Abstract
PCDH10 is a member of the protocadherin cell adhesion molecule family, which are frequently downregulated in cancers. This study aimed to characterize the methylation silencing of the PCDH10 gene in the full spectrum of cervical carcinogenesis and to clarify if a field effect of methylation might be a target for a diagnostic test from cervical scrapings. Methylation silencing of PCDH10 was found in four of five cervical cancers and one of two cervical precancerous cell lines, which could be reversed by demethylation treatment. The same methylation was detected in 85.7% (24/28) of invasive cancer tissues, 36.4% (4/11) of high‐grade squamous intraepithelial lesions, 20% (1/5) of low‐grade squamous intraepithelial lesions, and none (0/17) of the normal cervical tissues from non‐cancer subjects. In addition, methylation was also frequently found in histologically ‘normal’ cervical tissues adjacent to cancer lesions (7/13, 53.8%) and, less frequently, in vaginal and endometrial tissues (1/8, 12.5%). Further investigation of cervical scrapings revealed cancer‐specific methylation of PCDH10 with a methylation rate of 71% (22/31) in invasive cancer, 27.9% (12/43) in carcinoma in situ, and none in high‐grade squamous intraepithelial lesions excluding carcinoma in situ (n = 12), low‐grade squamous intraepithelial lesions (n = 27), and normal controls (n = 66) (P < 10−16). Compared to the high‐risk human papilloma virus test, PCDH10 methylation testing of cervical scrapings was more specific (92 vs 60%) but less sensitive (71 vs 96%) in detecting invasive cervical cancer. This study demonstrated field methylation of the PCDH10 gene specifically in the invasion stage of cervical carcinogenesis, which might be used to develop a highly specific diagnostic test for cervical scrapings. (Cancer Sci 2009)
Cervical cancer is one of the most common malignancies among women worldwide.( 1 ) High‐risk human papilloma virus (HPV) was detected in virtually all cervical cancer tissues and regarded as a necessary etiological cause of cervical cancer.( 2 ) However, HPV infection is clearly not a sufficient factor in cervical carcinogenesis as most HPV infections are transient and benign. Other genetic and/or epigenetic alterations are also critical in cervical carcinogenesis.
Members of the cadherin family function as tumor suppressors through maintenance of cell–cell adhesion and/or inhibition of the Wnt pathway by interaction with cytoplasmic β‐catenin.( 3 , 4 , 5 ) Tumor suppressor genes are frequently downregulated by DNA methylation in tumorigenesis, including cervical carcinogenesis.( 6 , 7 , 8 ) Genes of the cadherin family have been found to be downregulated in human malignancies, resulting in tumor progression, invasion, and metastasis.( 3 , 9 ) Protocadherin 10 (PCDH10), also known as OL‐protocadherin, is a newly identified protocadherin of the cadherin family and contains six extracellular repeats.( 10 ) PCDH10 is essential for brain development, especially in elongating striatal axons and guiding thalamocortical projections.( 11 ) The gene locus of PCDH10 at 4q28.3 is frequently deleted in hepatoma, colorectal cancer, prostate cancer, pancreatic cancer, and breast cancer.( 12 , 13 , 14 , 15 , 16 ) A recent study demonstrated a tumor suppressor role of PCDH10 with inhibition of the growth, migration, and invasion of nasopharyngeal cancer cells upon overexpression.( 17 ) Hypermethylation of the PCDH10 gene was found in nasopharyngeal, haematological, breast, and gastric cancers as well as in several other cancer cell lines.( 17 , 18 , 19 , 20 )
To clarify the clinical scenario (the specific step in carcinogenesis in which the PCDH10 gene is silenced by methylation), the present study surveyed the status of PCDH10 methylation in the full spectrum of cervical carcinogenesis, including low‐grade squamous intra‐epithelial lesion (LSIL or mild dysplasia) and high‐grade squamous intra‐epithelial lesion (HSIL) excluding carcinoma in situ (CIS) (moderate dysplasia and severe dysplasia), CIS, squamous cell carcinoma (SCC), and adenocarcinoma (AdenoCA). To know the extent of the methylation effect on the neoplastic cervix, we also tested the normal counterparts of the uterine cervix in three groups: (1) normal cervical tissue from the non‐cancer patient (normal); (2) normal tissues remote to the cervical cancer lesion (NCx); and (3) normal tissue adjacent to the cervical cancer lesion (NNC). The results showed highly specific field methylation of PCDH10 at the in situ‐to‐invasion stage of cervical carcinogenesis.
