Abstract
More than 10 G protein-coupled receptors (GPCRs) have been shown to act as coreceptors for infection of human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV). We have isolated HIV-1 variants infectious to primary brain-derived CD4-positive cells (BT-3 and BT-20/N) and U87/CD4 glioma cells that are resistant to T-cell line-tropic (T-tropic), macrophage-tropic (M-tropic), and T- and M-tropic (dualtropic) (X4, R5, and R5X4) HIV-1 strains. These primary brain-derived cells were also highly susceptible to HIV-2ROD, HIV-2SBL6669, and SIVmndGB-1. A factor or coreceptor that determines the susceptibility of these brain-derived cells to these HIV and SIV strains has not been fully identified. To identify this coreceptor, we examined amino acid sequences of all known HIV and SIV coreceptors and noticed that tyrosine residues are well conserved in their extracellular amino-terminal domains. By this criterion, we selected 18 GPCRs as candidates of coreceptors for HIV and SIV strains infectious to these brain-derived cells. mRNA expression of an orphan GPCR, RDC1, was detected in the brain-derived cells, the C8166 T-cell line, and peripheral blood lymphocytes, all of which are susceptible to HIV-1 variants, but not in macrophages, which are resistant to them. When a CD4-expressing cell line, NP-2/CD4, which shows strict resistance to infection not only with HIV-1 but also with HIV-2 or SIV, was transduced with the RDC1 gene, the cells became highly susceptible to HIV-2 and SIVmnd strains but to neither M- nor T-tropic HIV-1 strains. The cells also acquired a low susceptibility to the HIV-1 variants. These findings indicate that RDC1 is a novel coreceptor for several HIV-1, HIV-2, and SIV strains which infect brain-derived cells.
Human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus (SIV) enter target cells primarily in a CD4-dependent manner (12, 33). Several human or animal cell lines show resistance to infection with various HIV-1 strains, although the CD4 molecules are detected on the cell surface (7). These findings suggested that some factors other than the CD4 molecule are necessary for the HIV-1 infection process. Later, a CXC-chemokine receptor (CKR), CXCR4, was shown to mediate the fusion of the cells expressing the Env protein of T-cell line-tropic (T-tropic) HIV-1 strains, but not of macrophage tropic (M-tropic) HIV-1 strains, with CD4-positive cells (22).
CKRs, which belong to the G protein-coupled receptors (GPCRs), play important roles in the chemotaxis system in vivo in collaboration with their ligands, chemokines. GPCRs constitute a large family consisting of more than 100 receptor molecules, such as the receptors for chemoattractants, light, and peptide hormones (38). Four CC-CKRs (CCR2b, CCR3, CCR5, and CCR9) were shown to serve as specific coreceptors for M-tropic or primary T-tropic HIV-1 strains (1, 4, 5, 8, 9, 16, 22, 47, 49). Thus, several CKRs have abilities to act not only as coreceptors but also as determinants of the susceptibility of cells to HIV-1 strains. A cytomegalovirus-encoded CKR (US28), a CX3C-CKR (v28), and four orphan GPCRs (APJ, GPR1, GPR15, and STRL33) were also shown to act as coreceptors for several HIV-1, HIV-2, or SIV strains (8, 14, 19, 34, 35, 40).
We isolated three wild-type HIV-1 strains, GUN-1WT, GUN-4WT, and GUN-7WT, from peripheral blood lymphocytes (PBLs) of AIDS patients and found that they are T- and M-tropic (i.e., dualtropic) (R5-R3-X4) (46, 51, 52). From each viral preparation, variant strains GUN-1V, GUN-4V, and GUN-7V were isolated (46, 51, 52). These variants show another type of cell tropism: GUN-1V, GUN-4V, and GUN-7V strains infect both T-cell lines and brain-derived cells, such as the CD4-transduced glioma cell line U87/CD4, and BT-3 and BT-20/N cells, which are normal fibroblast-like cells probably originating in human brain blood vessels (51). T-tropic, M-tropic, and dualtropic HIV-1 strains (i.e., X4, R5, and X4R5 strains) cannot efficiently infect these brain-derived cells (51, 52). Recently, we found that a CC-CKR (CCR8/TER1) facilitates the infection of the dualtropic HIV-1 strains and the HIV-1 variants which are infectious to the brain-derived cells (31). However, a cell surface molecule that is expressed in the brain-derived cells and determines the susceptibility to the HIV-1 variants, HIV-2, or SIV infectious to the brain-derived cells has not been identified.
It has been shown that tyrosine residues are well conserved in the extracellular amino-terminal (N-terminal) domains of a few HIV/SIV coreceptors (8, 19, 20). Deletion of these tyrosines abolished the coreceptor function of CCR5 (20). Recently, it was shown that sulfation of these tyrosines in the N-terminal domain is critical for the coreceptor function of CCR5 (21). In this study, we show that mRNA for an orphan GPCR, RDC1, which also has tyrosines in the extracellular N-terminal domain, was detected in the human brain-derived cells, PBLs, and an established T-cell line and that it acted as a coreceptor for HIV-1, HIV-2, and SIV strains infectious to the brain-derived cells when it was transduced into HIV/SIV-resistant NP-2/CD4 cells.
MATERIALS AND METHODS
Cells.
A human T-cell line, C8166 (44), was mostly used to prepare the viral stocks of HIV-1, HIV-2, and SIV strains. NP-2/CD4, NP-2/RDC1, and NP-2/CD4/RDC1 cells were established by introducing CD4 and/or RDC1 genes into a human glioma cell line, NP-2 (31, 45), using retrovirus vectors as described elsewhere (31, 48). NP-2/CD4/CCR5 and NP-2/CD4/CXCR4 cells were also established by introducing the CCR5 or CXCR4 gene into NP-2/CD4 cells by using retrovirus vectors as described previously (31). These NP-2-derived cells and a CD4-transduced human glioma cell line, U87/CD4 (41, 51), were maintained in Eagle's minimum essential medium (Nissui Co., Ltd., Tokyo, Japan) supplemented with 10% fetal calf serum (FCS). C8166 cells were cultured in RPMI 1640 medium (Nissui) containing 10% FCS.
The brain-derived fibroblast-like BT-3 (51, 52) and BT-20/N (51, 52) cells, which have been thought to originate in brain blood vessels, were cultured in RPMI 1640 medium containing 10% FCS, 10 μg of endothelial cell growth supplement per ml, and 10 ng of epidermal growth factor per ml. BT-3 and BT-20/N cells were derived from surgically dissected human brain tissues of patients with meningioma and glioma, respectively. BT-3 and BT-20/N cells stop growing after 10 to 15 cell passages and have not shown any specifically differentiated cell markers so far tested (data not shown). Because monoclonal antibodies raised against BT-3 cells reacted with blood vessels, BT-3 and BT-20/N cells may be derived from the cells associated with blood vessels, although these brain-derived cells were negative for factor VIII-related antigens (endothelial cell marker) or glial fibrillary acid protein (astrocyte marker) (data not shown).
