Abstract
It is well known that S100A4 is overexpressed in many tumors and involved in tumor invasion and metastasis. But the regualtion of it is ill understood. We previously found that hypoxia mimicking cobalt chloride (CoCl2) enhanced the mRNA and protein expressions of the S100A4 gene in the gastric cancer cell line BGC823. In this study we found that S100A4 also displayed increased expression in BGC823 cells after exposure to real hypoxia (2.5% O2) as that by CoCl2 treatment. Moreover, S100A4 protein showed different subcellular distribution under real hypoxia compared with that by CoCl2 treatment or in normoxic conditions. To investigate the underlying molecular mechanism by which hypoxia regulates the expression of S100A4, we analyzed the regulatory sequences of the genes by bioinformatics and found a putative hypoxia responsive element (HRE) motif in the first intron of S1004. Furthermore, luciferase reporter assay showed that it is responsive to hypoxia. Electrophoretic mobility shift assay and chromatin immunoprecipitation assays demonstrated that hypoxia‐inducible factor 1 (HIF‐1) binds to the functional HRE in vitro and in vivo. The results provide evidence that S100A4 is a hypoxia‐inducible gene, whose transcription is stimulated at least partly through the interaction of HIF‐1 and HRE located at +329 to +334 of S100A4.
(Cancer Sci 2010; 101: 1141–1146)
S100A4 protein is a member of the S100 family of calcium‐binding proteins involved in the invasiveness and metastasis of tumors.( 1 , 2 , 3 , 4 ) S100A4 expression has been associated with metastasis in animal studies, and a number of studies have examined the utility of S100A4 expression as a prognostic marker in human cancers. Expression of S100A4 is significantly increased in tumors, such as thyroid, ovarian, lung, colorectal, pancreatic, and gastric cancers.( 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 ) It plays a key role in the process of metastasis by stimulating angiogenesis;( 13 ) promoting the migration of tumor cells,( 14 ) adhesion of cells to matrix, and escape of tumor cells from primary tumors; and inducing expression of protein hydrolase.( 15 , 16 ) However, the regulation of S100A4 gene expression is not well understood.
Hypoxia occurs in solid tumors due to uncoordinated tumor growth and angiogenesis. It was reported that hypoxia promoted tumor progression, invasion, metastasis, and resistance to chemotherapy, leading to poor patient prognosis.( 17 ) Whether S100A4 is regulated by hypoxia and mediates the effects of hypoxia in promoting the progression of cancer should be an interesting issue. In a previous study, we treated the gastric cancer cell line BGC823 with CoCl2 to mimic hypoxia and found that CoCl2 could up‐regulate S100A4 expression,( 18 ) but the effect of real hypoxia on S100A4 expression and underlying molecular mechanism are less well understood.
It is well known that hypoxia‐inducible factor 1 (HIF‐1) plays a critical role in cell response to hypoxia. HIF‐1 is a heterodimeric transcription factor which activates a wide range of genes encoding important proteins involved in the physiological adaptation to hypoxia.( 19 , 20 ) Most HIF‐1‐regulated genes contain an enhancer sequence termed the hypoxia responsive element (HRE).( 21 , 22 , 23 , 24 , 25 ) Hypoxia‐induced transcriptional activation of HIF‐1 involves, at least in part, a decrease in oxygen‐sensitive degradation of the HIF‐1α and an increase in its binding to the HIF‐1‐binding site (HBS) present in the HRE.( 26 ) The present study showed that S100A4 displayed increased expression and different subcellular distribution under real hypoxia (2.5% O2) conditions compared with that by hypoxia mimetics CoCl2‐treatment or in normoxia. We then explored the mechanism of transcriptional regulation of S100A4 by hypoxia. We found a putative HRE in the first intron of the S100A4 gene and demonstrated that HIF‐1 binds to the HRE in the S100A4 gene, by which S100A4 expression was regulated under hypoxic conditions.
Materials and Methods
Reagents. Cobalt chloride (CoCl2) was from Sigma‐Aldrich (St. Louis, MO, USA). The pGL3‐Basic, pRL‐TK luciferase reporter vectors and the Dual Luciferase Reporter assay system were obtained from Promega (Madison, WI, USA). The reverse transcription system was from Promega. Rabbit antibodies against HIF‐1α, and rabbit anti‐β‐actin, secondary antibodies were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA); rabbit anti‐S100A4 antibody was from Lab Vision & Neomarkers (Fremont, CA, USA). The DNA 3′‐end bio‐labeled kit was purchased from Pierce Biotechnology (Thermo Scientific, Rockford, IL, USA).
