Abstract
Gefitinib is an inhibitor of the tyrosine kinase activity of epidermal growth factor receptor (EGFR). Accumulating evidence suggests that gefitinib may provide a survival benefit to EGFR mutation‐positive non‐small lung cancer patients. We have established a clinical test that can detect EGFR mutations from cytological specimens or paraffin‐embedded tissue specimens that are contaminated by normal cells. This test is based on the peptide nucleic acid, locked nucleic acid polymerase chain reaction clamp method that can detect G719S, G719C, L858R, L861Q and seven different exon 19 deletions in the presence of 100–1000‐fold wild‐type alleles. Consequently, using a small aliquot of samples isolated to establish a cancer diagnosis, the EGFR mutation status is determined soon after the diagnosis of cancer is made. We investigated the EGFR mutation status in 86 patients using a variety of cytological specimens (59 bronchoscopy specimens, 16 pleural effusion, 9 sputum, and 2 pericardial effusion) and in 46 patients who had a disease relapse and paraffin‐embedded tissues were available. Forty‐five patients (34%) were positive for mutation (29 exon 19 deletions, 16 L858R and 1 L861Q). The sensitivity and the specificity of this test was 97% and 100%, respectively. EGFR mutation status thereby obtained was used to determine each patient's therapeutic regimen. This test is easily integrated into the normal clinical practice for lung cancer, while allowing the medical staff to select therapeutic regimen depending on the EGFR mutation status. (Cancer Sci 2007; 98: 246–252)
Abbreviations:
- EGFR
epidermal growth factor receptor
- PNA
peptide nucleic acid
- LNA
locked nucleic acid
Gefitinib is an inhibitor of the tyrosine kinase activity of epidermal growth factor receptor (EGFR) which competes with the adenosine triphosphate (ATP) for the ATP binding site of the receptor. Several initial clinical studies have elucidated that gefitinib shows a response in non‐small cell lung cancers, where a higher response is observed in women, in ‘never smokers’, in patients with adenocarcinoma and in Japanese.( 1 , 2 ) The genetic factors that predict responsiveness were not elucidated until 2004 when mutations in the EGFR gene were reported to be significantly correlated with the response to gefitinib.( 3 , 4 ) Mutations were located in exons 18, 19 or 21 which encode a segment of the kinase domain around the ATP binding site. These mutations are activating mutations. The mutant receptor changes the direction of signaling to Akt and STAT in order to stimulate cell survival, while the wild type mainly transmits the signal to Ras in order to promote cell proliferation. Gefitinib preferentially inhibits this survival signal from the mutant EGFR, thereby inducing apoptosis in cancer cells.( 3 , 5 )
Since the discovery of the relationship between EGFR mutations and the responsiveness to gefitinib, several retrospective and prospective studies have confirmed that gefitinib improves the response rate, the progression‐free survival time, and the overall survival time when administered to patients with EGFR mutations.( 6 , 7 , 8 , 9 , 10 ) EGFR mutations were easily examined from surgically resected tissue or biopsy tissue specimens that mostly consisted of cancer cells. In clinical practice, many of the cancers have been pathologically diagnosed using small biopsy specimens or cytological specimens that are contaminated by many normal cells. Therefore, the EGFR mutation test that is used in clinical practice should be able to examine cytological samples contaminated with normal cells as well as being rapid, sensitive and inexpensive to perform. We previously reported a rapid and sensitive EGFR detection method, called the peptide nucleic acid‐locked nucleic acid poloymerase chain reaction (PNA‐LNA PCR) clamp.( 11 ) Using this method, we designed a test that fulfills all these requirements.
