Abstract
CD98 is known as a cell surface antigen expressed in proliferating normal tissues and in almost all tumor cells. Although the function of CD98 is not yet fully elucidated, it is suggested that CD98 is concerned functionally in lymphocyte activation, cell proliferation, and malignant transformation. Monoclonal antibody against human CD98 heavy chain (h.c.), termed HBJ127, shows inhibition of lymphocyte activation and tumor cell growth in vitro. These observations suggest that the epitope recognized by HBJ127 may be crucial for CD98 function. In the present study, the authors investigated the epitope recognized by HBJ127 using a phage display random heptapeptide library. Approximately 2.4 × 104‐fold amplification of eluted phage titer was obtained after three rounds of panning of the phage library against HBJ127. Seven different heptapeptide sequences were isolated from 30 randomly selected clones of the post‐panning phage population. A homology search using ClustalW identified the peptide sequence corresponding to 442AFS444 of human CD98 h.c. It was also found that 443F is a human‐specific amino acid by comparing sequences of human, rat, and mouse origin. Reduced reactivity of HBJ127 was detected against the phenylalanine‐substituted peptide but not detected against the alanine or serine‐substituted one. It has been identified that HBJ127 reacts only with human species and the HBJ127 epitope position is predicted in 418–529 of human CD98 h.c. From these results and observations, it was estimated that 442AFS444 of human CD98 h.c. may be the HBJ127 epitope. Moreover, 443F may be critical for the binding of HBJ127 against human CD98 h.c. (Cancer Sci 2007; 98: 1696–1700)
CD98/GP125 is a heterodimeric protein with a relative molecular mass of 125‐kDa consisting of a glycosylated 85‐kDa heavy chain (h.c.) and a non‐glycosylated 40‐kDa light chain, which are disulfide‐linked.( 1 ) Analyses of human CD98 cDNA have revealed that CD98 is a type II transmembrane glycoprotein( 2 ) that is disulfide‐linked to a non‐glycosylated light chain of a member of the permease family.( 3 ) CD98 was identified originally as a cell surface antigen associated with lymphocyte activation defined by the 4F2 monoclonal antibody (mAb),( 1 ) and is expressed in proliferating normal tissues( 4 ) and in almost all tumor cells.( 5 ) These findings suggest that CD98 is involved functionally in lymphocyte activation, cell proliferation and malignant transformation. In fact, a mAb against CD98, termed HBJ127, which was originally raised against T24 human bladder cancer,( 5 ) inhibited tumor cell growth( 6 ) and lymphocyte proliferation.( 7 ) Furthermore, CD98 h.c. cDNA‐transfected murine fibroblasts showed various malignant phenotypes.( 8 )
Phage display, originally developed by Smith,( 9 ) is the procedure for expression of proteins or peptides of interest on the surface of filamentous phage M13 as a fusion protein with viral coat proteins. Biopanning, which is characteristic for phage display, made it easy to screen and select the positive clones because several rounds of reaction between the phage display library and the target molecule can concentrate the population of positive clones in the library. This technique is now widely used for isolation of recombinant antibody fragments,( 10 , 11 , 12 , 13 , 14 , 15 ) for identification of receptor ligands,( 16 , 17 , 18 ) and for epitope mapping of mAb.( 19 , 20 , 21 )
Because HBJ127 inhibits CD98 function, the epitope recognized by this mAb may play an important role in expressing CD98 function. In the present study, the authors tried to identify the epitope sequence recognized by HBJ127 using a phage display random peptide library. Three rounds of biopanning resulted in 2.4 × 104‐fold amplification of the HBJ127‐bound phage titer. Sequence analysis of 30 clones of the HBJ127‐bound phage revealed that these clones consisted of seven individual sequences. ClustalW analysis obtained seven different heptapeptide sequences and a human CD98 h.c. amino acid sequence revealed that the epitope recognized by HBJ127 might be 443AFS445 in CD98 h.c. The authors concluded that 444F might be crucial for HBJ127 binding to CD98 h.c. because the reactivity of HBJ127 against synthetic peptide with point mutation (444F to A) was greatly reduced compared with the peptide of the original sequence.
