Abstract
To assess the involvement of the TSLC cascade in hepatocarcinogenesis, we investigated the expression and DNA methylation patterns of the genes Tslc1 and Dal‐1 in hepatocellular carcinomas (HCC) induced using N‐nitrosodiethylamine (DEN) in rats. Six‐week‐old male F344 rats received a single intraperitoneal injection of DEN at a dose of 10 mg/kg body weight, followed by combined treatment with partial hepatectomy and colchicine to induce cell‐cycle disturbance and a selection procedure consisting of 2‐acetylaminofluorene and carbon tetrachloride. Total RNA was extracted from 10 HCC, and the expression levels of Tslc1 and Dal‐1 were measured using semiquantitative reverse transcription–polymerase chain reaction (RT‐PCR) analysis. Three of 10 HCC showed reduced expression of Tslc1, compared with normal liver tissues, but no changes in the expression level of Dal‐1 were found. For DNA methylation analysis, bisulfite sequencing was performed. The 5′ upstream region of Tslc1 was methylated in the three HCC in which its expression was reduced, but was unmethylated in normal liver tissue. Western blot analysis also revealed reduced expression of Tslc1 protein in the three HCC. These results suggest that alterations to the TSLC cascade might have a role in hepatocarcinogenesis using DEN in rats. (Cancer Sci 2007; 98: 943–948)
Abbreviations:
- DAL‐1
differentially expressed in adenocarcinoma of the lung
- DEN
N‐nitrosodiethylamine
- HCC
hepatocellular carcinoma
- NSCLC
non‐small‐cell lung cancer
- PCR
polymerase chain reaction
- RT
reverse transcription
- SDS
sodium dodecyl sulfate
- TGF
transforming growth factor
- TSLC1
tumor suppressor in lung cancer 1.
HCC is one of the most frequent and deadly human malignancies.( 1 , 2 , 3 , 4 , 5 ) Although infection with the hepatitis virus is a primary risk factor, environmental chemicals, such as aflatoxin, are also involved in the development of HCC.( 6 , 7 , 8 ) Furthermore, there are various nitroso‐compounds in the environment that possess hepatocarcinogenicity. To elucidate the molecular events underlying the development of HCC, animal models exposed to such nitroso‐compounds are very useful. In studies of one such animal model, rats treated with DEN, the authors have so far reported the occurrence of frequent mutations in the gene encoding β‐catenin,( 9 ) and alterations to Fhit;( 10 ) however, no alterations have been observed in genes related to the TGF‐β signaling pathway, such as those encoding Smad2, Smad4 and TGF‐β receptor II.( 11 )
TSLC1 has been identified as a tumor suppressor gene on chromosome 11q23.2 in NSCLC using functional complementation analyses.( 12 , 13 ) TSLC1 encodes a membrane glycoprotein that belongs to the immunoglobulin superfamily and participates in cell adhesion.( 13 , 14 ) DAL‐1/4.1B was initially identified as a candidate tumor suppressor gene on 18p11.3.( 15 ) DAL‐1 belongs to the 4.1 protein family, and is an anchoring protein connecting TSLC1 to the actin cytoskeleton.( 16 ) Recently, these TSLC cascade genes have been reported to be inactivated in several human cancers, and promoter methylation has been proposed as an important mechanism for inactivation of the genes encoding TSLC1 and DAL‐1.( 13 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 )
In a recent study by the authors, reduced expression of Tslc1 due to aberrant methylation in rat lung tumors induced by nitroso‐compounds was detected.( 24 ) Therefore, to assess the involvement of the TSLC cascade in hepatocarcinogenesis, we investigated the expression patterns of Tslc1 and Dal‐1, and their DNA methylation status, in HCC induced using DEN in rats.