Macrophages were isolated from PBLs of healthy blood donors as described previously (36). Macrophages were cultured in RPMI 1640 medium containing 10% FCS and 10% human serum. PBLs were stimulated with phytohemagglutinin and cultured in RPMI 1640 medium containing 10% FCS and 100 U of recombinant interleukin-2 per ml.
Virus strains.
Three HIV-1 strains infectious to T-cell lines and macrophages (GUN-1WT [52], GUN-4WT [46], and GUN-7WT [46]), three HIV-1 variants infectious to the brain-derived cells as well as T-cell lines (GUN-1V [52], GUN-4V [46], and GUN-7V [46]), one HIV-1 strain infectious to T-cell lines (IIIB [54]), two M-tropic HIV-1 strains (SF162 [6] and BaL [55]), two HIV-2 strains (ROD [25] and SBL6669 [25]), and three SIV strains (mac251 [13], mndGB-1 [53], and agmTYO-1 [23]) were used. GUN-4WT and GUN-7WT strains are primary HIV-1 isolates because their passage numbers are less than 10. The culture supernatants of C8166 cells infected with HIV-1, HIV-2, or SIV strains were harvested as the viral stocks when many syncytia were detected microscopically. The M-tropic HIV-1 strains SF162 and BaL were propagated in human PBLs.
GPCRs and PCR primers.
Oligonucleotide primers (Nihon Idenshi Kenkyujo, Co., Ltd., Miyagi, Japan) were synthesized to detect the expression of various GPCRs RNA by reverse transcription (RT)-PCR. Names of GPCRs, nucleotide sequences of PCR primers to detect the cDNA of GPCRs, orientations of the primers, and positions of the primers in the open reading frame (ORF) of GPCRs are as follows: APJ, 5′-ATGGAGGAAGGTGGTGATTTTGACAACTAC-3′ (sense, positions 1 to 30) and 5′-CTAGTCAACCACAAGGGTCTCCTGGCTGTAG-3′ (antisense, positions 1113 to 1143) (DDBJ/Genbank/EMBL accession no. U03642); C5a receptor, 5′-ATGAACTCCTTCAATTATACCACCCCTGAT-3′ (sense, positions 1 to 30) and 5′-CTACACTGCCTGGGTCTTCTGGGCCATAGTG-3′ (antisense, positions 1023 to 1,053) (M62505); CCR1, 5′-ATGGAAACTCAAAACACCACAGAGGACTATG-3′ (sense, positions 1 to 31) and 5′-TCAGAACCCAGCAGAGAGTTCATGCTCCCCTG-3′ (antisense, positions 1037 to 1,068) (L09230); CCR4, 5′-ATGAACCCCACGGATATAGCAGATACCACC-3′ (sense, positions 1 to 30) and 5′-CGTCGCATTCGCGGCCGCCTACAGAGCATCATGAAG-3′ (antisense, positions 1066 to 1083) (X85740); CCR7, 5′-CGCGTCCTTCTCATCAGCAAGCTGTCCTGTG-3′ (sense, positions 508 to 538) and 5′-GTGCCGACAGGAAGACCACTGCCGGAGCTG-3′ (antisense, positions 1051 to 1080) (X84702); CCR9/CCR10, 5′-ATCCCTGATATGGTCTTTGTACAGACACATG-3′ (sense, positions 529 to 559) and 5′-GCTGGATAATGAGGCCTGGGCAGTGCCAGG-3′ (antisense, positions 1002 to 1038) (U94888); CXCR1, 5′-GTAGGAGGTAACACGATGACGTGCCAAGAA-3′ (antisense, positions 988 to 1017); CXCR2, 5′-ATGGAAGATTTTAACATGGAGAGTGACAGC-3′ (sense, positions 1 to 30) and 5′-AAAGGAAGGCCTGCTGTCTTTGGGCAGGGA-3′ (antisense, positions 1015 to 1044) (M99412); CXCR3, 5′-CGGGGGCCCCCGGCCCGCGTGACCCTCACCTG-3′ (sense, positions 481 to 515) and 5′-CTGGAGCCCTCTCTGGTTGGGGCAGCCCAGGC-3′ (antisense, positions 1004 to 1035) (X95876); CXCR4, 5′-CCAAGGAAGCTGTTGGCTGAAAAGGTGGTCTA-3′ (sense, positions 439 to 470) and 5′-TCCACCTCGCTTTCCTTTGGAGAGGATCTT-3′ (antisense, positions 979 to 1008) (X71635); CXCR5/BLR1, 5′-GGGACCATCTGGCTGGTGGGCTTCCTCCTTG-3′ (sense, positions 511 to 546) and 5′-GAGACTGCTCCTGCGCCAGCTAGGGAAGAG-3′ (antisense, positions 1051 to 1080) (X68149); DEZ α, 5′-ATGAGAATGGAGGATGAAGATTACAACACTTC-3′ (sense, positions 1 to 32) and 5′-TCAAAGCATGCCGGTCTCCCTCTCATTCATAG-3′ (antisense, positions 1091 to 1122) (U79526); DEZ β, 5′-ATGGAGGATGAAGATTACAACACTTCCATC-3′ (sense, positions 1 to 30) and 5′-TCAAAGCATGCCGGTCTCCCTCTCATTCATAG-3′ (antisense, positions 1085 to 1116) (U79526); Duffy antigen, 5′-ATGGCCTCCTCTGGGTATGTCCTCCAGGCGGAG-3′ (sense, positions 1 to 33) and 5′-CTAGGATTTGCTTCCAAGGGTGTCCAGATGAG-3′ (antisense, positions 986 to 1017) (U01839); GPR-9-6, 5′-ATGGCTGATGACTATGGCTCTGAATCCACATC-3′ (sense, positions 1 to 32) and 5′-TCAGAGGGAGAGTGCTCCTGAGGTTGTCTCC-3′ (antisense, positions 1044 to 1074) (U45982); GPR5, 5′-ATGGAGTCCTCAGGCAACCCAGAGAGCACC-3′ (sense, positions 1 to 30) and 5′-TCAGTAGAAGGAGGCGCCCTCATAGGCGAAG-3′ (antisense, positions 972 to 1002) (L36149); GPR12, 5′-ATGAATGAAGACCTGAAGGTCAATTTAAGCGG-3′ (sense, positions 1 to 32) and 5′-CTACACATCACTGGGCGAGCGCGCTCTCTGGG-3′ (antisense, positions 974 to 1005) (U18548); GPR15, 5′-ATGGACCCAGAAGAAACTTCAGTTTATTTG-3′ (sense, positions 1 to 30) and 5′-TTAGAGTGACACAGACCTCTTCCTCCTCCTGG-3′ (antisense, positions 1052 to 1083) (U34806); GPR25, 5′-ATGGCCCCCACAGAGCCCTGGAGCCCCAGCCC-3′ (sense, positions 1 to 32) and 5′-CTACCAGGAGGCCGAGGCAGTGTTCGCGGCC-3′ (antisense, positions 1053 to 1083) (U91939); and RDC1, 5′-AAGAAGATGGTACGCCGTGTCGTCTGCATCCTG-3′ (sense, positions 469 to 501) and 5′-CTGCTGTGCTTCTCCTGGTCACTGGACGCCGAG-3′ (antisense, positions 717 to 749) (M64749).
