Abstract
We performed loss of heterozygosity (LOH) analysis on five chromosomal arms (1p, 3p, 9p, 10q, 17p) in hepatocellular carcinoma (HCC). Univariate analyses of 80 patients who underwent liver transplantation demonstrated significant correlations between cancer recurrence and the following variables: LOH on 3p26, LOH on 10q23, LOH on 17p13, tumor diameter ≥ 5 cm, number of tumors ≥ 4, histologic Grade 3, alpha‐fetoprotein (AFP) ≥ 400 ng/mL, American Joint Committee on Cancer (AJCC) pT classification, and portal invasion. Patients with LOH on 10q23 exhibited a significantly higher 3‐year recurrence rate (38.9%vs 11.9%, P = 0.0009). Multivariate analysis identified LOH on 10q23, histologic Grade 3, tumor nodules ≥ 4, and AFP ≥ 400 ng/mL as the risk factors of advanced HCC recurrence. These results suggest that LOH on 10q23 is associated with metastatic recurrence of HCC. (Cancer Sci 2009; 100: 520–528)
Loss of heterozygosity (LOH) has been documented in various human malignancies. Numerous studies have been conducted on hepatocellular carcinoma (HCC), and LOH has been reported on different chromosomes.( 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 ) Some studies have found a correlation between LOH and prognosis.( 5 , 6 , 7 ) However, many of these studies examined HCC patients who underwent hepatic resection,( 3 , 5 , 6 , 10 , 11 ) and HCC often recurs in patients who have undergone hepatic resection.( 12 ) Tumor recurrence complicates 70% of cases at 5 years, reflecting either intrahepatic metastases (true recurrences) or the development of de novo tumors.( 13 ) Furthermore, whether such recurrence represents de novo HCC or metastatic HCC is difficult to ascertain.( 13 ) Conversely, in HCC patients who have undergone liver transplantation (LT), the incidence of hepatic recurrence is low, and liver with chronic disease as a background factor for the onset of HCC is replaced at the time of LT. Almost all recurrences can thus be considered to represent metastasis. Studying patients with LT thus enables investigation of the chromosomal aberrations involved with metastatic recurrence of HCC. Another advantage can be seen in studying LT patients. In studies on patients who have undergone hepatic resection, the tumors can generally be assumed to be resectable, and the ratio of patients with early stage HCC within the patient cohort should thus be high. At our hospital, LT has been performed for HCC irrespective of tumor size and count since 1999.( 14 , 15 ) Subsequently, the ratio of patients with advanced HCC should be higher when compared to previous studies. With LT, the entire liver can be removed and the pathological and molecular biological characteristics of tumor tissue thoroughly ascertained.
The objective of this study was to investigate chromosomal aberrations involved with metastatic recurrence of HCC in LT patients, assess relationships between chromosomal aberrations and cumulative recurrence rate, and ascertain the clinical significance of chromosomal aberrations as predictors of recurrence.
Materials and Methods
Patients. Between February 12, 1999 and December 31, 2004, 340 adult patients over 18 years underwent primary living donor liver transplantation (LDLT) at Kyoto University Hospital. Of these, 117 had HCC, either diagnosed on preoperative evaluation or found on explant examination. Of these 117, eight patients with all necrotic nodules and 17 others who died within the first 3 months due to postoperative complications were excluded from the analysis. In addition, we excluded two patients with viable tumors that were smaller than 4 mm in size to avoid exhaustion of the tissue archives. The specimens of two patients were not available for genotype analysis after two rounds of independent microdissection and polymerase chain reaction (PCR). Eight patients did not provide informed consent. The remaining 80 patients were included in the analysis. The study comprised 53 men and 27 women ranging in age from 22 to 69 years (median, 55 years). Of the 80 patients, 62 patients (78%) had received preoperative treatments for HCC including transcatheter arterial embolization (n = 48), percutaneous ethanol injection or radiofrequency ablation (n = 40), or hepatic resection (n = 10). As of the end of April 2008, the median follow‐up was 54 months (3–109 months) from the time of operation. The clinical characteristics of study patients of the 80 patient with are shown in Table 1.
Table 1.
Clinical characteristics of patients (n = 80)
| No. of patients | |
|---|---|
| Age (years) | |
| Median | 54 |
| Range | 22–69 |
| Sex | |
| Male | 53 |
| Female | 27 |
| Background liver diseases | |
| Hepatitis C virus (HCV) | 44 |
| Hepatitis B virus (HBV) | 26 |
| Alcoholic liver cirrhosis | 2 |
| HBV + HCV | 2 |
| Primary biliary cirrhosis | 2 |
| None | 2 |
| Citrullinemia | 1 |
| HCV+ Biliary atresia | 1 |
Indication of LDLT for HCC. Unresectable HCC with poor liver function or with multiple tumor locations were included in the indication of LDLT regardless of the number or the diameters of the tumors. Exclusion criteria were extrahepatic tumor spread and/or major vessel involvement of the tumor in the liver detected by preoperative imaging.
Surgical procedure and follow‐up. Operative procedures are described elsewhere.( 16 ) Immunosuppressant included tacrolimus and low‐dose steroids.( 15 , 17 , 18 ) Steroids were gradually reduced and finally withdrawn during the first 3 months after the operation. There were no patients who received rapamycin. Patients were followed up with blood tests including tumor markers every month and computed tomography images every 3 months. Seventeen of 80 patients experienced HCC recurrence by the end of April 2008. The sites of metastatic recurrences are shown in Table 2.
Table 2.