Materials and Methods
Clinical samples. This study was approved by the institutional review board of Saitama Medical University and performed in accordance with the Declaration of Helsinki (1995, revised in Edinburgh 2000). The specimens were isolated from patients who gave their written informed consent. Cytology specimens were obtained from patients who visited Saitama Medical University Hospital from October 2004 to May 2006 and had a suspected diagnosis of cancer. Paraffin‐embedded specimens of surgically resected tumors were obtained from patients who had a disease relapse and required information on EGFR mutation to determine the regimen of therapy during the same period. The cytology specimens were divided into pathology samples (the main sample) and PNA‐LNA PCR clamp samples (a small aliquot). The cells in the PNA‐LNA PCR clamp sample were collected and stored in the AL buffer (a buffer containing protein denaturant: Qiagen, Hilden Germany). When the pathologist confirmed a pathology sample to contain cancer cells (i.e. rated as classes IV or V), DNA was purified from the PNA‐LNA PCR clamp samples using QIAamp DNA Micro Kit (Qiagen) and subjected to the analysis. The paraffin‐embedded tissue specimens were serially thin‐sliced: one slice was used to confirm the presence of cancer cells under microscopy, while the others were investigated by the PNA‐LNA PCR reaction after DNA was isolated using the QIAamp DNA Micro Kit.
PNA‐LNA PCR clamp. We have previously reported on the PNA‐LNA PCR clamp method and described its usefulness for detecting EGFR mutations in cell lines.( 11 ) The reaction preferentially amplifies and specifically detects mutant alleles. As a result, the mutant alleles can be detected in the presence of 100–1000‐fold background of the wild‐type alleles. In this study, we intentionally reduced the sensitivity that detects one mutant allele in the presence of 100‐fold wild‐type alleles (see Discussion). The PNA‐LNA PCR clamp is briefly described as follows. The PCR mixture contains a clamp primer that has the wild‐type sequence and fluorogenic detection probes that have mutant sequences. The clamp primer is a peptide nucleic acid (PNA) oligo, while the detection probes are usual nucleotide oligos with the locked nucleic acid (LNA) at the mutated residue and fluorogenic dyes on both ends. Because the annealing of PNA to DNA is highly specific to the matched complementary sequence, the PNA clamp primer specifically binds to the wild‐type sequence, thereby inhibiting its amplification. This allows only mutant sequences to be amplified. Because the annealing of LNA to DNA is also highly specific to the matched complementary sequence, the LNA detection probes specifically detect mutant sequences. An additional probe (the total probe) that is located on the flanking sequence monitors the amplification of the target DNA segment. To detect 11 different mutations (G719S, G719C, L858R, L861Q and 7 exon 19 deletions), we used different combinations of a clamp primer and detection probes in a total of five reactions as previously reported. For one reaction, 25 ng of genomic DNA was used. The reactions were performed on the Smart Cycler II (Cepheid, Sunnyvale, CA, USA).
To monitor the quality of DNA, we used the reaction that detects L858R. In the L858R reaction, the clamp primer does not completely inhibit the amplification of wild‐type sequence and the curve for the total probe for L858R fragment rises in late cycles even in the absence of the mutant sequence. We found this ‘leaky’ amplification to be reproducible, and thus it was used to monitor the progression of the PCR, and thus quality of DNA. Note that the total probe for exon 19 deletions completely suppress the amplification of wild‐type sequence, thereby enabling us to detect the presence of exon 19 deletions that are not detected by the detection probes (see Results). Control genomic DNA mixtures containing the mutant and the wild‐type EGFR gene at ratios of 1:1, 0.1:1.0, 0.01:1.00 and 0:0 were used to establish standards for the amplification curve.
Measurement of the cancer content in the specimens. Microscopic slides that had been confirmed to contain cancer cells in Step 1 were randomly selected to investigate the contents of the cancer cells. For each slide, at least five microscopic photos representing different areas of the slide were taken at a magnification of either ×100 or ×200. Normal cells and cancer cells were identified by at least two examiners, and the total number of both cells were counted using a computer.
DNA sequencing. The amplification product of the PNA‐LNA PCR clamp was purified using the Strataprep PCR purification kit (Stratagene, Palo Alto, CA, USA) and then it was subjected to DNA sequencing using an automatic DNA sequencer.
Results
System design. Fig. 1 is the overview of our system. The system consists of four steps. Step 1: Divide the clinical sample into the pathology sample (most parts of the sample) and the PNA‐LNA PCR clamp sample (a small aliquot). Step 2: Check the pathology sample for the presence of cancer cells. Step 3: Perform the PNA‐LNA PCR clamp reaction to detect EGFR mutations. Step 4: Perform direct sequencing using the amplification product by the PNA‐LNA PCR clamp, if the presence of a deletion is suspected that is not detected by the detection probes.