Cell culture. The gastric adenocarcinoma cell line BGC823 cells were cultured in RPMI‐1640 medium supplemented with 10% FCS. The cells were maintained at 37°C and 5% CO2 in a humidified incubator.
Hypoxia treatment. Cells were incubated in a temperature and humidity‐controlled environmental chamber IG750 (Jouan, St‐Herblain, France) in an atmosphere containing 2.5% O2, 5% CO2, and 92.5% N2. Hypoxia was also mimicked by treatment with CoCl2 (200 μm).
Reverse transcriptase–polymerase chain reaction (RT‐PCR). The total RNA of BGC823 cells was extracted using the Trizol reagent (Invitrogen, Carlsbad, CA, USA); 1 μg of RNA was reverse transcribed using the Promega RT system. The resulting cDNA was amplified by PCR. Primers specific for human S100A4 and β‐actin were designed as follows: S100A4 sense 5′‐GATGTGATGGTGTCCACCTT‐3′, antisense 5′‐ATTTCTTCCTGGGCTGCTTA‐3′; β‐actin sense 5′‐CTCTTCCAGCCTTCCTTCCT‐3′, antisense 5′‐CACCTTCACCGTTCCAGTTT‐3′. Aliquots of PCR products were checked by electrophoresis on a 1.5% agarose gel with the fragments visualized by ethidium bromide staining.
Immunofluorescence staining. The BGC823 cells were cultured on coverslip. Hypoxia treatment (2.5% O2) was carried out as described before. The cultured cells were rinsed in PBS and fixed in 10% formaldehyde for 15 min at room temperature. Fixed cells were incubated first with 0.1% Triton X‐100 for 10 min followed by incubation in blocking solution (5% BSA) for 30 min, then cells were incubated with a 1:500 dilution of rabbit anti‐S100A4 antibody for 2 h, and then incubated with a 1:2000 dilution of FITC‐conjugated mouse antirabbit IgG (Invitrogen) for 30 min at 37°C. Nuclei were stained with 4′,6‐diamidino‐2‐phenylindole (DAPI; Nakarai, Tokyo, Japan). The cells were observed under a fluorescence microscope.
Plasmid constructions. A 937 bp DNA fragment (937 wt) of S100A4 gene containing the putative HRE motif (from −387 to +550) was amplified by PCR using the human genomic DNA as a template and the primers were: 5′‐TCACCACATTTCCAGGGCA‐3′, 5′‐GCAAAGTGAGGTGGCAGAG‐3′. We employed overlap extension‐PCR to generate the mutated 937‐bp DNA fragment (937mu) by introducing the same mutations as those described in EMSA to the putative HRE motif using the following primer pairs: 5′‐TCACCACATTTCCAGGGCA‐3′, 5′‐GGCACATAATACCAACCTTTACTA‐3′, 5′‐TAGTAAAGGTTGGTATG TATGTGCC‐3′; 5′‐GCAAAGTGAGGTGGCAGAG‐3′, and the primers used in the generation of 937wt as described above. The fragments 937wt or 937mu were cloned into the KpnI/HindIII sites of pGL3‐Basic to generate pGL3‐937wt or pGL3‐937mu reporter constructs, respectively. All constructs were confirmed by sequencing.
Transient transfection and luciferase reporter assay. BGC823 cells were seeded at 1.5 × 104 cells per well (24‐well plate) and maintained at 37°C in a humidified 5% CO2 atmosphere. The DNA transfections with 0.8 μg of the pGL3‐937wt or pGL3‐937mu luciferase constructs with internal control vector phRL‐TK (50:1 ratio) were performed using Lipofectin (Invitrogen) 24 h after cell seeding. After incubation for 5 h, the medium was replaced with a fresh one, and these cells were incubated for 24 h under real hypoxic (2.5% O2) or normoxic (21% O2) conditions. The cells were then harvested, and the luciferase activities in the cell lysates were determined using dual luciferase assay reagents. The luciferase activities were normalized by the internal control values. The values were represented as the mean ± SD of triplicate samples.
