected to sequencing to obtain the entire map of γ‐synuclein CpG island. The genomic sequencing PCR product in the reaction tube was diluted 1:10, and 2 µL was used as DNA template for NMSP and real‐time Q‐MSP.
For NMSP, the methylated PCR was conducted for 34 cycles with the annealing temperature of 60°C, and the unmethylated PCR product was amplified at the annealing temperature of 59°C for 34 cycles. The PCR products were separated on 2% agarose gels and stained with ethidium bromide.
Real‐time Q‐MSP was carried out using SYBR Green Realtime PCR Master Mix (TOYOBO). The PCR was initiated at 95°C for 1 min and followed by 40 cycles of 95°C for 15 s, 59°C for 15 s, 72°C for 45 s, and an extra incubation step of 77°C for 30 s to overcome the primer dimer artifact as described previously.( 24 ) The ratio of amount of methylation gene (M) to unmethylation gene (U) was calculated to obtain the relative threshold cycle (ΔCtm/u). The percentage of unmethylated gene was calculated as the unmethylation index = U/(U + M) = 1/(1 + M/U) = 1/[1 + 2−(ΔCtm/u)] as described previously.( 18 , 25 ) An unconverted wild‐type version of the γ‐synuclein gene was amplified with wild‐type primers to estimate the completeness of bisulfite conversion. CpGenome Universal Methylated Control DNA and Unmethylated Control DNA (Chemicon/Millipore, Billerica, MA, USA) were used as positive controls, and H2O was used as a negative control for NMSP and real‐time Q‐MSP. Table 1 provides the sequences of the primers used in these studies.
Western blot analysis. We used an enhanced chemiluminescence western‐blotting analysis system (Amersham, UK) as previously described.( 23 ) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) was performed in 15% polyacrylamide gels. PAGE‐separated proteins were electroblotted onto Sequi‐blot polyvinylidene difluoride membranes (Bio‐Rad) and incubated with a 1:500 dilution of mouse anti‐γ‐synuclein monoclonal antibody (1H10D2, sc‐65979; Santa Cruz Biotechnology, Santa Cruz, CA, USA). A goat antimouse IgG‐HRP antibody (Santa Cruz) served as the secondary antibodies. GAPDH served as a loading control.
Immunohistochemistry (IHC) analysis. IHC was carried out as described previously,( 23 ) using mouse anti‐γ‐synuclein monoclonal antibody at a dilution of 1:100. Positive cases were defined by the presence of intracellular staining with red/brown color in epithelial cells. Negative cases were defined by the absence of specific intracellular staining as seen in the negative control. Samples were independently evaluated under light microscopy by two pathologists without prior knowledge of the patients’ clinical data.
Statistical analysis. Statistical analyses were performed using spss 11.0 software (at Shanghai Jiaotong, University School of Medicine, Shanghai, China). Fisher's exact test was used to analyze protein expression of γ‐synuclein in paired tumor and corresponding normal tissues, and to assess the relationship between the protein expression and the demethylation status of γ‐synuclein and clinicopathological characteristics. Wilcoxon's signed rank test was used to analyze mRNA expression and the unmethylation index of γ‐synuclein in paired tumor and normal tissues. Spearman's rank correlation coefficient and the Mann–Whitney U‐test were used to assess the relationship between the unmethylation index of γ‐synuclein and clinicopathological characteristics. P < 0.05 was considered statistically significant.
Results
Examination of γ‐synuclein mRNA and protein expression in CRC samples, NNAT samples, and five CRC cell lines. We examined γ‐synuclein mRNA expression in 30 pairs of tumor and normal tissues. The 30 tumors were diagnosed as CRC with 13 at TNM stage I/II and 17 at stage III/IV. Fig. 1(a) shows the results of RT‐PCR in 9/30 pairs. Fig. 1(b) shows the results of Q‐RT‐PCR in a total of 30 pairs. γ‐Synuclein mRNA expression in CRC samples ranged from 1.45 to 37.05 with a median value of 13.42, while in matched NNAT samples it ranged from 0.81 to 30.21 with a median value of 6.11. The γ‐synuclein mRNA expression levels in CRC samples were significantly higher than those in NNAT samples (P = 0.002). Of five CRC cell lines, HT‐29 and HCT116 cells displayed γ‐synuclein mRNA expression, whereas there was undetectable γ‐synuclein mRNA expression in COLO205, LoVo, and SW480 cells (Fig. 1c).
Figure 1.
