Abstract
The mutational status of the immunoglobulin variable region heavy chain genes (IGHV) is an important prognostic marker in chronic lymphocytic leukemia (CLL). The data accumulated in the literature has largely been derived from studies conducted on Caucasian Western populations. Little is known about Asian CLL patients. In this study the IGHV genes usage and somatic hypermutation status have been investigated in 87 Iranian CLL patients. Based on a cut‐off of 98% nucleotide sequence homology, 64.4% and 35.6% of the patients expressed mutated and unmutated IGHV genes, respectively, with most non‐progressive patients being in the mutated group (35/44 vs 19/40; P = 0.009). Progression‐free survival (PFS) and time to first treatment (TTFT) were significantly higher in our mutated and non‐progressive patients compared to unmutated and progressive subtypes, respectively. The most frequently used IGHV gene was IGHV3‐7 (12.6%) followed by IGHV3‐30 (11.4%), IGHV3‐48 (9.2%), IGHV4‐39 (6.9%), and IGHV1‐8 (6.9%) genes, which taken together comprised nearly half of the IGHV genes expressed in the Iranian CLL patients. Of the IGHV genes, IGHV3‐7 was significantly over‐represented in non‐progressive compared to progressive CLL patients (P = 0.036), whereas IGHV1‐69 and IGHV1‐2 were expressed at a higher frequency in unmutated compared to mutated CLL patients (P < 0.03). Comparison of IGHV gene usage in our patients with that of Western CLL patients revealed significant differences in expression of IGHV1‐69, IGHV3‐7, IGHV3‐21, and IGHV4‐34 genes. Analysis of the IGHV third complementary determining region (HCDR3) sequences revealed a high frequency use of certain HCDR3 motifs, such as YYYGMDV, in our samples. These findings imply contribution of antigen selection and regional (ethnic/geographic) parameters in the leukomogenesis of CLL. (Cancer Sci 2009; 100: 2346–2353)
Leukemic cells of chronic lymphocytic leukemia (CLL) are derived from clonally expanded and phenotypically mature B cells (CD5+/CD19+/sIglow) which accumulate in the bone marrow, blood, and lymphoid tissues.( 1 ) In recent years a number of biological prognostic factors have been identified in CLL.( 2 ) CD38 expression was the first marker that was found to correlate with disease progression.( 3 ) Intracytoplasmic detection of the ZAP‐70 enzyme in leukemic B cells was subsequently revealed to be correlated with CLL prognosis.( 4 ) Most patients with CD38 and ZAP‐70 expression on their leukemic cells tend to have a more aggressive disease. In addition to these markers, chromosomal abnormalities have also been documented in the majority of CLL patients and found to be associated with disease prognosis.( 5 )
Among the prognostic biomarkers, analysis of the immunoglobulin (Ig) genes has been of considerable interest, particularly in terms of somatic hypermutation status of Ig heavy chain variable region (IGHV) genes.( 6 , 7 , 8 ) This helped to define two disease subsets, mutated and unmutated, based on nucleotide sequence homology to the closest germline gene. Patients with unmutated leukemic cells tend to have a more aggressive clinical course relative to those with mutated IGHV.( 9 , 10 , 11 ) This categorization has led to the speculation that these two subtypes might represent two distinct entities: one derived from memory B cells that had undergone Ig somatic mutation, and the other derived from naïve or pre‐germinal center B cells.( 12 )
The results obtained from most previous studies which have been conducted in Western populations suggest that IGHV genes usage among CLL patients is not random; besides, their frequencies differ in mutated and unmutated subsets of CLL patients.( 3 , 6 , 7 , 8 , 13 , 14 , 15 , 16 , 17 , 18 , 19 ) Of the IGHV genes, IGHV1‐69, which is associated with unmutated subtype of CLL patients, is over‐represented in Western patients,( 18 , 20 ) predominantly recombined with IGHD3‐3 and IGHJ6 genes, encoding longer than average heavy chain complementarity determining region 3 (HCDR3) compared to other IGHV members.( 20 , 21 , 22 ) Other frequently used IGHV genes are IGHV3‐23, IGHV4‐34, and IGHV3‐7 ( 8 , 15 , 23 , 24 ) that usually belong to the mutated subtype.
In addition to the skewed representation of certain IGHV genes in mutated and unmutated subtypes of CLL, there is compelling evidence indicating differential expression of some IGHV genes in different geographic parts of Western countries. While IGHV3‐21 is frequently expressed in northern European (Scandinavian) CLL patients,( 20 , 25 ) there is no or a low frequency of usage of this gene in southern or central parts of European countries and the USA.( 15 , 26 , 27 )
All the findings of biased repertoire of IGHV genes, as well as similar amino acid motifs in HCDR3, have led to the hypothesis of involvement of an as yet undefined set of antigens (self or non‐self) with a restricted set of epitopes in the pathogenesis of CLL, implicating the role of B‐cell antigen receptor (BCR) in the etiopathogenesis of the disease.
As the current bulk of data on IGHV gene representation and mutational status in CLL is derived from studies conducted in Western populations with only two reports of Asian CLL patients from China and Japan, we investigated for the first time IGHV gene usage and mutational status of the leukemic B cells in an Iranian CLL population. Comparison of our results with those reported from the two other Asian populations, as well as Western CLL patients, strongly suggest contribution of regional parameters in shaping the IGHV repertoire of the leukemic CLL B cells.
Materials and Methods
Patients and controls. Twenty mL of heparinized peripheral blood was collected from 87 CLL patients attending the Hematology and Oncology Clinics of Vali‐Asr and Firozgar Hospitals, affiliated to Tehran University of Medical Sciences and Iran University of Medical Sciences, respectively. All patients were Iranians, living in Tehran city. Written consent was obtained from all patients and the study was approved by the Ethical Committee of Tehran University of Medical Sciences. The diagnosis was based on immunophenotypic analysis, blood cell count, cell morphology. and clinical symptoms, according to the World Health Organization classification of tumors.( 28 )
Immunophenotyping of leukemic cells by flow cytometry revealed expression of CD19 and CD5 molecules in all cases and CD23 in 77 cases. For the following reasons CD23‐negative patients were diagnosed as CLL and not mantle cell lymphoma (MCL): (a) as opposed to CLL, MCL is a very aggressive disease with a short overall survival and treatment free survival. Among these 10 patients, five patients displayed a non‐progressive course, of whom four patients did not require chemotherapy and the fifth patient responded very well to chemotherapy. (b) CLL usually displays a better response to conventional chemotherapy than MCL, with most MCL cases being refractory to chemotherapy. Five of our 10 CD23‐negative patients (four progressive and one non‐progressive) underwent chemotherapy. With the exception of one patient who died due to unrelated complications (sepsis), the remaining four patients responded quite well to therapy. (c) Surface Ig is usually expressed weakly in CLL as compared to MCL. Indeed, this has been the case in all our five non‐progressive patients (IgM+ leukemic cells, 7–26%; mean, 13.7%). In progressive patients, however, moderate expression of surface Ig was observed (IgM+ cells, 23–62%; mean, 44%).