Materials and Methods
Cell culture and DNA demethylation. Sixteen cell lines including five cervical cancer cells (SiHa, CaSki, HeLa, HeLaS3, C33A), two HPV16‐ and HPV18‐immortalized cervical epithelial cells displaying features of CIS (Z172 and Z183, respectively),( 21 ) one non‐HPV‐immortalized keratinocyte (HaCaT), two breast cancer cell lines (MCF7 and MDA‐MB 231), three lung cancer cell liness (A549, CL1‐0, and CL1‐5), two ovarian cancer cell lines (OVCAR3 and SKOV3), and one melanoma cell line (MDA‐MB 435) were studied. HeLa, HeLaS3, HaCaT, Z172, Z183, MDA‐MB 435, and breast cancer cells were cultured in DMEM medium. SiHa, CaSki, OVCAR3, SKOV3, and lung cancer cells were cultured in RPMI medium. C33A cells were cultured in MEM medium. Z172 and Z183 cells were supplemented with 10% Nu serum and 1% hydrocortisone, OVCAR3 cells with 20% FCS, and the rest of the cells with 10% FCS. For demethylation, cells were treated with 10 µM 5′‐aza‐2′‐deoxycytidine. The media containing 5′‐aza‐2′‐deoxycytidine was changed every 24 h for 4 days, and cells were harvested at day 5.
Study patients and controls. A hospital‐based cohort of cervical carcinogenesis was established at the Buddhist Tzu‐Chi General Hospital, Hualien, Taiwan from December 2005 to April 2009. Women with newly found abnormalities in Pap smears were invited. Exclusion criteria were acute or chronic non‐HPV infections, immune‐compromised status, pregnancy, history of previous cervical neoplasia, genital warts, other cancers, and previous surgeries of the uterine cervix. A standard guideline issued by National Health Research Institute, Taiwan, for screening, diagnosis, and management of cervical neoplasia (http://www.nhri.org.tw/NHRI_ADM/userfiles/file/tcog/gog_b.pdf) was followed in all subjects. At enrollment, all subjects were subjected to cervical scrapings, colposcopy with necessary biopsies, and venous blood collection. Two normal cervix‐control groups were recruited. Healthy women received a regular Pap smear with a ‘negative for cervical neoplasia’ result, and patients underwent hysterectomy on account of benign tumors of the uterine corpus with histologically proven normal cervix being categorized into ‘normal cervix from non‐cancer women (normal)’. Patients with gynecological cancers of a non‐cervical origin and a histologically proven normal cervix served as controls of ‘normal cervix from cancer patient’.
Clinical specimens. Surgical tissues were carefully procured with respect to their anatomical locations and the nature of the tumor lesions to prevent cross‐contamination. Primary and metastatic cervical tumors with different severities were procured during the operation. Grossly normal cervical tissues from gynecological malignancies of non‐cervix sites (NC) were also procured. Normal tissues with a different topological relationship to the cervical cancer lesions were procured. They included NNC and NCx, such as those from the vagina and endometrium. Tissue specimens were collected in multiple tubes by snap‐freezing in liquid nitrogen. The mean time of exposure of these specimens at room temperature was 19.9 ± 2 min. Cervical scrapings were collected with a cytobrush before colposcopy, cervical biopsy, or other cervical procedures. Typically, the same swab of cervix was collected after applying the Papanicolaou smear. Residual material on the swab was transferred into a universal tube containing 3 mL PBS. Following thorough agitation, it was then dispensed, snap‐frozen, and stored at –80°C. Overall, the study materials included: (1) cervical swabs of 27 SCC, four AdenoCA, 57 HSIL, 27 LSIL, and 66 normal controls; and (2) tumor tissues of 22 SCC, six AdenoCA, 11 HSIL, five LSIL, five metastatic lesions, and normal control tissues of 11 NN, 16 NNC, six NC, and eight NCx. The disease nature of the swabs and tissues were proved by cytology and histology, respectively. All specimens were procured under an Institutional Review Board‐approved project of Research Tissue Bank of Buddhist Tzu‐Chi General Hospital. This study was also approved by the Institutional Review Board with informed consent obtained from all subjects.
RT‐PCR. Total RNA was extracted using TRIzol Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. RNA was reverse‐transcribed to cDNA using the First Strand cDNA Synthesis kit (Roche, Mannheim, Germany). PCR was carried out with primers located at exon 1 (5′‐ACTGCTATCAGGTATGCCTG‐3′) and exon 2 (5′‐GTCTGT CAACTAGATAGCTG‐3′) of the PCDH10 gene.( 17 ) The β‐actin gene was amplified as an internal control, with primers 5′‐AACTCCATCATGAAGTGTGACG‐3′ and 5′‐GATCCACATCT GCTGGAAGG‐3′. PCR was carried out with initial denaturing at 95°C for 10 min, followed by 35 cycles of denaturing at 95°C for 1 min, annealing at 58°C for 1 min, and extension at 72°C for 1 min, with a final extension at 72°C for 7 min.