Detection of GPCR mRNA.
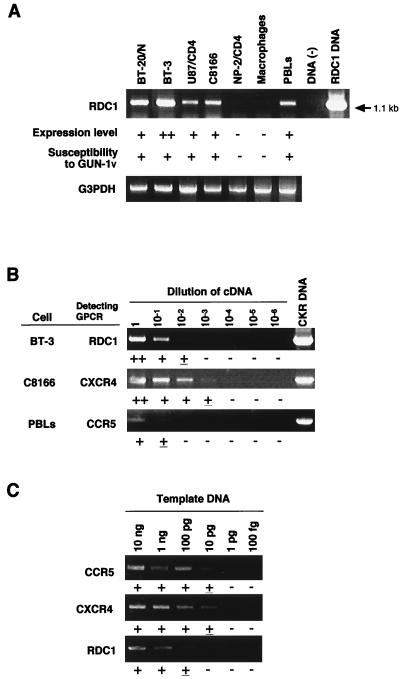
Total RNA was isolated from BT-3, BT-20/N, C8166, NP-2/CD4, and U87/CD4 cells as well as macrophages and PBLs by using an RNA extraction kit (SepaGene; Sanko-Junyaku Co., Ltd., Tokyo, Japan) in accordance with the manufacturer's protocol. cDNA of the total cellular RNA was constructed as described elsewhere (48). The GPCR sequences in the cDNA preparation were detected by PCR as described elsewhere (48), using the specific primers for each GPCR. As a control, the mRNA of glyceraldehyde-3-phosphate dehydrogenase was detected by RT-PCR. The efficiencies of PCR primer pairs to detect CCR5, CXCR4, and RDC1 RNA in cells were estimated by detection of serially diluted plasmid DNA containing each GPCR as PCR templates. The amplified cDNA was examined by 1% agarose gel electrophoresis.
Cloning of RDC1 gene.
The DNA fragment containing the ORF of the RDC1 gene was obtained by RT-PCR using RDC1-specific primers RDC1CN and RDC1CR. The cDNA was constructed from the total RNA which had been isolated from BT-3 cells. The ORF DNA of RDC1 was cloned into the EcoRV site of the expression plasmid pcDNA3 (Clontech Co., Palo Alto, Calif.), and the resultant plasmid was designated pcDNA3/RDC1. The DNA fragment containing the RDC1 ORF was separated from plasmid pcDNA3/RDC1 by digestion with the BamHI and NotI and subcloned into the expression plasmid pMX-puro. The RDC1 plasmid obtained was designated pMX-puro/RDC1. The cloned RDC1 gene was sequenced and found to be 99.2% homologous in amino acid sequence to the reported gene (50). Serine, glycine, and leucine at amino acid positions 130, 131, and 174 in the reported RDC1 were replaced with glycine, serine, and serine, respectively, in the RDC1 gene which we cloned. These changes were not localized in the extracellular regions.
Establishment of RDC1-expressing cells.
RDC1-transduced cell lines were established as follows. The plasmid harboring the genes of the receptor for ecotropic murine leukemia virus (MuLV) and hygromycin resistance was introduced into NP-2/CD4 cells by DNA transfection using LipofectAMINE (GIBCO BRL), and hygromycin-resistant cells were selected as described previously (31). These cells were infected with the ecotropic MuLV pseudotype, which had been produced by BOSC23 cells (ATCC CRL 11554) transfected with plasmid pMX-puro/RDC1 containing the puromycin resistance and RDC1 genes. The puromycin-resistant cells were selected by cultivation in medium containing 2 μg of puromycin per ml for 1 to 2 weeks. The obtained cell line was designated NP-2/CD4/RDC1. Over 80% of these RDC1-transduced cells were found to be positive for expression of CD4 molecules by flow cytometric analyses (data not shown). NP-2/RDC1 cells were established by transfection of pMX-puro/RDC1 DNA into NP-2 cells. NP-2/CD4/CCR5 and NP-2/CD4/CXCR4 cells were established as described previously (31).
Infection assay.
BT-20/N, NP-2/CD4, NP-2/CD4/CCR5, NP-2/CD4/CXCR4, NP-2/CD4/RDC1, and U87/CD4 cells (5 × 104) were seeded into 24-well culture plates 24 h prior to inoculation. These cells were inoculated with HIV-1, HIV-2, or SIV in an amount corresponding to 104 cpm of reverse transcriptase activity as described previously (30). The cells were passaged every 2 days.
Susceptibilities of BT-20/N, NP-2/CD4, NP-2/CD4/CCR5, NP-2/CD4/CXCR4, NP-2/CD4/RDC1, NP-2/RDC1, and U87/CD4 cells to HIV-1, HIV-2, and SIV strains were determined by an indirect immunofluorescence assay (IFA) which detected HIV or SIV antigens expressed in the infected cells as described previously (52). HIV-1-infected human serum or SIVmac-infected macaque serum was used as the first antibody. The infection was checked on days 2, 4, 6, and 8 after inoculation. The cells infected with HIV-1, HIV-2, or SIV were stained with Giemsa reagent after fixation with methanol on day 6 after infection, and syncytia induced were microscopically detected. Infection was also checked by PCR using specific primers for HIV-1, HIV-2, or SIV.
Phylogenetic analysis.
Multiple alignment of amino acid sequences for the conserved regions (amino acid positions 40 to 70, 76 to 96, 109 to 174, and 236 to 264 in the case of CXCR4) of 23 GPCRs (APJ, CCR1, CCR2b, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8/TER1, CCR9, CCR10, CXCR1, CXCR2, CXCR3, CXCR4, CXCR5/BLR1, Duffy antigen, GPR1, GPR15, RDC1, STRL33, US28, and v28) was carried out. The phylogenetic tree was constructed by the neighbor-joining (N-J) method (43).
RESULTS
Expression of candidate GPCRs for novel HIV/SIV coreceptors.
The four extracellular regions of HIV/SIV coreceptors (i.e., extracellular N-terminal domain and three loop structures) were reported to be involved in the interaction with the Env protein of HIV-1 (15, 39), and tyrosines in the extracellular N-terminal domains of human CCR5 and simian CCR5 were necessary for their function as an HIV/SIV coreceptor (19, 20). Therefore, we selected 18 GPCRs (APJ, C5a receptor, CCR1, CCR4, CCR7, CCR9/CCR10, CXCR1, CXCR2, CXCR3, CXCR5/BLR1, DEZ α, DEZ β, Duffy antigen, GPR5, GPR12, GPR25, GPR-9-6, and RDC1) from 96 GPCRs which were found in the DDBJ/EMBL/Genbank database as candidates for novel HIV/SIV coreceptors because they also have tyrosines in their extracellular N-terminal domains. APJ, DEZ β, and C5a receptor were included because they had not been identified as coreceptors when this study started. These candidates and newly identified coreceptors might also include a specific coreceptor for HIV-1, HIV-2, or SIV strains infectious to brain-derived cells. It was recently reported that sulfation of these tyrosines in the N-terminal domain is critical for the coreceptor function of CCR5 (21).