Materials and Methods
Monoclonal antibodies. HBJ127 (IgG1, κ)( 5 ) and 1–10 (IgG1, κ) recognize the human CD98 h.c. SER4 (IgG1, κ)( 22 ) recognizes the human c‐erb‐B2 proto‐oncogene product. HBJ127 was used as a target of epitope mapping. 1–10 and SER4 were used as negative controls for screening of HBJ127‐reactive phage clones.
Phage display random peptide library. The phage display random peptide library, Ph.D.‐7 (New England BioLabs, Ipswich, MA, USA), was used for the epitope mapping of HBJ127. The linear heptapeptide with random sequence was expressed as a fusion protein with the N‐terminal end of the minor coat protein of the M13 phage. This library contained 2.8 × 109 individual clones. The phage titer was checked before use, and found to be 2.81 × 1013 p.f.u./mL, which was almost equal to the manufacture's determination (2 × 1013 p.f.u./mL).
Panning of the phage library against HBJ127. The wells of Costar 3590 EIA/RIA plate were coated with HBJ127 (5 µg/well) overnight at 4°C. The wells were then blocked with Blockace (Dainippon Sumitomo Pharma, Osaka, Japan) for 1 h at 37°C. After discarding the blocking solution, the phage solution (2.8 × 1011 p.f.u./well) was added to the wells and incubated for 1 h at room temperature. After removal of the phage solution, the wells were washed 10 times with phosphate‐buffered saline (PBS; pH 7.4) containing 0.1% (v/v) polyoxyethylene sorbitan monolaurate (Tween 20; T‐PBS). HBJ127‐bound phages were then eluted with 0.1 M glycine‐hydrochrolide (pH 2.2). The eluate was neutralized using 2 M Tris. 1 µL of solution was used for determination of the eluted phage titer. The rest of the eluted solution was added to the exponentially growing Escherichia coli XL1‐Blue cells (Stratagene, La Jolla, CA, USA), and then culture was continued for 4.5 h at 37°C with vigorous shaking. The culture was transferred to a conical tube and centrifuged for 10 min at 13 000g at 4°C. Supernatant was added to 1/6 volume of 20% polyethylene glycol 8000 in 2.5 M sodium chloride solution (PEG/NaCl), then mix by inversion. The solution was left at 4°C overnight, and precipitated phages were collected using centrifugation for 15 min at 13 000g at 4°C. Phages were resuspended with PBS and re‐precipitated with PEG/NaCl. The phages were finally resuspended with 100 µL of PBS and used for the next round of panning. Panning was repeated three times in this experiment.
Screening of phage clones. The phages obtained after the third round of panning were infected with XL1‐Blue culture. After 5 min incubation at room temperature, LB top agar was added to the mixture and the solution was poured on to the LB agar plate containing 50 µg/mL IPTG and 50 µg/mL Xgal (LB/IPTG/Xgal plate). The plate was inverted and incubated overnight at 37°C. Thirty blue plaques on the plate were picked up and infected with 1:100 diluted XL1‐Blue overnight cultures in 15 mL PP tubes. After 4.5 h shaking at 37°C, cultures were transferred to 1.5 mL tubes and centrifuged for 10 min at 36 000g at 4°C. The cleared supernatants were used for testing the binding activity against HBJ127 by a phage ELISA and for making single‐stranded (ss)DNA to identify the heptapeptide sequence.
Phage ELISA. HBJ127, 1–10 and SER4 were coated onto a Costar 3690 EIA/RIA plate at 1 µg/mL overnight at 4°C. The wells were incubated with Blockace for 1 h at 37°C. After discarding the solution, phage solution was added to the wells and incubated for 1 h at 37°C. After the wells were washed five times with T‐PBS, horseradish peroxidase (HRP)‐labeled rabbit anti‐M13 (GE Healthcare, Tokyo, Japan; 1:3000 in T‐PBS) was added to the wells and incubated for 1 h at 37°C. The wells were washed in the same manner as described above, tetramethylbenzidine‐based substrate solution (ML‐1120T, Sumitomo Bakelite, Tokyo, Japan) was added to the wells and the color development was stopped by the addition of a sulfuric acid‐based stop solution. The absorbance at 450 nm was measured using a Model 550 Microplate Reader (Bio‐Rad, Tokyo, Japan).