Methylation‐specific PCR and bisulfite genomic sequencing. Genomic DNA was extracted from cervical scrapings and tissues using the DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Genomic DNA was modified with sodium bisulfite using the DNA Methylation Gold Kit (Zymo Research, Orange, CA, USA), which converted unmethylated cytosines to uracil while leaving methylated cytosines unchanged. Methylation‐specific PCR (MS‐PCR) designed by Ying et al.( 17 ) was used to detect CpG methylation of the PCDH10 gene, with methylation‐specific primers of 5′‐TCGTTAAATAG ATACGTTACGC‐3′ and 5′‐TAAAAACTAAAAACTTTCCGCG‐3′, and unmethylation‐specific primers of 5′‐GTTGTTAAATAGAT ATGTTATGT‐3′ and 5′‐CTAAAAACTAAAAACTTTCCACA‐3′. White blood cell (WBC) DNA treated with SssI (New England Biolab) was used as the methylated control, and untreated WBC DNA was used as the unmethylated control. All of the single MS‐PCR contained 10 ng bisulfate‐converted DNA template. The MS‐PCR products were visualized on a 2% agarose gel containing ethidium bromide and illuminated under UV light. For bisulfite genomic sequencing (BGS), the promoter region from –328 to +18 containing 27 CpG sites was PCR amplified with primers of 5′‐GTTGATGTAAATAGGGGAATT‐3′ and 5′‐CTTCAACCTCTAAACCTATAA‐3′. The PCR products of the bisulfate‐converted DNA were cloned and sequences of five randomly picked clones were determined.
HPV detection and typing. The presence of HPV DNA and the genotypes were determined by a highly sensitive assay of consensus PCR and reverse blot hybridization as previously described.( 22 , 23 ) Briefly, the MY11 (5′−ΓΧΜΧΑΓΓΓΩΧΑΤΑΑΨΑΑΤΓΓ−3′) and biotinylated GP6+(5′‐GAAAAAT AAACTGT AAATC AT ATTC‐3′) consensus primers were used to amplify a fragment of approximately 192 bp in the L1 open reading frame of the HPV genome for 40 cycles. The PCR products were then hybridized with an Easychip HPV Blot (King Car, I‐Lan, Taiwan), which included oligonucleotide probes of 39 different types of HPV (types 6, 11, 16, 18, 26, 31, 32, 33, 35, 37, 39, 42, 43, 44, 45, 51, 52, 53, 54, 55, 56, 58, 59, 61, 62, 66, 67, 68, 69, 70, 72, 74, 82, CP8061, CP8304, L1AE5, MM4, MM7, and MM8), and were visualized with biotinylated antibodies and alkaline phosphatase conjugation. The overall sensitivity of HPV detection of this assay was 1–50 copies of HPV genome equivalents, with no cross‐reactivity with amplicons of other HPV genotypes.( 24 )
Statistic analysis. Fisher's exact test was carried out using SPSS 15.0 (SPSS, Chicago, IL, USA).
Results
PCDH10 gene was methylation silenced in most cervical cancer cell lines. Among five cervical cancer cell lines (SiHa, CaSki, HeLa, HeLaS3, and C33A) and one immortalized keratinocyte (HaCaT) examined by RT‐PCR, only C33A expressed PCDH10. In two HPV‐immortalized human cervical epithelial cells displaying features of CIS (Z172 and Z183), only Z183 weakly expressed PCDH10. Also, PCDH10 seemed to be less expressed in the more invasive (MDA‐MB 231) than the less invasive (MCF7) breast cancer cells. It was also non‐expressed or weakly expressed in one of the three lung cancer cells, two ovarian cancer cells, and melanoma cells studied (Fig. 1a). After demethylation with 10 µM of 5′‐aza‐2′‐deoxycytidine, all four cervical cancer cell lines resumed expression of PCDH10 (Fig. 1b), with the methylation status confirmed by MS‐PCR (Fig. 1c). These results showed that PCDH10 was silenced by methylation in cervical cancer cells.
Figure 1.