The age range of patients was 39–84 years with a median of 63 years. Fifty‐four patients were males and 33 were females. The patients were classified into non‐progressive (n = 44) and progressive (n = 40) groups. Disease progression of three patients could not be identified. Disease progression was identified on the basis of either of the following criteria: lymphocyte count doubling time of <1 year; progression to a more advanced Rai stage; development of systemic symptoms; development of Richter’s syndrome; downward trend of hemoglobin (Hb) concentration or platelet count to below the normal range (Hb < 13.5 g/dL for males and <11.5 g/dL for females; platelet count <150 × 109/L) even when not meeting the criteria for stage III or IV disease (Hb <10 g/dL, platelet count <100 × 109/L). Possession of one of these characteristics was sufficient to qualify as progressive disease.( 29 ) Of 87 CLL patients, 47, five, 18, five, and 11 patients were in Rai stages 0, I, II, III, and IV, respectively. Of 44 non‐progressive CLL patients, 33, two, five, two, and one patient were in Rai stages 0, I, II, III, and IV, respectively. Staging of one non‐progressive patient could not be defined. Of 40 progressive CLL patients, 12, three, 12, three, and 10 patients were in Rai stages 0, I, II, III, and IV, respectively.
Patients with one or more of the following manifestations were treated: splenomegaly, anemia, thrombocytopenia, disfiguring lymphadenopathy, and constitutive symptoms not related to other syndromes. Thirty‐one (35.6%) patients were under therapy at the time of sample collection, whereas 48 (55.2%) patients received their first therapy after diagnosis. The remaining eight patients had stable disease and did not require treatment. The median time from diagnosis to first treatment (TTFT) for these 48 patients was 6 months with a minimum and maximum of 1 and 84 months, respectively. Of these 48 patients, 13 and 35 cases had non‐progressive and progressive diseases, respectively. The patients’ median TTFT time was 15 months for non‐progressive, 4 months for progressive, 12 months for mutated, and 4 months for unmutated groups of patients. The median follow‐up of patients was 49 months (range, 8–204 months). During this study, nine patients died, seven of whom due to CLL complications.
Progression‐free survival (PFS) was available for 33 patients with a median of 11 months. Median PFS for non‐progressive and progressive CLL patients was 24 and 8.5 months, respectively, whereas that of the mutated and unmutated patients was 24 and 8 months, respectively. The TTFT and PFS in our series of patients are lower than Western patients( 27 , 30 ) which could be due to genetic and/or environmental differences.
DNA, RNA isolation, and cDNA synthesis. Genomic DNA was extracted from heparinized peripheral blood leukocytes according to a standard protocol.( 31 ) Total cellular RNA was isolated from three million PBMCs of CLL patients using RNA‐Bee (BioSite, Taby, Sweden) based on the guanidine thiocyanate phenol chloroform extraction method. First strand cDNA was synthesized from 1–3 μg of total RNA (unfolded at 65°C for 5 min) using M‐MuLV reverse transcriptase (RT) enzyme (Fermentas Life Sciences, Ontario, Canada), random hexamer primers (20 pmol), and dNTPs (20 mm) (Roche, Mannheim, Germany). The mixture was incubated at 42°C for 45 min followed by 5 min at 90°C for inactivation of RT enzyme.
Determination of IGHV family gene expression by polymerase chain reaction (PCR). Serial dilutions of DNA and cDNA (1:10 to 1:2000) were prepared and PCR amplification was performed using IGHV family‐specific degenerative primers. The 5′ primers used for IGHV gene amplification were designed to anneal to the leader sequence of IGHV1‐7 families, and one degenerative IGHJ or a Cμ primer were used as 3′ primers. PCR reactions were performed as described.( 16 )
Cloning and sequencing of PCR products. Clonal PCR products were purified by excision using the QIAquick gel extraction kit (Qiagen, Hilden, Germany). Gel purified PCR products were cloned into pGEM‐T easy vector (Promega, Southampton, United Kingdom). Briefly, after transformation of chemically prepared JM109 competent cells, at least five colonies were selected randomly for sequencing. Sequencing was performed from both directions using the BigDye Terminator Cycle Sequencing Reaction Kit (Applied Biosystems, Foster City, CA, USA), T7, and SP6 primers.
Analysis of Ig gene sequences. For each sample IGHV, IGHD, and IGHJ genes were identified by matching to the closest known human germline gene using the ImMuno‐Gene Tics (IMGT) Database (http://imgt.cines.fr) and the IgBLast search (http://www.ncbi.nlm.nih.gov/igblast/). As the IMGT database is the most reliable and updated database, the results were reported based on the analysis of the sequences by this database. Classification of patients into mutated (n = 56) and unmutated (n = 31) subtypes was based on more than 98% homology in nucleotide sequence of immunoglobulin variable region heavy (IGHV) chain genes of the leukemic cells.( 13 ) The IGHV third complementary determining region (HCDR3) was determined by the IMGT database.
Antigenic selection and amino acid changes in IGHV. Distribution of replacement (R) and silent (S) mutations in the IGHV sequence of mutated CLL patients was evaluated by the multinomial distribution model proposed by Lossos et al. ( 32 )
Statistical analysis. Statistical analyses were performed using the Pearson χ2‐test, Fisher’s exact test, and Mann–Whitney U‐test. Kaplan–Meier methods and log‐rank tests were used to compare TTFT and PFS in mutated and unmutated as well as progressive and non‐progressive CLL patients. Analyses were conducted using the SPSS statistical package (SPSS, Chicago, IL, USA). P‐values of <0.05 were considered significant.
Results
IGHV mutation and disease outcome. Based on a cut‐off of 98% nucleotide sequence homology of isolated IGHV gene with that of the germline counterpart, 56 out of 87 (64.4%) IGHV sequences displayed somatic mutations, whereas the remaining 31 sequences (35.6%) were unmutated. In the mutated cases, the range of mutations varied from 2% to 19% with an average of 4.9%. The average of mutations was different among IGHV families with 6.1% for IGHV4, 5% for IGHV3, and 3.2% for IGHV1 families.
Of 44 non‐progressive patients, 35 cases had a mutated IGHV sequence, and from 40 progressive patients, 21 had an IGHV sequence with <98% homology to the corresponding germline IGHV sequence (P = 0.009).
Comparison of TTFT means between our CLL subtypes revealed a significantly higher TTFT in our non‐progressive and mutated (25.3 and 16.4 months) CLL patients compared to progressive and unmutated patients (6.6 and 7.4 months) (P = 0.001 and P = 0.03, respectively). A higher PFS mean was also observed in non‐progressive and mutated (44 and 37 months) CLL patients compared to progressive and unmutated (13 and 9 months) CLL patients (P = 0.006 and P = 0.0001, respectively). Representative TTFT and PFS results of the mutated and unmutated groups are illustrated in Figure 1. Overall survival was significantly lower in IGHV unmutated CLL patients compared to mutated patients (98%vs 84%, P = 0.04).
Figure 1.