We examined the mRNA expression of these 18 GPCRs in various cells, including the brain-derived cells, by RT-PCR using the primers specific for each GPCR (Table 1). The mRNA of an orphan GPCR, RDC1, was detected in the C8166 T-cell line, PBLs, and three brain-derived cell lines (BT-3, BT-20/N, and U87/CD4), all of which were susceptible to variants GUN-1V, GUN-4V, and GUN-7V (see Fig. 2B and C) (18, 51). Expression of RDC1 mRNA was not detected in NP-2/CD4 cells and macrophages (Table 1 and Fig. 1A), which were resistant to infection with these HIV-1 variants (Fig. 2E and data not shown). CCR4 mRNA was detected not only in BT-3, BT-20/N, and U87/CD4 cells brain-derived cells but also in NP-2/CD4 cells which were resistant to the HIV-1 variants (Table 1 and Fig. 2E). These results suggested that RDC1 but not CCR4 might act as a coreceptor for these variants.
TABLE 1.
mRNA expression of GPCRs containing tyrosines in the extracellular N-terminal domains in various cells
| GPCR | GPCR mRNA expressiona
|
||||||
|---|---|---|---|---|---|---|---|
| BT-20/N | BT-3 | U87/CD4 | C8166 | NP-2/CD4 | Macrophages | PBLs | |
| CCR1 | − | − | − | − | − | − | − |
| CCR4 | ± | + | ± | ++ | ± | − | ++ |
| CCR7 | − | + | − | ++ | − | + | ++ |
| CCR9/CCR10 | − | − | − | − | − | − | − |
| CXCR1 | − | ND | − | − | − | ND | ND |
| CXCR2 | − | ND | − | − | − | ND | ND |
| CXCR3 | − | − | − | − | − | − | ++ |
| CXCR5/BLR1 | − | + | − | − | − | + | ++ |
| APJ | − | − | − | − | − | − | − |
| C5a receptor | − | ++ | + | − | − | − | + |
| DEZ α | − | − | − | − | − | − | − |
| DEZ β | − | ± | − | − | − | − | − |
| Duffy antigen | − | − | − | − | − | − | − |
| GPR-9-6 | − | ± | − | − | − | ± | − |
| GPR5 | − | − | + | − | − | − | + |
| GPR12 | − | ± | − | ± | − | − | − |
| GPR25 | − | − | − | − | − | − | − |
| RDC1 | ++ | ++ | + | + | − | − | + |
Determined by the intensities of specific RT-PCR bands as shown in Fig. 1A. ±, faint; +, weak; ++, strong; −, not detected; ND, not done.
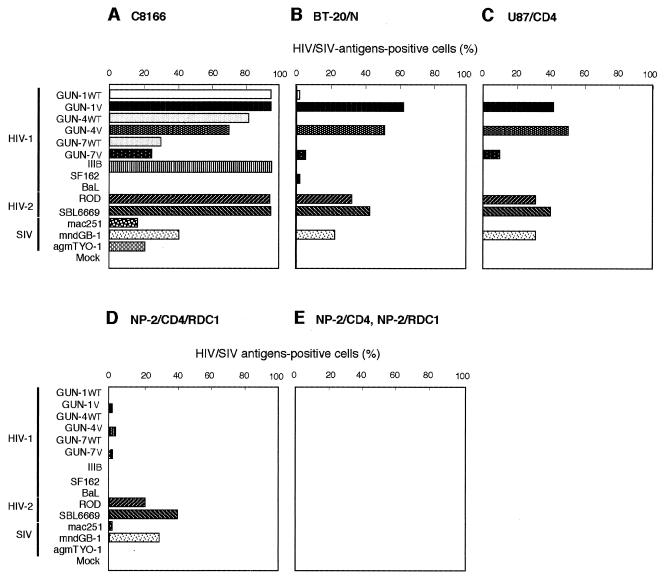
FIG. 2.
Susceptibilities of C8166 (A), BT-20/N (B), U87/CD4 (C), NP-2/CD4/RDC1 (D), NP-2/CD4 (E), and NP-2/RDC1 (F) cells to 14 HIV-1, HIV-2, and SIV strains. The results by IFA obtained 6 days after inoculation are shown. NP-2/CD4 and NP-2/RDC1 cells were resistant to all 14 viruses examined up to 8 days after infection. The experiments were repeated three times.
FIG. 1.
Detection of RDC1 mRNA expressed in several kinds of cells by RT-PCR. (A) RDC1 mRNA expression was detected in seven cell types (BT-3, BT-20/N, C8166, NP-2/CD4, and U87/CD4 cells, macrophages, and PBLs) by RT-PCR using primers specific for RDC1 (RDC1CN and RDC1CR). BT-3, BT-20/N, NP-2/CD4, and U87/CD4 cells originated in the human brain. The PCR primers amplify 1.1-kb DNA fragments when RDC1 mRNA is expressed in these cells. Expression levels (— to ++) were determined by the intensities of RT-PCR bands and are summarized in Table 1. As controls, the expression levels of glyceraldehyde-3-phosphate dehydrogenase (G3PDH) mRNA in each sample were determined by RT-PCR. The susceptibility of each cell to HIV-1 variant GUN-1V (Fig. 2) (40, 41) is shown by + or —. (B) The relative amount of the RDC1 mRNA in BT-3 cells was determined by comparison with that of CXCR4 in C8166 cells and CCR5 in PBLs. RT-PCR was done with 1:1, 1:10, 1:100, 1:1,000, and 1:10,000 dilutions of cDNA as templates. (C) Efficiencies of PCR primer sets to detect CCR5, CXCR4, and RDC1. The plasmid DNA containing each GPCR was serially diluted 10-fold and used as the template. PCR bands with similar intensities were detected at each dilution when CCR5 and CXCR4 were examined. Detection of RDC1 was 10-fold less efficient than that of CCR5 or CXCR4. Experiments were repeated three times. The intensity of the bands is indicated by +, ±, or — under each strip.
We determined the relative level of RDC1 mRNA expressed in BT-3 cells in comparison with those of CXCR4 or CCR5 in C8166 cells or PBLs (Fig. 1B). Reverse-transcribed cDNA was serially diluted 10-fold, and the level of RDC1 mRNA in the brain-derived BT-3 cells and levels of CCR5 and CXCR4 mRNA in C8166 cells and PBLs, respectively, were examined by RT-PCR under similar assay conditions. Serially 10-fold-diluted plasmids containing the CCR5, CXCR4, or RDC1 gene were used as PCR templates to detect corresponding genes. C8166 cells which abundantly express CD4 and CXCR4 have been used as cells highly susceptible to T-tropic HIV-1 strains (48). PBLs which express CCR3 and CCR5 are susceptible to M-tropic HIV-1 strains (9). From the results in Fig. 1B, the amount of the RDC1 mRNA expressed in BT-3 cells was estimated to be similar to that of CXCR4 mRNA expressed in C8166 cells and 100-fold more than that of CCR5 mRNA in PBLs when results shown in Fig. 1C were taken into account. These results suggest that RDC1 may be used as a coreceptor of HIV-1, HIV-2, or SIV strains which are susceptible to the brain-derived cells.