Clinicopathologic characteristics and LOH statuses in 80 patients
| Patient number | AJCC pT classification | Results of LOH analysis of chromosomal arm | Tumor diameter (maximum; cm) | Tumor number (nodules) | Grade (1–3) | AFP (ng/mL) | vp † | Recurrence free survival (months) | Recurrence | Sites of the recurrence | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1p | 3p | 9p | 10q | 17p | ||||||||||
| 1 | 1 | R | LOH | NI | R | LOH | 3.5 | 1 | 1 | 4 | 0 | 91 | No | |
| 2 | 1 | R | NI | R | R | R | 0.9 | 1 | 1 | 5 | 0 | 87 | No | |
| 3 | 1 | LOH | R | LOH | LOH | LOH | 2.3 | 1 | 2 | 4 | 0 | 78 | No | |
| 4 | 1 | LOH | R | NI | LOH | LOH | 0.9 | 1 | 2 | 3 | 0 | 77 | No | |
| 5 | 1 | LOH | LOH | NI | R | R | 0.9 | 1 | 1 | 60 | 0 | 77 | No | |
| 6 | 1 | R | R | LOH | R | R | 2.0 | 1 | 1 | 3 | 0 | 48 | No | |
| 7 | 1 | LOH | NI | R | R | LOH | 2.1 | 1 | 2 | 29 | 0 | 65 | No | |
| 8 | 1 | R | LOH | NI | NI | LOH | 2.7 | 1 | 1 | 32 | 0 | 109 | No | |
| 9 | 1 | NI | LOH | NI | LOH | LOH | 3.2 | 1 | 1 | 21 | 0 | 59 | No | |
| 10 | 1 | LOH | R | R | R | R | 1.2 | 1 | 2 | 10 | 0 | 92 | No | |
| 11 | 1 | LOH | R | R | R | R | 1.0 | 1 | 2 | 102 | 0 | 73 | No | |
| 12 | 1 | R | R | LOH | R | LOH | 1.8 | 1 | 2 | 6 | 0 | 22 | No | |
| 13 | 1 | R | LOH | R | R | R | 2.0 | 1 | 1 | 38 | 0 | 88 | No | |
| 14 | 1 | LOH | R | LOH | R | R | 1.0 | 1 | 2 | 29 | 0 | 71 | No | |
| 15 | 1 | NI | R | NI | R | R | 1.5 | 1 | 2 | 32 | 0 | 11 | No | |
| 16 | 1 | R | LOH | NI | R | R | 1.1 | 1 | 2 | 3 | 0 | 59 | No | |
| 17 | 1 | R | R | R | R | R | 2.0 | 2 | 1 | 11 | 0 | 40 | No | |
| 18 | 2 | LOH | R | LOH | R | NI | 1.5 | 2 | 2 | 8 | 0 | 89 | No | |
| 19 | 2 | R | R | R | LOH | LOH | 4.2 | 2 | 2 | 51 | 0 | 86 | No | |
| 20 | 2 | LOH | LOH | R | R | LOH | 1.5 | 3 | 1 | 11 | 0 | 86 | No | |
| 21 | 2 | R | R | LOH | LOH | NI | 6.0 | 1 | 3 | 3317 | 1 | 85 | No | |
| 22 | 2 | LOH | NI | R | LOH | LOH | 1.8 | 2 | 2 | 36 | 0 | 84 | No | |
| 23 | 2 | LOH | R | R | R | LOH | 3.3 | 5 | 2 | 16 | 0 | 82 | No | |
| 24 | 2 | NI | LOH | LOH | R | LOH | 4.2 | 10 | 2 | 1 | 1 | 81 | No | |
| 25 | 2 | R | R | LOH | LOH | LOH | 2.7 | 3 | 2 | 29 | 0 | 77 | No | |
| 26 | 2 | LOH | R | R | R | R | 2.1 | 1 | 1 | 434 | 1 | 75 | No | |
| 27 | 2 | LOH | R | LOH | LOH | LOH | 2.4 | 3 | 2 | 6 | 1 | 73 | No | |
| 28 | 2 | LOH | R | R | R | LOH | 3.1 | 5 | 3 | 37 | 1 | 72 | No | |
| 29 | 2 | R | R | R | R | R | 2.7 | 5 | 3 | 9 | 1 | 70 | No | |
| 30 | 2 | LOH | R | R | R | NI | 3.6 | 6 | 1 | 1490 | 1 | 55 | No | |
| 31 | 2 | LOH | LOH | NI | R | R | 3.0 | 2 | 3 | 3 | 0 | 72 | No | |
| 32 | 2 | LOH | R | LOH | R | LOH | 1.4 | 3 | 1 | 42 | 0 | 63 | No | |
| 33 | 2 | LOH | R | R | R | R | 2.8 | 6 | 2 | 12 | 0 | 51 | No | |
| 34 | 2 | R | R | NI | R | LOH | 2.5 | 16 | 2 | 34 | 1 | 39 | No | |
| 35 | 2 | R | R | LOH | LOH | LOH | 1.9 | 137 | 2 | 1276 | 1 | 49 | No | |
| 36 | 2 | LOH | R | R | R | R | 2.2 | 3 | 2 | 263 | 1 | 47 | No | |
| 37 | 2 | R | R | R | R | R | 2.5 | 10 | 2 | 7 | 0 | 47 | No | |
| 38 | 2 | LOH | LOH | R | R | R | 1.4 | 2 | 2 | 585 | 0 | 65 | No | |
| 39 | 2 | R | R | R | LOH | LOH | 2.1 | 3 | 2 | 11 | 0 | 62 | No | |
| 40 | 2 | LOH | LOH | R | R | LOH | 3.5 | 22 | 3 | 33 | 1 | 58 | No | |
| 41 | 2 | R | R | LOH | R | R | 2.1 | 1 | 2 | 19 | 0 | 53 | No | |
| 42 | 2 | LOH | R | R | R | LOH | 0.9 | 2 | 2 | 5 | 0 | 58 | No | |
| 43 | 2 | LOH | R | LOH | R | LOH | 1.8 | 4 | 2 | 14 | 0 | 46 | No | |
| 44 | 2 | LOH | R | LOH | LOH | NI | 2.0 | 7 | 1 | 224 | 0 | 92 | No | |
| 45 | 2 | R | NI | LOH | R | R | 2.0 | 50 | 2 | 410 | 1 | 79 | No | |
| 46 | 2 | LOH | R | NI | R | R | 2.1 | 9 | 2 | 3564 | 0 | 87 | No | |
| 47 | 2 | NI | LOH | NI | R | R | 1.5 | 2 | 2 | 24 | 0 | 82 | No | |
| 48 | 2 | R | R | R | R | R | 2.7 | 8 | 2 | 8 | 1 | 63 | No | |
| 49 | 2 | LOH | R | NI | R | NI | 2.5 | 3 | 2 | 96 | 1 | 43 | No | |
| 50 | 2 | R | R | NI | R | LOH | 1.5 | 2 | 2 | 19 | 0 | 42 | No | |
| 51 | 2 | R | NI | NI | LOH | LOH | 2.5 | 4 | 2 | 19 | 0 | 11 | No | |
| 52 | 2 | NI | R | R | R | NI | 2.5 | 3 | 1 | 4 | 0 | 46 | No | |
| 53 | 2 | NI | LOH | R | R | LOH | 2.4 | 8 | 2 | 27 | 1 | 46 | No | |
| 54 | 2 | LOH | R | R | R | R | 4.8 | 2 | 2 | 29 | 0 | 40 | No | |
| 55 | 2 | LOH | LOH | LOH | R | LOH | 3.0 | 5 | 2 | 2153 | 1 | 46 | No | |
| 56 | 2 | LOH | R | NI | R | LOH | 3.0 | 5 | 2 | 22 | 0 | 45 | No | |
| 57 | 2 | LOH | R | LOH | R | R | 5.0 | 1 | 2 | 23 | 1 | 7 | No | |
| 58 | 2 | LOH | LOH | NI | LOH | LOH | 5.0 | 5 | 2 | 3 | 1 | 26 | Yes | Lung |
| 59 | 2 | LOH | NI | NI | LOH | LOH | 4.0 | 4 | 3 | 1148 | 0 | 8 | Yes | Lung and diaphragm |
| 60 | 2 | LOH | R | LOH | LOH | LOH | 2.1 | 4 | 2 | 53 | 0 | 44 | Yes | Liver |
| 61 | 2 | LOH | LOH | R | LOH | LOH | 3.5 | 7 | 3 | 2363 | 1 | 3 | Yes | Lung |
| 62 | 2 | LOH | R | R | LOH | LOH | 4.9 | 2 | 3 | 779 | 0 | 6 | Yes | Lymph node |
| 63 | 2 | LOH | LOH | LOH | R | LOH | 1.8 | 96 | 2 | 12 727 | 0 | 16 | Yes | Liver, bone, and lung |
| 64 | 2 | LOH | LOH | LOH | LOH | LOH | 3.0 | 15 | 2 | 91 | 0 | 18 | Yes | Liver, lung, and bone |
| 65 | 2 | LOH | NI | R | LOH | LOH | 3.0 | 40 | 3 | 4699 | 2 | 6 | Yes | Lung and bone |
| 66 | 2 | R | LOH | LOH | LOH | LOH | 4.0 | 380 | 2 | 261 | 1 | 6 | Yes | Lung and bone |
| 67 | 2 | LOH | R | R | R | LOH | 4.0 | 3 | 3 | 6736 | 2 | 7 | Yes | Adrenal gland and bone |
| 68 | 3 | LOH | LOH | LOH | LOH | R | 7.0 | 2 | 3 | 529 | 0 | 96 | No | |
| 69 | 3 | LOH | R | LOH | R | R | 6.5 | 4 | 2 | 199 | 0 | 56 | No | |