Materials and Methods
Animals and treatment. A total of 12 male F344 rats were purchased at 5 weeks of age from Japan SLC Inc. (Shizuoka, Japan) and housed 3–5 per plastic cage with white flake bedding in an air‐conditioned room, with a constant temperature of 25°C and a 12‐h light–dark cycle. Food and water were given ad libitum throughout the study. After a 1‐week acclimation period on a basal diet in pellet form (CF‐2 diet; Clea Japan, Tokyo, Japan), 10 animals received DEN (Wako Pure Chemical Co. Ltd, Kyoto, Japan) i.p. at a dose of 10 mg/kg body weight, and, after 4 h, underwent partial hepatectomy performed using the method described by Higgins and Anderson.( 25 ) Colchicine (Sigma Chemical Co. St Louis, MO, USA), at a dose of 0.5 mg/kg body weight, was injected i.p. 3 days after DEN treatment. After an 11‐day recovery period, rats were placed on a selection regimen, which comprised feeding them a 0.02% 2‐acetylaminofluorene (Nacalai Tesque, Inc., Kyoto, Japan) diet for 2 weeks and a single intragastric administration of carbon tetrachloride (Nacalai Tesque, Inc.) at 1 mg/kg body weight, following the procedure described by Cayama et al.( 26 , 27 ) Rats were killed under ether anesthesia 42 weeks after the beginning of the experiment.( 28 , 29 )
Tissue preparation. After killing, livers were immediately excised and any grossly apparent tumors were dissected from the surrounding tissue. Samples were frozen in liquid nitrogen and stored at −80°C until analysis. Portions of the tumors were fixed in 10% neutrally buffered formalin at 4°C and routinely processed for H&E staining for histologic examination.
Semi‐quantitative RT‐PCR analysis of Tslc1 and Dal‐1 gene expression. Total RNA was extracted from frozen tissue using an RNeasy Mini Kit (Qiagen, Hilden, Germany) and first‐strand cDNA was synthesized from 0.2 µg of RNA using ready‐to‐go your‐prime first‐strand beads (Pharmacia Co. Ltd, Tokyo, Japan). To eliminate possible false positives caused by residual genomic DNA, all samples were treated with DNase.
Semi‐quantitative RT‐PCR analysis was performed as described previously.( 30 , 31 ) PCR amplification was carried out in a reaction volume of 20 µL containing 1 µM of each gene primer, 200 µM of each dNTP, 1 × PCR buffer (Perkin Elmer, Applied Biosystems Division, Foster City, CA, USA), 0.5 U of AmpliTaq Gold (Perkin Elmer), and 0.5 µL of synthesized cDNA mixture. Primer pairs for PCR amplification were designed against rat Tslc1 and Dal‐1 sequences (GenBank accession numbers AB257091 and NM_05392, respectively; Table 1). Rat Gapdh gene was used as an internal control gene.( 24 , 30 , 31 ) For each gene, multiple cycles of PCR amplification were tested. The cycle number at which the sample with the highest expression level reached an amplification plateau was determined and a cycle number smaller than this was adopted for the analysis. Amplified products were then separated on 2% agarose gels containing 0.05 µg/mL ethidium bromide.
Table 1.
The primer sequences used in this study
| Primer sequence | Annealing temperature (°C) | |
|---|---|---|
| Reverse transcription‐polymerase chain reaction | ||
| Tslc1 | 1F: 5′‐TTATCCTCTGCAAGGCCTAAC‐3′ | 62 |
| 1R: 5′‐TGTTGAGGCATTTCGTCATC‐3′ | ||
| Dal‐1 | 2F: 5′‐AGAGCCACAGAGGAATGACG‐3′ | 60 |
| 2R: 5′‐GGCACAGACCCCTAACATGA‐3′ | ||
| Bisulfite sequencing | ||
| Tslc1 | BS‐F: 5′‐GGGGAAGTAAAGGTTGAAATTTAA‐3′ | 60 |
| BS‐R: 5′‐AACAACACAATACTCACCATAT‐3′ | ||
| Dal‐1 | BS‐F: 5′‐TAGAGGATTTAGGTGTTTTAAAGA‐3′ | 56 |
| BS‐R: 5′‐CTCAAAACCCCTCAACCACCAA‐3′ | ||
Bisulfite sequencing. Bisulfite treatment of genomic DNA was performed as previously described.( 24 , 31 ) Briefly, genomic DNA was extracted from frozen tissue using a DNeasy Tissue Kit (Qiagen) and 500 ng of each sample was denatured in 0.3 N NaOH, followed by 2.9 M sodium bisulfite (Sigma), and 0.5 mM hydroquinone (Sigma) was added and the mixture underwent 15 cycles of 30 s denaturation at 95°C and 15 min incubation at 50°C. Samples were then desalted using the Wizard DNA cleanup system (Promega, Madison, WI, USA) and desulfonated by treatment with 0.3 N NaOH at room temperature for 5 min. After ethanol precipitation with ammonium acetate, DNA was dissolved in distilled water.