RDC1-mediated infection.
To examine the activity of RDC1 as a coreceptor for HIV-1, HIV-2, or SIV, NP-2/CD4/RDC1 and NP-2/RDC1 cells as well as four different types of cells (BT-20/N, C8166, NP-2/CD4, and U87/CD4) were infected with nine HIV-1 strains (GUN-1WT, GUN-1V, GUN-4WT, GUN-4V, GUN-7WT, GUN-7V, IIIB, SF162, and BaL), two HIV-2 strains (ROD and SBL6669), and three SIV strains (mac251, mndGB-1, and agmTYO-1). C8166 cells were highly susceptible to HIV-1 strains GUN-1WT, GUN-1V, GUN-4WT, GUN-4V, GUN-7WT, and GUN-7V and to all HIV-2 and SIV strains, while C8166 cells were not susceptible to the M-tropic HIV-1 strains SF162 and BaL (Fig. 2A). The brain-derived cells BT-20/N and U87/CD4 were susceptible to GUN-1V, GUN-4V, and GUN-7V strains (Fig. 2B and C) as reported previously (46). Moreover, BT-20/N and U87/CD4 cells were also susceptible to HIV-2ROD, HIV-2SBL6669, and SIVmndGB-1 strains: 20 to 40% of the cells were IFA positive 6 days after inoculation (Fig. 2B and C). These three HIV-1 variants, two HIV-2 strains, and SIVmndGB-1 strain induced numerous syncytia in the brain-derived cells (data not shown). SIVmac251 and SIVagmTYO-1 strains infected BT-20/N and U87/CD4 cells but inefficiently: less than 2% of the cells were IFA positive on day 6 after infection.
As control experiments, NP-2/CD4/CXCR4 (31) and NP-2/CD4/CCR5 (31) cells were also infected with these HIV and SIV strains. NP-2/CD4 cells were highly resistant to infection with all virus strains tested (Fig. 2E). NP-2/CD4/CXCR4 and NP-2/CD4/CCR5 cells were infected with the 14 HIV and SIV strains. Seven HIV-1 strains (GUN-1WT, GUN-1V, GUN-4WT, GUN-4V, GUN-7WT, GUN-7V, and IIIB), two HIV-2 strains (ROD and SBL6669), and SIVmndGB-1 efficiently plated onto NP-2/CD4/CXCR4 cells, while seven HIV-1 strains (GUN-1WT, GUN-1V, GUN-4WT, GUN-4V, GUN-7WT, GUN-7V, and SF162), HIV-2SBL6669, and SIVmac251 efficiently infected NP-2/CD4/CCR5 cells: more than 50% of the cells were IFA positive on day 6 after infection (data not shown). These findings indicate that all HIV and SIV strains were highly infectious to compatible target cells as previously reported.
NP-2/CD4/RDC1 cells were susceptible to HIV-2ROD, HIV-2SBL6669, and SIVmndGB-1 (Fig. 2D): 20 to 40% of the cells were IFA positive on day 6 after infection. HIV-2ROD and HIV-2SBL6669 induced large syncytia in NP-2/CD4/RDC1 cells on day 6 after infection, although no syncytia were detected in NP-2/CD4 cells infected with these strains (data not shown). SIVmndGB-1 also induced syncytia in NP-2/CD4/RDC1 cells on day 6 after infection (data not shown). NP-2/CD4/RDC1 cells showed low susceptibilities to variant strains GUN-1V, GUN-4V, and GUN-7V and to SIVmac251. Only 2 to 5% of the cells were IFA positive on day 6, and large syncytia could not be detected (data not shown). The infections of NP-2/CD4/RDC1 cells with GUN-1V, GUN-4V, and GUN-7V were also confirmed by PCR assay using primers specific for HIV-1 (data not shown). The infection of these HIV and SIV strains to NP-2/CD4/RDC1 cells spread slowly, and less than 10% of the cells became IFA positive after cultivation for up to 8 days (data not shown). NP-2/CD4/RDC1 cells were resistant to infection with GUN-1WT, GUN-4WT, GUN-7WT, IIIB, SF162, BaL, and SIVagmTYO-1. Recently, we noticed that NP-2/CD4/RDC1 cells have been infected with amphotropic MuLV. However, NP-2/CD4 and NP-2/RDC1 cells infected with amphotropic MuLV were highly resistant to infection with HIV and SIV strains (data not shown). This result indicates that the susceptibilities of NP-2/CD4/RDC1 cells to several HIV and SIV strains are CD4 and RDC1 dependent. Thus, RDC1 can facilitate the cell entry of HIV-1, HIV-2, and SIV strains infectious to the brain-derived BT-20/N and U87/CD4 cells (Fig. 2B and C). RDC1 functions as a novel coreceptor for several HIV-1, HIV-2, and SIV strains, supporting the entry of cell-free viruses and cell fusion after infection.
CD4 dependency.
It was reported that HIV-2ROD/B and neurotropic SIVmac174 strains can enter CD4-negative cells and that their entry is directly mediated through coreceptors CXCR4 and CCR5, respectively (3, 17, 42). To examine the CD4 dependency of the infection of NP-2/CD4/RDC1 cells with HIV-1, HIV-2, or SIVmndGB-1, CD4-negative NP-2/RDC1 cells were infected with these strains. The expression of CD4 molecules was detected on the surface of NP-2/CD4 cells but not of NP-2/RDC1 cells by flow cytometry using anti-CD4 monoclonal antibodies (data not shown). Figure 2E shows that NP-2/RDC1 cells were resistant to infection with all HIV-1, HIV-2, and SIV strains examined. HIV-2ROD, HIV-2SBL6669, and SIVmndGB-1, which efficiently infected NP-2/CD4/RDC1 cells, did not induce syncytia in NP-2/RDC1 cells (data not shown). These results indicate that RDC1 serves as a coreceptor for several HIV-1, HIV-2, and SIV strains in a CD4-dependent manner.
Molecular phylogeny of RDC1.