Western blot analysis. BGC823 cells were treated with CoCl2 (200 μm) or real hypoxia (2.5% O2), and harvested at different time points for the detection of HIF‐1α or S100A4 protein expression, respectively. Then CoCl2 treatment or real hypoxia treatment as well as untreated BGC823 cells were lysed with lysis buffer. Protein concentrations were measured using a modified Lowry procedure with bovine serum albumin (BSA) as the standard. Equal amounts of proteins in whole cell lysates were separated on 12% SDS‐polyacrylamide gels, transferred onto PVDF membranes (Millipore, Bedford, MA, USA), and blocked with TBST supplemented with 5% nonfat milk. Membranes were incubated with the rabbit anti‐HIF‐1α, S100A4, or anti‐actin antibodies at 1:1000 dilution for 16 h. After extensive washing with TBST, the membranes were incubated for 1 h with horseradish peroxidase‐conjugated goat antirabbit IgG (1:5000) in blocking solution. The blots were visualized using the Amersham ECL system.
Electrophoretic mobility shift assay (EMSA). The nuclear extracts were prepared from BGC823 cells maintained under normoxic or hypoxic conditions for 24 h using the NE‐PER Nuclear Extraction Reagent Kit (Pierce Biotechnology). Wild‐type and mutated oligonucleotides corresponding to the S100A4‐HRE region were designed according to the S100A4 gene sequence (GenBank accession no. 6275). The sequences of the sense strands used for EMSA are shown in Table 1. The probes were labeled using the Biotin 3′ End DNA Labeling Kit according to the manufacturer’s instructions. The binding reactions were carried out with 15 μg of nuclear extracts and bio‐labeled S100A4‐HREwt, S100A4‐HREmu, and consensus probes (Epo‐HRE). The mixtures were incubated for 1 h at room temperature. The supershift analysis was performed using 1 μg of rabbit antibody against HIF‐1α. For the oligonucleotide competition experiments, unlabeled S100A4‐HREwt was used. The complexes were resolved in a 6% nondenaturing polyacrylamide gel in 1× Tris‐Borate‐EDTA electrophoresis buffer.
Table 1.
Oligonucleotides used for EMSA
| S100A4‐HREwt | 5′‐ CACACGCTGTTGCTA TAGTACGTGTTGGTATGTA‐3′ |
| Erythropoietin (Epo‐HRE) | 5′‐CTATAGTACGTGTTGGTA‐3′ |
| S100A4‐HREmu | 5′‐CACACGCTGTTGCTATAGTAAAGGTTGGTATGTA‐3′ |
Core sequences of the S100A4‐HRE are shown in bold and mutant bases are underlined. HRE, hypoxia responsive element.
Chromatin immunoprecipitation (ChIP). BGC823 cells were treated with CoCl2 (200 μm) for 24 h to induce the expression of HIF‐1, with the untreated cells as control. DNA and proteins were cross‐linked by the addition of 1% formaldehyde for 20 min before harvesting. Cells were washed twice with ice cold PBS, scraped off the plates, resuspended in 300 μL of SDS lysis buffer (1% SDS, 10 mm EDTA, 50 mm Tris‐HCl [pH8.0], and a protease inhibitor mixture), then sonicated to generate 100–1000 bp fragments. After centrifugation, the cleared supernatants were diluted 10‐fold with immunoprecipitation buffer (50 mm Tris‐HCl [pH 8], 150 mm NaCl, 5 mm EDTA, 0.5% Nonidet P‐40). The cell lysates were precleared by incubation at 4°C with 15 μL protein G beads pre‐adsorbed with sonicated single‐stranded DNA and BSA. The cleared lysates were incubated overnight with anti‐HIF‐1α antibody or without any antibodies. Immune complexes were precipitated with 30 μL protein G beads pre‐absorbed with sonicated single‐stranded DNA and BSA. After centrifugation, the beads were washed, and the antigen was eluted with 1% SDS, 100 mm sodium carbonate. DNA‐protein cross‐links were reversed by heating at 65°C for 4–5 h, and DNA was phenol‐extracted and analyzed by PCR with Taq DNA Polymerase (Promega) using the following primers: forward 5′‐ATGTGGAAGACAGCATAGACC‐3′ and reverse 5′‐AGAGGCAAAGAGGA AAACCACCA‐3′.
PCR and sequencing. DNA was extracted from 25 cases of gastric carcinoma tissues. The fragment of the S100A4 gene containing the core HRE motif was analyzed by the combined PCR and sequencing techniques. The PCR primers were: forward 5′‐TTCTTGGTTTGGTGAGTTGTG‐3′ and reverse 5′‐GCAAAGTGAGGTGGCA GAG‐3′.
Results
S100A4 expression was up‐regulated and showed distinct subcellular distribution under real hypoxic conditions. RT‐PCR showed that the level of S100A4 mRNA at 24 h after exposure to hypoxia was the highest among the time points observed (Fig. 1a). Therefore, we analyzed S100A4 protein expression at 24 h after exposure to hypoxia by western blotting and immunofluorescence staining, which showed up‐regulation of S100A4 protein (Fig. 1b,c). Moreover, S100A4 protein displayed distinct subcellular distribution after real hypoxia exposure. High nuclear S100A4 protein expression was observed in BGC823 cells under real hypoxic conditions (Fig. 1c).