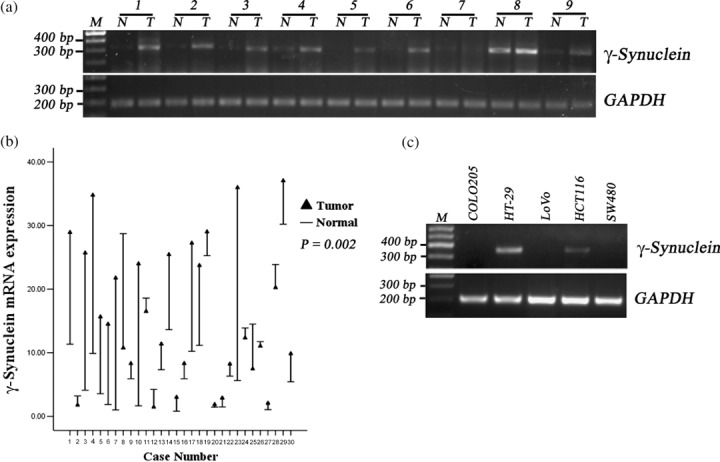
Detection of γ‐synuclein mRNA in colorectal cancer (CRC) samples (T), non‐neoplastic adjacent tissues (NNAT) samples (N), and CRC cell lines. M, 100 bp Maker. (a) The results of reverse transcription–polymerase chain reaction (RT‐PCR) in nine of 30 pairs of samples. (b) Drop‐line graph compares γ‐synuclein mRNA expression detected by quantitative RT‐PCR in CRC versus NNAT samples (P = 0.002). (c) The results of RT‐PCR in CRC cells.
The results of western blot analysis demonstrated that γ‐synuclein protein was detected in 19/30 (63.3%) CRC samples and 8/30 (26.7%) NNAT samples (representative four pairs, Fig. 2a). γ‐Synuclein protein expression was up‐regulated in CRC samples compared to NNAT samples (P = 0.009, Fig. 2b). γ‐Synuclein protein was also detected in γ‐synuclein mRNA‐positive CRC cells, HT‐29, and HCT116 (Fig. 2c).
Figure 2.
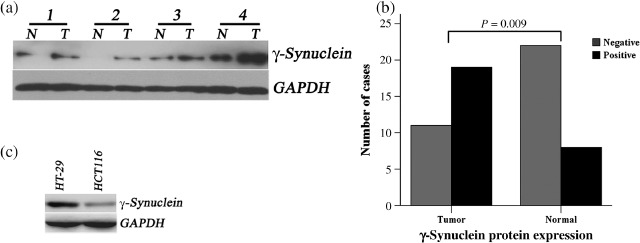
Detection of γ‐synuclein protein in colorectal cancer (CRC) samples (T), non‐neoplastic adjacent tissues (NNAT) samples (N), HT‐29, and HCT116 cells. (a) The results of western blot analysis in four of 30 pairs of samples. (b) The bar graph compares γ‐synuclein protein expression in CRC versus NNAT samples (P = 0.009). (c) The results of western blot analysis in γ‐synuclein mRNA‐positive cells, HT‐29, and HCT116.
Immunohistochemical staining of γ‐synuclein protein in CRC samples and NNAT samples. γ‐Synuclein protein expression in the 30 pairs of tissues was also evaluated by IHC. The subcellular localization of γ‐synuclein protein was cytoplasmic in the epithelial cells of tumor or normal tissues, and nuclear staining was sometimes observed in strong immunostaining tumors. Negative or weak immunostaining for γ‐synuclein protein was observed primarily in NNAT sections. In contrast, most CRC sections showed a variety of immunoreactivity (Fig. 3). Eighteen of 30 (60%) CRC samples and 9/30 (30%) NNAT samples were positive for γ‐synuclein protein. The frequency of positive immunostaining was similar to the frequency of detectable γ‐synuclein protein expression by western blot analysis.
Figure 3.
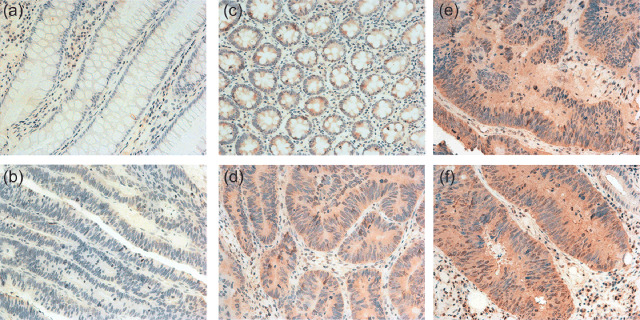
γ‐Synuclein protein expression detected by immunohistochemistry. (a,b). A pair of colorectal cancer (CRC) and non‐neoplastic adjacent tissues (NNAT) without γ‐synuclein protein expression. (c) Weak immunostaining in NNAT. (d). Weak immunostaining in a CRC tissue at stage II. (e,f). Strong immunostaining in CRC tissues at stage III and IV (original magnification ×400).
To investigate the clinical significance of aberrant γ‐synuclein expression in CRC, we chose other 37 tumor samples for IHC. The results demonstrated that γ‐synuclein protein expression was detected in 37 of a total 67 samples. The frequency of positive immunostaining for individual pathological stages was 7/17 (41.18%) for stage I, 8/19 (42.11%) for stage II, 17/26 (65.38%) for stage III, and 5/5 (100%) for stage IV. The γ‐synuclein protein expression significantly correlated with the clinical stage (P = 0.047, Table 2). Similarly, there was a significant correlation between the γ‐synuclein protein expression and lymph node involvement (P = 0.047, Table 2). Although the significant correlation was not reached, there was a tendency that tumors with γ‐synuclein expression were more likely to distantly metastasize compared with γ‐synuclein‐negative tumors (P = 0.060, Table 2).