For bisulfite sequencing, primer pairs were designed against the 5′ upstream regions of rat Tslc1 and Dal‐1 (GenBank accession numbers AB257090 and NC_005108, respectively; Table 1). Although there is a CpG site within the primer sequence of Tslc1 BS‐R, the PCR products showed a clear single band in 1% agarose gels. PCR products were subcloned using a TOPO TA cloning kit (Invitrogen, CA, USA) and sequenced using a BigDye terminator v3.0 cycle sequencing ready reaction kit (Applied Biosystems) and an ABI PRISM 310 genetic analyzer (Applied Biosystems). For each sample, eight clones were sequenced.
Western blot analysis. Proteins were extracted from frozen tissues in a buffer solution containing 50 mM Tris‐HCl (pH 8.0), 150 mM NaCl, 1% Triton X‐100, and 1 mM phenylmethylsulfonyl fluoride.( 32 ) Briefly, the extracted proteins were electrophoresed on 8% SDS‐polyacrylamide gels, transferred to Hybond ECL (Amersham Biosciences Corp., NJ, USA) and probed with a human polyclonal anti‐TSLC1 antibody (Santa Cruz Biotechnology Inc., CA, USA). Mouse monoclonal antiactin antibody (Santa Cruz Biotechnology) was used as an internal control. The membranes were developed with an ECL advance western blotting detection kit (Amersham Biosciences). Densitometric and quantitative analysis of signals was performed using Scion Image (Scion Corporation, Frederick, MD, USA).( 33 )
Results
A total of 10 of the HCC obtained were histologically well‐differentiated. To assess the expression levels of Tslc1 and Dal‐1 in HCC, semiquantitative RT‐PCR analysis on the 10 HCC and tissue from two normal livers was performed. Representative results are shown in Fig. 1. In three of the 10 HCC (30%) the expression level of Tslc1 was reduced compared with that in normal liver tissues. By contrast, no changes in the expression level of Dal‐1 were found in any of the 10 HCC.
Figure 1.
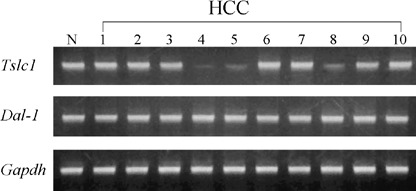
Representative results of the semiquantitative reverse transcription‐polymerase chain reaction analysis of the expressions of Tslc1 and Dal‐1 in one normal (N) liver tissue and 10 hepatocellular carcinomas (HCC).
Next, we measured the DNA methylation status of the 5′ upstream region of Tslc1 (between nucleotides‐273 and 20), which contains 23 CpG sites, and that of Dal‐1 (between nucleotides ‐200 and 107), which contains 33 CpG sites. Representative results of the bisulfite sequencing analysis of Tslc1 and Dal‐1 are shown in 2, 3. Methylation was observed in the 5′ upstream region of Tslc1 in all three HCC in which the expression level of Tslc1 was reduced. By contrast, in normal liver tissues and HCC, in which Tslc1 expression was not reduced, this region was unmethylated. The 5′ upstream region of Dal‐1 was unmethylated in normal liver tissues and HCC.
Figure 2.