Figure 1.
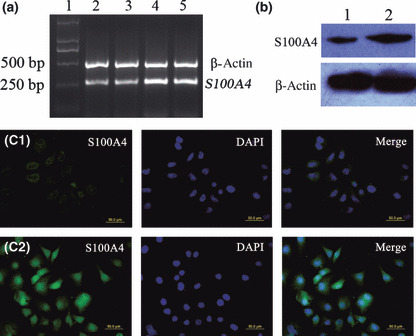
Effects of real hypoxia (2.5% O2) on S100A4 expression and subcecullar distribution of S100A4 protein. BGC823 gastric cancer cells were cultured under hypoxic conditions (2.5% O2), and S100A4 mRNA and protein expression were analyzed by RT‐PCR (a), western blot analysis (b), and immunofluorescence staining (c) as described in the Materials and Methods. (a) Lane 1: M, DNA marker – DL2000; Lane 2: sample from untreated cells; Lanes 3 to 5: samples representing 6, 24, and 48 h after exposure to hypoxia, respectively. (b) Lane 1: sample from untreated cells; Lane 2: sample representing 24 h after exposure to hypoxia. (c) C1, C2: samples representing untreated cells, 24 h after exposure to hypoxia, respectively. The primary antibody used in this experiment was anti‐S100A4 antibody (green). DAPI was used to visualize nuclear staining (blue). Merged images of green and blue signals are shown in the right panels.
Identification of the putative HRE motif in the first intron of the human S100A4 gene. Bioinformatic analysis of the promoter region and the first intron sequences of the S100A4 gene showed that there was HIF‐1 binding site, named HRE motif, located at +329 to +334 (Fig. 2) in the first intron, suggesting that it may have an oxygen‐dependent regulation. Potential HRE motif was identified using the programs at URL: http://www.gene‐regulation.com.
Figure 2.

Prediction of the hypoxia responsive element (HRE) motif in the first intron of the S100A4 gene. The sequence containing the entire first exon and part of the first intron in the S100A4 gene is shown, in which the putative HRE motif is indicated.
Transcriptional regulation of the region harboring the putative HRE in the S100A4 gene by hypoxia. The promoter activity of pGL3‐973wt increased by 2.6‐fold upon exposure to hypoxia compared with normoxia, and activity was markedly abrogated by the mutation of the HRE (pGL3‐973mu) (Fig. 3). The data indicated that the HRE motif was directly involved in the activation of S100A4 transcription under hypoxia.
Figure 3.
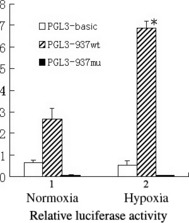
Luciferase reporter assay demonstrating that the putative hypoxia responsive element (HRE) is responsible for the transcriptional activation of S100A4 under hypoxia. Each bar represents the relative luciferase activity under hypoxic and normoxic conditions. The relative luciferase activity is represented by the ratio of luciferase activity of firefly (from the various pGL3 vectors) to that of Renilla (from phRL‐TK construct) as an internal control. The data represent the mean ± SD from three experiments (*P < 0.05).
HIF‐1 protein expression in BGC823 cells at different time points after exposure to CoCl2. Western blot showed up‐regulation of HIF‐1 protein after CoCl2 exposure compared with nontreated cells. The up‐regulation of HIF‐1 protein in BGC823 cells occurred 6 h after cells were exposed to CoCl2, reached a peak at 24 h, and the overexpression lasted for 48 h (Fig. 4). Therefore, we treated BGC823 cells with 200 μm CoCl2 for 24 h for EMSA and ChIP.
Figure 4.
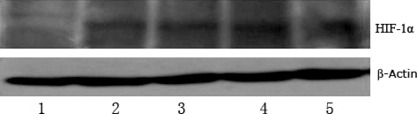
Hypoxia‐inducible factor 1 (HIF‐1) protein expression at different time points after exposure to CoCl2 in BGC823 cells as measured by western blot analysis. Lane 1: sample from untreated cells; Lanes 2 to 5: samples representing 6, 12, 24, and 48 h after exposure to CoCl2.