Table 2.
Correlation between γ‐synuclein protein expression and clinicopathological factors of colorectal cancer patients
| Variable | No | γ‐Synuclein protein expression | P‐value* | |
|---|---|---|---|---|
| Positive | Negative | |||
| Age | ||||
| ≥60 | 34 | 19 | 15 | 1.000 |
| <60 | 33 | 18 | 15 | |
| Sex | ||||
| Male | 37 | 21 | 16 | 0.809 |
| Female | 30 | 16 | 14 | |
| Histological types | ||||
| Differentiated | 62 | 33 | 29 | 0.370 |
| Undifferentiated | 5 | 4 | 1 | |
| Stage | ||||
| I | 17 | 7 | 10 | 0.047 |
| II | 19 | 8 | 11 | |
| III | 26 | 17 | 9 | |
| IV | 5 | 5 | 0 | |
| Lymph node invasion | ||||
| Positive | 30 | 21 | 9 | 0.047 |
| Negative | 37 | 16 | 21 | |
| Distant metastasis | ||||
| Positive | 5 | 5 | 0 | 0.060 |
| Negative | 62 | 32 | 30 | |
Statistical significance was determined with Fisher's exact test.
Re‐expression of γ‐synuclein by 5‐aza‐C treatment. To test whether the molecular mechanism of CpG island demethylation was underlying aberrant γ‐synuclein expression in CRC, we chose γ‐synuclein‐negative COLO205, LoVo, and SW480 cells, and treated them with the demethylating agent 5 µM 5‐aza‐C. After 3–6 days treatment, we isolated mRNA and DNA from the cells, and then detected γ‐synuclein mRNA re‐expression in these cells using RT‐PCR assay (Fig. 4a). Then, NMSP assay was used to analyze the methylation status of γ‐synuclein before and after the treatment. NMSP assay indicated that treatment of COLO205, LoVo, and SW480 cells with 5‐µM 5‐aza‐C induced a time‐dependent demethylation of γ‐synuclein (Fig. 4b). To obtain the detailed methylation status of the entire γ‐synuclein CpG island with 15 CpG sites, we conducted genomic sequencing; the results of which showed that the density of methylated CpG sites gradually lowered in the three cells after treatment (Fig. 4c).
Figure 4.
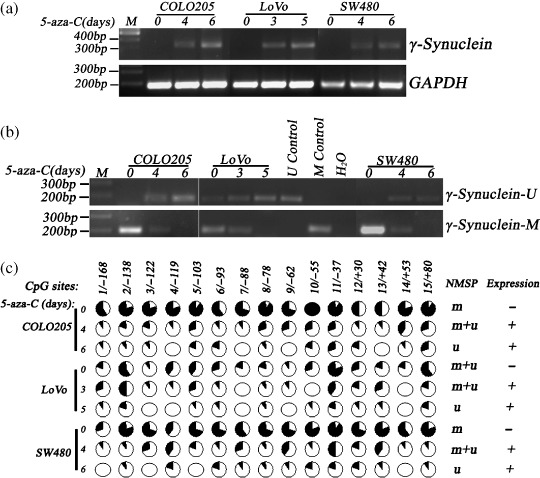
Re‐expression of γ‐synuclein mRNA and changes in methylation status of γ‐synuclein CpG island in COLO205, LoVo, and SW480 cells after 5‐aza‐C treatment. M control, methylated control; m + u, partially unmethylated; M, 100 bp Maker; m, completely methylated; U control, unmethylated control; u, completely unmethylated; γ‐synuclein‐M, methylated polymerase chain reaction (PCR) product; γ‐Synuclein‐U, unmethylated PCR product. (a) reverse transcription–polymerase chain reaction (RT‐PCR) shows re‐expression of γ‐synuclein mRNA after the treatment. (b) Nested methylation‐specific PCR (NMSP) shows demethylation of γ‐synuclein after the treatment. (c) The results of genomic bisulfite DNA sequencing before and after the treatment. Each circle in the figure represents a single CpG site. For each DNA sample, the percentage of unmethylation at a single CpG site is calculated from the sequencing results of 10 independent clones.  , 100% unmethylation;
, 100% unmethylation;  , 0% unmethylation.
, 0% unmethylation.
Examination of γ‐synuclein methylation status in CRC samples, NNAT samples, and CRC cell lines. We further examined the methylation status of γ‐synuclein in the 30 pairs of tumor and normal tissues by NMSP and real‐time Q‐MSP. The unmethylated PCR product was detected in 24/30 (80%) tumor samples and 15/30 (50%) normal samples by NMSP. In 14/30 (46.67%) tumor samples and 6/30 (20%) normal samples, both alleles became completely unmethylated, and in 10/30 (33.33%) tumor samples and 9/30 (30%) normal samples contained both methylated and unmethylated alleles (partial unmethylation). Completely methylated alleles were detected in the remaining 6/30 (20%) tumor samples and 15/30 (50%) normal samples (representative eight pairs, Fig. 5a). Real‐time Q‐MSP assay indicated that the median (range) unmethylation index of tumor samples was 76.34% (84.84%), and the median (range) unmethylation index of normal samples was 19.68% (84.03%). The demethylated status of CRC samples was significantly higher than that of matched NNAT samples (P = 0.001, Fig. 5b). γ‐Synuclein methylation status of the five cells was also examined by NMSP. The results demonstrated that HT‐29 and HCT116 cells showed complete unmethylation, COLO205 and SW480 cells showed complete methylation, and LoVo cell displayed partial unmethylation.