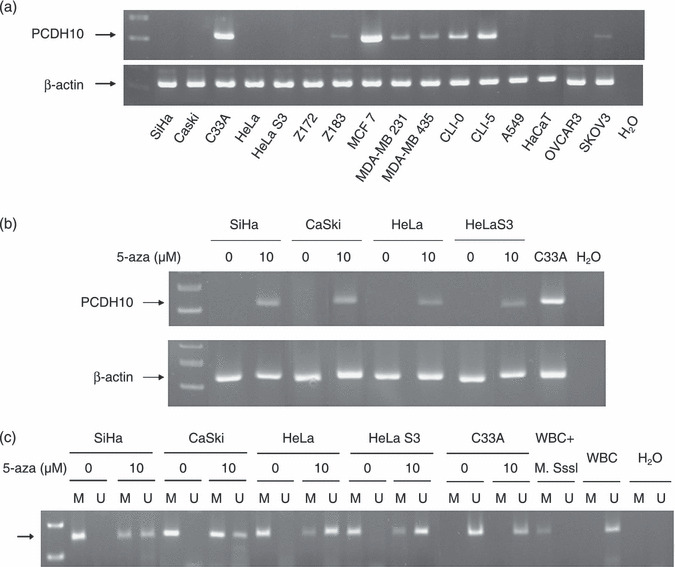
PCDH10 was silenced by hypermethylation in cervical cancer cells. (a) Among five cervical cancer cell lines (SiHa, CaSki, HeLa, HeLa3, and C33A), two human papillomavirus (HPV)‐immortalized human cervical epithelial cell lines (Z172 and Z183), and one immortalized keratinocyte (HaCaT) were examined by RT‐PCR. Only C33A expressed a significant amount of PCDH10. PCDH10 was weakly expressed or non‐expressed in the more invasive breast cancer cell line (MDA‐MB 231), melanoma cells (MDA‐MB 435), one of the three lung cancer cell lines (A549), and two ovarian cancer cell lines (OVCAR3 and SKOV3). (b) After treatment with 10 µM 5‐aza‐dC, PCDH10 was re‐expressed in SiHa, Caski, HeLa, and HeLaS3 cells. (c) Methylation‐specific‐PCR (MS‐PCR) confirmed the demethylation treatment. In contrast to little unmethylated (U) PCDH10 before treatment, both methylated (M) and U forms were found in these four cancer cells after demethylation treatment. Untreated white blood cell (WBC) DNA and H2O were used as negative controls. The arrow signified the expected location of the PCR product.
Methylation of PCDH10 specifically occurred in cervical cancer but not normal cervix. The methylation status of PCDH10 in tissue specimens including normal cervix, cervical dysplasia, CIS, and invasive carcinoma was examined. Methylation of PCDH10 was found in 85.7% (24/28) of invasive cervical cancers including 22 of 23 SCC and two of five AdenoCA, 80% (4/5) of metastatic cervical lesions, 36.4% (4/11) of CIS or moderate/severe dysplasia (including weakly positive in each of the moderate dysplasia and severe dysplasia and strong positive in 2 of 9 CIS), and 20% (1/5) of mild dysplasia, but not in 17 normal cervical tissues from normal control subjects (Fig. 2a; Table 1).
Figure 2.
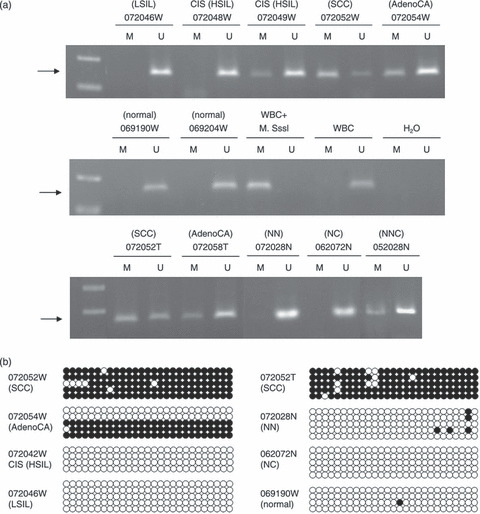
Methylation of PCDH10 promoter was closely related to cervical carcinogenesis. (a) Representive methylation‐specific‐PCR (MS‐PCR) results are shown indicating that only unmethylated (U) PCDH10 was noted in cervical scrapings from the normal control and patients with low‐grade squamous intra‐epithelial lesion (LSIL), but both U and methylated (M) PCDH10 were noted from patients with carcinoma in situ (CIS) (high‐grade squamous intra‐epithelial lesion; HSIL), squamous cell carcinoma (SCC), and adenocarcinoma (AdenoCA). White blood cell (WBC) treated with SssI methylase served as the methylation control, and untreated WBC served as the U control. (b) The detailed methylation status of the 27 CpG sites (indicated by circles) located at the promoter of the PCDH10 gene were determined in representative samples. Both cervical scrapings (W) and tissues (T) of SCC and AdenoCA showed extensive methylation of almost all CpG sites, with the proportion of methylation higher in SCC than AdenoCA. M and U CpG sites are shown by closed and open circles respectively. The arrow signified the expected location of the PCR product. NC, normal tissue from non‐cancer organ of cancer patient; NN, normal tissue from non‐cancer patient; NNC, normal tissue next to cervical cancer.
Table 1.