Analysis of binding of nuclear protein HIF‐1 to the predicted HRE by EMSA and ChIP. To determine whether the HRE identified within the first intron of the S100A4 gene binds the HIF‐1 protein, EMSA was performed with the nuclear extracts obtained from CoCl2‐treated BGC823 cells. As shown in Figure 5, a specific complex with retarded migration appeared exclusively when CoCl2‐treated nuclear extracts were incubated together with the labeled probe containing the intact HRE motif. No complex was visualized when mutated oligonucleotides were used as the probe. In addition, the specific binding of CoCl2‐treated nuclear extracts to the wild‐type probe was eliminated by competition with an excess of homologous unlabeled probe. These results demonstrate that HIF‐1 binds to the consensus HRE. To identify proteins involved in the formation of these complexes, we performed supershift assays which showed that a HIF‐1α antibody decreased the formation of the hypoxia‐inducible complex and caused the appearance of a supershifted complex. These results indicate that HIF‐1α was present in these hypoxia‐induced complexes.
Figure 5.
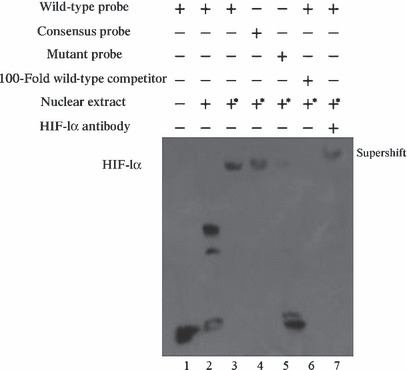
Analysis of hypoxia‐inducible factor 1 (HIF‐1) binding to the hypoxia responsive element (HRE) motifs in the S100A4 gene in vitro using EMSA. BGC823 cells were exposed to normoxic or hypoxic mimicking conditions for 24 h prior to preparation of nuclear extracts. Various probes were incubated with nuclear extracts from CoCl2‐treated or nontreated BGC823 cells. Lane 1: labeled S100A4‐HREwt probe without nuclear extract; Lane 2: labeled S100A4‐HREwt probe incubated with nuclear extract from nontreated cells formed no specific complex; Lane 3: labeled S100A4‐HREwt incubated with the nuclear extracts from CoCl2‐treated cells forming a complex; Lane 4: labeled Epo‐HREwt as consensus oligonucleotide probe incubated with the nuclear extracts from CoCl2‐treated cells forming a complex; Lane 5: labeled S100A4‐HREmu probe did not form a complex; Lane 6: competition experiments performed with the S100A4‐HREwt probe using a 100‐fold molar excess of unlabeled S100A4‐HREwt oligonucleotides; Lane 7: supershift analysis of HIF‐1 bound to the S100A4‐HREwt probe performed using anti‐HIF‐1α antibody; *nuclear extracts from the CoCl2‐treated cells.
ChIP experiments were performed to check the binds of HIF‐1 proteins and HRE motifs in vivo, using HIF‐1α antibody, with the IgG antibody as a negative control. Positive signals were found when the HIF‐1α antibody but not the IgG antibody was used in BGC823 cells subjected to hypoxic conditions, demonstrating that HIF‐1 interacts with the S100A4 gene in vivo (Fig. 6). Significantly weaker signals were found in cells exposed to normoxic compared to hypoxic conditions, due to the up‐regulation of HIF‐1 under hypoxic conditions.
Figure 6.
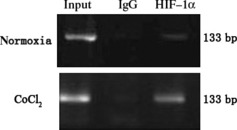
Analysis of the hypoxia‐inducible factor 1 (HIF‐1) binding to the hypoxia responsive element (HRE) motif of the S100A4 gene in vivo by ChIP assay. BGC823 cells were grown under normoxic or hypoxic conditions for 24 h. Lysates of BGC823 cells were immunoprecipitated with HIF‐1α antibody or normal rabbit IgG. Subsequently, a DNA segment of the first intron of the S1004 gene containing the HRE was amplified by PCR. Lysates without immunoprecipitation were used as the control for DNA input in each sample. Binding of HIF‐1α was increased by CoCl2 treatment compared with untreated samples.
Sequence analysis of fragments harboring HRE of S100A4 in gastric carcinoma tissues. The combined PCR and sequencing techniques were used to analyze the fragment of the S100A4 gene (+44 bp to +507 bp) containing the core HRE motif in 25 gastric carcinomas, but no variation was detected among all the samples.