Figure 5.
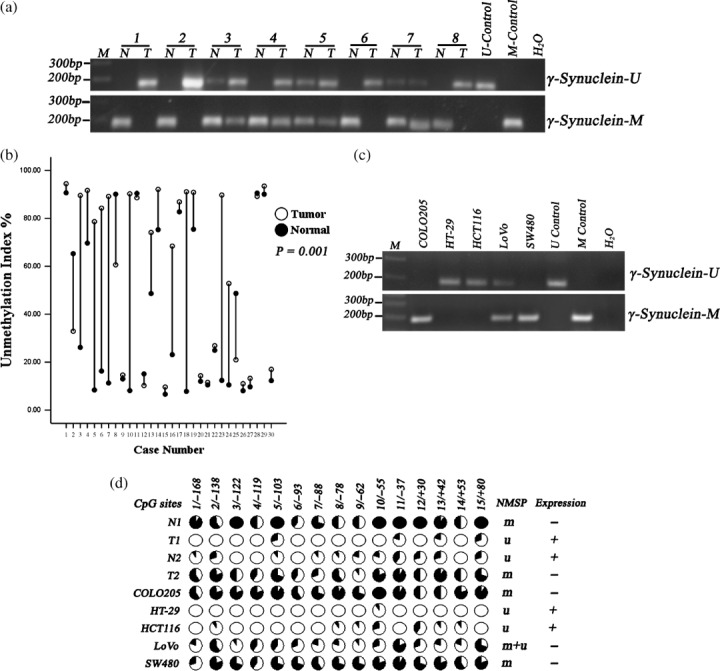
Detection of methylation status of the γ‐synuclein CpG island in colorectal cancer (CRC) samples (T), non‐neoplastic adjacent tissues (NNAT) samples (N), and CRC cell lines. M control, methylated control; m + u, partially unmethylated; M, 100 bp Maker; m, completely methylated; U control, unmethylated control; u, completely unmethylated; γ‐synuclein‐M, methylated polymerase chain reaction (PCR) product; γ‐Synuclein‐U, unmethylated PCR product. (a) The results of NMSP in nine of 30 pairs of samples. (b) Drop‐line graph compares unmethylation index of γ‐synuclein detected by real‐time quantitative methylation‐specific PCR (real‐time Q‐MSP) in CRC versus NNAT samples (P = 0.001). (c) The results of nested methylation‐specific PCR (NMSP) in CRC cells. (d) The results of genomic bisulfite DNA sequencing.
Genomic sequencing was also conducted in two pairs of representative tissues and the five cells (Fig. 5d). Normal tissue (N1) with negative γ‐synuclein expression showed densely methylated CpG sites in the CpG island, and only scattered methylated CpG sites were detected in matched tumor tissue (T1) that expressed γ‐synuclein protein. By contrast, normal tissue (N2) with γ‐synuclein expression showed scattered methylated CpG sites, and matched tumor tissue (T2) with negative γ‐synuclein expression showed densely methylated CpG sites. Densely methylated CpG sites were detected in COLO205 and SW480 cells, only scattered methylated CpG sites were detected in HT‐29 and HCT116 cells, and LoVo cells displayed medium density of methylated CpG sites. Bisulfite genome sequencing analysis confirmed the results of NMSP.
Correlation between demethylation status of γ‐synuclein and clinicopathological factors of CRC. To investigate clinical significance of aberrant γ‐synuclein demethylation in CRC, we also examined the demethylation status of γ‐synuclein in the other 37 CRC samples using MSP. The results demonstrated that of the total 67 tumor samples, 51 samples displayed unmethylation including 30 samples that were completely unmethylated and 21 samples that were partially unmethylated, and the remaining 16 samples were completely methylated. Furthermore, the demethylation tendency was more prominent in later‐stage tumors than in early stage tumors (P = 0.003, Table 3), in lymph node‐positive tumors than in lymph node‐negative tumors (P = 0.025, Table 3), and in distant metastasis‐positive tumors than in distant metastasis‐negative ones (P = 0.049, Table 3). We further performed real‐time Q‐MSP, the results of which showed that the median (range) unmethylation indices for individual pathological stages were 15.01% (82.52%) for stage I, 42.55% (83.65%) for stage II, 85.9% (85.31%) for stage III, and 88.69% (5.900%) for stage IV. The unmethylation index of γ‐synuclein was significantly correlated with clinical stage (P < 0.0001, Table 3). Similarly, there was a significant correlation between the unmethylation index of γ‐synuclein and lymph node involvement and distant metastasis (P = 0.009, P = 0.039, Table 3). Furthermore, it seemed more sensitive to predict advanced stage, lymph node invasion, and distant metastasis by detection of the demethylation status than by detection of the protein expression. To interpret the interesting phenomenon, we compared the demethylation status with the protein expression in total 67 samples (Table 4). We found that, of total 30 γ‐synuclein protein‐negative samples, 17 samples were completely or partially unmethylated; however, of total 16 completely methylated samples, only three samples expressed γ‐synuclein protein. Twenty‐five of 30 (83.33%) lymph node‐positive samples, 43/50 (86%) stage II~IV samples, and 5/5 (100%) distant metastasis samples were completely or partially unmethylated. However, the corresponding ratios of samples that expressed γ‐synuclein protein were only 21/30 (70%), 30/50 (60%), and 5/5 (100%), respectively.