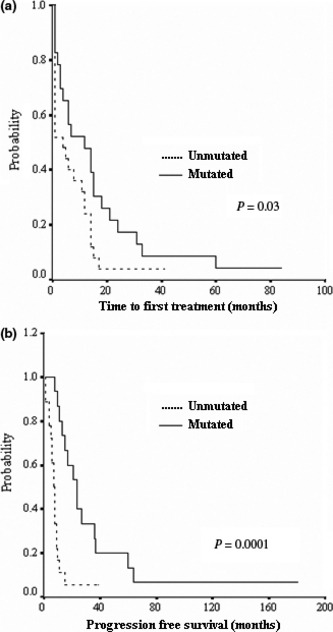
Kaplan–Meier plot comparing time from diagnosis to first treatment (a) and progression‐free survival (b) between immunoglobulin variable region heavy chain (IGHV) mutated and unmutated chronic lymphocytic leukemia (CLL) patients.
IGHV gene usage in CLL patients. The frequencies of the expressed IGHV gene families were determined in our CLL patients. Cumulative results are summarized in Table 1. Overall, the most frequently expressed IGHV family was IGHV3 (n = 49, 56.3%), followed by IGHV4 (n = 18, 20.7%), IGHV1 (n = 16, 18.4%), IGHV6 (n = 3, 3.4%), and IGHV5 (n = 1, 1.1%), with no expression of IGHV2 and IGHV7 families.
Table 1.
Frequency of IGHV gene families expressed in leukemic cells of Iranian CLL patients
| Patients | IGHV family | Total | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||
| Non‐progressive | 6 (13.6) | 0 | 30 (68.2) | 8 (18.2) | 0 | 0 | 0 | 44 |
| Progressive | 9 (22.5) | 0 | 17 (42.5) | 10 (25) | 1 (2.5) | 3 (7.5) | 0 | 40 |
| Newly diagnosed | 1 (33.3) | 0 | 2 (66.7) | 0 | 0 | 0 | 0 | 3 |
| Mutated | 6 (10.7) | 0 | 32 (57.1) | 14 (25) | 1 (1.8) | 3 (5.4) | 0 | 56 |
| Unmutated | 10 (32.3) | 0 | 17 (54.8 | 4 (12.9) | 0 | 0 | 0 | 31 |
| Total | 16 (18.4) | 0 | 49 (56.3)* | 18 (20.7) | 1 (1.1) | 3 (3.4) | 0 | 87 |
| †Normal adult B cells | 778 (28.3) | 104 (3.8) | 971 (35.3) | 770 (28) | 94 (3.4) | 33 (1.2) | 0 | 2750 |
| ‡Normal CD5+/IgM+ B cells | 27 (18.8) | 3 (5.9) | 81 (56.3) | 26 (18.1) | 2 (1.4) | 2 (1.4) | 3 (2.1) | 144 |
| ¶Known functional genes per family | 11 (21.6) | 3 (5.9) | 22 (43.1) | 11 (21.6) | 2 (3.9) | 1 (2) | 1 (2) | 51 |
Data are presented as number (%) of patients expressing the corresponding immunoglobulin variable region heavy chain (IGHV) family; All significant differences between groups are shown in bold. *Comparison between the frequency of expressed gene family in the Iranian chronic lymphocytic leukemia (CLL) population and the frequency of gene family expression in normal adult B cells (P < 0.0001). †The frequency of IGHV gene family expression in normal adult B cells represents IGHV clones identified in IgM cDNA libraries of unstimulated peripheral blood lymphocytes from healthy adults.( 41 ) ‡The frequency of expressed IGHV family genes in human peripheral CD5 + /IgM + B cells from two healthy adult donors.( 42 ) ¶The percent of functional IGHV genes per family observed as IGHV recombinants were obtained from the V BASE sequence directory.
The IGHV3 family was more represented in non‐progressive patients. Of the IGHV3 family members, only IGHV3‐7 was preferentially expressed in mutated and non‐progressive CLL subtypes, but significant difference was observed only between non‐progressive and progressive groups of patients (20.5%vs 5%; P = 0.05) (Table 2).
Table 2.
Frequency of IGHV genes expressed in leukemic cells of Iranian CLL patients
| No | IGHV | Total | Mutated (n = 56) | Unmutated (n = 31) | Non‐progressive (n = 44) | Progressive (n = 40) |
|---|---|---|---|---|---|---|
| 1 | IGHV3‐7 | 11 (12.6) | 9 (16.1) | 2 (6.5) | 9 (20.5) | 2 (5) |
| 2 | IGHV3‐30 | 10 (11.5) | 5 (8.9) | 5 (16.1) | 5 (11.4) | 5 (12.5) |
| 3 | IGHV3‐48 | 8 (9.2) | 6 (10.7) | 2 (6.5) | 5 (11.4) | 3 (7.5) |
| 4 | IGHV4‐39 | 6 (6.9) | 5 (8.9) | 1 (3.2) | 3 (6.8) | 3 (7.5) |
| 5 | IGHV1‐8 | 6 (6.9) | 5 (8.9) | 1 (3.2) | 2 (4.5) | 3 (7.5) |
| 6 | IGHV4‐34 | 5 (5.7) | 5 (8.9) | 0 | 3 (6.8) | 2 (5) |
| 7 | IGHV3‐33 | 5 (5.7) | 2 (3.6) | 3 (9.7) | 3 (6.8) | 2 (5) |
| 8 | IGHV1‐69 | 5 (5.7) | 1 (1.8) | 4 (12.9) | 3 (6.8) | 2 (5) |
| 9 | IGHV3‐23 | 4 (4.6) | 3 (5.4) | 1 (3.2) | 4 (9.1) | 0 |
| 10 | IGHV1‐2 | 3 (3.4) | 0 | 3 (9.7) | 0 | 3 (7.5) |
| 11 | IGHV3‐11 | 3 (3.4) | 2 (3.6) | 1 (3.2) | 1 (2.3) | 2 (5) |
| 12 | IGHV3‐64 | 3 (3.4) | 3 (5.4) | 0 | 2 (4.5) | 0 |
| 13 | IGHV6‐1 | 3 (3.4) | 3 (5.4) | 0 | 0 | 3 (7.5) |
| 14 | IGHV1‐46 | 2 (2.3) | 0 | 2 (6.5) | 1 (2.3) | 1 (2.5) |
| 15 | IGHV3‐15 | 2 (2.3) | 2 (3.6) | 0 | 0 | 1 (2.5) |
| 16 | IGHV3‐49 | 2 (2.3) | 0 | 2 (6.5) | 0 | 2 (5) |
| 17 | IGHV4‐30 | 2 (2.3) | 1 (1.8) | 1 (3.2) | 0 | 2 (5) |
| 18 | IGHV4‐4 | 2 (2.3) | 2 (3.6) | 0 | 1 (2.3) | 1 (2.5) |
| 19 | IGHV4‐b | 2 (2.3) | 0 | 2 (6.5) | 1 (2.3) | 1 (2.5) |
| 20 | *Others | 3 (3.4) | 2 (3.6) | 1 (3.2) | 1 (2.3) | 2 (5) |
Data are presented as number (%) of patients expressing the corresponding immunoglobulin variable region heavy chain (IGHV) member; All significant (P < 0.05) differences between groups are shown in bold. *Others represent IGHV3‐66, IGHV4‐59, and IGHV5‐a genes, each expressed in one patient. CLL, chronic lymphocytic leukemia.