Discussion
Our previous study demonstrated that the S100A4 gene was induced during chemical induction of hypoxia in BGC823 cells.( 18 ) In the present study, we found that real hypoxia (2.5% O2) could also up‐regulate S100A4 expression at both mRNA and protein levels in BGC823 cells. Moreover, the subcellular distribution of S100A4 by real hypoxia was different from that by hypoxia mimicking CoCl2 treatment in our previous study. Increased S100A4 protein occurred mainly in the cytoplasm of BGC823 cells after CoCl2 treatment, whereas it occurred mainly in nuclear after real hypoxia exposure. The different subcellular distribution of S100A4 protein maybe due to different properties of hypoxia mimics and real hypoxia, although they share some commonality. It seems that nuclear overexpression of S100A4 under a real hypoxic condition maybe an interesting issue. Maelandsmo et al. ( 6 ) reported that nuclear S100A4 expression in tumor cells was associated with more aggressive ovarian carcinomas. Furthermore, it has been reported that S100A4 regulated the expression of many genes.( 27 ) So we can speculate that hypoxia induces S100A4 expression and translocation to the nucleus, where it regulates many gene expressions and promotes the progression of cancer.
Our previous and present studies showed that real hypoxia and CoCl2 are consistent in promoting the expression of S100A4. So we wanted to explore the mechanism by which hypoxia regulates S100A4 expression. One possible mechanism by which hypoxia regulates S100A4 expression is through the transcriptional factor HIF‐1. Previous studies have shown several regulatory elements such as kB‐related, specificity protein 1 (Sp1), core‐binding factor (Ap‐1), core‐binding factor (CBF), and κ recognition component (KRC) binding sites etc., in the first intron of S100A4,( 28 , 29 , 30 ) which suggest that the first intron is an important region for gene regulation. In the present study, bioinformatics analysis was used to analyze the promoter region and sequences downstream of the transcription initiation site, including the first intron of the S100A4 gene in the search for HREs. The core sequence of the potential HRE motif was found in the first intron of the S100A4 gene, located at +329 to +334 (TACGTG) which conforms to the consensus sequence of the HIF‐1‐binding site 5′‐RCGTG‐3′ found in other genes such as erythropoietin (EPO), vascular endothelial growth factor (VEGF), endothelin‐1, endothelial nitric oxide synthase (eNOS), etc.( 22 , 23 , 24 , 25 ) It might be essential to the regulation of S100A4 under hypoxic conditions in BGC823 cells.
To identify whether the putative HRE motif found in the first intron of S100A4 gene was indeed responsible for the transcriptional activation under hypoxia, we performed luciferase reporter assays. The results showed that transfection of pGL3‐937wt containing the regulatory fragment with the wild‐type HRE motif enhanced the reporter gene’s activity under hypoxia compared with that in normoxia. Meanwhile, the pGL3‐937mu construct with the mutated HRE exhibited far lower luciferase activity than the pGL3‐937wt construct in hypoxia, and no significant difference in the regulatory activity of pGL3‐937mu construct was found between normoxic and hypoxic conditions, indicating that the putative HRE motif in S100A4 is essential for the regulatory activity under hypoxia. It is interesting that the regulatory activity of the pGL3‐937mu construct decreased dramatically compared to the pGL3‐937wt and basic pGL3 construct even under normoxia. But no known human regulatory elements were found in the mutated HRE motif and its neighboring sequences by bioinformatics analysis. Therefore, we speculated that wild‐type HRE may antagonize the effect of a potential repressor element in S100A4. When it was mutated, not only was the induction of HRE disrupted and abolished under hypoxia, but also the activity of related repressor element(s) increased and caused the reduced luciferase activity from the pGL3‐937mu construct even in normoxic conditions. However, the exact molecular mechanism remains to be clarified.
It is known that CoCl2 can also induce HIF‐1α expression in many different kinds of cells.( 31 ) We used 200 μm CoCl2 to mimic the hypoxic conditions and detected the HIF‐1α protein expression in BGC823 cells at different time points after exposure to CoCl2. We determined that the peak expression of the HIF‐1α protein occurred at 24 h after exposure to CoCl2. We therefore treated cells with CoCl2 for 24 h in subsequent studies.
To identify the binding of nuclear protein HIF‐1 to the HRE motif, EMSA and ChIP were performed. EMSA showed that a specific DNA‐protein complex, absent with control extracts from normoxic cells, was formed with nuclear extracts from CoCl2‐treated gastric cancer cells. Mutant oligonucleotides failed to form the complex with nuclear extracts from CoCl2‐treated gastric cancer cells as well. Additionally, the complex was prevented by an excess of cold S100A4‐HREwt probes. Anti‐HIF‐1α was able to “supershift” the hypoxia‐induced binding activity, showing that HIF‐1α could bind the HRE motif in vitro. The ChIP assay results also showed that HIF‐1 could bind to the HRE of the S100A4 gene in vivo and that CoCl2 treatment could enhance their interactions. Collectively, these data indicate that HIF‐1 can bind the HRE motif of S100A4 both in vivo and in vitro.