DNA sequencing. The ssDNA of M13 phage clones was isolated and purified from supernatants by QIAprep Spin M13 kit (Qiagen, Tokyo, Japan) according to the manufacturer's instructions. Nucleic acid sequencing was carried out on an ABI PRISM 310 Genetic Analyzer (Applied Biosystems, Tokyo, Japan) using a BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems). The primer of ‐96gIII (5′‐CCC TCA TAG TTA GCG TAA CG‐3′), hybridizing to the (–) strand of the 96 base‐downstream of the 3′‐end of random heptamer coding sequence, was used for sequencing of the random heptamer.
Western blot. Selected phage clones were amplified and concentrated using PEG/NaCl precipitation. The amount of obtained phage was determined using sandwich ELISA with labeled and non‐labeled anti‐M13 antibody. Immunologically equal amounts of phages were separated using 12.5% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) under a non‐reducing condition, and transferred to polyvinylidene fluoride (PVDF) membrane (Immobilon‐N, Millipore, Tokyo, Japan) using semi‐dry blotter (Transblot SD Cell, Bio‐Rad). The blotted membrane was blocked with Blockace, then successively treated with HBJ127 (10 µg/mL), biotin‐labeled antimouse IgG, and peroxidase labeled‐ABC (Vector Laboratories, Burlingame, CA, USA). The immunoreactive bands were visualized by treatment of 4‐chloro‐1‐naphtol‐based substrate solution (HRP color development kit, Bio‐Rad).
Peptide synthesis. A peptide with the sequence of the predicted HBJ127 epitope (LLHGDFHAFSAG‐GGGC, GGGC: linker), epitope‐related peptides (LLHGDFH‐GGGC, FHAFSAG‐GGGC, FHGFSAG‐GGGC, FHAASAG‐GGGC, FHAFAAG‐GGGC) and a control peptide (AHNQVRQVPLQR‐GGGC) were chemically synthesized (Torey Research Center, Tokyo, Japan). Peptide–bovine serum albumin (BSA) conjugates were prepared by reaction of the cysteine residue at the C‐terminal end of the peptide and maleimide‐activated BSA (Imject Maleimide Activated BSA, Pierce, Rockford, IL, USA). The protein content of the BSA conjugates was determined using BCA Protein Assay (Pierce), and used as an antigen for testing of reactivity against HBJ127.
Antibody ELISA. The wells of the Costar 3690 EIA/RIA plate were coated with the sequentially diluted peptide–BSA conjugates and incubated overnight at 4°C. After discarding the antigen solution, the wells were filled with 1% BSA‐PBS and incubated for 1 h at 37°C. After discarding the blocking solution, the wells were successively treated with HBJ127 (5 µg/mL), an alkaline phosphatase‐labeled rabbit antimouse IgG Fc portion (Jackson ImmunoResearch, West Grove, PA, USA), and a p‐nitrophenyl phosphate (4‐nitrophenyl phosphate disodium salt hexahydrate 5 mg tablet, Sigma‐Aldrich, Tokyo, Japan)‐based substrate solution. The absorbance of resultant p‐nitrophenol was determined at 405 nm using a Model 550 Microplate Reader.
Results
Panning of the library and screening of the HBJ127‐reactive clones. To identify the sequence mimicking the epitope recognized by HBJ127, three rounds of biopanning of the Ph.D.‐7 phage display random peptide library against HBJ127 were performed. The eluted phage titer was increased with each successive round of panning, indicating the presence of phage clones bearing the peptide sequence reactive with HBJ127. The phage titer increased from 1 × 105 p.f.u./mL (first round) to 3.6 × 107 p.f.u./mL (second round), and finally 2.9 × 109 p.f.u./mL (third round). Consequently, the library was concentrated to 2.4 × 104‐fold after three round of biopanning against HBJ127. Thirty blue color plaques were randomly picked up from the plate of the eluted phages after the third round of panning. Each clone was amplified by infection with XL1‐Blue, and then served for sequencing and testing reactivity against HBJ127. All tested clones showed reactivity to HBJ127 and consisted of seven different sequences (frequency of appearance): HPMHFPS (11); YPRWQIP (6); SVFWMIP (6); YPQLPFT (4); HPMHFPC (1); HHYAFSV (1); SPNVNAN (1).