The expression of the IGHV1 gene family was more represented in progressive and unmutated patients compared to non‐progressive and mutated groups of patients, but the difference was significant only between mutated and unmutated patients (10.7%vs 32.3%; P = 0.02) (Table 1). Among the IGHV1 family members, IGHV1‐69 (12.9%vs 1.8%; P = 0.05) and IGHV1‐2 (7.9%vs 0%; P = 0.04) genes were expressed at significantly higher frequency in unmutated compared to mutated patients (Table 2).
The most predominantly used IGHV4 members were IGHV4‐39 (n = 6) and IGHV4‐34 (n = 5) genes representing 61% (11/18) of the total IGHV4 family. All these genes, with the exception of one, were mutated (Table 2).
Significant differences were observed neither for the frequency of IGHV family members, nor for their mutational status between male and female CLL patients (data not shown).
Association of rearranged IGHV genes with disease outcome. Analysis of the association of TTFT and PFS with the usage of IGHV families revealed no significant association with any of the IGHV gene families. Of the IGHV gene members a higher TTFT was observed in patients expressing IGHV3‐7 compared to those expressing other IGHV family members (29 vs 10 months, P = 0.08).
HCDR3 length and motifs. The median length of the HCDR3 region was 17 amino acids (aa) in our CLL patients. There was a significantly higher mean length of HCDR3 in unmutated CLL patients compared to the mutated group (20 vs 15 aa, P < 0.0001). Comparing non‐progressive and progressive patients, there was also a significantly higher mean length of HCDR3 in progressive CLL patients (18 vs 15 aa, P = 0.008).
When HCDR3 mean length was analyzed in relation to the rearranged IGHV gene family, the length of IGHV1‐expressing samples (19 aa) was greater than IGHV3‐ (16 aa) and IGHV4‐ (15 aa) expressing samples. When HCDR3 length was compared among the most frequently used gene members of these families, IGHV3‐7 was associated with an average length of 16 aa, IGHV3‐30 of 20, IGHV3‐48 of 19, IGHV4‐39 of 15, IGHV1‐8 of 19, IGHV4‐34 of 19, IGHV3‐33 of 20, and IGHV1‐69 of 21 amino acids.
Composition of the HCDR3 region expressed in our CLL samples revealed similarities between some samples. We found the motif (Y or H) YYYGMDV in 13 (15%) samples who all used IGHJ6 gene in their HCDR3 together with IGHD6‐13 in six samples, IGHD3‐22 in two, and IGHD1‐26, IGHD5‐5, IGHD4‐17, IGHD3‐3, and IGHD2‐2 each in one sample. Seven of these patients used the IGHV3‐30 gene, while IGHV4‐34 and IGHV3‐48 were used each in two patients, and IGHV3‐49 and IGHV1‐69 were used each by one patient. Eight of these patients had an unmutated IGHV gene.
The motif ‘ARDQDGGWGVDY’ was found in three (3.5%) patients with IGHV3‐64, IGHD6, and IGHJ4 rearrangement, all with mutated IGHV sequence. The HCDR3 sequence ‘GRVIYSSSERNLDF’ was also observed in two (2.3%) patients, both with IGHV3‐48/ IGHD6/ IGHJ4 genes.
Discussion
The observation of highly significant differences in disease outcome between mutated and unmutated CLL subtypes has prompted extensive investigations on the mutational status and VH/VL gene usage in CLL over the last few years. These studies have demonstrated a biased use of certain IGHV gene family members, preferentially higher mutation rate in certain IGHV gene members as well as higher frequency of closely related (stereotyped) HCDR3 sequences in patients sharing the same or homologous IGHV gene members.( 33 , 34 ) These findings imply selection and clonal expansion of leukemic B cells by certain unidentified foreign or self antigens. As such, environmental and/or ethnic variations may affect IGHV gene usage in CLL.
Most previous studies have been conducted in Western populations with similar geographic and/or ethnic backgrounds. Nevertheless, such variations have been reported; perhaps the most frequent one is the use of IGHV3‐21 with a frequency of approximately 11.7% in the Scandinavian region,( 20 ) 7.9% in Ireland,( 35 ) and 2–3% in the Mediterranean region.( 6 ) Such analysis in CLL patients from disparate ethnic and geographic origins could expand our understanding of CLL and deserves special consideration.
Little is known about Asian CLL patients. Analysis of IGHV gene family usage in Asian CLL patients has so far been reported only in small cohorts of Japanese,( 7 , 36 ) Chinese,( 37 ) and Iranian( 38 ) patients. This could partly be due to the low incidence of CLL in Asian populations( 39 , 40 ) compared to the Western countries. Our previous study, which was performed in a small number of Iranian CLL patients to determine the frequency of the expressed IGHV gene family, revealed significant differences in the expression of IGHV1 and IGHV4 gene families between Iranian and Western patients.( 38 )
In the present study we investigated the frequency of the expressed IGHV gene family members and the prognostic significance of somatic hypermutation of IGHV genes in Iranian CLL patients. We also analysed the HCDR3 sequence in leukemic B cells to find out homologous motifs in our CLL patients. In general, the most frequently used IGHV family by the leukemic B cells of our CLL patients was IGHV3 followed by IGHV4, IGHV1, IGHV6, and IGHV5, with no expression of the IGHV2 and IGHV7 families. Although the IGHV3 family was predominantly expressed in non‐progressive (30/49, 61.2%) and mutated CLL subtypes (32/49, 65.3%), the IGHV1 family was more represented in progressive (9/16, 56.3%) and unmutated (10/16, 62.5%) CLL patients (Table 1). Our results on the frequency of IGHV families are similar to those obtained in Japanese( 7 , 36 ) and Chinese( 37 ) CLL patients. Comparing the frequency of IGHV family expression in CLL patients with normal adult B cells( 41 ) revealed a higher representation of the IGHV3 family in our CLL patients, but no differences with their normal CD5+/IgM+ B cell counterparts,( 42 ) which is also in agreement with the Japanese CLL patients.( 7 , 36 ) Our results did not confirm the results of Fais et al.,( 13 ) who reported a significant decrease in IGHV3 family expression in their CLL patients compared to normal CD5+/IgM+ B cells. The IGHV1 family is predominantly used after the IGHV3 family in Western CLL patients,( 15 , 16 , 25 , 43 ) while the use of IGHV4 family is more predominant after IGHV3 in our patients and those from China( 37 ) and Japan.( 7 , 36 )
The most frequently represented IGHV genes in our cohort of CLL patients were IGHV3‐7 (12.6%), IGHV3‐30 (11.4%), IGHV3‐48 (9.2%), IGHV4‐39 (6.9%), and IGHV1‐8 (6.9%). Collectively, these five IGHV genes represent nearly half (47%) of all Iranian CLL cases, whereas they are expressed in about 20% of Western, 21.3% of Japanese,( 36 ) and 14.3% of Chinese( 37 ) CLL patients. While IGHV3‐7 is the most frequent IGHV gene in Iranian CLL patients, IGHV4‐34 is most frequently expressed in Chinese( 37 ) and Japanese( 36 ) CLL patients. IGHV3‐7 was predominantly expressed in our mutated and non‐progressive CLL patients. Higher frequency usage of this gene in mutated CLL patients has also been observed in other studies.( 16 , 19 , 25 ) Comparing the expression frequency of IGHV3‐7 in Iranian CLL patients with Western CLL patients revealed a significantly higher representation of this gene in Iranian CLL patients (Iranian vs Western, P = 0.005; Iranian vs American, P = 0.03; Iranian vs European, P = 0.004), but no differences with Chinese and Japanese CLL patients as Asian populations (Table 3).