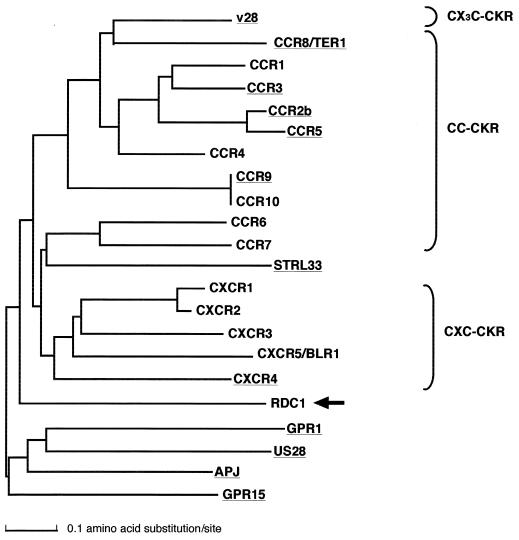
A natural ligand for RDC1 has not been identified, although RDC1 has the characteristic amino acid sequence of GPCRs (18). Amino acid sequences required for GPCRs to function as HIV or SIV coreceptors have not been clearly identified. To determine common properties of the HIV and SIV coreceptors among many GPCRs, we analyzed the evolutional relationship of the novel HIV/SIV coreceptor RDC1 with other reported HIV and SIV coreceptors. We constructed a phylogenetic tree for 10 CKRs (CCR1, CCR4, CCR6, CCR7, CCR9, CCR10, CXCR1, CXCR2, CXCR3, and CXCR5/BLR1) and 12 HIV/SIV coreceptors (APJ, CCR2b, CCR3, CCR5, CCR8, CCR9, CXCR4, GPR1, GPR15, STRL33, v28, US28, and RDC1) by the N-J method (43) (Fig. 3). In this tree, CXC-CKRs, including CXCR1, CXCR2, CXCR3, CXCR4, and CXCR5, and CC-CKRs, including CCR1, CCR2b, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR9 and CCR10, were clustered separately. This result suggests that the CC-CKRs and CXC-CKRs have evolved independently of each other. An orphan GPCR, STRL33, was located in the cluster containing CC-CKRs. Four orphan GPCRs (APJ, GPR1, GPR15, and RDC1) were located outside the clusters containing CC-CKRs and CXC-CKRs. The data in Fig. 3 suggest that RDC1 may not be a CX3C-CKR, CC-CKR, or CXC-CKR; that is, GPCRs other than CKRs can function as HIV and SIV coreceptors.
FIG. 3.
Phylogenetic tree of CKRs and the HIV/SIV coreceptors, including RDC1. This tree was constructed by the N-J method using multiple alignment of amino acids for their conserved regions as described in Materials and Methods. Reported HIV/SIV coreceptors are underlined. The branch of RDC1 is indicated by the arrow.
DISCUSSION
We showed that an orphan GPCR, RDC1, which is expressed in brain-derived cells, PBLs, and T-cell lines, acted as a novel coreceptor for several HIV-1, HIV-2, and SIV strains. Tyrosines are conservatively contained in the extracellular N-terminal domains of known HIV and SIV coreceptors (20, 21). This property was used as a criterion to select 18 candidates for a novel coreceptor of HIV and SIV strains infectious to T-cell lines, PBLs, and brain-derived cells. Among them, RDC1 was chosen as a possible GPCR for the coreceptor because it was expressed in cells susceptible to these HIV and SIV strains but not in cells resistant to them (Table 1 and Fig. 1A). Recently, it was shown that sulfation of these tyrosines of CCR5 is necessary for its coreceptor functions (21).
NP-2/CD4/RDC1 cells were much more susceptible to HIV-2 strains HIV-2ROD and HIV-2SBL6669 than to HIV-1 variants GUN-1V, GUN-4V, and GUN-7V and to SIVmndGB-1, all of which infected the brain-derived cells (Fig. 2D). Therefore, the susceptibilities of the brain-derived cells to HIV-1, SIV, and, in particular, HIV-2 strains may partly be explained by their use of RDC1 as a coreceptor. RDC1 is obviously used by HIV-1 variants GUN-1V, GUN-4V, and GUN-7V as their coreceptor even though the levels were much lower than those found for infection of HIV-2ROD and HIV-2SBL6669 strains. Thus, the RDC1 may not be a major coreceptor used by GUN-1V, GUN-4V, and GUN-7V variants. The primary brain-derived BT-3 cells were estimated to express 10-fold more RDC1 mRNA relative to that of CXCR4 in C8166 cells and that of CCR5 in PBLs (Fig. 1B).
It was reported that some primary HIV-1 isolates as well as HIV-2 and SIV strains infect U87/CD4 cells, which are not transduced with any CKR genes (2, 10). The cell tropism of these HIV-1 isolates may be similar to that of the HIV-1 variants described here. The HIV-1 variants which we used have glycine-serine-glycine-arginine (GSGR), GT(threonine)GR, or GA(alanine)GR sequence at the tip of the V3 region of gp120, while most HIV-1 strains have GP(proline)GR at this position. It was reported that a small percentage of HIV-1 isolates from patients have the sequence GSGR (32). This report suggests that HIV-1 strains which may show cell tropism similar to that of the HIV-1 variants described here are present in vivo. As for SIVs, we examined only three strains (SIVagmTYO-1, SIVmac251, and SIVmndGB-1). It is thought that there are genetic variations among these SIV strains. Although SIVmac251 and SIVagmTYO-1 did not use RDC1 as an efficient coreceptor, other strains of SIVmac and SIVagm may do so.
We recently noticed that NP-2/CD4/RDC1 cells harbor amphotropic MuLV. We also detected expression of amphotropic MuLV in PA317/CD4 packaging cells (37), HOS.T4 cells (28), and some U87/CD4 cells. Nucleotide sequences of the env gene were partially determined after RT-PCR of cellular RNA of these cells: the sequence of MuLV expressed in NP-2/CD4/RDC1 cells was identical to that of PA317/CD4 cells but slightly different from that of HOS.T4 cells. These findings suggest that PA317/CD4 cells may have produced competent MuLV, which might have infected NP-2 cells when CD4 was transduced.
RDC1 has been isolated as a human homologue of the canine vasoactive intestinal peptide receptor (18). However, a natural ligand for RDC1 has not been identified (11). Phylogenetic analysis for CKRs and HIV/SIV coreceptors including RDC1 revealed that RDC1 is located in the clusters of neither CC-CKRs nor CXC-CKRs, suggesting that a ligand for RDC1 may not be a typical chemokine (Fig. 3). This result may make it easier to identify a natural ligand for RDC1.
The RDC1 mRNA was reported to be detected in the lung, testis, and kidney but not in the brain (18). However, we detected the RDC1 mRNA in the human glioma cell line U87/CD4 and the primary brain-derived cell lines BT-20/N and BT-3 (Table 1 and Fig. 1A), which may have originated in cells associated with the brain blood vessels, such as smooth muscle cells or pericytes. The blood-brain barrier consists mainly of astrocytes, endothelial cells, and pericytes which affect the function of endothelial cells (27). Dysfunction of the blood-brain barrier may result from damage of the brain blood vessels. Infection of these cells with HIV-1 may promote the development of neurological disorders such as HIV encephalitis and AIDS dementia complex, which have frequently been observed in AIDS patients (24). The expression of RDC1 in different types of the cells of the brain has not been well elucidated. The role of RDC1 in the transmission and pathogenesis of HIV and SIV remains to be studied.