Reactivity of the phage clones against HBJ127. Representative clones with a distinct sequence (MP‐1: YPRWQIP; MP‐3: SVFWMIP; MP‐6: HPMHFPS, MP‐9: HHYAFSV; MP‐10: YPQLPFT; MP‐16: SPNVNAN; MP‐17: HPMHFPC) were selected and characterized using phage ELISA and western blot. Before the experiments, the phage amount to be tested was adjusted using sandwich phage ELISA with an anti‐M13 antibody. The reactivity of epitope‐mimicking (mimotope) phages against HBJ127 and isotype‐matched other antibodies (1–10: anti‐CD98 h.c. reactive with the epitope different from HBJ127, SER4: anti‐c‐erbB‐2) were determined using phage ELISA. MP‐1, ‐6, ‐9, ‐10, and ‐17 showed the predominant reactivity against HBJ127, with slight cross‐reactivity to 1–10 and SER4 (Fig. 1). MP‐3 and ‐16 showed reactivity against 1–10 at the same extent to HBJ127 (Fig. 1). To confirm this result, mimotope phages blotted on the PVDF membrane were detected using HBJ127. An approximately 50‐kDa band of peptide‐pIII fusion protein was detected in MP‐1, ‐6, ‐9, ‐10, and ‐17, but not in MP‐3 and ‐16 (Fig. 2). MP‐3 and ‐16 showed reactivity to HBJ127 from phage ELISA, but not from western blot. Because, in general, higher affinity may be required for the binding of blotted antigens compared with plastic adsorbed antigens, MP‐3 and ‐16 were found to be less reactive with HBJ127 than the other clones.
Figure 1.
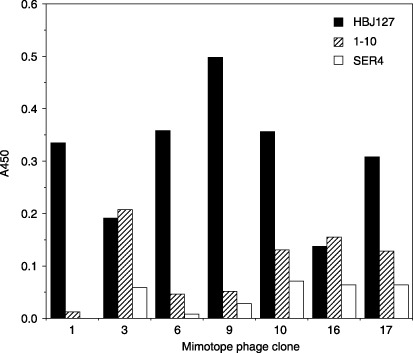
Reactivity of mimotope phage clones against monoclonal antibodies. Reactivity of mimotope phages against plate‐coated monoclonal antibodies were determined using phage ELISA as described in the Materials and Methods.
Figure 2.
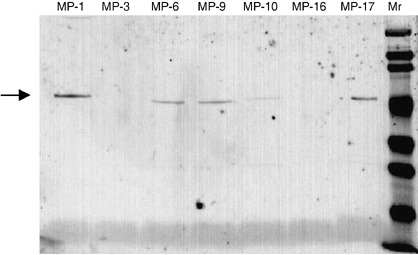
Western blot of phage particles bearing HBJ127‐reactive mimotopes. Mimotope‐bearing phages blotted on the polyvinylidene fluoride membrane were stained with HBJ127 as described in the Materials and Methods. The arrowhead shows the position (~50 kDa) of the mimotope‐pIII fusion protein. Mr, molecular weight marker.
ClustalW analysis of homology between isolated heptapeptides and human CD98 heavy chain. Epitope analysis was performed by searching the homologous region between the human CD98 h.c. amino acid sequence and the mimotope phages using ClustalW software on the web of DNA Data Bank of Japan (Mishima, Shizuoka, Japan). MP‐1, ‐3, ‐6/‐17, ‐9, and ‐10 showed the homology at 378–399, 393–399, 381–387, 437–445, and 395–407 of CD98 h.c., respectively (Fig. 3). All clones other than MP‐9 was located in the region of 378–407 of CD98 h.c. Although CD98 h.c. is known as a highly conserved molecule between human and rodents (mouse and rat), HBJ127 shows reactivity only against the human CD98 h.c. At the region of 378–407, human‐specific 395F and 396P were found but solely existed in the matched region of MP‐3 and ‐10, respectively (Fig. 3). In contrast, at the region of 437–445, three consecutive amino acids identical to human CD98 h.c. including human‐specific 443F were found in the matched region of MP‐9 (Fig. 3). The HBJ127 epitope has been predicted to be between 418 and 529 of the human CD98 h.c.( 23 ) From the present results and observation, MP‐9 may represent the epitope recognized by HBJ127.