| 70 | 3 | LOH | LOH | NI | LOH | R | 3.7 | 2 | 2 | 188 | 3 | 104 | No | |
| 71 | 3 | R | LOH | R | R | LOH | 8.0 | 15 | 2 | 5 | 1 | 11 | No | |
| 72 | 3 | LOH | LOH | LOH | R | NI | 6.0 | 2 | 2 | 341 | 3 | 24 | No | |
| 73 | 3 | LOH | LOH | NI | LOH | R | 6.0 | 2 | 2 | 233 | 1 | 41 | No | |
| 74 | 3 | LOH | LOH | NI | LOH | LOH | 5.5 | 15 | 3 | 173 | 1 | 3 | Yes | Liver, adrenal gland, and brain |
| 75 | 3 | R | LOH | R | R | LOH | 9.0 | 9 | 2 | 2093 | 1 | 22 | Yes | Liver and luns |
| 76 | 3 | LOH | R | LOH | LOH | LOH | 8.0 | 5 | 3 | 3209 | 3 | 10 | Yes | Lymph node |
| 77 | 3 | LOH | R | LOH | R | R | 9.1 | 2 | 2 | 20 362 | 0 | 30 | Yes | Right subphrenic space |
| 78 | 3 | NI | LOH | NI | LOH | R | 22.5 | 23 | 3 | 3 | 1 | 8 | Yes | Diaphragm |
| 79 | 3 | LOH | R | NI | R | R | 5.2 | 4 | 2 | 2 | 2 | 14 | Yes | Liver |
| 80 | 3 | LOH | LOH | R | R | NI | 8.5 | 50 | 3 | 212 220 | 3 | 9 | Yes | Systemic |
| † HCC invasion into the portal vein was classified as vp0 (none), vp1 (microscopic), vp2 (into the segmental branch), and vp3 (into the lobar vein or the main trunk). | ||||||||||||||
| AFP, alpha‐fetoprotein; AJCC, American Joint Committee on Cancer; LOH, loss of heterozygosity; R, retention of heterozigosity; NI, not informative. | ||||||||||||||
Histopathologic diagnosis of the tumor. Histologic diagnosis of HCC in explanted liver was performed by a pathologist (H.H.) according to the guidelines of the World Health Organization.( 19 ) The sizes of the tumors, location of the tumors, number of tumors, and vascular invasion were evaluated by sections of explanted livers at 1‐cm intervals. HCC were classified by gross and microscopic examination. Pathological T (pT) classification was assigned in accordance with the American Joint Committee on Cancer (AJCC) sixth edition.( 20 ) Definitions are as follows: pT1, a solitary tumor without vascular invasion; pT2, a solitary tumor with vascular invasion, or multiple tumors, none larger than 5 cm; pT3, multiple tumors larger than 5 cm or a tumor involving a major branch of the portal or hepatic vein(s); and pT4, tumor(s) with direct invasion of adjacent organs other than the gallbladder or with perforation of the visceral peritoneum. Several staging systems exist for HCC. In LT, background liver diseases are also removed. Thus, the present study employed the AJCC staging system, as this system is based only on tumor progression, not the combination of severity of cirrhosis and tumor progression. While the Liver Cancer Study Group of Japan (LCSGJ) system resembles the AJCC system, the AJCC is currently more accessible around the world, and Minagawa et al. conducted a multicenter study on 13 773 hepatic resection patients and reported that compared to the AJCC system, the LCSGJ system might make it possible to classify patients with early stage HCC more precisely than with AJCC system. The present study therefore utilized the AJCC system. Histologic grading was performed as described in another report.( 21 )
DNA extraction and PCR. Microdissection‐based allelotyping was performed using paraffin‐embedded specimens.( 7 , 22 ) Serial 4‐µm‐thick sections were cut from the embedded tissue. The slide with the paraffin sections was placed in a jar filled with xylene for 8 h and then in a jar with 100% ethanol and stained with 1% pH 2.0 toluidine blue. Cancer tissue and non‐cancerous tissue were separately obtained by manual microdissection under the Nikon BH microscope. The largest nodule was microdissected at two sites, and up to two additional samples were microdissected in different nodules when there were multiple lesions.( 7 , 22 ) Because our goal was to analyze the LOH statuses in patients including advanced HCC, the purpose of this multiple sampling was to maximize the sensitivity of LOH in heterogenous HCC. Microdissected cancer tissue fragments were approximately 4 mm in diameter, and non‐cancerous fragments were smaller than the corresponding cancer tissue.( 7 , 22 ) These tissue fragments were placed on 50 µL of a reaction buffer (TE solution pH 8.0, 3% Tween 20; 1 mg/mL proteinase K enzyme),( 23 ) and incubated at 55°C for 16 h. DNA was extracted according to the conventional phenol‐chloroform method. The DNA pellet was diluted in 20 µL of TE and 1‐µL aliquot was used as a template for PCR amplification. PCR was carried out in 10‐µL reaction mixture containing 1 µL of supplied buffer, 0.75 µL of supplied dNTP mixture (2.5 mM each), 0.5 µM primers (final concentration), and 0.25 units of TaKaRa Ex Taq (Takara Bio, Otsu, Japan). Ten microsatellite markers were available as polymorphic probes in reference to previous studies.( 7 ),2 The chromosomal regions and primer sequences are as follows:
D1S407, 1p36, 5′‐TGCTAACCACATGGAGAGG‐3′, 5′‐GGGCGGGGGATAGAAGGA‐3′;
MYCL1, 1p34, 5′‐TGGCGAGACTCCATCAAAG‐3′, 5′‐CCTTTTAAGCTGCAACAATTTC;
D3S1539, 3p26, 5′‐CTTTCCATTACTCTCTCCATA‐3′, 5′‐CCAGTGCTGTTTTAGCTTC‐3′;
D3S2303, 3p25, 5′‐TTTCTGCCCTGCCTACATG‐3′, 5′‐TCAGAATCACCCACAAGGG‐3′;
D9S254, 9p23, 5′‐TCCTGGGTAATAACTGCCG‐3′, 5′‐CACTCACACACACGCTCAG‐3′;
D9S251, 9p21, 5′‐CCTGTGTTGAAATTTTGACTG‐3′, 5′‐ATTTCA GACTTCCTTGTGTTC‐3′;
D10S520, 10q23, 5′‐GCACTCCAGCCTATGCAAC‐3′, 5′‐GTCCTTGTGAGAAACTGGATG‐3′;
D10S1173, 10q23, 5′‐CATGCCAAGACTGAAACTCC‐3′, 5′‐AAACCCCAATGCCATAATGG‐3′;
TP53, 17p13, 5′‐GAATCCGGGAGGAGGTTG‐3′, 5′‐AACAGCTCCTTTAATGGCAG‐3′;
D17S1289, 17p13, 5′‐CATGGTCTTTTTCCATTCC‐3′, 5′‐CTTCAGAACTTACTGCCTC‐3′.