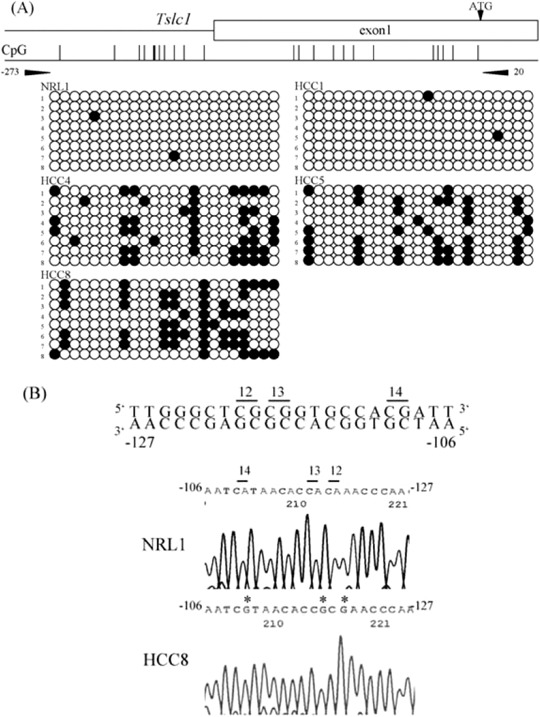
Results of the methylation analysis of the 5′ upstream region of rat Tslc1 (tumor suppressor in lung cancer 1) gene. (a) For each sample, eight clones were sequenced. The primer pair for bisulfite sequencing is shown (arrowheads). There is a CpG site within the primer sequence of BS‐R. Methylated CpG sites are shown by closed circles and unmethylated CpG sites are shown by open circles. Reduced expression of Tslc1 was seen in HCC4, 5 and 8, but not in NRL1 and HCC1. NRL, normal liver tissue, HCC, hepatocellular carcinoma. (b) Bisulfite sequencing of the 5′ upstream region of rat Tslc1 gene. Sequence traces in NRL1 and HCC8 correspond to the genomic sequence (–127 to‐106 bp from the transcription initiation site) shown in the top line. CpG sites were numbers as 12, 13 and 14. Asterisks indicate methylated CpG sites.
Figure 3.
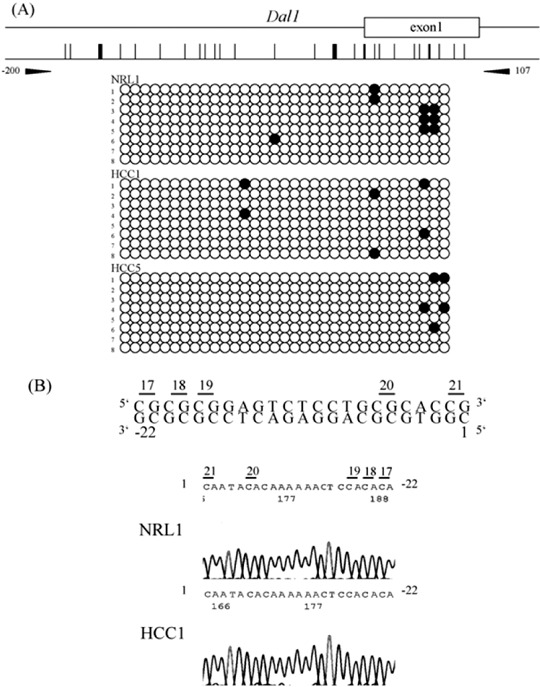
Results of the methylation analysis of the 5′ upstream region of rat Dal‐1 (differentially expressed in adenocarcinoma of the lung) gene. (a) For each sample, eight clones were sequenced. The primer pair for bisulfite sequencing is shown (arrow heads). Methylated CpG sites are shown by closed circles and unmethylated CpG sites are shown by open circles. NRL, normal liver tissue, HCC, hepatocellular carcinoma. (b) Bisulfite sequencing of the 5′ upstream region of rat Dal‐1 gene. Sequence traces in NRL1 and HCC1 correspond to the genomic sequence (–22–1 bp from the first nucleotide of exon 1) shown in the top line. CpG sites were numbers as 17, 18, 19, 20 and 21.
Western blot analysis also revealed reduced expression of Tslc1 protein in the three HCC, correlating with reduced expression of the Tslc1 mRNA and its DNA methylation status. Representative results are shown in Fig. 4.
Figure 4.