Figure 3.
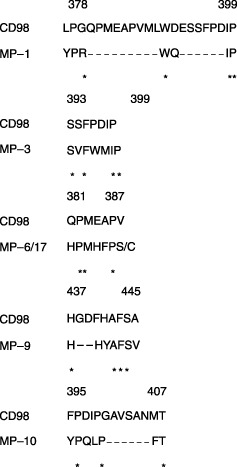
Identification of mimotope‐matched regions in human CD98 heavy chain (h.c.). The homologous region between the mimotopes and the CD98 h.c. were searched using ClustalW. The human‐specific amino acid is shown in bold typeface. The dashed line shows the gap in the sequence. The asterisk shows the amino acid that is identical to the CD98 h.c. sequence.
Reactivity of HBJ127 against synthetic peptides derived from the expected epitope sequence. To confirm that the sequence of 442AFS444 in human CD98 h.c. is the epitope recognized by HBJ127, various synthetic peptides including the expected epitope sequence were prepared and the reactivity against HBJ127 were tested using ELISA. Based on the amino acid sequence of human CD98 h.c., 435LLHGDFHAFSAG446 dodecapeptide was synthesized, and a linker sequence (GGGC) was introduced into the C‐terminal end of the peptide to prepare the peptide–BSA conjugate. HBJ127 showed reactivity against the LLHGDFHAFSAG peptide in a concentration‐dependent manner with no‐cross reactivity to an unrelated but the same chain length peptide–BSA conjugate (Fig. 4). Next, N‐terminal (LLHGDFH) and C‐terminal (FHAFSAG) heptapeptides were synthesized, and tested for reactivity against HBJ127 (Fig. 5). FHAFSAG showed the highest reactivity against HBJ127. LLHGDFH also showed reactivity against HBJ127, but was weaker than FHAFSAG and LLHGDFHAFSAG. These results suggest that the epitope recognized by HBJ127 may be located in the sequence of FHAFSAG. Finally, alanine (alanine to glycine)‐substituted peptides (FH GFSAG, FHAA SAG, FHAFA AG) were synthesized and tested for reactivity against HBJ127 (Fig. 6). Reduced reactivity was detected against the phenylalanine‐substituted peptide. Other peptides showed reactivity in the same extent to that against the FHAFSAG peptide. These results imply that 443F in human CD98 h.c. may be critical for binding HBJ127 against the human CD98 h.c.
Figure 4.
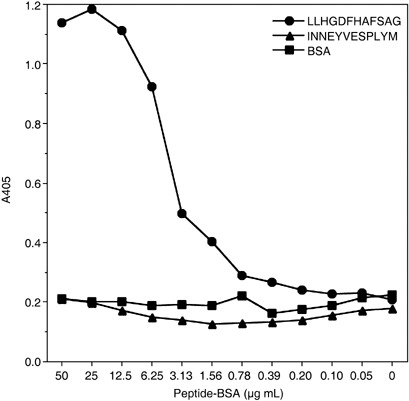
Reactivity of HBJ127 against dodecapeptide corresponding to 435–446 of the human CD98 heavy chain (h.c.). The reactivity of HBJ127 against plate‐coated peptide–bovine serum albumin (BSA) was determined using antibody ELISA as described in the Materials and Methods.
Figure 5.
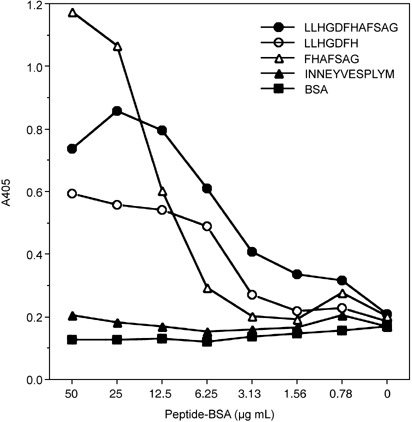
Reactivity of HBJ127 against N‐ and C‐terminal heptapeptides. Reactivity of HBJ127 against plate‐coated peptide–bovine serum albumin (BSA) was determined using antibody ELISA as described in the Materials and Methods.
Figure 6.