Figure 1.
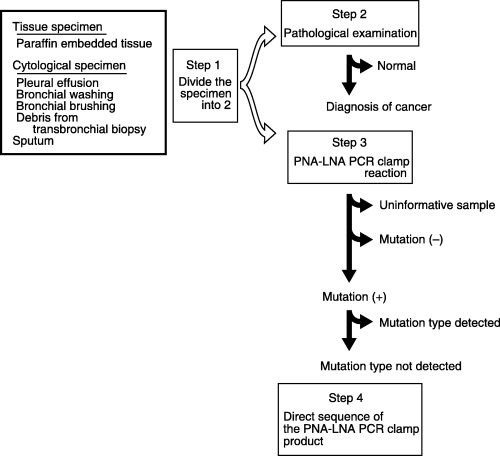
An overview of the peptide nucleic acid‐locked nucleic acid poloymerase chain reaction (PNA‐LNA PCR) clamp system. A small aliquot of the samples that were confirmed to contain cancer cells were examined for the epidermal growth factor receptor (EGFR) mutations by the PNA‐LNA PCR clamp. Samples that contain an insufficient amount of DNA or insufficient quality of DNA for PCR amplification were judged to be uninformative. The PNA‐LNA PCR clamp amplification product was subjected to direct sequencing when the reaction suggests the presence of exon 19 deletions that cannot be detected by the detection probes.
Steps 1 and 2 establish the diagnosis of cancer as well as confirming that the PNA‐LNA PCR clamp sample contains cancer cells.
Step 3 detects EGFR mutations by the PNA‐LNA PCR clamp. Fig. 2A is a schematic presentation of the reaction. We limited the detection range to avoid any false positives by a small amount of contaminating DNA in the sample (Fig. 2B). We judged the sample to be positive for mutation when more than 1% of the EGFR alleles were mutant. This corresponds to the peak of the second derivative of the amplification curve that occurs at less than 35 cycles (Fig. 2B). The detection probes detect all EGFR mutations that were initially reported (Fig. 2C).( 3 , 4 ) However, five representative reports from Japan in which direct sequencing was used to study EGFR mutations( 7 , 12 , 13 , 14 , 15 ) indicate that our detection probe set is unable to detect 10% of all occurrences of exon 19 deletions (Fig. 2C). Nevertheless, most exon 19 deletions are detected as a positive signal from the total probe contained in the reaction that detects exon 19 deletions, because the clamp primer does not inhibit the amplification of the fragments that does not have a completely matched sequence.
Figure 2.
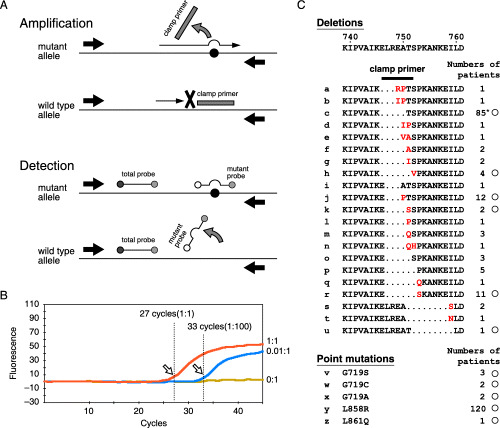
The peptide nucleic acid‐locked nucleic acid polymerase chain reaction (PNA‐LNA PCR) clamp reaction. (A) A schematic presentation of the reaction. (B) A representative amplification curve. The amount of target molecules can be determined from the position of the peak of the 2nd derivative of the amplification curve (arrows) which is calculated using the Smart Cycler program. 1:1, 0.1:1.0, and 0:0: the molar ratios of the mutant and the wild‐type epidermal growth factor receptor (EGFR) sequences. (C) A list of the EGFR mutations that are suggested to be linked to the responsiveness of gefitinib and their occurrences are summarized based on a review of the literature. 7 , 12 , 13 , 14 , 15 ) The inserted amino acids are written in red. The position of the clamp primer used to detect the deletions in this study is shown by a bar. The mutations that can be detected by our detection probes are marked by circles. *: the occurrences of two deletions (2235–2249delGGAATTAAGAGAAGC and 2236–2250delGAATTAAGAGAAGCA) that give the same amino acid change were combined.