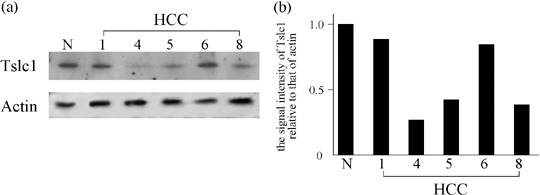
(a) Representative results of western blot analysis for the Tslc1 protein. N, normal liver tissue, HCC, hepatocellular carcinoma. (b) The signal intensity of Tslc1 relative to that of actin for each HCC case.
There were no differences in the pathologic appearance and proliferation capacity between the tumors expressing Tslc1 and those lacking Tslc1 using DNA methylation. All results are summarized in Table 2.
Table 2.
Summary for the detected alterations of the Tslc1 (tumor suppressor in lung cancer 1) and Dal‐1 (differentially expressed in adenocarcinoma of the lung) genes
| Samples | Tslc1 | Dal‐1 | |||
|---|---|---|---|---|---|
| mRNA expression | DNA methylation | Protein abnormality | mRNA expression | DNA methylation | |
| HCC1 | Expressed | Unmethylated | ND | Expressed | Unmethylated |
| HCC2 | Expressed | – | – | Expressed | – |
| HCC3 | Expressed | – | – | Expressed | – |
| HCC4 | Reduced | Methylated | Reduced | Expressed | – |
| HCC5 | Reduced | Methylated | Reduced | Expressed | Unmethylated |
| HCC6 | Expressed | – | ND | Expressed | – |
| HCC7 | Expressed | – | – | Expressed | – |
| HCC8 | Reduced | Methylated | Reduced | Expressed | – |
| HCC9 | Expressed | – | – | Expressed | – |
| HCC10 | Expressed | – | – | Expressed | – |
–, not evaluated; HCC, hepatocellular carcinoma; ND, not detected.
Discussion
TSLC1 and DAL‐1 are components of the TSLC cascade, which participates in organizing the actin cytoskeleton and constructing stable cell‐to‐cell adhesion.( 16 ) It is thought that disturbances in the cell adhesion system are critical factors in the growth, invasion and metastasis of cancer cells.( 34 ) Indeed, apoptosis and inhibition of tumor cell growth can be induced by re‐expression of TSLC1 in lung cancer cell lines.( 35 , 36 ) Moreover, re‐expression of DAL‐1 also increases cellular adhesion and induces apoptosis in a breast cancer cell line.( 37 , 38 )
Recently, it was reported that the genes encoding several of the components of the TSLC cascade are inactivated in several human cancers.( 13 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 ) As mutations in TSLC1 and DAL‐1 are rare in human cancers, aberrant promoter methylation might be a major mechanism of silencing of these genes.( 13 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 ) Aberrant promoter methylation of TSLC1 was detected in 44% of non‐small cell lung cancers, 32% of prostate cancers, and 27% of pancreatic cancers, in association with loss or reduced expression.( 13 , 17 , 18 , 19 , 20 , 21 , 22 ) In contrast, aberrant methylation of DAL‐1 has also been observed in 55% of primary NSCLC, 33% of NSCLC cell lines, 9% of small cell lung cancer cell lines, and 45% of renal cell carcinomas, and this methylation was also associated with a loss of, or reduction in DAL‐1 expression.( 16 , 21 , 22 , 23 ) Although to our knowledge there are few reports of DAL‐1 alterations in HCC, aberrant promoter methylation of TSLC1 has been detected in 29% of HCC.( 13 , 17 ) Thus, in the present study we investigated the expression levels of Tslc1 and Dal‐1 and their DNA methylation status in rat HCC, and showed reduced expression of Tslc1 due to aberrant methylation in 30% of HCC, but no changes in Dal‐1 expression. It has been suggested that inactivation of one of these two genes, TSLC1 and DAL‐1, might disrupt cell adhesion and links between the membrane and the cytoskeleton, resulting in gain of invasion and metastasis of cancer cells.( 22 ) Furthermore, aberrant methylation of at least one of these two genes occurred at a statistically significant higher frequency in patients with advanced stage disease compared with patients with early stage disease.( 22 ) Therefore, the present study suggests that alterations in the TSLC cascade might have a role in rat hepatocarcinogenesis caused by DEN. Although promoter methylation is often accompanied by complete loss of the TSLC1 expression in cancer cell lines,( 13 , 16 , 17 , 18 , 20 , 22 ) a low amount of Tslc1 expression was observed in three HCC. It may be derived from contaminated non‐cancerous tissues.