Furthermore, we hypothesized that there might be sequence variations in the HRE of the S100A4 gene which may affect the S100A4 hypoxic response in gastric carcinoma tissues. To test this possibility, we screened 25 primary gastric carcinoma tissues by amplifying and sequencing a fragment of approximately 500 bp containing the HRE of the S100A4 gene. However, no sequence variations were found in the fragments in any of the gastric carcinoma tissues investigated. The results did not seem to support our hypothesis. However, the sample number in our study was small; further study by increasing sample numbers is needed to clarify this.
In summary, our data showed that real hypoxia (2.5% O2) induced S100A4 expression at both the mRNA and protein level. Moreover, real hypoxia led to nuclear overexpression of S100A4, which is different from subcellular distribution of S100A4 by CoCl2 treatment or in normoxia. Our data also showed that HIF‐1 could bind the HRE at position +329 to +334 of the S100A4 gene in vivo and in vitro. The results provide important insights into the molecular mechanism by which human S100A4 gene expression is induced under hypoxic conditions.
Acknowledgment
This project was supported by a grant from the National Natural Science Foundation of China (no. 30570848).
References
- 1. Zimmer DB, Cornwall EH, Landar A, Song W. The S100 protein family: history, function, and expression. Brain Res Bull 1995; 37: 417–29. [DOI] [PubMed] [Google Scholar]
- 2. Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: from evolution to function and pathology (including an update of the nomenclature). Biochem Biophys Res Commun 2004; 322(4): 1111–22. [DOI] [PubMed] [Google Scholar]
- 3. Ebralidze A, Tulchinsky E, Grigorian M et al. Isolation and characterization of a gene specifically expressed in different metastatic cells and whose deduced gene product has a high degree of homology to a Ca2+‐binding protein family. Genes Dev 1989; 3: 1086–93. [DOI] [PubMed] [Google Scholar]
- 4. Davies MPA, Rudland PS, Robertaon L, Parry EW. Expression of the calcium‐binding protein S100A4 (p9Ka) in MMTV‐neu transgenic mice induces metastasis of mammary tumors. Oncogene 1996; 13: 1631–7. [PubMed] [Google Scholar]
- 5. Min HS, Choe G, Kim SW et al. S100A4 expression is associated with lymph node metastasis in papillary microcarcinoma of the thyroid. Mod Pathol 2008; 21: 748–55. [DOI] [PubMed] [Google Scholar]
- 6. Mælandsmo GM, Flørenes VA, Nguyen MT. Different Expression and Clinical Role of S100A4 in Serous Ovarian Carcinoma at Different Anatomic Sites. Tumour Biol 2009; 30: 15–25. [DOI] [PubMed] [Google Scholar]
- 7. Yoon CS, Hyung WJ, Lee JH et al. Expression of S100A4, E‐cadherin, alpha‐ and beta‐catenin in gastric adenocarcinoma. Hepatogastroenterology 2008; 55: 1916–20. [PubMed] [Google Scholar]
- 8. Cho YG, Nam SW, Kim TY et al. Overexpression of S100A4 is closely related to the aggressiveness of gastric cancer. APMIS 2003; 111: 539–45. [DOI] [PubMed] [Google Scholar]
- 9. Miyazaki N, Abe Y, Oida Y. Poor outcome of patients with pulmonary adenocarcinoma showing decreased E‐cadherin combined with increased S100A4 expression. Int J Oncol 2006; 28: 1369–74. [PubMed] [Google Scholar]
- 10. Hemandas AK, Salto‐Tellez M, Maricar SH. Metastasis‐associated protein S100A4: a potential prognostic marker for colorectal cancer. J Surg Oncol 2006; 93: 498–503. [DOI] [PubMed] [Google Scholar]
- 11. De Silva Rudland S, Martin L, Roshanlall C et al. Association of S100A4 and osteopontin with specific prognostic factors and survival of patients with minimally invasive breast cancer. Clin Cancer Res 2006; 12: 1192–200. [DOI] [PubMed] [Google Scholar]
- 12. Sato N, Maitra A, Fukushima N et al. Frequent hypomethylation of multiple genes overexpressed in pancreatic ductal adenocarcinoma. Cancer Res 2003; 63: 4158–66. [PubMed] [Google Scholar]