Table 3.
Comparison of IGHV gene usage in Iranian CLL patients with other studies
| Country | No of patients | IGHV member | Reference | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1–2 | 1–8 | 1–18 | 1–69 | 3–7 | 3–21 | 3–23 | 3–30 | 4–34 | 3–48 | |||
| USA | 83 | 6 | 0 | 4.8 | 7.1 | 12.1 | 0 | 4.8 | 3.6 | 18.1 | 2.5 | 13 |
| USA | 34 | 3 | 3 | 0 | 5.9 | 3 | 5.9 | 8.8 | 5.9 | 11.8 | 0 | 26 |
| USA | 172 | 6.4 | 0.6 | 2.3 | 10.5 | 8.2 | 1.2 | 6.4 | 4.7 | 14.5 | 2.9 | 17 |
| USA | 356 | 4.8 | 0.5 | 2.4 | 18 | 4.3 | 1.6 | 7 | 4.8 | 7 | 4.3 | 19 |
| USA | 190 | 4.7 | NI | NI | 11 | 4.7 | NI | 5.8 | 4.7 | 14.2 | 7.9 | 45 |
| England | 84 | 7.1 | 0 | 1.2 | 11.9 | 7.1 | 1.2 | 11.9 | 11.9 | 11.9 | 4.8 | 15 |
| England | 154 | 6.3 | 0 | 1.3 | 11.3 | 5 | 3.5 | 8.2 | 8.8 | 7.1 | 5.7 | 16 |
| Sweden | 346 | 4 | NI | NI | 14 | 3 | 9 | 5 | 6 | 7 | NI | 25 |
| Sweden | 265 | NI | NI | NI | 17.4 | 3.8 | 11.7 | 4.9 | 5.7 | NI | NI | 20 |
| Italy | 85 | 5.7 | 5.7 | NI | 9.1 | 5.7 | 1.1 | 8 | 9.1 | 8 | NI | 8, 43 |
| Spain | 56 | 1.8 | 0 | 3.6 | 21.5 | 1.8 | 1.8 | 5.4 | 3.6 | 7.1 | 3.6 | 4 |
| Mediterranean area | 553 | 4.2 | 1 | 2.8 | 10.7 | 5.4 | 3 | 9.2 | 6.7 | 10.7 | 2.2 | 6 |
| Ukraine | 189 | 5.3 | NI | NI | 21.7 | 4.8 | 5.8 | 1.6 | 3.7 | 7.4 | 4.2 | 44 |
| Ireland | 228 | 7.9 | NI | NI | 12.3 | NI | 7.9 | NI | NI | 13.5 | 3.5 | 35 |
| Japan | 44 | NI | NI | NI | 2.3 | NI | 7 | 14 | 7 | 20.9 | NI | 7 |
| Japan | 80 | 1.25 | 3.75 | 2.5 | 1.25 | 5 | 7.5 | 8.75 | 6.25 | 27.5 | 3.75 | 36 |
| China | 65 | 0 | 0 | 0 | 1.5 | 4.6 | 3 | 7.7 | 6.2 | 12.3 | 1.5 | 37 |
| Iran | 87 | 3.4 | 6.9 | 0 | 5.7 | 12.6 | 0 | 4.6 | 11.4 | 5.7 | 9.2 | Present study |
The results represent the percent of each immunoglobulin variable region heavy chain (IGHV) member. CLL, chronic lymphocytic leukemia; NI, not identified.
IGHV1‐69 is the most frequently used IGHV gene in Western CLL patients reported by many investigators, generally with unmutated phenotype (Table 3). The frequency of this gene varies in different populations based on their geographic origin. The most frequent usage of this gene has been reported in Ukrainian( 44 ) and Spanish( 4 ) CLL patients (∼21.5%) of whom 98% displayed an unmutated sequence. The frequency of this gene is significantly higher in our unmutated CLL patients compared to mutated ones (12.9%vs 1.8%, P = 0.03) (Table 2) confirming the results obtained in Western patients.( 4 , 6 , 8 ) However, the frequency of this gene in Iranian patients is significantly lower than that in Western CLL patients (Iranian vs Western, P = 0.02; Iranian vs American, P = 0.04; Iranian vs European, P = 0.02), with no significant difference compared to other Asian (Japanese and Chinese) CLL populations. Taken together, representation of IGHV1‐69 is significantly lower in Asian CLL patients (Asian vs Western, P < 0.00001, Table 3). This difference is the most striking feature of Asian compared to Western CLL patients.
While IGHV1‐2 and IGHV1‐69 genes in our patients carry a few or no mutations, IGHV1‐8, IGHV3‐7, IGHV3‐23, and IGHV3‐48 genes showed a high load of mutations in their sequences. Our results are in agreement with other studies,( 1 , 6 , 13 ) showing a few mutations in the IGHV1‐69 gene and a higher frequency of mutations in IGHV3‐23 and IGHV3‐7 genes (Fig. 2).
Figure 2.
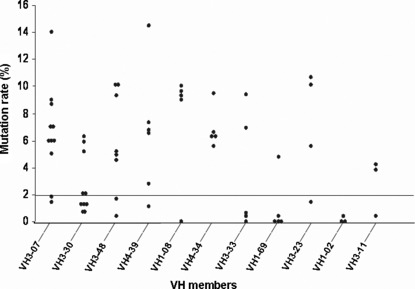
Distribution of mutations in the most frequently expressed immunoglobulin variable region heavy chain (IGHV) genes in chronic lymphocytic leukemia (CLL) patients. Baseline denotes a cut‐off mutation value (2%) assigned to differentiate between mutated and unmutated IGHV sequences.
In contrast to Western CLL patients, especially of those Scandinavian origin,( 20 , 25 ) leukemic B cells of our CLL patients did not use the IGHV3‐21 gene. Over‐representation of IGHV3‐21 has also been reported in other Western CLL populations from the USA,( 13 ) Italy,( 8 , 43 ) and England,( 15 ) though contradicting results have also been reported.( 6 , 17 , 19 , 26 ) The differences in the frequency of this gene in patients with the same ethnic origin might be due to the small number of patients studied or more importantly because of their geographic origin. The important role of geographic origin of patients for the expression of IGHV3‐21 by leukemic B cells of CLL patients has been verified in Scandinavian patients.( 20 , 25 ) This possibility is highlighted by differential usage of IGHV3‐21 and IGHV1‐69 genes between Asian (especially Iranian) and Western CLL patients. For further insight into this issue, the normal IGHV repertoire should also be investigated in healthy subjects of the same ethnicity living in the same geographic areas, to be able to find out associations, if any, between the over‐expressed IGHV genes and ethnic/geographic parameters.