As epigenetic silencing of TSLC1 and DAL‐1 are frequently detected in human lung cancers, it is suggested that alterations in the TSLC cascade are particularly important for lung carcinogenesis.( 16 , 22 ) Recently, the authors reported that the expression of Tslc1 is significantly reduced in rat lung cancers induced by nitroso‐compounds, and that this reduction is associated with aberrant methylation.( 24 ) Moreover, the authors have also detected reduced expression of Dal‐1 due to aberrant methylation in the same tumor series of rats.( 39 ) By contrast, alterations of TSLC cascade genes were relatively less frequent in rat HCC than in rat lung tumors. Therefore, it seems that the biologic significance of the TSLC cascade in carcinogenesis might depend on organ and tumor type in rats as well as in humans.
It is well known that liver tumors can be induced by feeding rats a choline‐deficient diet without the need for exposure to any chemical carcinogens.( 40 ) This model has advantages for investigating the mechanisms underlying hepatocarcinogenesis due to endogenous factors.( 40 ) So far, we have revealed the possibility that different genetic pathways underlie DEN‐induced and choline‐deficient diet‐induced hepatocarcinogenesis in rats.( 9 ) Therefore, to better understand the involvement of the TSLC cascade in hepatocarcinogenesis, further studies should investigate the alterations to Tslc1 and Dal‐1 in rat HCC induced by a choline‐deficient diet.
Acknowledgments
This study was supported in part by the Foundation for Promotion of Cancer Research in Japan, and by a grant (RK17‐027) from the Faculty of Science and Engineering, Kinki University.
References
- 1. Simonetti RG, Camma C, Firelo F, Politi F, D’Amico G, Pagliaro L. Hepatocellular carcinoma. A worldwide problem and the major risk factors. Dig Dis Sci 1991; 36: 962–72. [DOI] [PubMed] [Google Scholar]
- 2. Tsuda H, Oda T, Sakamoto M, Hirohashi S. Different pattern of chromosomal allele loss in multiple hepatocellular carcinomas as evidence of their multifocal origin. Cancer Res 1992; 52: 1504–9. [PubMed] [Google Scholar]
- 3. Nagasue N, Uchida M, Makino Y et al. Incidence and factors associated with intrahepatic recurrence following resection of hepatocellular carcinoma. Gastroenterology 1993; 105: 488–94. [DOI] [PubMed] [Google Scholar]
- 4. Takenaka K, Adachi E, Nishizaki T et al. Possible multicentric occurrence of hepatocellular carcinoma: a clinicopathological study. Hepatology 1994; 19: 889–94. [PubMed] [Google Scholar]
- 5. Kumada T, Nakano S, Takeda I et al. Patterns of recurrence after initial treatment in patients with small hepatocellular carcinoma. Hepatology 1997; 25: 87–92. [DOI] [PubMed] [Google Scholar]
- 6. Saito I, Miyamura T, Ohbayashi A et al. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA 1990; 87: 6547–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tanaka K, Hirohata T, Koga S et al. Hepatitis C and hepatitis B in the etiology of hepatocellular carcinoma in the Japanese population. Cance Res 1991; 51: 2842–7. [PubMed] [Google Scholar]
- 8. Qian GS, Ross RK, Yu MC et al. A following‐up study of urinary markers of aflatoxin exposure and liver cancer risk in Shanghai, People's Republic of China. Cancer Epidemiol Biomarkers Prev 1994; 3: 3–10. [PubMed] [Google Scholar]