- 13. Ambartsumian N, Klingelhöfer J, Grigorian M et al. The metastasis‐associated Mts1 (S100A4) protein could act as an angiogenic factor. Oncogene 2001; 20: 4685–95. [DOI] [PubMed] [Google Scholar]
- 14. Oslejskova L, Grigorian M, Gay S, Neidhart M, Senolt L. The metastasis associated protein S100A4: a potential novel link to inflammation and consequent aggressive behavior of rheumatoid arthritis synovial fibroblasts. Ann Rheum Dis 2008; 67: 1499–504. [DOI] [PubMed] [Google Scholar]
- 15. Schmidt‐Hansen B, Klingelhöfer J, Grum‐Schwensen B. Functional significance of metastasis‐inducing S100A4 (Mts1) in tumor‐stroma interplay. J Biol Chem 2004; 279: 24498–504. [DOI] [PubMed] [Google Scholar]
- 16. Novitskaya V, Grigorian M, Kriajevska M et al. Oligomeric forms of the metastasis‐related Mts1 (S100A4) protein stimulate neuronal differentiation in cultures of rat hippocampal neurons. J Biol Chem 2000; 275: 41278–86. [DOI] [PubMed] [Google Scholar]
- 17. Ferrara N, Kerbel RS. Angiogenesis as a therapeutic target. Nature 2005; 438: 967–74. [DOI] [PubMed] [Google Scholar]
- 18. Hua J, Fu H, Zhang RX et al. Effect of cobalt chloride on S100A4 expression in human gastric cancer cells BGC823. Yi Chuan 2008; 30: 1563–6. [DOI] [PubMed] [Google Scholar]
- 19. Rankin EB, Giaccia AJ. The role of hypoxia inducible factors in tumorigenesis. Cell Death Differ 2008; 15: 678–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med 2003; 9: 677–84. [DOI] [PubMed] [Google Scholar]
- 21. Semenza G, Wang G. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol 1992; 12: 5447–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Forsythe JA, Jiang BH, Iyer NV et al. Activation of vascular endothelial growth factor gene transcription by hypoxia‐inducible factor 1. Mol Cell Biol 1996; 16: 4604–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hayashi M, Sakata M, Takeda T et al. Induction of glucose transporter 1 expression through hypoxia‐inducible factor 1alpha under hypoxic conditions in trophoblast‐derived cells. J Endocrinol 2004; 183: 145–54. [DOI] [PubMed] [Google Scholar]
- 24. Coulet F, Nadaud S, Agrapart M, Soubrier F. Identification of hypoxia‐response element in the human endothelial nitric‐oxide synthase gene promoter. J Biol Chem 2003; 278: 46230–40. [DOI] [PubMed] [Google Scholar]
- 25. Lok CN, Ponka P. Identification of a hypoxia response element in the transferrin receptor gene. J Biol Chem 1999; 274: 24147–52. [DOI] [PubMed] [Google Scholar]
- 26. Wang GL, Semenza GL. General involvement of hypoxia‐inducible factor 1 in transcriptional response to hypoxia. Proc Natl Acad Sci U S A 1993; 90: 4304–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saleem M, Kweon MH, Johnson JJ et al. S100A4 accelerates tumorigenesis and invasion of human prostate cancer through the transcriptional regulation of matrix metalloproteinase 9. Proc Natl Acad Sci U S A 2006; 103: 14825–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tulchinsky E, Ford HL, Kramerov D et al. Transcriptional analysis of the mts1 gene with specific reference to 5′ flanking sequences. Proc Natl Acad Sci U S A 1992; 89: 9146–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tulchinsky E, Prokhortchouk E, Georgiev G, Lukanidin E. A κB‐related binding site is an integral part of the mts1 gene composite enhancer element located in the first intron of the gene. J Biol Chem 1997; 272: 4828–35. [DOI] [PubMed] [Google Scholar]
- 30. Cohn MA, Hjelmsø I, Wu LC, Guldberg P, Lukanidin EM, Tulchinsky EM. Characterization of Sp1, AP‐1, CBF and KRC binding sites and minisatellite DNA as functional elements of the metastasis‐associated mts1/S100A4 gene intronic enhancer. Nucleic Acids Res 2001; 29: 3335–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ardyanto TD, Osaki M, Nagahama Y et al. Down‐regulation of cobalt‐induced HIF‐1alpha expression correlates with cell proliferation and apoptosis in human gastric carcinoma cells. Oncol Rep 2008; 19: 339–43. [PubMed] [Google Scholar]