Because our goal was to analyze the factors to predict metastatic recurrence after LT, we analyzed the microsatellites according to the reports from University of Pittsburgh which analyzed the LOHs in transplant patients.( 7 , 22 ) They were tetranucleotide or pentanucleotide repeat markers.( 7 , 22 ) Advantages of such repeat markers are objective identification of peaks by uncomplicated method and reduction of stutter bands. In fact, Finkelstein et al. have reported many microdissection‐based studies using these microsatellites.( 22 , 24 , 25 , 26 ) Our samples were formalin‐fixed, paraffin‐embedded tissue blocks that are not optimal for experiments including PCR and LOH analyses. For these reasons, we selected microsatellite markers according to Finkelstein et al.( 22 , 24 , 25 , 26 ) Sequences obtained from Entrez UniSTS (http://www.ncbi.nlm.nih.gov/) or modified sequences were used as primer sequences. PCRs were performed using the TAKARA Dice TP600 (Takara Bio, Otsu, Japan) with 10‐min initial denaturation at 95°C followed by 35 cycles of 40 s at 94°C, 40 s at 56°C (D3S1539, D9S251, D17S1289) or 58°C (D1S407, MYCL1, D3S2303, D9S254, D10S520, D10S1179, TP53), and 60 s at 72°C and a 10‐min extension at 74°C. These microsatellite markers are located adjacent or within known or putative tumor‐suppressor genes as follows: TP73 (tumor protein p73) on 1p36.3; VHL (von Hippel‐Lindau tumor suppressor) on 3p26‐p25; CDKN2A (cyclin‐dependent kinase inhibitor 2A) on 9p21; TP53 (tumor protein p53) on 17p13.1; and PTEN (phosphatase and tensin homolog) on 10q23.3. The information of the genetic locations was obtained from Entrez Gene (http://www.ncbi.nlm.nih.gov/).
LOH analysis. The amplified products were separated using an Agilent 2100 Bioanalyzer system (Agilent Technologies, Palo Alto, CA, USA). When the PCR product of a given microsatellite marker in non‐cancerous tissue demonstrated only a single peak, that patient was designated as non‐informative for that microsatellite. When the PCR products in non‐cancerous tissue demonstrated two allele peaks of the target of interest, the patient was designated as informative. On informative loci, the allele ratios were calculated by the peak heights of the individual allele. Only those specimens with a ratio of (allele ratio in tumor sample)/(allele ratio in normal sample) out of a range between 0.5 and 2.0 were determined as having LOH.( 27 ) LOH on an individual chromosomal arm was determined positive when one or more microsatellite markers on each chromosomal arm detected LOH. LOH in an individual patient was determined positive when LOH in one or more tumor nodule was determined as positive.
Statistical analysis. Cumulative HCC recurrence rates were estimated according to the Kaplan–Meier method and compared at the univariate analysis using the log‐rank test. The variables analyzed were maximum tumor diameter (≥5 cm vs <5 cm), number of viable tumor nodules (≥4 vs 3 ≥), HCC invasion into portal vein(s) (presence vs absence), histologic Grade (1, 2, vs 3), AFP (alpha‐fetoprotein) ≥ 400 ng/mL, and LOH status (positive vs negative on 1p, 3p, 9p, 10q, and 17p loci). For multivariate analysis, variables were included when there was statistical significance in univariate analysis. Multivariate analysis of factors contributing to HCC recurrence was performed by means of the Cox proportional hazard model and the variables were chosen by Akaike information criterion,( 28 ) in a forward–backward stepwise selection. Statistical significance was assumed at P < 0.05.
Cohen's kappa values were calculated to evaluate interrelationships between dichotomous variables. The Kruskal–Wallis test was used to evaluate the association between AJCC pT classification (pT1, pT2, and pT3) and LOH statuses (positive vs negative). All statistical analyses were performed using R version 2.4.1.( 29 )
Approval by the ethical committee of Kyoto University. This study was approved by the ethical committee of Kyoto University, and informed consent was obtained from the patients or their relatives. No donor organs were obtained from executed prisoners or other institutionalized persons.
Results
Frequency of LOH in the HCC. In this study, we found LOH in 67% (49/73) of the patients on 1p, in 38% (28/73) on 3p, in 45% (26/58) on 9p, 33% (26/79) on 10q, and in 57% (41/72) on 17p. Detailed results are shown in Table 2. Frequencies of LOH at each of the loci were as follows: 72% (42/58) at D1S407, 46% (20/44) at MYCL1, 19% (12/63) at D3S2303, 43% (20/47) at D3S1539, 38% (12/32) at D9S254, 45% (20/44) at D9S251, 31% (22/70) at D10S1173, 26% (15/58) at D10S520, 54% (33/61) at D17S1289, and 57% (21/37) at TP53. Representative electropherograms of PCR products are shown in Fig. 1(a–c).
Figure 1.
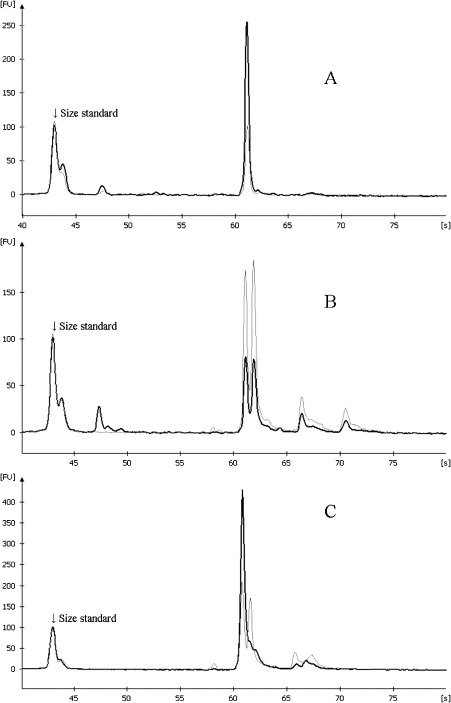
Representative electropherograms demonstrating the allelic profiles of D10S520 locus. Darker curves and lighter curves indicate polymerase chain reaction products from cancer tissue and non‐cancerous liver tissue, respectively. The x‐ and y‐axes indicate the migration time (second) and fluorescence unit (FU), respectively. (a) Non‐informative status (patient no. 8), (b) allelic retention (No. 9), and (c) loss of heterozygosity (LOH; no. 44). Patient no. 9 showed allelic retention on D10S520 but LOH on D10S1173 so this patient was interpreted as positive for LOH on 10q23.
Risk factors of HCC recurrence. Univariate analyses demonstrated significant associations between cancer recurrence and the following variables: LOH on 3p26, LOH on 10q23, LOH on 17p13, maximum of the tumors ≥ 5 cm, number of the tumors ≥ 4, and histologic Grade 3, AFP ≥ 400 ng/mL, presence of HCC invasion into the portal vein, and AJCC pT classification (Table 3). Fig. 2 illustrates the cumulative recurrence curves of patients stratified by the LOH status on 10q23. Next, a multivariate analysis of the above‐identified genotypic and pathological factors was performed. However, the AJCC pT classification was not included in the multivariate analysis because tumor diameter, tumor count, and portal invasion were included among the factors constituting the AJCC pT classification system.( 20 ) The analysis identified LOH on 10q23, histologic Grade 3, tumor nodules ≥ 4, and AFP ≥ 400 ng/mL as the risk factors for advanced HCC recurrence (Table 4).