Step 4 confirms the deletions that are not detected by our detection probes. Only when we noticed a signal from the total probe contained in the reaction that detects exon 19 deletions without any signals from the detection probes, do we then proceed to Step 4 and perform direct sequencing of the PNA‐LNA PCR clamp product in which the mutant fragment is enriched.
Results of the mutation detection. Using this system, we investigated 132 cases of non‐small cell lung cancer using a variety of sample sources (1, 2, 3). In 126 cases we could obtain information on EGFR mutations. Six uninformative cases included two bronchial washing specimens which contained too few cells to isolate a sufficient amount of DNA, and four paraffin‐embedded tissues in which the DNA had degraded and PCR failed.
Table 1.
Epidermal growth factor receptor mutations classified according to the patients’ sex and histology of the samples
| Total | Mutation | Uninformative positive | Negative | |
|---|---|---|---|---|
| Sex | ||||
| Male | 80 | 19 | 60 | 1 |
| Female | 52 | 26 | 21 | 5 |
| Total | 132 | 45 | 81 | 6 |
| Histology | ||||
| Adenocarcinoma | 102 | 43 | 55 | 4 |
| Squamous cell carcinoma | 15 | 1 | 14 | 0 |
| Adeno‐squamous cell carcinoma | 4 | 1 | 3 | 0 |
| Large cell carcinoma | 2 | 0 | 2 | 0 |
| Undifferentiated/unclassified | 9 | 0 | 7 | 2 |
| Total | 132 | 45 | 81 | 6 |
Table 2.
Source of the clinical specimens and pidermal growth factor receptor mutations
| No. of samples | Mutation | Uninformative | ||
|---|---|---|---|---|
| Positive | Negative | |||
| Cytolotical specimens | ||||
| Bronchial washing/bronchial brushing | 50 | 16 | 32 | 2 |
| Pleural effusion | 16 | 7 | 9 | |
| Debris of bronchial biopsy specimen | 9 | 3 | 6 | |
| Sputum | 9 | 3 | 6 | |
| Pericardial effusion | 2 | 0 | 2 | |
| Subtotal | 86 | 29 | 55 | 2 |
| Tissue specimens | ||||
| Paraffin embedded tissue | 46 | 16 | 26 | 4 |
| Total | 132 | 45 | 81 | 6 |
Table 3.
Mutations of the epidermal growth factor receptor gene. The change in amino acid sequence and the change in the nucleotide sequence are shown for each mutation. A letter before each sequence corresponds to that shown in Fig. 2C
| No. | No. | ||
|---|---|---|---|
| Exon 19 deletions | 29 | ||
| c | E746‐A750del | 13 | |
| 2235–2249delGGAATTAAGAGAAGC | |||
| c | E746‐A750del | 7 | |
| 2236–2250delGAATTAAGAGAAGCA | |||
| h | L747‐S752del E746V | 1 | |
| 2238–2255delATTAAGAGAAGCAACATC, 2237 A > T | |||
| j | L747‐E749del A750P | 2 | |
| 2239–2247delTTAAGAGAA, 2248G > C | |||
| k | L747‐A750del T751S | 1 | |
| 2240–2251delTAAGAGAAGCAA | |||
| r | L747‐S752del P753S | 3 | |
| 2240–2257delTAAGAGAAGCAACATCTC | |||
| L747‐T751del | 1 † | ||
| 2238–2252delATTAAGAGAAGCAAC | |||
| L747‐T753del ins A | 1 ‡ | ||
| 2239–2258delTTAAGAGAAGCAACA, ins GCT | |||
| Exon 21 point mutations | 16 | ||
| y | L858R | ||
| 2573T > G | 15 | ||
| z | L861Q | 1 | |
| 2582T > A | |||
Mutations determined by the direct sequencing of the peptide nucleic acid‐locked nucleic acid polymerase chain reaction clamp product.