- 9. Tsujiuchi T, Tsutsumoto M, Sasaki Y, Takahama M, Konishi Y. Different frequencies and patterns of β‐catenin mutations in hepatocellular carcinomas induced by N‐nitrosodiethylamine and a choline‐deficient 1‐amino acid‐defined diet in rats. Cancer Res 1999; 59: 3904–7. [PubMed] [Google Scholar]
- 10. Tsujiuchi T, Sasaki Y, Kubozoe T, Tsutsumoto M, Konishi Y, Nakae D. Alterations of the Fhit gene in hepatocellular carcinomas induced by N‐nitrosodiethylamine in rats. Mol Carcinog 2002; 34: 19–24. [DOI] [PubMed] [Google Scholar]
- 11. Sasaki Y, Tsujiuchi T, Murata N, Tsutsumoto M, Konishi Y, Nakae D. Alterations of the transforming growth factor‐β signaling pathway in hepatocellular carcinomas induced endogenously and exogenously in rats. Jpn J Cancer Res 2001; 92: 16–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Murakami Y, Nobukuni T, Tamura K et al. Localization of tumor suppressor activity important in non‐small cell lung carcinoma on chromosome 11q. Proc Natl Acad Sci USA 1998; 95: 8153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuramochi M, Fukuhara H, Nobukuni T et al. TSLC1 is a tumor suppressor gene in human non‐small‐cell lung cancer. Nat Genet 2001; 27: 427–30. [DOI] [PubMed] [Google Scholar]
- 14. Gomyo H, Arai Y, Tanigami A et al. A 2‐Mb sequence‐ready contig map and a novel immunoglobulin superfamily gene IGSF4 in the LOH region of chromosome 11q23.2. Genomics 1999; 62: 139–46. [DOI] [PubMed] [Google Scholar]
- 15. Tran YK, Bogler O, Gorse KM, Wieland I, Green MR, Newsham IF. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res 1999; 59: 35–43. [PubMed] [Google Scholar]
- 16. Yageta M, Kuramochi M, Masuda M et al. Direct association of TSLC1 and DAL‐1, two distinct tumor suppressor proteins, in lung cancer. Cancer Res 2002; 62: 5129–33. [PubMed] [Google Scholar]
- 17. Murakami Y. Functional cloning of a tumor suppressor gene, TSLC1, in human non‐small cell lung cancer. Oncogene 2002; 21: 6936–48. [DOI] [PubMed] [Google Scholar]
- 18. Fukami F, Fukuhara H, Kuramochi M et al. Promoter methylation of the TSLC1 gene in advanced lung tumors and various cancer cell lines. Int J Cancer 2003; 107: 53–9. [DOI] [PubMed] [Google Scholar]
- 19. Fukuhara H, Kuramochi M, Fukumi T et al. Promoter methylation of the TSLC1 and tumor suppression by its gene product in human prostate cancer. Jpn J Cancer Res 2002; 93: 605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jansen M, Fukushima N, Rosty C et al. Aberrant methylation of the 5′‐CpG island of TSLC1 is common in pancreatic ductal adenocarcinoma and is first manifest in high‐grade PanlNs. Cancer Biol Ther 2002; 1: 293–6. [DOI] [PubMed] [Google Scholar]
- 21. Kikuchi S, Yamada D, Fukami T et al. Promoter methylation of DAL‐1/4.1B predicts poor prognosis in non‐small cell lung cancer. Clin Cancer Res 2005; 11: 2954–61. [DOI] [PubMed] [Google Scholar]
- 22. Heller G, Fong KM, Girard L et al. Expression and methylation pattern of TSLC1 cascade genes in lung carcinomas. Oncogene 2006; 25: 959–68. [DOI] [PubMed] [Google Scholar]
- 23. Yamada D, Kikuchi S, Williams YN et al. Promoter hypermethylation of the potential tumor suppressor DAL‐1/4.1B gene in renal clear cell carcinoma. Int J Cancer 2006; 118: 916–23. [DOI] [PubMed] [Google Scholar]
- 24. Shimizu K, Itsuzaki Y, Onishi M, Fujii H, Honoki K, Tsujiuchi T. Reduced expression of the Tslc1 gene and its aberrant DNA methylation in rat lung tumors. Biochem Biophys Res Commun 2006; 347: 358–62. [DOI] [PubMed] [Google Scholar]