Table 3.
Univariate analysis of HCC recurrence
| Variables | n | 3‐year cumulative recurrence rate | Standard error | P‐values | |
|---|---|---|---|---|---|
| LOH on 1p (7) † | Negative | 24 | 8.7% | 5.9 | 0.0631 |
| Positive | 49 | 27.0% | 6.4 | ||
| LOH on 3p (7) † | Negative | 45 | 11.4% | 4.8 | 0.0464* |
| Positive | 28 | 33.0% | 9.0 | ||
| LOH on 9p (22) † | Negative | 32 | 18.9% | 6.9 | 0.7520 |
| Positive | 26 | 20.3% | 8.1 | ||
| LOH on 10q (1) † | Negative | 53 | 11.9% | 4.6 | 0.0009* |
| Positive | 26 | 38.9% | 9.7 | ||
| LOH on 17p (8) † | Negative | 31 | 10.2% | 5.6 | 0.0255* |
| Positive | 41 | 30.0% | 7.3 | ||
| Number of the tumor nodule | <4 | 45 | 6.9% | 3.9 | 0.0003* |
| ≥4 | 35 | 38.3% | 8.4 | ||
| Maximum diameter of tumor | <5 cm | 65 | 12.4% | 4.1 | 0.0002* |
| ≥5 | 15 | 62.8% | 14.1 | ||
| Histologic grade | Grade 1–2 | 65 | 11.5% | 4.1 | 0.0000* |
| Grade 3 | 15 | 60.0% | 12.7 | ||
| AFP (ng/mL) | <400 | 61 | 10.3% | 4.0 | 0.0001* |
| ≥400 | 19 | 52.6% | 11.5 | ||
| Portal vein invasion | Absent | 48 | 10.8% | 4.6 | 0.0100* |
| Present | 32 | 35.7% | 8.7 | ||
| AJCC T classification | pT1 | 17 | Not computable ‡ | 0.0008* | |
| pT2 | 50 | 18.3% | 5.5 | ||
| pT3 | 13 | 58.5% | 14.7 | ||
P < 0.05.
Number of patients who had non‐informative alleles.
Not computable because no recurrence was observed in this group.
AFP, alpha‐fetoprotein; AJCC, American Joint Committee on Cancer; HCC, hepatocellular carcinoma; LOH, loss of heterozygosity.
Figure 2.
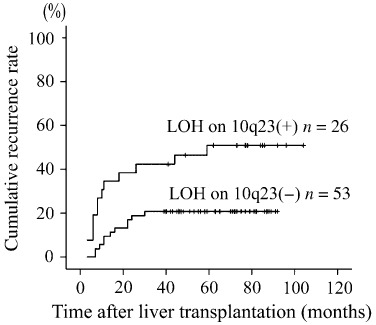
Cumulative recurrence rates of patients with 10q23‐LOH (loss of heterozygosity) and those without 10q23‐LOH.
Table 4.
Multivariate analysis of HCC recurrence (n = 64 † )
| Hazard ratio | 95% confidence interval | P‐values | |
|---|---|---|---|
| LOH on 10q (+) | 5.06 | 1.566–16.33 | 0.0067* |
| Tumor number = 4 | 7.45 | 1.858–29.92 | 0.0046* |
| Grade 3 | 3.21 | 1.008–10.19 | 0.0480* |
| AFP = 400 ng/mL | 3.10 | 1.04–9.26 | 0.0420* |
| LOH on 3p (+) | 2.23 | 0.728–6.85 | 0.1600 |
P < 0.05.
Of the 80 patients 16 patients were excluded for multivariate analysis because either of 10q or 3p was non‐informative.
AFP, alpha‐fetoprotein; HCC, hepatocellular carcinoma; LOH, loss of heterozygosity.
The relations between AJCC pT classification and LOH status, and pT classification and clinicopathologic variables. Relationships between HCC progression and LOH, and the progression and clinicopathologic variables are shown in Table 5. Regarding LOH, a significant correlation existed only for LOH on 3p. A significant correlation existed for a few clinicopathologic variables (Grade 3 and AFP ≥ 400 ng/mL).
Table 5.
Distribution of patients according to the AJCC pT classification
| AJCC pT | Number and % of patients positive for each category | P‐values | |
|---|---|---|---|
| LOH on 1p | pT1 | 7/15 (47%) | 0.1143 |
| pT2 | 32/46 (70%) | ||
| pT3 | 10/12 (83%) | ||
| Total | 73 | ||
| LOH on 3p | pT1 | 6/15 (40%) | 0.0323* |
| pT2 | 13/45 (29%) | ||
| pT3 | 9/13 (69%) | ||
| Total | 73 | ||
| LOH on 9p | pT1 | 4/10 (40%) | 0.5567 |
| pT2 | 17/40 (43%) | ||
| pT3 | 5/8 (63%) | ||
| Total | 58 | ||
| LOH on 10q | pT1 | 3/16 (19%) | 0.2893 |
| pT2 | 17/50 (34%) | ||
| pT3 | 6/13 (46%) | ||
| Total | 79 | ||
| LOH on 17p | pT1 | 7/17 (41%) | 0.0548 |
| pT2 | 30/44 (68%) | ||
| pT3 | 4/11 (36%) | ||
| Total | 72 | ||
| Grade 3 | pT1 | 0/17 (0%) | 0.0273* |
| pT2 | 10/50 (20%) | ||
| pT3 | 5/13 (38%) | ||
| Total | 80 | ||
| AFP ≥ 400 | pT1 | 0/17 (0%) | 0.0266* |
| pT2 | 14/50 (28%) | ||
| pT3 | 5/13 (38%) | ||
| Total | 80 |
P < 0.05.
Tumor diameter, tumor count, and portal invasion were not analyzed because these are included among the factors constituting the AJCC system.
AFP, alpha‐fetoprotein, AJCC, American Joint Committee on Cancer; LOH, loss of heterozygosity.
Intervariable relationships. We anticipated that there would be a strong intervariable correlation. Thus, Cohen's kappa statistics of the pairs of the variables were calculated and ranked. The result revealed that pairs including LOH status tended to be ranked lower than those without LOH status. As expected, there were various degrees of intervariable concordance, with kappa values of 0.36–0.77. (Table 6)
Table 6.