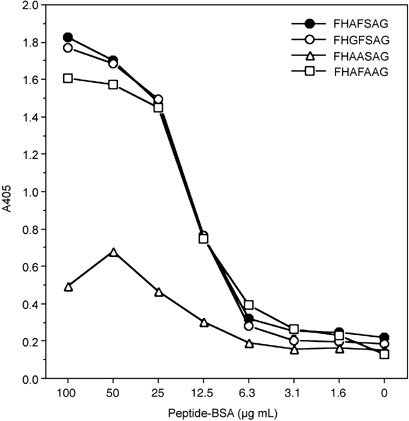
Reactivity of HBJ127 against mutation‐introduced C‐terminal heptapeptides. Reactivity of HBJ127 against plate‐coated peptide–bovine serum albumin (BSA) was determined using antibody ELISA as described in the Materials and Methods.
Discussion
In the present study, the phage display random peptide library was used for the identification of the epitope recognized by HBJ127. In general, immunoprecipitation with a specific mAb followed by liquid chromatography–mass spectrometry (LC‐MS) analysis has been used for the identification of a recognized antigen. The epitope may be determined by an oligo‐peptide array constructed using sequence information of the antigen obtained as above. These procedures seem laborious and time consuming. As for the epitope mapping using the phage display random peptide library, panning of the library against the antibody of interest followed by screening of phage clones from post‐panning the library will give phage clones with the antibody binding activity. Sequencing of the peptide‐coding region in phage DNA can identify the consensus sequence of antibody‐binding clones.
Although the phage display approach is actually simple and easy, this procedure has some disadvantages. Identification of the epitope may be difficult if the sequence information of the antigen of interest is not available, because the homology search of the obtained sequence and the antigen sequence is indispensable for identification of the epitope region on the antigen. The peptide sequence of the most frequent or of the strongest binding to antibody may not be the epitope sequence. In fact, in the present study, the expected epitope‐bearing sequence was found in only one of 30 tested clones. Moreover, if the antibody recognizes the conformation‐dependent epitope, the actual epitope sequence cannot be identified using this procedure. Combined use of phage display and immunological techniques makes it possible to identify the epitope of the antigen, easily, simply and accurately.
The epitope of HBJ127 was estimated by comparing the isolated heptapeptide sequences and the human CD98 h.c. amino acid sequence using ClustalW. The epitope of HBJ127 lies in between 418 and 529 in the human CD98 h.c. from the study using deletion mutants of the human CD98 h.c. expressed on mouse NIH 3T3 cells.( 23 ) HBJ127 reacts with the human CD98 h.c., not with mouse or rat molecule.( 5 ) The authors’ predicted epitope position (442AFS444) is within 418–529 of the human CD98 h.c. One of the constituents, 443F, is human sequence‐specific (mouse and rat sequences have 443L). Decreased reactivity of HBJ127 against the point mutation (F to A)‐introduced epitope peptide–BSA conjugate was shown. From these observations and the present findings, we propose that the epitope would be 442AFS444 in human CD98 h.c., and that 443F may be crucial for HBJ127‐binding to human CD98 h.c. Binder et al. reported that the epitope of anti‐CD25 mAb, dacrizumab, was found to be overlapped with basiliximab, which is also an anti‐CD25 mAb, using the mutation‐introduced epitope peptides prepared based on the sequences isolated from the phage display peptide library panned against basiliximab.( 24 ) The present procedure for determination of the HBJ127 epitope may be appropriate because a similar approach to the above was used: identification of the epitope‐related sequences using the phage display peptide library and confirmation of the predicted epitope sequence using mutation‐introduced synthetic peptides.