Methylation of PCDH10 in tissue specimens
| Age ± SE (years) | Methylation (%) | P † | |
|---|---|---|---|
| Normal | 50.8 ± 3.3 | 0/17 (0) | |
| NN | 0/11 (0) | ||
| NC | 0/6 (0) | ||
| NNC | 52.3 ± 3.3 | 7/13 (53.8) | |
| LSIL | 38.8 ± 8.0 | 1/5 (20) | |
| HSIL | 49.5 ± 4.1 | 4/11 (36.4) | |
| SCC/AdenoCA | 56.7 ± 2.3 | 24/28 (85.7) | |
| Metastatic CA | 53.4 ± 8.8 | 4/5 (80) | 1.48E‐08 |
Fisher's exact test. AdenoCA, adenocarcinoma; CA, carcinoma; HSIL, high‐grade squamous intra‐epithelial lesion; LSIL, low‐grade squamous intra‐epithelial lesion; NC, normal tissue from non‐cancer organ of cancer patient; NN, normal tissue from non‐cancer patient; NNC, normal tissue next to cervical cancer; SCC, squamous cell carcinoma.
Field effect of PCDH10 methylation in cervical cancer. Various topological controls of normal tissues from cervical or non‐cervical cancers were tested for the extent of field methylation and its specificity. Methylation of PCDH10 was found in 53.8% (7/13) of NNC but in only 12.5% (1/8) of NCx (Table 2), indicating the limit of the field methylation. Methylation of PCDH10 was not found in six NC (Table 2).
Table 2.
Topological controls used to test the field effect of PCDH10 methylation
| ID | Primary cancer | Topological control specimens | ||||
|---|---|---|---|---|---|---|
| Diagnosis | Site | Methylation | Procured site | Type | Methylation | |
| 052028 | SCC of cervix, stage 2a | Cervix | Positive | Vagina | MC | Negative |
| 062037 | SCC of cervix, stage 4b | Cervix | Positive † | Vagina | MC | Positive |
| 062020 | Adenocarcinoma of cervix, stage 2a | Cervix | Positive | Pelvic LN | MC | Positive |
| 062043 | SCC of cervix, stage 3a at least | Cervix | Negative † | Vagina | MC | Positive |
| 062049 | Recurrent SCC of cervix, with lymph node metastasis | Cervix | N/A | Common iliac LN | MC | Positive |
| 052014 | SCC of cervix, stage 2a | Cervix | Positive | Cervix | NNC | Positive |
| 052028 | SCC of cervix, stage 2a | Cervix | Positive | Cervix | NNC | Positive |
| Vagina | NNC | Positive | ||||
| 062011 | SCC of cervix, stage 2a | Cervix | Positive | Cervix | NNC | Positive |
| 062045 | SCC of cervix, stage 2a | Cervix | Positive | Cervix | NNC | Positive |
| Vagina | NNC | Positive | ||||
| 072052 | SCC of cervix, stage 1b | Cervix | Positive | Cervix | NNC | Positive |
| 072094 | SCC of cervix, stage 1b2 | Cervix | Positive | Cervix | NNC | Positive |
| Stroma under SCC | NNC | Positive | ||||
| 062048 | SCC of cervix, stage 2a | Cervix | Positive | Cervix | NNC | Negative |
| 072058 | Adenocarcinoma of cervix, stage 1b | Cervix | Positive | Cervix | NNC | Negative |
| 082012 | Villoglandular adenocarcinoma of endocervix | Cervix | Negative † | Cervix | NNC | Negative |
| 072081 | CIS of cervix | Cervix | Negative † | Cervix | NNC | Positive |
| 082016 | CIS of cervix | Cervix | Negative † | Cervix | NNC | Negative |
| 082071 | Poorly differentiated adenosquamous Ca of cervix, stage 1b1 | Cervix | Negative | Vagina | NNC | Negative |
| 082085 | SCC of cervix, stage 1a | Cervix | Negative | Cervix | NNC | Negative |
| 062063 | Adenocarcinoma of endometrium, stage 3c | Endometrium | Positive | Cervix | NC | Negative |
| 062072 | Adenocarcinoma of endometrium, stage 1b | Endometrium | Negative | Cervix | NC | Negative |
| 062091 | Endometroid Ca of oviduct, stage 2 | Fallopian tube | Negative | Cervix | NC | Negative |
| 072098 | Adenocarcinoma of endometrium, stage 1a | Endometrium | Negative | Cervix | NC | Negative |
| 082031 | Adenocarcinoma of endometrium, stage 1a | Endometrium | Negative | Cervix | NC | Negative |
| 082069 | Serous cyst adenocarcinoma of ovary, stabe 3c | Ovary | Negative | Cervix | NC | Negative |
| 062048 | SCC of cervix, stage 2a | Cervix | Positive | Endometrium | NCx | Positive |
| 072094 | SCC of cervix, stage 1b2 | Cervix | Positive | Ovary | NCx | Negative |
| Pelvic LN | NCx | Negative | ||||
| 082001 | Adenocarcinoma of cervix, stage 1b2 | Cervix | Negative | Common iliac LN | NCx | Negative |
| 082012 | Villoglandular adenocarcinoma of endocervix | Cervix | Negative † | Endometrium | NCx | Negative |
| 082016 | CIS of cervix | Cervix | Negative † | Endometrium | NCx | Negative |
| 082065 | Neuroendocrine differentiated carcinoma of cervix, stage 4b | Cervix | Negative | Vagina | NCx | Negative |
| Uterus | NCx | Negative | ||||
Data were collected from swab specimens. Ca, carcinoma; LN, lymph node; MC, metastatic cancer; N/A, not available; NC, normal cervix from cancer patient; NCx, non‐cervical tissue from cervical cancer patient; NNC, normal tissue next to cancer; SCC, squamous cell carcinoma.