Chromatogram of the direct sequence is shown in Fig. 3B
Two samples proceeded to Step 4 (Fig. 3A). Both samples were found to have deletions. Figure 3B shows the advantage of the PNA‐LNA PCR clamp product for direct sequencing in which the mutant fragment is enriched. The product of the PNA‐LNA PCR clamp clearly presented the mutant sequence, while the product of the conventional PCR presented only the wild type sequence.
Figure 3.
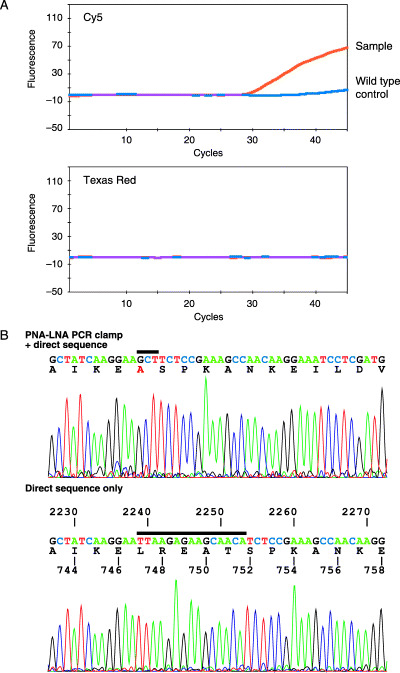
Detection of the deletions at Step 4. (A) The presence of deletion is suggested by a positive signal from the total probe (Cy5 channel) in the absence of signals from the detection probes (Texas red channel is shown as an example). (B) (Upper panel) Direct sequencing of the peptide nucleic acid‐locked nucleic acid polymerase chain reaction (PNA‐LNA PCR) clamp product. Because the mutated sequence is preferentially amplified, only the mutant sequence is observed. An inserted sequence is indicated by a bar. (Lower panel) The direct sequencing of the same sample amplified by the conventional PCR. The wild‐type sequence predominates and the deletion is difficult to visualize. The sequence indicated by a bar is replaced by ‘GCT’ in the mutant shown in the upper panel.
Sensitivity and specificity of the system. Our system is designed to determine the EGFR mutation status of cancer cells in clinical samples that are pathologically confirmed to contain cancer cells. In this context, we evaluated the sensitivity and specificity of the system (Table 4). At first, we evaluated the sensitivity and specificity of the PNA‐LNA PCR clamp method itself. As mutation‐negative controls, we used genomic DNA isolated from the peripheral blood of 10 normal volunteers from whom DNA sequences were confirmed to be wild type. As mutation‐positive controls, we used 11 mutant EGFR sequences( 11 ) synthesized by the mega‐primer method( 16 ) and diluted by the normal genomic DNA at a ratio of mutant:wild type = 1:100. More than 200 PNA‐LNA PCR clamp reactions performed on negative controls gave negative results. More than 200 reactions performed on positive controls gave positive results (data not shown). Therefore, we concluded that the specificity and sensitivity of the PNA‐LNA PCR clamp are 100% as long as the mutation is detectable by the detection probes.
Table 4.
Sensitivity and specificity of the system
| Sensitivity | Specificity | |
|---|---|---|
| PNA‐LNA PCR clamp for mutations with detection probes | 1.00 | 1.00 ‡ |
| PNA‐LNA PCR clamp for all mutations | >0.98 † | 1.00 ‡ |
| Cancer cell content is more than 1% | 0.99 § | 1.00 ¶ |
| Overall | >0.97 | 1.00 |
Our system detects 267 out of 269 mutation occurrences listed in Fig. 2C. Step 3 detects 240 occurrences (89%). Step 4 detects additional 27 occurrences (10%). Two occurrences which failed to be detected are those of G719A.
‡ More than 200 controls did not demonstrate any false results.
At Step 2, 59 of 59 specimens had a cancer content of more than 1% (see Fig. 4).