ACKNOWLEDGMENTS
We thank T. J. Schall and K. Matsushima for kindly supplying CKR-expressing pcDNA3 plasmids and CXCR4 plasmid, respectively. We also thank T. Kumanishi, P. R. Clapham, B. Chesebro, W. S. Pear, and G. P. Nolan for kindly providing NP-2 glioma cells, U87 glioma cells, PA317/CD4 cells, BOSC23 cells, and phiNX-A cells, respectively.
This work was supported in part by grants-in-aid from the Ministry of Education, Science and Culture and the Ministry of Health and Welfare of Japan.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Bjorndal A, Deng H, Jansson M, Fiore J R, Colognesi C, Karlesson A, Albert J, Scarlatti G, Littman D R, Fenyo E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1998;10:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bron R, Klasse P J, Wilkinson D, Clapham P R, Matthews A P, Power C, Wells T N, Kim J, Peiper S C, Hoxie J A, Marsh M. Promiscuous use of CC and CXC chemokine receptors in cell-to-cell fusion mediated by a human immunodeficiency virus type 2 envelope protein. J Virol. 1997;71:8405–8415. doi: 10.1128/jvi.71.11.8405-8415.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen Z, Gettie A, Ho D D, Mark P A. Primary SIVsm isolates use the CCR5 coreceptor from sooty mangabeys naturally infected in west Africa: a comparison of coreceptor usage of primary SIVsm, HIV-2, and SIVmac. Virology. 1998;246:113–124. doi: 10.1006/viro.1998.9174. [DOI] [PubMed] [Google Scholar]
- 5.Chen Z, Zhou P, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. Viral determinants of human immunodeficiency virus type 1 T-cell or macrophage tropism, cytopathyogenicity, and CD4 antigen modulation. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chesebro B, Buller R, Portis J, Wehrly K. Failure of human immunodeficiency virus entry and infection in CD4-positive human brain and skin cells. J Virol. 1990;64:215–221. doi: 10.1128/jvi.64.1.215-221.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor APJ supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 10.Clapham P R, Blanc D, Weiss R A. Specific cell surface requirements for the infection of CD4-positive cells by human immunodeficiency virus types 1 and 2 and by simian immunodeficiency virus. Virology. 1991;181:703–715. doi: 10.1016/0042-6822(91)90904-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook J S, Wolsing D H, Lameh J, Olson C A, Correa P E, Sadee W, Blumenthal E M, Rosenbaum J S. Characterization of the RDC1 gene which encodes the canine homolog of a proposed human VIP receptor. Expression does not correlate with an increase in VIP binding sites. FEBS Lett. 1992;300:149–152. doi: 10.1016/0014-5793(92)80184-i. [DOI] [PubMed] [Google Scholar]
- 12.Dalgleish A G, Beverley P C, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 13.Daniel M D, Letvin N L, King N W, Kanniga M, Sehgal P K, Hunt R D, Kanki P J, Essex M, Desrosiers R C. Isolation of a T-cell tropic HTLV-III-like retrovirus from macaques. Science. 1985;228:1201–1204. doi: 10.1126/science.3159089. [DOI] [PubMed] [Google Scholar]
- 14.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 15.Doranz B J, Lu Z H, Rucker J, Zhang T Y, Sharron M, Cen Y H, Wang Z X, Guo H H, Du J G, Accavitti M A, Doms R W, Peiper S C. Two distinct CCR5 domains can mediate coreceptor usage by human immunodeficiency virus type 1. J Virol. 1997;71:6305–6314. doi: 10.1128/jvi.71.9.6305-6314.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the beta chemokine receptors CKR-5, CKR-3, and CKR-2b as fusin cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 17.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by a neurovirulent simian immunodeficiency virus strain. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eva C, Sprengel R. A novel putative G protein-coupled receptor highly expressed in lung and testis. DNA Cell Biol. 1993;12:393–399. doi: 10.1089/dna.1993.12.393. [DOI] [PubMed] [Google Scholar]
- 19.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farzan M, Choe H, Vaca L, Martin K, Sun Y, Desjardins E, Ruffing N, Wu L, Wyatt R, Gerard N, Gerard C, Sodroski J. Tyrosine-rich region in the N terminus of CCR5 is important for human immunodeficiency virus type 1 entry and mediates an association between gp120 and CCR5. J Virol. 1998;72:1160–1164. doi: 10.1128/jvi.72.2.1160-1164.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farzan M, Mizabekov T, Kolchinsky P, Wyatt R, Cayabyab M, Gerard N P, Gerard C, Sodroski J, Choe H. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96:667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 22.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 23.Fukasawa M, Miura T, Hasegawa A, Morikawa S, Tsujimoto H, Miki K, Kitamura T, Hayami M. Sequence of simian immunodeficiency virus from African green monkey, a new member of the HIV/SIV group. Nature. 1988;333:457–461. doi: 10.1038/333457a0. [DOI] [PubMed] [Google Scholar]
- 24.Glass J D, Johnson R T. Human immunodeficiency virus and the brain. Annu Rev Neurosci. 1996;19:1–26. doi: 10.1146/annurev.ne.19.030196.000245. [DOI] [PubMed] [Google Scholar]
- 25.Guyader M, Emerman M, Sonigo P, Clavel F, Montagnier L, Alizon M. Genome organization and transactivation of the human immunodeficiency virus type 2. Nature. 1987;326:662–669. doi: 10.1038/326662a0. [DOI] [PubMed] [Google Scholar]
- 26.Haraguchi Y, Takeuchi Y, Hoshino H. Inhibition of plating of human T cell leukemia virus type 1 and syncytium-inducing types of human immunodeficiency virus type 1 by polycations. AIDS Res Hum Retroviruses. 1997;13:1517–1523. doi: 10.1089/aid.1997.13.1517. [DOI] [PubMed] [Google Scholar]
- 27.Hasan M, Glees P. The fine structure of human cerebral perivascular pericytes and juxtavascular phagocytes: their possible role in hydrocephalic edema resolution. J Hirnforsch. 1990;31:237–249. [PubMed] [Google Scholar]