- 25. Higgins GM, Anderson RM. Experimental pathology of the liver. 1. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol Lab Med 1931; 12: 1186–202. [Google Scholar]
- 26. Cayama E, Tsuda H, Sarma DSR, Farber E. Initiation of chemical carcinogenesis requires cell proliferation. Nature (Lond) 1978; 275: 60–2. [DOI] [PubMed] [Google Scholar]
- 27. Tsuda H, Lee G, Farber E. Induction of resistant hepatocytes as a new principle for a possible short‐term in vivo test for carcinogens. Cancer Res 1980; 40: 1157–64. [PubMed] [Google Scholar]
- 28. Ohashi K, Tsutsumi M, Tsujiuchi T et al. Enhancement of N‐nitrosodiethylamine‐initiated hepatocarcinogenesis caused by a colchicines‐induced cell cycle disturbance in partially hepatectomized rats. Cancer Res 1996; 56: 3473–9. [PubMed] [Google Scholar]
- 29. Tsutsumi M, Ohashi K, Tsujiuchi T et al. Disturbance of the cell cycle with colchicine enhances the growth advantage of diethylnitrosamine‐initiated hepatocytes in rats. Jpn J Cancer Res 1996; 87: 5–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shimizu K, Kato A, Hinotsume D et al. Reduced expressions of Foxp1 and Rassfla genes in lung adenocarcinomas induced by N‐nitrosobis (2‐hydroxypropyl) amine in rats. Cancer Lett 2006; 236: 186–90. [DOI] [PubMed] [Google Scholar]
- 31. Shimizu K, Kato A, Shigemura M, Fujii H, Honoki H, Tsujiuchi T. Aberrant methylation patterns of the Rassfla gene in rat lung adenocarcinomas induced by N‐nitrosobis (2‐hydroxypropyl) amine. Mol Carcinog 2006; 45: 112–17. [DOI] [PubMed] [Google Scholar]
- 32. Uchino K, Ito A, Wakayama T et al. Clinical implication and prognostic significance of the tumor suppressor TSLC1 gene detected in adenocarcinoma of the lung. Cancer 2003; 98: 1002–7. [DOI] [PubMed] [Google Scholar]
- 33. Fukushima N, Morita Y. Actomysin‐dependent microtubule rearrangement in lysophosphatidic acid‐induced neurite remodeling of young cortical neurons. Brain Res 2006; 1094: 65–75. [DOI] [PubMed] [Google Scholar]
- 34. Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci 2003; 94: 575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mao X, Seidlitz E, Ghosh K, Murakami Y, Ghosh HP. The cytoplasmic domain is critical to the tumor suppressor activity of TSLC1 in non‐small cell lung cancer. Cancer Res 2003; 63: 7979–85. [PubMed] [Google Scholar]
- 36. Mao X, Seidlitz E, Truant R, Hitt M, Ghosh HP. Re‐expression of TSLC1 in a non‐small‐cell lung cancer cell line induced apoptosis and inhibits tumor growth. Oncogene 2004; 23: 5632–42. [DOI] [PubMed] [Google Scholar]
- 37. Charboneau AL, Singh VYuT, Newsham IF. Suppression of growth and increased cellular attachment after expression of DAL‐1 in MCF‐7 breast cancer cells. Int J Cancer 2002; 100: 181–8. [DOI] [PubMed] [Google Scholar]
- 38. Jiang W, Newsham IF. The tumor suppressor DAL‐1/4.1B and protein methylation cooperate in inducing apoptosis in MCF‐7 breast cancer cells. Mol Cancer 2006; 5: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsujiuchi T, Masaoka T, Sugata E et al. Hypermethylation of the Dal‐1 gene in lung adenocarcinomas induced by N‐nitrosobis (2‐hydroxypropyl) amine in rats. Mol Carcinog 2007. (in press). [DOI] [PubMed]
- 40. Nakae D, Yoshiji H, Mizumoto Y et al. High incidence of hepatocellular carcinomas induced by a choline deficient 1‐amino acid defined diet in rats. Cancer Res 1992; 52: 5042–5. [PubMed] [Google Scholar]