¶ Specificity of the pathological examination is assumed to be 1.0 in this study. PNA‐LNA PCR, peptide nucleic acid‐locked nucleic acid polymerase chain reaction.
Second, sensitivity and specificity of the whole system (Fig. 1) was evaluated. One of the possible causes of false negatives is samples in which the cancer content is less than 1%. Because we limited the detection range of the PNA‐LNA PCR clamp, such samples may always be judged as negative for mutations. We checked 59 samples randomly chosen out of 136 samples and measured the cancer content. As shown in Figure 4, it was more than 1% for all samples. This indicates that the limitations of the detection range did not increase the false negative rate. Another source of false negatives is the presence of mutations that are not detected by our detection probes. We added Step 4 to deal with this problem. We synthesized fragments containing the individual deletions shown in Figure 2C by the mega‐primer method,( 16 ) and investigated whether they could be detected in the presence of 100‐fold wild‐type alleles. All deletions were clearly detected as shown in Figure 3, thus indicating that almost all of the exon 19 deletions would be detected by our system.
Figure 4.
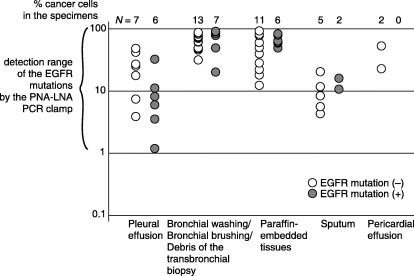
Cancer content in the samples. The number of cancer cells and normal cells was counted for the microscopic slides used in Step 1. The detection range of the peptide nucleic acid‐locked nucleic acid polymerase chain reaction clamp in Step 3, which is one mutant allele in the presence of 100 wild‐type alleles, is shown.
In summary, the sensitivity and the specificity of the PNA‐LNA PCR clamp itself are 100% for mutations detected by the detection probes. Samples that will give false negative results are: (i) samples that contain cancer cells less than 1% and still pathologically diagnosed as containing cancer cells; and (ii) samples that contain cancer cells with EGFR mutations that are neither exon 19 deletions nor are detected by the PNA‐LNA PCR clamp probe set.
Discussion
Approximately 60 000 patients died of advanced lung cancer in Japan in 2004.( 17 ) Accumulating evidence suggests that gefitinib provides a survival benefit to patients with non‐small cell lung cancers harboring EGFR mutations. Therefore, the screening for EGFR mutations in all advanced non‐small cell lung cancer patients is expected to become a routine procedure. To examine a large number of samples and to deal with the variety of clinical settings where the samples are isolated, an EGFR mutation test that is inexpensive, rapid, sensitive and integrated into the standard procedures for cancer diagnosis is needed. Our system fulfills all these requirements. The PNA‐LNA PCR clamp reaction is inexpensive (consumables cost about US$20 per sample: unpublished data). It can also be completed within 40 min. It can detect one mutant allele in the presence of 100 wild‐type alleles. The test uses a small aliquot of the samples isolated to establish the diagnosis of cancer, and therefore, patients do not need to undergo additional procedures. In our hospital, information on EGFR mutations is therefore available at the time we determine the treatment regimen for each patient, and we have found this information to be very useful. As of September 2006, we have been examining approximately 65 fresh cancer cases per month using our system.
Regardless of how high the sensitivity of the method may be, the test is useless if the samples do not contain cancer cells. We confirm the presence of cancer cells in Step 1. We use 25 ng of DNA per reaction, that is, the amount of 8000 haploid genomes. If we set up the PNA‐LNA PCR reaction at a sensitivity of one mutant to 1000 wild types, it would thus detect eight copies of mutant DNA. Some of the apparatuses used for the diagnosis of cancer are not disposable, for example bronchoscopy or bronchoscopy forceps. We considered that an excessively high sensitivity would make the system vulnerable to the accidental contamination of DNA adhering to these apparatuses, and thus would be more harmful than beneficial. For this reason, we intentionally reduced the sensitivity to one mutant to 100 wild types. Pathologists often request re‐sampling when a sample contains very few abnormal cells and thus they cannot make a conclusive diagnosis. The result shown in Fig. 4 suggests that only samples that have a cancer content more than 1% are acknowledged by the pathologist and proceed to Step 3. Moreover, in some of the mutant cells, mutant alleles have been reported to be amplified, thus increasing the proportion of mutant alleles to a level higher than expected based on the cancer content.( 7 , 18 ) Therefore, the limitations of the sensitivity do not reduce the performance of the PNA‐LNA PCR clamp method.