Methylation of PCDH10 in cervical scrapings was more specific than high‐risk HPV DNA test. The field effect of PCDH10 methylation was anticipated to develop a feasible test for cervical carcinogenesis in cervical scraping specimens. Indeed, methylation of the PCDH10 gene was noted in cervical swabs of 71% (22/31) of SCC or AdenoCA (including 20 of 27 SCC, and two of four AdenoCA), 27.9% (12/43) of CIS, and 0 of 12 CIN2 or 3, 27 LSIL, and 66 normal (Table 3). The specific methylation of PCDH10 in cervical cancer was further confirmed by BGS, where the methylation status of the 27 CpG sites around the promoter was determined in representative specimens. Extensive methylation of almost all CpG sites were noted in SCC and AdenoCA, but not seen in CIS, LSIL, and normal (Fig. 2b). On the other hand, high‐risk HPV DNA was detected in 21 of 22 (95.4%) SCC or AdenoCA, 31 of 31 (100%) HSIL, 9 of 12 (75%) LSIL, and 3 of 65 (4.6%) normal (Fig. 2a; Table 3). The sensitivity, specificity, positive predictive value, and negative predictive value of PCDH10 methylation in the diagnosis of SCC or AdenoCA of the cervix in this hospital‐based cohort was 71, 91.9, 64.7, and 93.8% respectively. On the other hand, the performances of the HPV test were 95.5, 60.2, 32.8, and 98.5%, respectively (Table 4).
Table 3.
Methylation of PCDH10 and human papilloma virus (HPV) typing in cervical swab specimens
| Age ± SE (years) | Methylation (%) | P † | Age ± SE (years) | High‐risk HPV (%) | P † | |
|---|---|---|---|---|---|---|
| Normal | 53.2 ± 1.8 | 0/66 (0) | 53.2 ± 1.8 | 3/65 (4.6) | ||
| LSIL | 46.8 ± 2.1 | 0/27 (0) | 49.2 ± 2.2 | 9/12 (75) | ||
| HSIL exclude CIS | 46.6 ± 5.4 | 0/12 (0) | 52.9 ± 6.7 | 8/8 (100) | ||
| CIS | 49.4 ± 2.4 | 12/43 (27.9) | 52.4 ± 3.6 | 23/23 (100) | ||
| SCC/AdenoCA | 58.9 ± 2.4 | 22/31 (71.0) | 2.31E‐17 | 57.1 ± 2.9 | 21/22 (95.5) | 6.02E‐30 |
Fisher's exact test. AdenoCA, adenocarcinoma; CIS, carcinoma in situ; HSIL, high‐grade squamous intra‐epithelial lesion; LSIL, low‐grade squamous intra‐epithelial lesion; SCC, squamous cell carcinoma.
Table 4.
Comparison of sensitivity, specificity, positive, and negative predictive value in a hospital‐based study
| Methylation of PCDH10 | High‐risk HPV | |||||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity % | Specificity % | PPV % | NPV % | Sensitivity % | Specificity % | PPV % | NPV % | |
| Normal/LSIL | ||||||||
| HSIL/SCC/AdenoCA | 39.5 | 100 | 100 | 64.1 | 98.1 | 84.4 | 81.3 | 98.5 |
| Normal/SIL exclude CIS | ||||||||
| CIS/SCC/AdenoCA | 45.9 | 100 | 100 | 72.4 | 97.8 | 76.5 | 68.8 | 98.5 |
| Normal/SIL | ||||||||
| SCC/AdenoCA | 71 | 91.9 | 64.7 | 93.8 | 95.5 | 60.2 | 32.8 | 98.5 |
AdenoCA, adenocarcinoma; CIS, carcinoma in situ; HPV, human papilloma virus; HSIL, high‐grade squamous intra‐epithelial lesion; LSIL, low‐grade squamous intra‐epithelial lesion; NPV, negative predictive value; PPV, positive predictive value; SCC, squamous cell carcinoma; SIL, squamous intraepithelial lesion.