Antibody medicines are widely used for the treatment of tumors, autoimmune disorders, and infectious diseases. As for tumor therapy, the representatives are transtuzumab and rituximab, which are used for the treatment of c‐erb‐B2‐positive breast cancers and CD20‐positive B‐cell lymphoma, respectively. As for rheumatoid arthritis therapy, the anti‐tumor necrosis factor (TNF)‐α mAb, infliximab, is used for treatment in rheumatrex‐unresponsive patients. Although antibody medicines show excellent efficacy in the therapy of such diseases as above, they have some disadvantages, for example, repeated injection may be necessary for maintaining efficacy, there is a risk of anaphylaxis, there can be decreased effectiveness due to the anti‐antibody response in the hosts, and they are very expensive medicines. The development of new medicines with an efficacy equivalent to the antibody medicines without the shown drawbacks is expected. One candidate is the peptide vaccine prepared based on the epitope sequence of antibody medicines or antibodies with biological activities. Induction of protective immunity would be expected by active immunization of the host with epitope vaccines. Mimotope peptides obtained using the phage display peptide library from cetuximab,( 25 ) rituximab,( 26 ) transtuzumab,( 27 ) and anti‐GD2 mAb,( 28 , 29 ) successfully induced protective immunity in the hosts. Evaluation of the usefulness of a HBJ127‐derived epitope peptide immunization for induction of CD98‐targeting immunity in the hosts is now in progress.
Acknowledgments
This work is supported in part by a Grant‐in‐aid for Scientific Research (C) from The Japan Society for Promotion of Science (17590467).
References
- 1. Haynes BF, Hemler ME, Mann DL et al . Characterization of a monoclonal antibody (4F2) that binds to human monocytes and to a subset of activated lymphocytes. J Immunol 1981; 126: 1409–14. [PubMed] [Google Scholar]
- 2. Broer S, Broer A, Hamprecht B. The 4F2hc surface antigen is necessary for expression of system 1‐like neutral amino acid‐transport activity in C6‐BU‐1 rat glioma cells: evidence from expression studies in Xenopus laevis oocytes. Biochem J 1995; 312: 863–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kanai Y, Segawa H, Miyamoto K, Uchino H, Takeda E, Endou H. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J Biol Chem 1998; 273: 23 629–32. [DOI] [PubMed] [Google Scholar]
- 4. Hashimoto Y, Masuko T, Yagita H, Endo N, Kanazawa J, Tazawa J. A proliferation‐associated rat cell surface antigen recognized by a murine monoclonal antibody. Gann 1983; 74: 819–21. [PubMed] [Google Scholar]
- 5. Masuko T, Abe J, Yagita H, Hashimoto Y. Human bladder cancer cell‐surface antigens recognized by murine monoclonal antibodies raised against T24 bladder cancer cells. Jpn J Cancer Res 1985; 76: 386–94. [PubMed] [Google Scholar]
- 6. Yagita H, Masuko T, Hashimoto Y. Inhibition of tumor cell growth in vitro by murine monoclonal antibodies that recognize a proliferation‐associated cell surface antigen system in rats and humans. Cancer Res 1986; 46: 1478–84. [PubMed] [Google Scholar]
- 7. Yagita H, Masuko T, Takahashi N, Hashimoto Y. Monoclonal antibodies that inhibit activation and proliferation of lymphocytes. I. Expression of the antigen on monocytes and activated lymphocytes. J Immunol 1986; 136: 2055–61. [PubMed] [Google Scholar]
- 8. Hara K, Kudoh H, Enomoto T, Hashimoto Y, Masuko T. Malignant transformation of NIH3T3 cells by overexpression of early lymphocyte activation antigen CD98. Biochem Biophys Res Commun 1999; 262: 720–5. [DOI] [PubMed] [Google Scholar]
- 9. Smith GP. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science 1985; 228: 1315–17. [DOI] [PubMed] [Google Scholar]
- 10. Burton DR, Barbas CF, 3rd. Human antibodies from combinatorial libraries. Adv Immunol 1994; 57: 191–280. [DOI] [PubMed] [Google Scholar]
- 11. Winter G, Griffiths AD, Hawkins RE, Hoogenboom HR. Making antibodies by phage display technology. Annu Rev Immunol 1994; 12: 433–55. [DOI] [PubMed] [Google Scholar]