Analysis of intervariable relationships between major risk factors for HCC recurrence
| Rank | Risk factors for HCC recurrence | Cohen's kappa | ||
|---|---|---|---|---|
| 1 | Grade 3 | by | AFP ≥ 400 ng/mL | 0.77 |
| 2 | Tumor size ≥ 5 cm | by | Grade 3 | 0.76 |
| 3 | Tumor size ≥ 5 cm | by | AFP ≥ 400 ng/mL | 0.72 |
| 4 | LOH on 10q | by | Grade 3 | 0.69 † |
| 5 | Tumor size ≥ 5 cm | by | Portal vein invasion | 0.68 |
| 6 | Tumor number ≥ 4 | by | Portal vein invasion | 0.68 |
| 7 | LOH on 10q | by | Tumor size ≥ 5 cm | 0.67 † |
| 8 | Grade 3 | by | Portal vein invasion | 0.67 |
| 9 | LOH on 10q | by | AFP ≥ 400 ng/mL | 0.62 † |
| 10 | LOH on 3p | by | Tumor size ≥ 5 cm | 0.61 † |
| 11 | LOH on 3p | by | Grade 3 | 0.60 † |
| 12 | AFP ≥ 400 ng/mL | by | Portal vein invasion | 0.59 |
| 13 | Tumor number ≥ 4 | by | Tumor size ≥ 5 cm | 0.59 |
| 14 | Tumor number ≥ 4 | by | Grade 3 | 0.58 |
| 15 | LOH on 17p | by | LOH on 10q | 0.57 † |
| 16 | LOH on 3p | by | AFP ≥ 400 ng/mL | 0.56 † |
| 17 | LOH on 3p | by | Portal vein invasion | 0.56 † |
| 18 | LOH on 3p | by | LOH on 10q | 0.56 † |
| 19 | LOH on 10q | by | Portal vein invasion | 0.56 † |
| 20 | Tumor number ≥ 4 | by | AFP ≥ 400 ng/mL | 0.54 |
| 21 | LOH on 17p | by | Tumor number ≥ 4 | 0.53 † |
| 22 | LOH on 3p | by | Tumor number ≥ 4 | 0.49 † |
| 23 | LOH on 10q | by | Tumor number ≥ 4 | 0.46 † |
| 24 | LOH on 17p | by | Grade 3 | 0.45 † |
| 25 | LOH on 17p | by | Portal vein invasion | 0.44 † |
| 26 | LOH on 17p | by | AFP ≥ 400 ng/mL | 0.44 † |
| 27 | LOH on 3p | by | LOH on 17p | 0.40 † |
| 28 | LOH on 17p | by | Tumor size ≥ 5 cm | 0.36 † |
Two‐variable combinations which include one or two LOH statuses.
AFP, alpha‐fetoprotein; HCC, hepatocellular carcinoma; LOH, loss of heterozygosity.
Discussion
According to a study of patients who underwent hepatic resection, patients generally have resectable tumors, and the ratio of patients with advanced HCC in a patient cohort should be low. Furthermore, as various risk factors for HCC have been identified in recent years, hepatic resection is more often performed on patients with small HCC.( 30 ) Minagawa et al. conducted a large‐scale study and reported that of 14 992 patients who underwent hepatic resection, 13 772 underwent curative resection. Using the AJCC pT classification, the curative‐hepatic‐resection‐N0M0‐cohort included 8457 T1 cases (67.5%), 2888 T2 cases (23%), and 1189 T3 cases (9.5%).( 30 ) Varotti et al. studied 393 patients who underwent curative resection, with 173 stage I (T1N0M0) cases (44%), 200 stage II (T2N0M0) cases (51%), 19 stage IIIa (T3N0M0) cases (5%), and one stage IIIc (any T1N1M0) case.( 31 ) Therefore, most studies on hepatic resection have shown a higher ratio of pT1 cases than the present study. Okabe et al.,( 3 ) Fujiwara et al.,( 8 ) Zhang et al.,( 2 ) Wong et al.,( 4 ) and Nagai et al.( 32 ) studied LOH, but did not employ pT classification as described in version 6 of the AJCC classification. Nishida et al. studied 49 patients who underwent hepatic resection at our hospital and described pathological information in great detail.( 6 ) When classifying cases using the AJCC system based on this information described in the paper, that study included 19 pT1 cases, 20 pT2 cases, and 10 pT3 cases (39%, 41%, and 20%, respectively). The present study included 17 pT1 cases, 50 pT2 cases, and 13 pT3 cases (21%, 63%, and 16%, respectively), showing a high proportion of pT2/3 cases. The present study of LT patients thus included more patients with advanced HCC when compared to past studies on hepatic resection.
LOH was confirmed in all chromosomes examined in this study. Microsatellite analysis and CGH (comparative genomic hybridization) have identified various LOHs in human malignancies. Certain chromosomal aberrations have been linked to survival. In hepatocellular carcinoma, LOH has been documented on 1p, 3p, 4q, 5q, 6q, 8p, 9p, 10q, 11p, 13q, 14q, 16p, 16q, and 17p.( 1 , 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 ) The present results do not contradict the results of previous studies.
Furthermore, few studies have investigated relationships between LOH and HCC prognosis. According to Nishida et al. 16q and 17p are correlated to metastasis‐free survival,( 6 ) and according to the analysis by Kusano et al. using CGH, LOH on 13q correlates to recurrence following hepatic resection.( 5 ) These studies also analyzed patients who underwent hepatic resection. To the best of our knowledge, reports from University of Pittsburgh and the validation study have been the only ones to investigate relationships between LOH and prognosis following LT.( 7 , 22 , 33 ) As far as past studies of LOH on 10q in HCC are concerned, Fujimori et al.,( 34 ) Nagai et al.,( 32 ) and Piao et al.( 35 ) performed microsatellite analyses, and Kusano et al.( 5 , 36 ) performed CGH studies. Fujiwara et al. reported LOH on 10q.23. However, no studies have documented the relationship of LOH on 10q23 and HCC recurrence following hepatic resection.( 8 ) Finkelstein et al. and Marsh et al. documented relationships between various microsatellite markers and tumor‐free survival following transplantation. However, none of the microsatellite markers on 10q23.3 exhibited significant correlations (P = 0.01). Finkelstein et al. discussed only part of the relationship between tumor‐free survival and LOH at certain loci.( 22 ) Marsh et al. did not conduct multivariate analysis because their objective was to combine with neural networks. Furthermore, only microsatellites with P < 0.001 were listed, and markers on 10q were not included.( 7 ) The present study was the first to clarify relationships between 10q23 LOH and recurrent HCC following LT. Nevertheless, whether this correlation is specific for only patients who underwent LT is yet unclear. Further investigation is required on this point. If the correlation between LOH on 10q23 and extrahepatic metastatic recurrence is observed in patients who underwent hepatic resection, LOH on 10q23 would prove more generally involved in metastatic recurrence of HCC.
The microsatellite markers used in the present study (D10S1173 and D10S520) are located on 10q23.3 near the phosphatase and tensin homology gene (PTEN). PTEN converts PIP3 (phosphatidylinositol [3,4,5]‐trisphosphate) into PIP2 (phosphatidylinositol 4,5‐biphosphate) by dephosphorylation. Since PIP3 serves as a second messenger in activation of Akt, PTEN down‐regulates Akt. One of direct targets of Akt is m‐TOR (the mammalian target of rapamycin) which promotes cell proliferation by regulating p70 S6 kinase and 4E‐BP1 (eukaryotic initiation factor 4E binding protein‐1).( 37 ) An in vitro study revealed the antineoplastic effect of rapamycin in HCC,( 38 ) and early clinical studies with rapamycin analogs (e.g. everolimus, temsirolimus) are in progress.( 13 ) Analysis of LOH on 10q23 might have predictive value in identifying patients suitable for rapamycin analogs. While LOHs on 10q23 are frequently observed in HCC, mutations of PTEN are rare.( 8 , 9 , 39 ) Although correlation between LOH and PTEN expression is still unclear, one of possible mechanisms of PTEN inactivation could be haploinsufficiency. There is also a possibility that LOH is the event that precedes PTEN mutation leading to inactivation of both alleles during cancer progression. Alternatively there might be another tumor suppressor gene.