- 28.He J, Landau N R. Use of a novel human immunodeficiency virus type 1 reporter virus expressing human placental alkaline phosphatase to detect an alternative virus receptor. J Virol. 1995;69:4587–4592. doi: 10.1128/jvi.69.7.4587-4592.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill C M, Deng H, Unutmaz D, Kewalramani V N, Bastiani L, Gorny M K, Zolla-Pazner S, Littman D R. Envelope glycoproteins from human immunodeficiency virus types 1 and 2 and simian immunodeficiency virus can use human CCR5 as a coreceptor for viral entry and make direct CD4-dependent interactions with this chemokine receptor. J Virol. 1997;71:6296–6304. doi: 10.1128/jvi.71.9.6296-6304.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoshino H, Esumi H, Miwa M, Shimoyama M, Minato K, Tobinai K, Hirose M, Watanabe S, Inada N, Kinoshita K, Kamihira S, Ichimaru M, Sugimura T. Establishment and characterization of 10 cell lines derived from patients with adult T-cell leukemia. Proc Natl Acad Sci USA. 1983;80:6061–6065. doi: 10.1073/pnas.80.19.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jinno A, Shimizu N, Soda Y, Haraguchi Y, Kitamura T, Hoshino H. Identification of the chemokine receptor TER1/CCR8 expressed in brain-derived cells and T cells as a new coreceptor for HIV-1 infection. Biochem Biophys Res Commun. 1998;243:497–502. doi: 10.1006/bbrc.1998.8130. [DOI] [PubMed] [Google Scholar]
- 32.LaRosa G J, Davide J P, Weinhold K, Waterbury J A, Profy A T, Lewis J A, Langlois A J, Dreesman G R, Boswell R N, Shadduck P. Conserved sequence and structural elements in the HIV-1 principal neutralizing determinant. Science. 1991;249:932–935. doi: 10.1126/science.2392685. [DOI] [PubMed] [Google Scholar]
- 33.Lasky L A, Nakamura G, Smith D H, Fennie C, Shimasaki C, Patzer E, Berman P, Gregory T, Capon D J. Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with the CD4 receptor. Cell. 1987;50:975–985. doi: 10.1016/0092-8674(87)90524-1. [DOI] [PubMed] [Google Scholar]
- 34.Liao F, Alkhatib G, Peden K W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchese A, Docherty J M, Nguyen T, Heiber M, Cheng R, Heng H H, Tsui L C, Shi X, George S R, O'Dowd B F. Cloning of human genes encoding noble G protein-coupled receptors. Genomics. 1994;23:609–618. doi: 10.1006/geno.1994.1549. [DOI] [PubMed] [Google Scholar]
- 36.McKeating J A, Gow J, Goudsmit J, Pearl J H, Mulder C, Weiss R A. Characterization of HIV-1 neutralization escape mutants. AIDS. 1989;3:777–784. doi: 10.1097/00002030-198912000-00001. [DOI] [PubMed] [Google Scholar]
- 37.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murphy P M. The molecular biology of leukocyte chemoattractant receptors. Annu Rev Immunol. 1994;12:593–633. doi: 10.1146/annurev.iy.12.040194.003113. [DOI] [PubMed] [Google Scholar]
- 39.Picard L, Simmons G, Power C A, Meyer A, Weiss R A, Clapham P R. Multiple extracellular domains of CCR-5 contribute to human immunodeficiency virus type 1 entry and fusion. J Virol. 1997;71:5003–5011. doi: 10.1128/jvi.71.7.5003-5011.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pleskoff O, Treboute C, Brelot A, Heveker N, Seman M, Alizon M. Identification of a chemokine receptor encoded by human cytomegalovirus as a cofactor for HIV-1 entry. Science. 1997;276:1874–1878. doi: 10.1126/science.276.5320.1874. [DOI] [PubMed] [Google Scholar]
- 41.Ponten J, Macintyre E H. Long term culture of normal and neoplastic human glia. Acta Pathol Microbiol Scand. 1968;74:465–486. doi: 10.1111/j.1699-0463.1968.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 42.Potempa S, Picard L, Reeves J D, Wilkinson D, Weiss R A, Talbot S J. CD4-independent infection by human immunodeficiency virus type 2 strain ROD/B: the role of the N-terminal domain of CXCR-4 in fusion and entry. J Virol. 1997;71:4419–4424. doi: 10.1128/jvi.71.6.4419-4424.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 44.Salahuddin S Z, Markham P D, Wang-Staal F, Franchini G, Kalyanaraman V S, Gallo R C. Restricted expression of human T-cell leukemia-lymphoma virus HTLV in transformed human umbilical cord blood lymphocytes. Virology. 1983;129:51–54. doi: 10.1016/0042-6822(83)90395-1. [DOI] [PubMed] [Google Scholar]
- 45.Shimizu N, Kobayashi M, Liu H-Y, Kido H, Hoshino H. Detection of tryptase TL2 and CD26 antigen in brain-derived cells nonpermissive to T-cell line-tropic human immunodeficiency virus type 1. FEBS Lett. 1995;358:48–52. doi: 10.1016/0014-5793(94)01394-g. [DOI] [PubMed] [Google Scholar]
- 46.Shimizu S N, Shimizu N G, Takeuchi Y, Hoshino H. Isolation and characterization of human immunodeficiency virus type 1 variants infectious to brain-derived cells: detection of common point mutations in the V3 region of the env gene of the variants. J Virol. 1994;68:6130–6135. doi: 10.1128/jvi.68.9.6130-6135.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soda Y, Shimizu N, Jinno A, Liu H-Y, Kanbe K, Kitamura T, Hoshino H. Establishment of a new system for determination of coreceptor usages of HIV based on the human glioma NP-2 cell line. Biochem Biophys Res Commun. 1999;258:313–321. doi: 10.1006/bbrc.1999.0633. [DOI] [PubMed] [Google Scholar]
- 49.Sol N, Ferchal F, Braun J, Pleskoff O, Treboute C, Ansart I, Alizon M. Usage of the coreceptors CCR-5, CCR-3, and CXCR-4 by primary and cell line-adapted human immunodeficiency virus type 2. J Virol. 1997;71:8237–8244. doi: 10.1128/jvi.71.11.8237-8244.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sreedharan S P, Robichon A, Peterson K E, Goetzl E J. Cloning and expression of the human vasoactive intestinal peptide receptor. Proc Natl Acad Sci USA. 1991;88:4986–4990. doi: 10.1073/pnas.88.11.4986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Takeuchi Y, Akutsu M, Murayama K, Shimizu N, Hoshino H. Host range mutant of human immunodeficiency virus type 1: modification of cell tropism by a single point mutation at the neutralization epitope in the env gene. J Virol. 1989;65:1710–1718. doi: 10.1128/jvi.65.4.1710-1718.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takeuchi Y, Inagaki M, Kobayashi N, Hoshino H. Isolation of human immunodeficiency virus from a Japanese hemophilia B patient with AIDS. Jpn J Cancer Res. 1989;78:11–15. [PubMed] [Google Scholar]
- 53.Tsujimoto H, Cooper R W, Kodama T, Fukasawa M, Miura T, Ohta Y, Ishikawa K, Nakai M, Frost E, Roelants G E. Isolation and characterization of simian immunodeficiency virus from mandrills in Africa and its relationship to other human and simian immunodeficiency viruses. J Virol. 1988;62:4044–4050. doi: 10.1128/jvi.62.11.4044-4050.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wong-Staal F, Gallo R C, Chang N, Ghrayeb J, Papas T S, Lautenberger J A, Pearson M L, Petteway S R, Jr, Ivanoff L, Baumeister K, Whitehorn E A, Rafalski J A, Doran E R, Josephs S J, Starcich B, Livak K J, Patarca R, Haseltine W A, Ratner L. Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature. 1985;313:277–284. doi: 10.1038/313277a0. [DOI] [PubMed] [Google Scholar]
- 55.Yamaguchi K, Byrn R A. Clinical isolates of HIV-1 contain few pre-existing proteinase inhibitor resistance conferring mutations. Biochim Biophys Acta. 1995;1253:136–140. doi: 10.1016/0167-4838(95)00167-1. [DOI] [PubMed] [Google Scholar]