Discussion
PCDH10 was shown to be an important tumor suppressor gene, inhibiting the growth, migration, and invasion of nasopharyngeal cancer cells.( 17 ) Promoter methylation and transcription silencing of PCDH10 were frequently found in nasopharyngeal, esophageal, breast, colorectal, cervical, lung, and hepatocellular carcinoma cell lines.( 17 ) Our study confirmed the methylation silencing of PCDH10 in cervical cancer cell lines, and disclosed frequent methylation of PCDH10 in cervical cancer lesions but not the normal controls. Although the chromosomal locus 4q28.3, where PCDH10 resides, was reported to be frequently deleted in many cancers,( 12 , 13 , 14 , 15 , 16 ) we did not find alleleic deletion in microsatellite markers adjacent to the PCDH10 gene (Supporting information Fig. 1).
The present study surveyed clinical specimens across the full spectrum of cervical carcinogenesis, from normal cervix, LSIL, HSIL‐CIN2 or 3, HSIL‐CIS to invasive carcinoma. Methylation of PCDH10 was only found in in situ and invasive carcinoma (Fig. 2; Table 1), indicating that methylation silencing occurs predominantly at the CIS stage of carcinogenesis. In addition to PCDH10, E‐cadherin was downregulated at early invasion of breast and prostate cancers.( 24 , 25 ) These results suggest that cancer cells may evolve agglomerative silencing of cadherin genes by methylation( 26 ) to acquire plasticity and mobility before invasion.
Methylation of PCDH10 was found in the cancer tissues as well as the adjacent, histologically normal cervical tissue, but not in remote tissues in the vagina or endometrium. This suggests a field effect of hypermethylation occurring before the invasion of cervical cancer. It takes at least 10 years for the HSIL to become invasive cervical cancer. Long‐term epigenetic modification of the in situ cancer may be required for cancer cells to acquire the plasticity or a state of epithelial–mesenchymal transition for invasion, presumably by dedifferentiation into a more stemness state.( 27 ) Our study indicates that this epigenetic effect seems not to be confined to cancer cells, rather they may act in a field, most probably through interaction with the underlying stroma.( 28 )
The field hypermethylation of PCDH10 was made feasible to develop a sensitive and specific test using cervical scrapings to detect cervical cancer. In the present study, a single PCR test of cervical scraping for PCDH10 methylation achieved a sensitivity of 71% and specificity of 91.9% in identifying invasive cervical cancer, which was better than any current diagnostic test in practice, including Pap smear and HPV test. In a head‐to‐head comparison with a highly performed high‐risk HPV DNA chip test on the same cervical scrapings, the methylation test was much more specific than the HPV test. Due to its high prevalence in normal and LSIL women, testing of high‐risk HPV has a low (32.8%) positive predictive value in diagnosing cervical cancer, which can be even lower when conducted in a general population. In addition, our study also showed that cancers of adjacent organs, that is, the endometrium, rarely reveal field methylation of PCDH10 in normal‐appearing uterine cervix, thus adding to the specificity of the test for the origin of cancer. This novel methylation marker may thus be used in screening for cervical cancer and the triage of ambiguous or low‐grade smear results, with more certainty of positivity than the HPV test.
Supporting information
Fig. S1. Absence of loss of heterozygosity (LOH) of microsatellite marker adjacent to the PCDH10 gene.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
References
- 1. Parkin DM. The global health burden of infection‐associated cancers in the year 2002. Int J Cancer 2006; 118: 3030–44. [DOI] [PubMed] [Google Scholar]
- 2. Walboomers JM, Jacobs MV, Manos MM et al . Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 1999; 189: 12–19. [DOI] [PubMed] [Google Scholar]
- 3. Jeanes A, Gottardi CJ, Yap AS. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 2008; 27: 6920–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel SD, Chen CP, Bahna F, Honig B, Shapiro L. Cadherin‐mediated cell–cell adhesion: sticking together as a family. Curr Opin Struct Biol 2003; 13: 690–8. [DOI] [PubMed] [Google Scholar]
- 5. Wheelock MJ, Johnson KR. Cadherin‐mediated cellular signaling. Curr Opin Cell Biol 2003; 15: 509–14. [DOI] [PubMed] [Google Scholar]