The frequency of IGHV4‐34 has been reported to be high in Americans( 13 , 17 , 26 , 45 ) and the two Asian, Japanese and Chinese,( 7 , 36 , 37 ) CLL patients, but only a small number (5.7%) of our Iranian CLL patients expressed this gene. The frequency of IGHV4‐34 gene usage in our patients is comparable to that of European patients, but significantly lower than the American CLL patients (P = 0.003). Over‐representation of this gene has also been observed in Japanese and Chinese patients (Japanese–Chinese vs Iranian, P = 0.003; Japanese–Chinese vs European, P < 0.0001). Together with some similarities between Asian populations, the lower frequency of the IGHV4‐34 gene in Iranian patients compared to that of Japanese and Chinese CLL patients may also reflect the differences in genetic background or environmental factors between these two distinct populations.
Collectively four IGHV genes, including IGHV1‐69, IGHV3‐7, IGHV3‐21, and IGHV4‐34 among all other IGHV genes showed significant differences in different ethnic/geographic CLL populations. This is a very important finding for our understanding of CLL etiopathogenesis. The apparent geographic/ethnic bias for the frequency of defined IGHV genes shown in our study and by others in different ethnic populations may theoretically be due to specific genetic background, depending on variations in germline composition of the IGHV locus, or to the effects of a potential environmental variable less frequently encountered in different regions as proposed by Ghia et al. ( 6 ) The lack of information for the frequency of IGHV genes in normal B cells and especially CD5+ B cells in Iranian and also other ethnic populations such as Scandinavian and Asian populations is the main gap in determining the exact biased expression of IGHV families and members in our CLL patients. The biased usage of IGHV genes and the presence of shared antigen‐binding motifs in HCDR3 regions of neoplastic B cells in our series of CLL patients is important evidence indicating that CLL may be sustained by an antigen‐driven process.( 1 ) It has been shown recently that BCR stimulation of B cells from CLL patients results in over‐expression of genes involved in signal transduction and cell cycle regulation, especially in unmutated CLL patients which resulted in extended survival of CLL cells.( 46 ) These results imply that antigenic stimulation of BCR by a self or foreign antigen plays a pivotal role in disease progression and/or initiation of malignant transformation of B cells.
The sequence ([Y or H] YYYGMDV) was the most predominant HCDR3 motif found in 13 (15%) of our patients. This sequence is largely encoded by the IGHJ6 gene which was found to be rearranged in all these patients. The motif ‘ARDQDGGWGVDY’ was found in three patients with the IGHV3‐64/IGHD6/IGHJ4 combination, all with mutated IGHV sequences, and ‘GRVIYSSSERNLDF’ was used in two patients with IGHV3‐48/IGHD6/IGHJ4 genes. This information is exclusive to our report with no previous precedent.
The motifs ‘DIVVVPA’, ‘DFWSGYY’,‘RFLEWL’, and ‘YYYGMDV’, which have previously been reported in CLL patients,( 8 , 18 , 22 , 47 ) were each found in HCDR3 of only one of our patients.
Comparison of the HCDR3 sequences of our CLL patients with archetypes clustering of HCDR3 recently reported by Messmer et al. ( 33 ) revealed that only one of our patients has the same sequence as the archetype no. 31 (CARDQYYYDSSGYYSGYFDYW) rearranging the same IGHV1‐46/ IGHD3‐22/IGHJ4 combination with no mutation in the IGHV sequence. Based on either 70% or 60% sequence homology with 67 represented HCDR3 archetypes clustered by Messmer et al.,( 33 ) seven and 31 patients within our samples could be classified within 12 and 27 archetypes, respectively. The small sample size of our patients and the fact that these archetypes are based on HCDR3 sequences from a large cohort of Western CLL patients, may explain the low level of similarities of HCDR3 observed in our patients compared to those reported by others.( 33 , 48 ) To determine the real archetypes of Iranian or Asian CLL patients, analysis of a large series of CLL patients is necessary.
The length of HCDR3 in our unmutated CLL patients was significantly longer than that in mutated cases (P < 0.0001). This finding was also reported by others( 8 , 18 , 22 , 49 ) and confirms that the IGHV sequence of unmutated leukemic B cells in the germline configuration might be derived from a more immature B cell than that of mutated CLL patients.( 8 , 18 ) The differences in HCDR3 length was not due to the selected germline IGHD genes, since for instance IGHD3‐3 (overused in unmutated group) and IGHD3‐22 (overused in mutated group) are both 31 nucleotides long. Furthermore, in the mutated group rearrangements involving the overused IGHV3‐7 gene were associated with a shorter HCDR3 (average 16 aa) than those of the IGHV3‐7 gene used in the unmutated group (18 aa). Our results are in agreement with the study of Duke et al. ( 16 ) who reported a significantly longer HCDR3 length in the unmutated group with overused IGHV1‐69 gene compared to the IGHV1‐69 gene used in the mutated CLL patients.
The present data on the similarity of HCDR3 in a subset of CLL patients strongly suggests the role of antigen selection in these patients. Since the ratio of R/S mutations in Ig CDRs may reflect the role of antigenic selection in the mutated CLL patients, nearly half of our mutated CLL patients (46.5%) displayed this pattern, in agreement with other studies.( 50 , 51 ) Together, the presence of specific HCDR3 motifs, biased expression of certain IGHV genes, and the ratio of R/S mutations in leukemic cells of our CLL patients, suggest involvement of an as yet undefined pathogen or autoantigen in the pathogenesis or initiation of CLL.
Although it seems that patients with CLL have some defects in their immune responses (especially T‐cell defects),( 52 ) the better prognosis of patients with mutated IGHV genes could be due to induction of autologous T‐cell response to the mutated epitopes (idiotopes) located on the IGHV region of the leukemic cells. We are currently embarking on elucidation of this issue by analysis of the autologous T‐cell response to peptides resembling the mutated epitopes of the IGHV region of our CLL patients.
In conclusion, it seems that skewed representation of IGHV genes and the presence of similar HCDR3 sequences in leukemic B cells of mutated and unmutated CLL patients from different ethnic populations suggest a role for an as yet undefined set of pathogens or autoantigens in the initiation of CLL. Some types of lymphomas such as marginal zone lymphoma have been proposed to be associated with chronic stimulation by persistent microbial antigens and/or autoantigens.( 53 ) The role of pathogens in malignancies may be direct through their oncogenes or indirect by promoting favorable conditions for lymphocyte transformation, such as increased proliferation or decreased apoptosis of lymphoid cells.( 53 , 54 ) Regional differences observed between our patients and those from other ethnic and/or geographic origins may imply contribution of different antigenic stimuli for selection of the neoplastic B cells. Extension of this study in a large cohort of Iranian and other Asian populations, particularly Chinese and Japanese patients with completely diverse ethnic and geographic backgrounds, as well as other ethnic populations, especially African and Australian CLL patients, may reveal some new aspects of the etiopathogenesis of CLL.