Of the various clinicopathologic variables and LOH, AJCC pT classification exhibited a significant correlation to LOH on 3p, Grade 3, and AFP ≥ 400 ng/mL. Univariate analysis showed a significant correlation between LOH on 3p and recurrence, but no significant correlation was seen by multivariate analysis. The present study clarified that LOH on 10q is a risk factor for recurrence and is a factor for metastatic recurrence of HCC, but no significant correlation with AJCC pT classification was seen. This indicates that LOH on 10q is not a substitute factor for disease staging.
The present study investigated interrelationships among recurrence risk factors using Cohen's kappa values. In the past, Pawlik et al. studied 1073 HCC patients who underwent hepatic resection and reported the high incidence of occult vascular invasion and advanced histologic grade in HCC tumors larger than 5 cm.( 40 ) The present study documented that Grade 3 and vascular invasion that cannot be assessed by diagnostic imaging before transplantation exhibited a high degree of correlation to tumor size ≥ 5 cm (kappa: 0.76 and 0.68, respectively). A high degree of correlation existed between LOH on 10q and Grade 3 (kappa: 0.69), but both LOH on 10q and Grade 3 were selected by multivariate analysis. LOH on 10q is a potent risk factor for metastatic recurrence.
A previous report of a multicenter study in Japan demonstrated that portal invasion has proved to be a significant tumor recurrence factor after LT.( 41 ) Our study also proved that portal invasion is a significant risk factor for metastatic recurrence by univariate analysis. (Table 3) Mitsunobu et al. demonstrated that the main drainage vessels of HCC are portal vein by radiopaque medium injection into the resected liver. They showed the correlation between vascular invasion and occurrence of daughter nodules.( 42 ) Our results also suggested the substantial concordance between portal invasion and tumor count. Nevertheless, the metastatic routes of HCC should not be the portal vein in transplant patients. The remaining possible origins of recurrence after LT are micrometastasis and cancer cells in the circulation. It remains unclear whether there is a direct association between these possible origins of metastatic recurrence between the risk factors investigated in this study. Moreover, there were various degrees of intervariable concordance (Table 6). In such a situation, it is difficult to illustrate causal links among these risk factors and metastatic recurrence.
In the present study, all experiments were conducted by a pathologist registered with the Japanese Society of Pathology (T.O.). As expected, microscopic grading is more convenient than LOH analysis. In the present study, multivariate analysis identified LOH on 10q, ≥ 4 tumors, Grade 3, and AFP ≥ 400 ng/mL as risk factors for recurrence. In clinical settings, when predicting recurrence before transplantation, an accurate count of the number of tumors by diagnostic imaging and measurement of AFP are important. When predicting recurrence after transplantation, the pathologist needs to accurately grade the tumor. LOH on 10q23.3 may help to predict recurrence. In addition, the present study clarified LOH on 10q23.3 as a chromosomal aberration involved with metastatic recurrence of HCC.
In conclusion, LOH on 10q23.3 where the PTEN gene is encoded is one of the risk factors for recurrent HCC following transplantation.
Acknowledgments
This work was supported in part by a Grant‐in‐Aid for Scientific Research and targeted support for creating world‐standard research and education bases (Center of Excellence) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
E‐mails for all authors: Tomoko Okuno, tokuno@legal.med.kyoto-u.ac.jp; Tatsuaki Tsuruyama, tsuruyam@path1.med.kyoto-u.ac.jp; Hironori Haga, haga@kuhp.kyoto-u.ac.jp; Yasutsugu Takada, takaday@kuhp.kyoto-u.ac.jp; Yoji Maetani, mbo@kuhp.kyoto-u.ac.jp; Koichi Tanaka, k-tanaka@fbri.org; Toshiaki; Manabe, manabet@kuhp.kyoto-u.ac.jp; Tamaki Keiji, ktamaki@legal.med.kyoto-u.ac.jp; Shinji Uemoto, uemoto@kuhp.kyoto-u.ac.jp. For pathological findings contact Dr Tatsuaki Tsuruyama.
References
- 1. Nishida N, Nishimura T, Ito T, Komeda T, Fukuda Y, Nakao K. Chromosomal instability and human hepatocarcinogenesis. Histol Histopathol 2003; 18: 897–909. [DOI] [PubMed] [Google Scholar]
- 2. Zhang SH, Cong WM, Xian ZH, Wu MC. Clinicopathological significance of loss of heterozygosity and microsatellite instability in hepatocellular carcinoma in China. World J Gastroenterol 2005; 11: 3034–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Okabe H, Ikai I, Matsuo K et al . Comprehensive allelotype study of hepatocellular carcinoma: potential differences in pathways to hepatocellular carcinoma between hepatitis B virus‐positive and ‐negative tumors. Hepatology 2000; 31: 1073–9. [DOI] [PubMed] [Google Scholar]
- 4. Wong N, Lai P, Lee SW et al . Assessment of genetic changes in hepatocellular carcinoma by comparative genomic hybridization analysis: relationship to disease stage, tumor size, and cirrhosis. Am J Pathol 1999; 154: 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kusano N, Okita K, Shirahashi H et al . Chromosomal imbalances detected by comparative genomic hybridization are associated with outcome of patients with hepatocellular carcinoma. Cancer 2002; 94: 746–51. [DOI] [PubMed] [Google Scholar]
- 6. Nishida N, Fukuda Y, Komeda T et al . Prognostic impact of multiple allelic losses on metastatic recurrence in hepatocellular carcinoma after curative resection. Oncology 2002; 62: 141–8. [DOI] [PubMed] [Google Scholar]
- 7. Marsh JW, Finkelstein SD, Demetris AJ et al . Genotyping of hepatocellular carcinoma in liver transplant recipients adds predictive power for determining recurrence‐free survival. Liver Transpl 2003; 9: 664–71. [DOI] [PubMed] [Google Scholar]
- 8. Fujiwara Y, Hoon DS, Yamada T et al . PTEN / MMAC1 mutation and frequent loss of heterozygosity identified in chromosome 10q in a subset of hepatocellular carcinomas. Jpn J Cancer Res 2000; 91: 287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bae JJ, Rho JW, Lee TJ et al . Loss of heterozygosity on chromosome 10q23 and mutation of the phosphatase and tensin homolog deleted from chromosome 10 tumor suppressor gene in Korean hepatocellular carcinoma patients. Oncol Rep 2007; 18: 1007–13. [PubMed] [Google Scholar]
- 10. Tamura S, Nakamori S, Kuroki T et al . Association of cumulative allelic losses with tumor aggressiveness in hepatocellular carcinoma. J Hepatol 1997; 27: 669–76. [DOI] [PubMed] [Google Scholar]
- 11. Nishimura T, Nishida N, Itoh T et al . Comprehensive allelotyping of well‐differentiated human hepatocellular carcinoma with semiquantitative determination of chromosomal gain or loss. Genes Chromosomes Cancer 2002; 35: 329–39. [DOI] [PubMed] [Google Scholar]