- 6. Duenas‐Gonzalez A, Lizano M, Candelaria M, Cetina L, Arce C, Cervera E. Epigenetics of cervical cancer. An overview and therapeutic perspectives. Mol Cancer 2005; 4: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3: 415–28. [DOI] [PubMed] [Google Scholar]
- 8. Knudson AG. Two genetic hits (more or less) to cancer. Nat Rev Cancer 2001; 1: 157–62. [DOI] [PubMed] [Google Scholar]
- 9. Stemmler MP. Cadherins in development and cancer. Mol Biosyst 2008; 4: 835–50. [DOI] [PubMed] [Google Scholar]
- 10. Wolverton T, Lalande M. Identification and characterization of three members of a novel subclass of protocadherins. Genomics 2001; 76: 66–72. [DOI] [PubMed] [Google Scholar]
- 11. Uemura M, Nakao S, Suzuki ST, Takeichi M, Hirano S. OL‐Protocadherin is essential for growth of striatal axons and thalamocortical projections. Nat Neurosci 2007; 10: 1151–9. [DOI] [PubMed] [Google Scholar]
- 12. Hammond C, Jeffers L, Carr BI, Simon D. Multiple genetic alterations, 4q28, a new suppressor region, and potential gender differences in human hepatocellular carcinoma. Hepatology 1999; 29: 1479–85. [DOI] [PubMed] [Google Scholar]
- 13. Knosel T, Schluns K, Stein U et al . Chromosomal alterations during lymphatic and liver metastasis formation of colorectal cancer. Neoplasia 2004; 6: 23–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Matsui S, LaDuca J, Rossi MR, Nowak NJ, Cowell JK. Molecular characterization of a consistent 4.5‐megabase deletion at 4q28 in prostate cancer cells. Cancer Genet Cytogenet 2005; 159: 18–26. [DOI] [PubMed] [Google Scholar]
- 15. Nowak NJ, Gaile D, Conroy JM et al . Genome‐wide aberrations in pancreatic adenocarcinoma. Cancer Genet Cytogenet 2005; 161: 36–50. [DOI] [PubMed] [Google Scholar]
- 16. Price GR, Armes JE, Ramus SJ et al . Phenotype‐directed analysis of genotype in early‐onset, familial breast cancers. Pathology 2006; 38: 520–7. [DOI] [PubMed] [Google Scholar]
- 17. Ying J, Li H, Seng TJ et al . Functional epigenetics identifies a protocadherin PCDH10 as a candidate tumor suppressor for nasopharyngeal, esophageal and multiple other carcinomas with frequent methylation. Oncogene 2006; 25: 1070–80. [DOI] [PubMed] [Google Scholar]
- 18. Miyamoto K, Fukutomi T, Akashi‐Tanaka S et al . Identification of 20 genes aberrantly methylated in human breast cancers. Int J Cancer 2005; 116: 407–14. [DOI] [PubMed] [Google Scholar]
- 19. Ying J, Gao Z, Li H et al . Frequent epigenetic silencing of protocadherin 10 by methylation in multiple haematologic malignancies. Br J Haematol 2007; 136: 829–32. [DOI] [PubMed] [Google Scholar]
- 20. Yu J, Cheng YY, Tao Q et al . Methylation of protocadherin 10, a novel tumor suppressor, is associated with poor prognosis in patients with gastric cancer. Gastroenterology 2008; 136: 640–51. [DOI] [PubMed] [Google Scholar]
- 21. Pecoraro G, Lee M, Morgan D, Defendi V. Evolution of in vitro transformation and tumorigenesis of HPV16 and HPV18 immortalized primary cervical epithelial cells. Am J Pathol 1991; 138: 1–8. [PMC free article] [PubMed] [Google Scholar]
- 22. Huang SL, Chao A, Hsueh S et al . Comparison between the Hybrid Capture II Test and an SPF1/GP6 + PCR‐based assay for detection of human papillomavirus DNA in cervical swab samples. J Clin Microbiol 2006; 44: 1733–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lin CY, Chen HC, Lin RW et al . Quality assurance of genotyping array for detection and typing of human papillomavirus. J Virol Methods 2007; 140: 1–9. [DOI] [PubMed] [Google Scholar]
- 24. Berx G, Van Roy F. The E‐cadherin/catenin complex: an important gatekeeper in breast cancer tumorigenesis and malignant progression. Breast Cancer Res 2001; 3: 289–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Paul R, Ewing CM, Jarrard DF, Isaacs WB. The cadherin cell–cell adhesion pathway in prostate cancer progression. Br J Urol 1997; 79(Suppl 1): 37–43. [DOI] [PubMed] [Google Scholar]
- 26. Novak P, Jensen T, Oshiro MM, Watts GS, Kim CJ, Futscher BW. Agglomerative epigenetic aberrations are a common event in human breast cancer. Cancer Res 2008; 68: 8616–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Li F, Tiede B, Massague J, Kang Y. Beyond tumorigenesis: cancer stem cells in metastasis. Cell Res 2007; 17: 3–14. [DOI] [PubMed] [Google Scholar]
- 28. Kopfstein L, Christofori G. Metastasis: cell‐autonomous mechanisms versus contributions by the tumor microenvironment. Cell Mol Life Sci 2006; 63: 449–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Absence of loss of heterozygosity (LOH) of microsatellite marker adjacent to the PCDH10 gene.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