Table 3.
Correlation between methylation status of γ‐synuclein and clinicopathological factors of colorectal cancer patients
| Variable | n | Unmethylation Index percentage | P‐value | Methylation status (n) | P‐value | |||
|---|---|---|---|---|---|---|---|---|
Mean (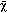 ± s)
¶ ± s)
¶
|
Median (range) | u ¶ | m + u ¶ | m ¶ | ||||
| Age | ||||||||
| ≥ 60 | 34 | 52.84 ± 34.50 | 56.69 (84.32) | 0.340 † | 13 | 11 | 10 | 0.393 § |
| < 60 | 33 | 59.34 ± 34.47 | 82.31 (87.01) | 17 | 10 | 6 | ||
| Sex | ||||||||
| Male | 37 | 54.50 ± 36.19 | 68.38 (86.93) | 0.612 † | 16 | 11 | 10 | 0.401 § |
| Female | 30 | 57.93 ± 32.51 | 68.48 (83.45) | 14 | 10 | 6 | ||
| Histological types | ||||||||
| Differentiated | 62 | 54.65 ± 34.58 | 61.73 (87.01) | 0.298 † | 27 | 19 | 16 | 0.301 § |
| Undifferentiated | 5 | 73.24 ± 29.49 | 86.84 (70.11) | 3 | 2 | 0 | ||
| Stage | ||||||||
| I | 17 | 35.27 ± 32.86 | 15.01 (82.52) | < 0.0001 ‡ | 3 | 5 | 9 | 0.003 § |
| II | 19 | 52.40 ± 31.96 | 42.55 (83.65) | 7 | 10 | 2 | ||
| III | 26 | 66.25 ± 32.95 | 85.90 (85.31) | 15 | 6 | 5 | ||
| IV | 5 | 89.35 ± 2.410 | 88.69 (5.900) | 5 | 0 | 0 | ||
| Lymph node invasion | ||||||||
| Positive | 30 | 68.88 ± 31.32 | 87.25 (84.84) | 0.009 † | 19 | 6 | 5 | 0.025 § |
| Negative | 37 | 45.62 ± 33.58 | 28.98 (85.33) | 11 | 15 | 11 | ||
| Distant metastasis | ||||||||
| Positive | 5 | 89.35 ± 2.410 | 88.69 (5.900) | 0.039 † | 5 | 0 | 0 | 0.049 § |
| Negative | 62 | 53.35 ± 34.35 | 56.69 (87.01) | 25 | 21 | 16 | ||
Calculated by the Mann–Whitney U‐test.
‡ Calculated by Spearman's correlation analysis.
§ Calculated by Fisher's exact test.
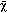 ± s: mean ± standard deviation. m: completely methylated; m + u: partially unmethylated or partially methylated; u, completely unmethylated.
± s: mean ± standard deviation. m: completely methylated; m + u: partially unmethylated or partially methylated; u, completely unmethylated.
Table 4.
Comparison between protein expression and methylation status in 67 tumor samples
| Unmethylation † (n = 51) | Methylation (n = 16) | |||
|---|---|---|---|---|
| Protein expression positive (n = 34) | Protein expression negative (n = 17) | Protein expression positive (n = 3) | Protein expression negative (n = 13) | |
| Stage II~IV (n = 50) | 29 | 14 | 1 | 6 |
| Lymph node invasion (n = 30) | 20 | 5 | 1 | 4 |
| Distant metastasis (n = 5) | 5 | 0 | 0 | 0 |
Unmethylation here means complete or partial unmethylation.
Discussion
Although there has been a report that showed down‐regulation of γ‐synuclein in human ESCC,( 22 ) more studies support the statement of overexpression of γ‐synuclein in more types of cancer. Furthermore, the overexpression is primarily due to loss of methylation control in a CpG island of the γ‐synuclein gene in those cancers.( 15 , 16 , 17 , 18 ) Altered expression and demethylation status of γ‐synuclein have been reported in breast, ovarian, and gastric cancer as a useful biomarker for evaluation of disease progression and patient prognosis.( 13 , 15 , 16 , 17 , 26 ) On the basis of previous findings, in the present study on CRC, NNAT, and five CRC cell lines, we investigated γ‐synuclein expression by RT‐PCR, western blot analysis, and IHC methods, and methylation status by MSP and genomic bisulfite DNA sequencing analyses.