- 12. Poon RT. Liver transplantation for solitary hepatocellular carcinoma less than 3 cm in diameter in Child A cirrhosis. Dig Dis 2007; 25: 334–40. [DOI] [PubMed] [Google Scholar]
- 13. Llovet JM, Bruix J. Novel advancements in the management of hepatocellular carcinoma in 2008. J Hepatol 48 Suppl 2008; 1: S20–37. [DOI] [PubMed] [Google Scholar]
- 14. Kaihara S, Kiuchi T, Ueda M et al . Living‐donor liver transplantation for hepatocellular carcinoma. Transplantation 2003; 75: S37–40. [DOI] [PubMed] [Google Scholar]
- 15. Takada Y, Ueda M, Ito T et al . Living donor liver transplantation as a second‐line therapeutic strategy for patients with hepatocellular carcinoma. Liver Transpl 2006; 12: 912–19. [DOI] [PubMed] [Google Scholar]
- 16. Inomata Y, Uemoto S, Asonuma K, Egawa H. Right lobe graft in living donor liver transplantation. Transplantation 2000; 69: 258–64. [DOI] [PubMed] [Google Scholar]
- 17. Ito T, Takada Y, Ueda M et al . Expansion of selection criteria for patients with hepatocellular carcinoma in living donor liver transplantation. Liver Transpl 2007; 13: 1637–44. [DOI] [PubMed] [Google Scholar]
- 18. Inomata Y, Tanaka K, Egawa H et al . The evolution of immunosuppression with FK506 in pediatric living‐related liver transplantation. Transplantation 1996; 61: 247–52. [DOI] [PubMed] [Google Scholar]
- 19. Hirohashi T, Ishak KG, Kojiro M et al . Hepatocellular carcinoma. In: Hamilton SR, Aaltonen LA (eds) World Health Organization Classification of Tumors. Pathology and Genetic Tumors of the Digestive System. Lyon: IAPS Press, 2000: 157–72. [Google Scholar]
- 20. Frederick L, Greene David L, Page Irvin DF et al . AJCC Cancer Staging Manual/American Joint Committee on Cancer . Cancer Staging Manual, 6th edn. New York: Springer‐Verlag, 2002: 131–44. [Google Scholar]
- 21. Lauwers GY, Terris B, Balis UJ et al . Prognostic histologic indicators of curatively resected hepatocellular carcinomas: a multi‐institutional analysis of 425 patients with definition of a histologic prognostic index. Am J Surg Pathol 2002; 26: 25–34. [DOI] [PubMed] [Google Scholar]
- 22. Finkelstein SD, Marsh W, Demetris AJ et al . Microdissection‐based allelotyping discriminates de novo tumor from intrahepatic spread in hepatocellular carcinoma. Hepatology 2003; 37: 871–9. [DOI] [PubMed] [Google Scholar]
- 23. Heinmoller E, Liu Q, Sun Y et al . Toward efficient analysis of mutations in single cells from ethanol‐fixed, paraffin‐embedded, and immunohistochemically stained tissues. Laboratory Invest 2002; 82: 443–53. [DOI] [PubMed] [Google Scholar]
- 24. Sasatomi E, Finkelstein SD, Woods JD et al . Comparison of accumulated allele loss between primary tumor and lymph node metastasis in stage II non‐small cell lung carcinoma: implications for the timing of lymph node metastasis and prognostic value. Cancer Res 2002; 62: 2681–9. [PubMed] [Google Scholar]
- 25. Lee JY, Finkelstein S, Hamilton RL, Rekha R, King JT Jr, Omalu B. Loss of heterozygosity analysis of benign, atypical, and anaplastic meningiomas. Neurosurgery 2004; 55: 1163–73. [DOI] [PubMed] [Google Scholar]
- 26. Khalid A, Pal R, Sasatomi E et al . Use of microsatellite marker loss of heterozygosity in accurate diagnosis of pancreaticobiliary malignancy from brush cytology samples. Gut 2004; 53: 1860–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheung TH, Lo KW, Yim SF et al . Clinicopathologic significance of loss of heterozygosity on chromosome 1 in cervical cancer. Gynecol Oncol 2005; 96: 510–15. [DOI] [PubMed] [Google Scholar]
- 28. Akaike H. A new look at the statistical model identification. IEEE Trans Automat Cotr 1974; AC‐19: 716–23. [Google Scholar]
- 29. R Development Core Team . A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Cited 30 April 2008] Available from URL: http://www.R‐project.org. [Google Scholar]
- 30. Minagawa M, Ikai I, Matsuyama Y, Yamaoka Y, Makuuchi M. Staging of hepatocellular carcinoma: assessment of the Japanese TNM and AJCC/UICC TNM systems in a cohort of 13 772 patients in Japan. Ann Surg 2007; 245: 909–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Varotti G, Ramacciato G, Ercolani G et al . Comparison between the fifth and 6th edns of the AJCC/UICC TNM staging systems for hepatocellular carcinoma: multicentric study on 393 cirrhotic resected patients. Eur J Surg Oncol 2005, 31, 760–7. [DOI] [PubMed] [Google Scholar]
- 32. Nagai H, Pineau P, Tiollais P, Buendia MA, Dejean A. Comprehensive allelotyping of human hepatocellular carcinoma. Oncogene 1997; 14: 2927–33. [DOI] [PubMed] [Google Scholar]
- 33. Rodriguez Luna H, Vargas HE, Byrne T, Rakela J. Artificial neural network and tissue genotyping of hepatocellular carcinoma in liver‐transplant recipients: prediction of recurrence. Transplantation 2005; 79: 1737–40. [DOI] [PubMed] [Google Scholar]
- 34. Fujimori M, Tokino T, Hino O et al . Allelotype study of primary hepatocellular carcinoma. Cancer Res 1991; 51: 89–93. [PubMed] [Google Scholar]
- 35. Piao Z, Park C, Park JH, Kim H. Allelotype analysis of hepatocellular carcinoma. Int J Cancer 1998; 75: 29–33. [DOI] [PubMed] [Google Scholar]
- 36. Kusano N, Shiraishi K, Kubo K, Oga A, Okita K, Sasaki K. Genetic aberrations detected by comparative genomic hybridization in hepatocellular carcinomas: their relationship to clinicopathological features. Hepatology 1999; 29: 1858–62. [DOI] [PubMed] [Google Scholar]
- 37. Vivanco I, Sawyers CL. The phosphatidylinositol 3‐Kinase AKT pathway in human cancer. Nat Rev Cancer 2002; 2: 489–501. [DOI] [PubMed] [Google Scholar]
- 38. Zhang JF, Liu JJ, Lu MQ et al . Rapamycin inhibits cell growth by induction of apoptosis on hepatocellular carcinoma cells in vitro. Transpl Immunol 2007; 17: 162–8. [DOI] [PubMed] [Google Scholar]
- 39. Kawamura N, Nagai H, Bando K et al . PTEN/MMAC1 mutations in hepatocellular carcinomas: somatic inactivation of both alleles in tumors. Jpn J Cancer Res 1999; 90: 413–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pawlik TM, Delman KA, Vauthey JN et al . Tumor size predicts vascular invasion and histologic grade: Implications for selection of surgical treatment for hepatocellular carcinoma. Liver Transpl 2005; 11: 1086–92. [DOI] [PubMed] [Google Scholar]
- 41. Todo S, Furukawa H. Living donor liver transplantation for adult patients with hepatocellular carcinoma: experience in Japan. Ann Surg 2004; 240: 451–9, discussion. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mitsunobu M, Toyosaka A, Oriyama T, Okamoto E, Nakao N. Intrahepatic metastases in hepatocellular carcinoma: the role of the portal vein as an efferent vessel. Clin Exp Metastasis 1996; 14: 520–9. [DOI] [PubMed] [Google Scholar]