Acknowledgments
This study was supported by a grant from the Nanotechnology Network of the Ministry of Health and Medical Education of Iran. We are grateful to Joseph Lawrence for his technical assistance in nucleotide sequencing.
References
- 1. Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med 2005; 352: 804–15. [DOI] [PubMed] [Google Scholar]
- 2. Oscier DG, Gardiner AC, Mould SJ et al. Multivariate analysis of prognostic factors in CLL: clinical stage, IGVH gene mutational status, and loss or mutation of the p53 gene are independent prognostic factors. Blood 2002; 100: 1177–84. [PubMed] [Google Scholar]
- 3. Damle RN, Wasil T, Fais F et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood 1999; 94: 1840–7. [PubMed] [Google Scholar]
- 4. Crespo M, Bosch F, Villamor N et al. ZAP‐70 expression as a surrogate for immunoglobulin‐variable‐region mutations in chronic lymphocytic leukemia. N Engl J Med 2003; 348: 1764–75. [DOI] [PubMed] [Google Scholar]
- 5. Caporaso N, Goldin L, Plass C et al. Chronic lymphocytic leukaemia genetics overview. Br J Hematol 2007; 139: 630–4. [DOI] [PubMed] [Google Scholar]
- 6. Ghia P, Stamatopoulos K, Belessi C et al. Geographic patterns and pathogenetic implications of IGHV gene usage in chronic lymphocytic leukemia: the lesson of the IGHV3‐21 gene. Blood 2005; 105: 1678–85. [DOI] [PubMed] [Google Scholar]
- 7. Koiso H, Yamane A, Mitsui T et al. Distinctive immunoglobulin VH gene usage in Japanese patients with chronic lymphocytic leukemia. Leuk Res 2006; 30: 272–6. [DOI] [PubMed] [Google Scholar]
- 8. Donisi PM, Di Lorenzo N, Riccardi M et al. Pattern and distribution of immunoglobulin VH gene usage in a cohort of B‐CLL patients from a Northeastern region of Italy. Diagn Mol Pathol 2006; 15: 206–15. [DOI] [PubMed] [Google Scholar]
- 9. Rassenti LZ, Huynh L, Toy TL et al. ZAP‐70 compared with immunoglobulin heavy‐chain gene mutation status as a predictor of disease progression in chronic lymphocytic leukemia. N Engl J Med 2004; 351: 893–901. [DOI] [PubMed] [Google Scholar]
- 10. Montillo M, Hamblin T, Hallek M et al. Chronic lymphocytic leukemia: novel prognostic factors and their relevance for risk‐adapted therapeutic strategies. Haematologica 2005; 90: 391–9. [PubMed] [Google Scholar]
- 11. Vasconcelos Y, Davi F, Levy V et al. Binet’s staging system and VH genes are independent but complementary prognostic indicators in chronic lymphocytic leukemia. J Clin Oncol 2003; 21: 3928–32. [DOI] [PubMed] [Google Scholar]
- 12. Maloum K, Davi F, Merle‐Beral H et al. Expression of unmutated VH genes is a detrimental prognostic factor in chronic lymphocytic leukemia. Blood 2000; 96: 377–9. [PubMed] [Google Scholar]
- 13. Fais F, Ghiotto F, Hashimoto S et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest 1998; 102: 1515–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rosenquist R, Lindstrom A, Holmberg D et al. V(H) gene family utilization in different B‐cell lymphoma subgroups. Eur J Hematol 1999; 62: 123–8. [DOI] [PubMed] [Google Scholar]
- 15. Hamblin TJ, Davis Z, Gardiner A et al. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood 1999; 94: 1848–54. [PubMed] [Google Scholar]
- 16. Duke VM, Gandini D, Sherrington PD et al. V(H) gene usage differs in germline and mutated B‐cell chronic lymphocytic leukemia. Haematologica 2003; 88: 1259–71. [PubMed] [Google Scholar]
- 17. Messmer BT, Albesiano E, Messmer D et al. The pattern and distribution of immunoglobulin VH gene mutations in chronic lymphocytic leukemia B cells are consistent with the canonical somatic hypermutation process. Blood 2004; 103: 3490–5. [DOI] [PubMed] [Google Scholar]
- 18. Widhopf GF II, Rassenti LZ, Toy TL et al. Chronic lymphocytic leukemia B cells of more than 1% of patients express virtually identical immunoglobulins. Blood 2004; 104: 2499–504. [DOI] [PubMed] [Google Scholar]
- 19. Mauerer K, Zahrieh D, Gorgun G et al. Immunoglobulin gene segment usage, location and immunogenicity in mutated and unmutated chronic lymphocytic leukaemia. Br J Hematol 2005; 129: 499–510. [DOI] [PubMed] [Google Scholar]
- 20. Tobin G, Thunberg U, Johnson A et al. Chronic lymphocytic leukemias utilizing the VH3‐21 gene display highly restricted Vlambda2‐14 gene use and homologous CDR3s: implicating recognition of a common antigen epitope. Blood 2003; 101: 4952–7. [DOI] [PubMed] [Google Scholar]
- 21. Widhopf GF II, Kipps TJ. Normal B cells express 51p1‐encoded Ig heavy chains that are distinct from those expressed by chronic lymphocytic leukemia B cells. J Immunol 2001; 166: 95–102. [DOI] [PubMed] [Google Scholar]
- 22. Potter KN, Orchard J, Critchley E et al. Features of the overexpressed V1‐69 genes in the unmutated subset of chronic lymphocytic leukemia are distinct from those in the healthy elderly repertoire. Blood 2003; 101: 3082–4. [DOI] [PubMed] [Google Scholar]
- 23. Kipps TJ, Tomhave E, Pratt LF et al. Developmentally restricted immunoglobulin heavy chain variable region gene expressed at high frequency in chronic lymphocytic leukemia. Proc Natl Acad Sci USA 1989; 86: 5913–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schroeder HW Jr, Dighiero G. The pathogenesis of chronic lymphocytic leukemia: analysis of the antibody repertoire. Immunol Today 1994; 15: 288–94. [DOI] [PubMed] [Google Scholar]
- 25. Tobin G, Thunberg U, Karlsson K et al. Subsets with restricted immunoglobulin gene rearrangement features indicate a role for antigen selection in the development of chronic lymphocytic leukemia. Blood 2004; 104: 2879–85. [DOI] [PubMed] [Google Scholar]
- 26. Pasqualucci L, Neri A, Baldini L et al. BCL‐6 mutations are associated with immunoglobulin variable heavy chain mutations in B‐cell chronic lymphocytic leukemia. Cancer Res 2000; 60: 5644–8. [PubMed] [Google Scholar]
- 27. Ghia EM, Jain S, Widhopf GF II et al. Use of IGHV3‐21 in chronic lymphocytic leukemia is associated with high‐risk disease and reflects antigen‐driven, post‐germinal center leukemogenic selection. Blood 2008; 111: 5101–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jaffe ES, Harris NL, Stein H, Vardiman JW, eds. World Health Organization Classification of Tumours. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC Press, 2001. [Google Scholar]