Our results showed that γ‐synuclein mRNA and protein expression were up‐regulated in CRC samples and HT‐29, HCT116 cells, compared to those in matched NNAT samples (P < 0.05). γ‐Synuclein protein expression was significantly correlated with clinical stage and lymph node involvement of CRC. Although no significant correlation was reached, there was a tendency that tumors with γ‐synuclein expression were more likely to distantly metastasize compared with γ‐synuclein‐negative tumors, and a significant correlation might be reached in a study with large quantities of samples. In general, these results provided strong clinical evidence that supported a positive role of γ‐synuclein in promoting disease progression and tumor metastasis, keeping in line with the data from previous study, in which overexpression of γ‐synuclein in MDA‐MB 435 breast cancer cells led to a significant increase in motility and invasiveness in cell culture and to a profound augmentation of metastasis in nude mice.( 14 ) These results were also parallel to previous report on diversified types of cancer including 20 colon cancer tissue samples.( 18 ) However, γ‐synuclein expression status was different in CRC cell lines. There was undetectable γ‐synuclein expression in LoVo cells with strong metastatic potentiality, but there was high expression in HT‐29 cells with relatively weak metastatic potentiality. The reason might be due to only five CRC cell lines having been chosen; other cells with strong metastatic potentiality may have high γ‐synuclein expression. In addition, γ‐synuclein may show other biological effects in CRC cells. The effects of γ‐synuclein on cell survival and antiapoptosis have been investigated in breast and ovarian cancer cells.( 27 , 28 , 29 , 30 , 31 , 32 , 33 ) The role that γ‐synuclein plays in CRC and how it is implicated in CRC remain unknown.
After treatment with 5‐aza‐C, demethylation of the CpG island was accompanied by re‐expression of γ‐synuclein in γ‐synuclein‐negative CRC cells, COLO205, LoVo, and SW480. This result led us further investigate the methylation status in tissues and total five cells. We then detected unmethylated γ‐synuclein alleles in HT‐29, HCT116, and LoVo cells, and significantly higher demethylation status in CRC samples than in matched NNAT samples (P < 0.05). The results of genomic sequencing approved the specificity of MSP to faithfully detect unmethylated alleles in tissues and cells. In addition, unmethylated alleles were also detected in NNAT samples without γ‐synuclein expression, and a similar observation has been reported in a previous study on benign and malignant colon neoplasms.( 34 ) This raises the interesting question of whether the demethylation event actually occurs in premalignant stages of CRC. Further studies are required to investigate the demethylation status of γ‐synuclein in preneoplastic lesions of the colorectum.
When associating the demethylation status of γ‐synuclein with clinicopathological factors, we found a stage‐specific pattern for demethylation status in CRC, which was parallel to previous findings for gastric cancer.( 26 ) Furthermore, the demethylation status seemed more sensitive compared to the protein expression for evaluation of the disease progression. To interpret the phenomenon, we compared the demethylation status with protein expression in CRC tissues and cells. We found that 17/30 (56.7%) of γ‐synuclein expression‐negative tumors were completely or partially unmethylated, and LoVo cells did not express γ‐synuclein but showed partially unmethylated γ‐synuclein alleles. Possibly this is because the demethylation event indirectly works upstream and protein directly performs downstream. The demethylation event of γ‐synuclein precedes the γ‐synuclein expression in some CRC tissues, especially in advanced‐stage tumors, such as a distant metastasis tumor with a metastatic tumor nodule in the left supraclavicular region, from which a LoVo cell was initiated in 1971.( 35 )
In addition to lack of γ‐synuclein expression in some tumor tissues with unmethylated genes, we also found lack of unmethylated genes in some tumor tissues with γ‐synuclein expression. We tentatively interpret this discrepancy thus: The unmethylated or methylated genes could be derived from other cell types such as fat cells, fibroblasts, or mononuclear leukocytes, in which γ‐synuclein expression is regulated by other transcriptional mechanisms. To minimize the impact of contamination by non‐tumor cells, we purified tumor tissues by washing them with RNase‐Free 0.9% NaCL after surgery, and then by trimming off non‐tumor portions from the tumor tissues as far as possible to ensure that there was a greater proportion of tumor cells in the selected areas for DNA extraction. We also quantified the demethylated status to minimize potential errors. However, two other possibilities cannot be excluded. First, it is possible that methylation in some key γ‐synuclein CpG sites is sufficient to block its expression, and some demethylated CpG sites are sufficient for transcriptional activation in CRC.( 17 ) Second, the demethylation of the CpG island is not completely responsible but is primarily responsible for aberrant γ‐synuclein expression, and other mechanisms of transcriptional activation could account for aberrant γ‐synuclein expression in CRC, keeping in line with previous findings in breast cancer cells, in which cellular content of transcription activators and repressors such as AP‐1, SP1, and CRE binding protein that interact with the cis‐regulatory sequences present in the intron 1 contribute prominently to the expression of γ‐synuclein.( 36 )
In conclusion, we provide strong evidence suggesting that γ‐synuclein expression is up‐regulated in CRC, which is primarily attributed to the demethylation of the CpG island in exon 1 of the γ‐synuclein gene, and that aberrant expression and demethylation of γ‐synuclein closely correlate with advanced clinical stage, lymph node involvement, and distant metastasis. The demethylation status is more sensitive than the protein expression for evaluation of disease progression. Further study is needed to prove the value of γ‐synuclein as a tumor marker in large quantities of CRC tissue samples as well as in serum or stool samples from patients with CRC, and as a factor in patient prognosis evaluation.