There have been several reports that lung cancers may be heterogeneous for EGFR mutation.( 11 , 19 , 20 ) In some patients, a small number of cancer cells with mutant alleles may exist among a large number of cancer cells with the wild‐type alleles. We limited the sensitivity of our system, and thus may be unable to detect small numbers of mutants when heterogeneity exists. It is not clear whether killing a small number of mutant cells can provide any clinical benefit while a large number of wild‐type cells survive. We placed a higher priority on avoiding contamination than dealing with the possibility of heterogeneity.
It is clear that the T790M mutation which occurs in the cells that have a preexisting mutation confers to the cancer cells gefitinib resistance.( 21 , 22 ) Our system is easily expanded and, after completing this study, we integrated the reaction for T790M into our system and it is now working effectively.
It is of interest that the EGFR mutations detected by our system can predict the response to gefitinib. To address this issue, we completed a phase II study in which 107 non‐small cell lung cancer patients were examined for EGFR mutations by our system; gefitinib was administered, and responses were compared between the mutation‐positive and mutation‐negative patients.( 23 ) The response rate was 78% for mutation‐positive patients, while it was 14% for mutation‐negative patients. The median survival time after the initiation of gefitinib treatment was 15.4 months and 11.1 months, respectively. These data support the usefulness of our system for predicting the effect of gefitinib.
In this study, we reported a PNA‐LNA PCR clamp‐based test for the detection of EGFR mutations and investigated its performance. Our system is easily integrated into clinical practice and it is expected to help medical specialists utilize the latest research results in order to select the optimal treatment strategy for cancer patients.
Acknowledgments
We thank Ms. Akemi Yokote for technical assistance. This work was supported in part by a grant‐in‐aid for scientific research (No. 16390236) from the Japan Society for the Promotion of Science and in part by a grant‐in‐aid for scientific research from the Japanese Foundation for the Multidisciplinary Treatment of Cancer.
References
- 1. Fukuoka M, Yano S, Giaccone G et al. Multi‐institutional randomized phase II trial of gefitinib for previously treated patients with advanced non‐small‐cell lung cancer (The IDEAL 1 Trial). J Clin Oncol 2003; 21: 2237 – 46. [DOI] [PubMed] [Google Scholar]
- 2. Kris MG, Natale RB, Herbst RS et al. Efficacy of gefitinib, an inhibitor of the epidermal growth factor receptor tyrosine kinase, in symptomatic patients with non‐small cell lung cancer: a randomized trial. Jama 2003; 290: 2149 – 58. [DOI] [PubMed] [Google Scholar]
- 3. Lynch TJ, Bell DW, Sordella R et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2004; 350: 2129 – 39. [DOI] [PubMed] [Google Scholar]
- 4. Paez JG, Janne PA, Lee JC et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science 2004; 304: 1497 – 500. [DOI] [PubMed] [Google Scholar]
- 5. Sordella R, Bell DW, Haber DA, Settleman J. Gefitinib‐sensitizing EGFR mutations in lung cancer activate anti‐apoptotic pathways. Science 2004; 305: 1163 – 7. [DOI] [PubMed] [Google Scholar]
- 6. Chou TY, Chiu CH, Li LH et al. Mutation in the tyrosine kinase domain of epidermal growth factor receptor is a predictive and prognostic factor for gefitinib treatment in patients with non‐small cell lung cancer. Clin Cancer Res 2005; 11: 3750 – 7. [DOI] [PubMed] [Google Scholar]