- 12. Itoh K, Nakagomi O, Suzuki K, Inoue K, Tada H, Suzuki T. Recombinant human monoclonal Fab fragments against rotavirus from phage display combinatorial libraries. J Biochem (Tokyo) 1999; 125: 123–9. [DOI] [PubMed] [Google Scholar]
- 13. Hoogenboom HR. Overview of antibody phage‐display technology and its applications. Meth Mol Biol 2002; 178: 1–37. [DOI] [PubMed] [Google Scholar]
- 14. Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat Biotechnol 2005; 23: 1105–16. [DOI] [PubMed] [Google Scholar]
- 15. Dong L, Masaki Y, Takegami T et al . Cloning and expression of two human recombinant monoclonal Fab fragments specific for EBV viral capsid antigen. Int Immunol 2007; 19: 331–6. [DOI] [PubMed] [Google Scholar]
- 16. McLafferty MA, Kent RB, Ladner RC, Markland W. M13 bacteriophage displaying disulfide‐constrained microproteins. Gene 1993; 128: 29–36. [DOI] [PubMed] [Google Scholar]
- 17. Sparks AB, Quilliam LA, Der Thorn JMCJ, Kay BK. Identification and characterization of Src SH3 ligands from phage‐displayed random peptide libraries. J Biol Chem 1994; 269: 23 853–6. [PubMed] [Google Scholar]
- 18. Healy JM, Murayama O, Maeda T, Yoshino K, Sekiguchi K, Kikuchi M. Peptide ligands for integrin alpha v beta 3 selected from random phage display libraries. Biochemistry 1995; 34: 3948–55. [DOI] [PubMed] [Google Scholar]
- 19. Christian RB, Zuckermann RN, Kerr JM, Wang L, Malcolm BA. Simplified methods for construction, assessment and rapid screening of peptide libraries in bacteriophage. J Mol Biol 1992; 227: 711–18. [DOI] [PubMed] [Google Scholar]
- 20. Orlandi R, Menard S, Colnaghi MI, Boyer CM, Felici F. Antigenic and immunogenic mimicry of the HER2/neu oncoprotein by phage‐displayed peptides. Eur J Immunol 1994; 24: 2868–73. [DOI] [PubMed] [Google Scholar]
- 21. Bottger V, Bottger A, Lane EB, Spruce BA. Comprehensive epitope analysis of monoclonal anti‐proenkephalin antibodies using phage display libraries and synthetic peptides: revelation of antibody fine specificities caused by somatic mutations in the variable region genes. J Mol Biol 1995; 247: 932–46. [DOI] [PubMed] [Google Scholar]
- 22. Masuko T, Sugahara K, Kozono M et al . A murine monoclonal antibody that recognizes an extracellular domain of the human c‐erbB‐2 protooncogene product. Jpn J Cancer Res 1989; 80: 10–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hara K, Kudoh H, Enomoto T, Hashimoto Y, Masuko T. Enhanced tumorigenicity caused by truncation of the extracellular domain of GP125/CD98 heavy chain. Oncogene 2000; 19: 6209–15. [DOI] [PubMed] [Google Scholar]
- 24. Binder M, Vogtle FN, Michelfelder S et al . Identification of their epitope reveals the structural basis for the mechanism of action of the immunosuppressive antibodies basiliximab and daclizumab. Cancer Res 2007; 67: 3518–23. [DOI] [PubMed] [Google Scholar]
- 25. Riemer AB, Kurz H, Klinger M, Scheiner O, Zielinski CC, Jensen‐Jarolim E. Vaccination with cetuximab mimotopes and biological properties of induced anti‐epidermal growth factor receptor antibodies. J Natl Cancer Inst 2005; 97: 1663–70. [DOI] [PubMed] [Google Scholar]
- 26. Li M, Yan Z, Han W, Zhang Y. Mimotope vaccination for epitope‐specific induction of anti‐CD20 antibodies. Cell Immunol 2006; 239: 136–43. [DOI] [PubMed] [Google Scholar]
- 27. Riemer AB, Klinger M, Wagner S et al . Generation of Peptide mimics of the epitope recognized by trastuzumab on the oncogenic protein Her‐2/neu. J Immunol 2004; 173: 394–401. [DOI] [PubMed] [Google Scholar]
- 28. Riemer AB, Forster‐Waldl E, Bramswig KH et al . Induction of IgG antibodies against the GD2 carbohydrate tumor antigen by vaccination with peptide mimotopes. Eur J Immunol 2006; 36: 1267–74. [DOI] [PubMed] [Google Scholar]
- 29. Bolesta E, Kowalczyk A, Wierzbicki A et al . DNA vaccine expressing the mimotope of GD2 ganglioside induces protective GD2 cross‐reactive antibody responses. Cancer Res 2005; 65: 3410–18. [DOI] [PubMed] [Google Scholar]