Acknowledgments
This work was funded by the Terry Fox Foundation for Cancer Research, and was supported in part by Shanghai Biosune Biological Technology for genomic sequencing. We thank Dr Wen‐Jie Dong (Department of Digestive Medicine, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai, China) for his helpful discussions on MSP and genomic bisulfite DNA sequencing methods. We thank Dr Yuan‐Fei Peng, Dr Feng Zhang, and Dr Fei Yue (Institute of Digestive Surgery, Ruijin Hospital) for excellent technical assistance and advice.
References
- 1. Jemal A, Thomas A, Murray T, Thun M. Cancer statistics, 2002. CA Cancer J Clin 2002; 52: 23–47. [DOI] [PubMed] [Google Scholar]
- 2. Samoha S, Arber N. Cyclooxygenase‐2 inhibition prevents colorectal cancer: from the bench to the bed side. Oncology 2005; 69 (Suppl 1): 33–7. [DOI] [PubMed] [Google Scholar]
- 3. McGartland LP, Mulcahy MF, Benson AB III Pre‐ and postoperative adjuvant therapy for. locally advanced rectal cancer. Clin Adv Hematol Oncol 2004; 2: 806–14. [PubMed] [Google Scholar]
- 4. Lavedan C, Leroy E, Dehejia A et al . Identification, localization and characterization of the human γ‐Synuclein gene. Human Genet 1998; 103: 106–12. [DOI] [PubMed] [Google Scholar]
- 5. Lavedan C. The synuclein family. Genome Res 1998; 8: 871–80. [DOI] [PubMed] [Google Scholar]
- 6. Clayton DF, George JM. The synucleins: a family of proteins involved in synaptic function, plasticity, neurodegeneration and disease. Trends Neurosci 1998; 21: 249–54. [DOI] [PubMed] [Google Scholar]
- 7. Ahmad M, Attoub S, Singh MN, Martin FL, El‐Agnaf OM. γ‐Synuclein and the progression of cancer. FASEB J 2007; 21: 3419–30. [DOI] [PubMed] [Google Scholar]
- 8. Spillantini M, Schmidt M, Lee V, Trojanowski J, Jakes R, Goedert M. α‐Synuclein in Lewy bodies. Nature 1997; 38: 839–40. [DOI] [PubMed] [Google Scholar]
- 9. Takeda A, Mallory M, Sundsmo M, Honer W, Hansen L, Masliah E. Abnormal accumulation of NACP/α‐Synuclein in neurodegenerative disorders. Am J Pathol 1998; 152: 367–72. [PMC free article] [PubMed] [Google Scholar]
- 10. Ueda K, Fukushima H, Masliah E et al . Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci 1993; 90: 11 282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yoshimoto M, Iwai A, Kang D, Otero DA, Xia Y, Saitoh T. NACP, the precursor protein of the non‐amyloid β/A4 protein (Aβ) component of Alzheimer disease amyloid, binds Aβ and stimulates Aβ aggregation. Proc Natl Acad Sci 1995; 92: 9141–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ji H, Liu Y, Jia T et al . Identification of a breast cancer‐specific gene, BCSG1, by direct differential cDNA sequencing. Cancer Res 1997; 57: 759–64. [PubMed] [Google Scholar]
- 13. Wu K, Weng Z, Tao Q et al . Stage‐specific expression of breast cancer‐specific gene γ‐Synuclein. Cancer Epidemiol Biomarkers Prev 2003; 12: 920–5. [PubMed] [Google Scholar]
- 14. Jia T, Liu YE, Liu J, Shi YE. Stimulation of breast cancer invasion and metastasis by synuclein‐γ. Cancer Res 1999; 59: 742–7. [PubMed] [Google Scholar]
- 15. Guo J, Shou C, Meng L et al . Neuronal protein synuclein gamma predicts poor clinical outcome in breast cancer. Int J Cancer 2007; 121: 1296–305. [DOI] [PubMed] [Google Scholar]
- 16. Wu K, Quan Z, Weng Z et al . Expression of neuronal protein synuclein gamma gene as a novel marker for breast cancer prognosis. Breast Cancer Res Treat 2007; 101: 259–67. [DOI] [PubMed] [Google Scholar]
- 17. Gupta A, Godwin AK, Vanderveer L, Lu A, Liu J. Hypomethylation of the synuclein‐γ gene CpG island promotes its aberrant expression in breast carcinoma and ovarian carcinoma. Cancer Res 2003; 63: 664–73. [PubMed] [Google Scholar]
- 18. Liu H, Liu W, Wu Y et al . Loss of epigenetic control of synuclein‐γ gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res 2005; 65: 7635–43. [DOI] [PubMed] [Google Scholar]