- 29. Cheson BD, Bennett JM, Grever M et al. National Cancer Institute‐sponsored Working Group guidelines for chronic lymphocytic leukemia: revised guidelines for diagnosis and treatment. Blood 1996; 87: 4990–7. [PubMed] [Google Scholar]
- 30. Jelinek DF, Tschumper RC, Geyer SM et al. Analysis of clonal B‐cell CD38 and immunoglobulin variable region sequence status in relation to clinical outcome for B‐chronic lymphocytic leukaemia. Br J Hematol 2001; 115: 854–61. [DOI] [PubMed] [Google Scholar]
- 31. Olerup O, Zetterquist H. HLA‐DR typing by PCR amplification with sequence‐specific primers (PCR‐SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor–recipient matching in cadaveric transplantation. Tissue Antigens 1992; 39: 225–35. [DOI] [PubMed] [Google Scholar]
- 32. Lossos IS, Tibshirani R, Narasimhan B et al. The inference of antigen selection on Ig genes. J Immunol 2000; 165: 5122–6. [DOI] [PubMed] [Google Scholar]
- 33. Messmer BT, Raphael BJ, Aerni SJ et al. Computational identification of CDR3 sequence archetypes among immunoglobulin sequences in chronic lymphocytic leukemia. Leuk Res 2009; 33: 368–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Murray F, Darzentas N, Hadzidimitriou A et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood 2008; 111: 1524–33. [DOI] [PubMed] [Google Scholar]
- 35. Matthews C, Catherwood MA, Morris TC et al. V(H)3‐48 and V(H)3‐53, as well as V(H)3‐21, gene rearrangements define unique subgroups in CLL and are associated with biased lambda light chain restriction, homologous LCDR3 sequences and poor prognosis. Leuk Res 2007; 31: 231–4. [DOI] [PubMed] [Google Scholar]
- 36. Nakahashi H, Tsukamoto N, Hashimoto Y et al. Characterization of immunoglobulin heavy and light chain gene expression in chronic lymphocytic leukemia and related disorders. Cancer Sci 2009; 100: 671–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen L, Zhang Y, Zheng W et al. Distinctive IgVH gene segments usage and mutation status in Chinese patients with chronic lymphocytic leukemia. Leuk Res 2008; 32: 1491–8. [DOI] [PubMed] [Google Scholar]
- 38. Farsangi MH, Jeddi‐Tehrani M, Sharifian RA et al. Analysis of the immunoglobulin heavy chain variable region gene expression in Iranian patients with chronic lymphocytic leukemia. Leuk Lymphoma 2007; 48: 109–16. [DOI] [PubMed] [Google Scholar]
- 39. Moazzeni SM, Amirzargar AA, Shokri F. HLA antigens in Iranian patients with B‐cell chronic lymphocytic leukemia. Pathol Oncol Res 1999; 5: 142–5. [DOI] [PubMed] [Google Scholar]
- 40. Tamura K, Sawada H, Izumi Y et al. Chronic lymphocytic leukemia (CLL) is rare, but the proportion of T‐CLL is high in Japan. Eur J Hematol 2001; 67: 152–7. [DOI] [PubMed] [Google Scholar]
- 41. Kraj P, Friedman DF, Stevenson F et al. Evidence for the overexpression of the VH4‐34 (VH4.21) Ig gene segment in the normal adult human peripheral blood B cell repertoire. J Immunol 1995; 154: 6406–20. [PubMed] [Google Scholar]
- 42. Brezinschek HP, Foster SJ, Brezinschek RI et al. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(−)/IgM+ B cells. J Clin Invest 1997; 99: 2488–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nardini E, Neri F, Vicenzi E et al. Thymic function and immunoglobulin mutation genotype in B‐cell chronic lymphocytic leukemia patients. Int J Cancer 2003; 107: 958–61. [DOI] [PubMed] [Google Scholar]
- 44. Abramenko I, Bilous N, Kryachok I et al. IGHV3‐21 gene expression in patients with B‐cell chronic lymphocytic leukemia in Ukraine. Exp Oncol 2007; 29: 226–30. [PubMed] [Google Scholar]
- 45. Weinberg JB, Volkheimer AD, Chen Y et al. Clinical and molecular predictors of disease severity and survival in chronic lymphocytic leukemia. Am J Hematol 2007; 82: 1063–70. [DOI] [PubMed] [Google Scholar]
- 46. Guarini A, Chiaretti S, Tavolaro S et al. BCR ligation induced by IgM stimulation results in gene expression and functional changes only in IgV H unmutated chronic lymphocytic leukemia (CLL) cells. Blood 2008; 112: 782–92. [DOI] [PubMed] [Google Scholar]
- 47. Rosenwald A, Alizadeh AA, Widhopf G et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med 2001; 194: 1639–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stamatopoulos K, Belessi C, Moreno C et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: pathogenetic implications and clinical correlations. Blood 2007; 109: 259–70. [DOI] [PubMed] [Google Scholar]
- 49. Tobin G, Thunberg U, Johnson A et al. Somatically mutated Ig V(H)3‐21 genes characterize a new subset of chronic lymphocytic leukemia. Blood 2002; 99: 2262–4. [DOI] [PubMed] [Google Scholar]
- 50. Degan M, Rupolo M, Bo MD et al. Mutational status of IgVH genes consistent with antigen‐driven selection but not percent of mutations has prognostic impact in B‐cell chronic lymphocytic leukemia. Clin Lymphoma 2004; 5: 123–6. [DOI] [PubMed] [Google Scholar]
- 51. Degan M, Bomben R, Bo MD et al. Analysis of IgV gene mutations in B cell chronic lymphocytic leukaemia according to antigen‐driven selection identifies subgroups with different prognosis and usage of the canonical somatic hypermutation machinery. Br J Hematol 2004; 126: 29–42. [DOI] [PubMed] [Google Scholar]
- 52. Gallego A, Vargas JA, Castejon R et al. Production of intracellular IL‐2, TNF‐alpha, and IFN‐gamma by T cells in B‐CLL. Cytometry B Clin Cytom 2003; 56: 23–9. [DOI] [PubMed] [Google Scholar]
- 53. Suarez F, Lortholary O, Hermine O et al. Infection‐associated lymphomas derived from marginal zone B cells: a model of antigen‐driven lymphoproliferation. Blood 2006; 107: 3034–44. [DOI] [PubMed] [Google Scholar]
- 54. Li HC, Stoicov C, Rogers AB et al. Stem cells and cancer: evidence for bone marrow stem cells in epithelial cancers. World J Gastroenterol 2006; 12: 363–71. [DOI] [PMC free article] [PubMed] [Google Scholar]