- 7. Takano T, Ohe Y, Sakamoto H et al. Epidermal growth factor receptor gene mutations and increased copy numbers predict gefitinib sensitivity in patients with recurrent non‐small‐cell lung cancer. J Clin Oncol 2005; 23: 6829 – 37. [DOI] [PubMed] [Google Scholar]
- 8. Riely GJ, Pao W, Pham D et al. Clinical course of patients with non‐small cell lung cancer and epidermal growth factor receptor exon 19 and exon 21 mutations treated with gefitinib or erlotinib. Clin Cancer Res 2006; 12: 839 – 44. [DOI] [PubMed] [Google Scholar]
- 9. Inoue A, Suzuki T, Fukuhara T et al. Prospective phase II study of gefitinib for chemotherapy‐naive patients with advanced non‐small‐cell lung cancer with epidermal growth factor receptor gene mutations. J Clin Oncol 2006; 24: 3340 – 6. [DOI] [PubMed] [Google Scholar]
- 10. Uramoto H, Sugio K, Oyama T et al. Epidermal growth factor receptor mutations are associated with gefitinib sensitivity in non‐small cell lung cancer in Japanese. Lung Cancer 2006; 51: 71 – 7. [DOI] [PubMed] [Google Scholar]
- 11. Nagai Y, Miyazawa H. Huqun et al. Genetic heterogeneity of the epidermal growth factor receptor in non‐small cell lung cancer cell lines revealed by a rapid and sensitive detection system, the peptide nucleic acid‐locked nucleic acid PCR clamp. Cancer Res 2005; 65: 7276 – 82. [DOI] [PubMed] [Google Scholar]
- 12. Kosaka T, Yatabe Y, Endoh H, Kuwano H, Takahashi T, Mitsudomi T. Mutations of the epidermal growth factor receptor gene in lung cancer: biological and clinical implications. Cancer Res 2004; 64: 8919 – 23. [DOI] [PubMed] [Google Scholar]
- 13. Sasaki H, Shimizu S, Endo K et al. EGFR and erbB2 mutation status in Japanese lung cancer patients. Int J Cancer 2006; 118: 180 – 4. [DOI] [PubMed] [Google Scholar]
- 14. Sonobe M, Manabe T, Wada H, Tanaka F. Mutations in the epidermal growth factor receptor gene are linked to smoking‐independent, lung adenocarcinoma. Br J Cancer 2005; 93: 355 – 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tokumo M, Toyooka S, Kiura K et al. The relationship between epidermal growth factor receptor mutations and clinicopathologic features in non‐small cell lung cancers. Clin Cancer Res 2005; 11: 1167 – 73. [PubMed] [Google Scholar]
- 16. Sarkar G, Sommer SS. The ‘megaprimer’ method of site‐directed mutagenesis. Biotechniques 1990; 8: 404 – 7. [PubMed] [Google Scholar]
- 17. Vital statistics of population. Ministry of Health, Labour and Welfare, Japan.
- 18. Yokoyama T, Kondo M, Goto Y et al. EGFR point mutation in non‐small cell lung cancer is occasionally accompanied by a second mutation or amplification. Cancer Sci 2006; 97: 753 – 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Huang SF, Liu HP, Li LH et al. High frequency of epidermal growth factor receptor mutations with complex patterns in non‐small cell lung cancers related to gefitinib responsiveness in Taiwan. Clin Cancer Res 2004; 10: 8195 – 203. [DOI] [PubMed] [Google Scholar]
- 20. Inukai M, Toyooka S, Ito S et al. Presence of Epidermal Growth Factor Receptor Gene T790M Mutation as a Minor Clone in Non‐Small Cell Lung Cancer. Cancer Res 2006; 66: 7854 – 8. [DOI] [PubMed] [Google Scholar]
- 21. Kobayashi S, Boggon TJ, Dayaram T et al. EGFR mutation and resistance of non‐small‐cell lung cancer to gefitinib. N Engl J Med 2005; 352: 786 – 92. [DOI] [PubMed] [Google Scholar]
- 22. Pao W, Miller VA, Politi KA et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med 2005; 2: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sutani A, Nagai Y, Udagawa K et al. Gefitinib for non‐small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) gene mutations screened by Peptide Nucleic Acid‐Locked Nucleic Acid PCR clamp. Br J Cancer in press. [DOI] [PMC free article] [PubMed]