- 19. Bruening W, Giasson BI, Klein‐Szanto AJ, Lee VM, Trojanowski JQ, Godwin AK. Synucleins are expressed in the majority of breast and ovarian carcinomas and in preneoplastic lesions of the ovary. Cancer 2000; 88: 2154–63. [PubMed] [Google Scholar]
- 20. Li Z, Sclabas GM, Peng B et al . Overexpression of synuclein‐γ in pancreatic adenocarcinoma. Cancer 2004; 101: 58–65. [DOI] [PubMed] [Google Scholar]
- 21. Iwaki H, Kageyama S, Isono T et al . Diagnostic potential in bladder cancer of a panel of tumor markers (calreticulin, γ‐synuclein, and catechol‐o‐methyltransferase) identified by proteomic analysis. Cancer Sci 2004; 95: 955–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou CQ, Liu S, Xue LY et al . Down‐regulation of γ‐synuclein in human esophageal squamous cell carcinoma. World J Gastroenterol 2003; 9: 1900–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feng B, Xu WB, Zheng MH et al . Clinical significance of human kallikrein 10 gene expression in colorectal cancer and gastric cancer. J Gastroenterol Hepatol 2006; 21: 1596–603. [DOI] [PubMed] [Google Scholar]
- 24. Chan MW, Chu ES, To KF, Leung WK. Quantitative detection of methylated SOCS‐1, a tumor suppressor gene, by a modified protocol of quantitative real time methylation‐specific PCR using SYBR green and its use in early gastric cancer detection. Biotechnol Lett 2004; 26: 1289–93. [DOI] [PubMed] [Google Scholar]
- 25. Lo YM, Wong IH, Zhang J, Tein MS, Ng MH, Hjelm NM. Quantitative analysis of aberrant p16 methylation using real‐time quantitative methylation‐specific polymerase chain reaction. Cancer Res 1999; 59: 3899–903. [PubMed] [Google Scholar]
- 26. Yanagawa N, Tamura G, Honda T, Endoh M, Nishizuka S, Motoyama T. Demethylation of the synuclein‐γ gene CpG island in primary gastric cancers and gastric cancer cell lines. Clin Cancer Res 2004; 10: 2447–51. [DOI] [PubMed] [Google Scholar]
- 27. Gupta A, Inaba S, Wong OK, Fang G, Liu J. Breast cancer‐specific gene 1 interacts with the mitotic checkpoint kinase BubR1. Oncogene 2003; 22: 7593–9. [DOI] [PubMed] [Google Scholar]
- 28. Inaba S, Li C, Shi YE, Song DQ, Jiang JD, Liu J. Synuclein gamma inhibits the mitotic checkpoint function and promotes chromosomal instability of breast cancer cells. Breast Cancer Res Treat 2005; 94: 25–35. [DOI] [PubMed] [Google Scholar]
- 29. Jiang Y, Liu YE, Lu A et al . Stimulation of Estrogen Receptor Signaling by Synuclein‐γ. Cancer Res 2003; 63: 3899–903. [PubMed] [Google Scholar]
- 30. Liu YE, Pu W, Jiang Y, Shi D, Dackour R, Shi YE. Chaperoning of estrogen receptor and induction of mammary gland proliferation by neuronal protein synuclein gamma. Oncogene 2007; 26: 2115–25. [DOI] [PubMed] [Google Scholar]
- 31. Jiang Y, Liu YE, Goldberg ID, Shi YE. γ‐synuclein, a novel heat‐shock protein‐associated chaperone, stimulates ligand‐dependent estrogen receptor α signaling and mammary tumorigenesis. Cancer Res 2004; 64: 4539–46. [DOI] [PubMed] [Google Scholar]
- 32. Pan ZZ, Bruening W, Godwin AK. Involvement of RHO GTPases and ERK in synuclein‐γ enhanced cancer cell motility. Int J Oncol 2006; 29: 1201–5. [PubMed] [Google Scholar]
- 33. Pan ZZ, Bruening W, Giasson BI, Lee VM, Godwin AK. γ‐synuclein promotes cancer cell survival and inhibits stress‐ and chemotherapy drug‐induced apoptosis by modulating MAPK pathways. J Biol Chem 2002; 277: 35 050–60. [DOI] [PubMed] [Google Scholar]
- 34. Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 1985; 228: 187–90. [DOI] [PubMed] [Google Scholar]
- 35. Drewinko B, Romsdahl MM, Yang LY, Ahearn MJ, Trujillo JM. Establishment of a human carcinoembryonic antigen‐producing colon adenocarcinoma cell line. Cancer Res 1976; 36: 467–75. [PubMed] [Google Scholar]
- 36. Lu A, Gupta A, Li C et al . Molecular mechanisrns for aberrant expression of the human breast cancer specific gene 1 in breast cancer cells: control of transcription by DNA methylation and intronic sequences. Oncogene 2001; 20: 5173–85. [DOI] [PubMed] [Google Scholar]


