Abstract
Many growth factors and cytokines are immobilized on the extracellular matrix (ECM) by binding to glycosaminoglycans and are stored in an inactive form in the cellular microenvironment. However, the mechanisms of ECM‐bound growth factor or cytokine activation have not been well documented. We showed that the insulin‐like growth factor type‐1 receptor (IGF‐1R) was rapidly phosphorylated after the addition of matrix metalloproteinase (MMP)‐7 to a serum‐starved human colon cancer cell line (HT29) and that phosphorylation was completely inhibited by an IGF‐II neutralizing antibody. In the ECM of this cell line, IGF‐II and IGF binding protein (BP)‐2 coexisted, but IGFBP‐2 disappeared from the ECM fraction after treatment with MMP‐7 or heparinase III. On the other hand, in a cell line in which IGF‐1R was overexpressed, IGF‐1R was phosphorylated by supernatant from the MMP‐7‐treated ECM fraction of HT29 but not by that from a heparinase‐III‐treated ECM fraction. We also demonstrated that MMP‐7 degrades IGFBP‐2 in vitro at three cleavage sites (peptide bonds E151–L152, G175–L176 and K181–L182), which have not been documented previously. Taken together, these results demonstrate that MMP‐7 generates bioactive IGF‐II by degrading the IGF‐II/IGFBP‐2 complex binding to heparan sulfate proteoglycan in the ECM, resulting in IGF‐II‐induced signal transduction. This evidence indicates that some ECM‐associated growth factors enhance their ability to bind to their receptors by some proteases in the tumor microenvironment. This mechanism of action (‘protease‐triggered matricrine’) represents an attractive model for understanding ECM–tumor interactions. (Cancer Sci 2007; 98: 685–691)
The potential roles of insulin‐like growth factors (IGFs) in cancer growth and survival have been extensively investigated.( 1 , 2 , 3 , 4 ) IGFs are expressed ubiquitously and function as endocrine, paracrine and autocrine growth factors. In most tissues, they are synthesized together with six binding proteins (IGFBP‐1 to IGFBP‐6). Limited proteolysis of IGFBPs is essential for regulation of IGF bioavailability in the bloodstream and at cellular level.( 5 , 6 ) IGFBP‐2, ‐3 and ‐5 can also associate with the extracellular matrix (ECM),( 7 ) and may concentrate IGF near the IGF type‐1 receptor (IGF‐1R). In the case of IGFBP‐5, binding to the ECM decreases IGFBP affinity for IGF and potentiates IGF action.( 8 ) The consequence for cell function of binding of IGFBP‐2 to the ECM is unknown. In the only report about membrane‐associated IGFBP‐2, it was shown that IGFs bind to high levels of membrane‐associated IGFBP‐2 in small‐cell lung carcinoma, which does not respond to IGFs even though IGF‐1R is present.( 9 ) This suggests that membrane‐associated IGFBP‐2 may compete with IGF‐1R for ligand and regulate IGF responsiveness in lung carcinoma tissue. How cancer cells use the membrane‐ or ECM‐bound IGF/IGFBP complex as an active growth factor is still unclear. Remacle‐Bonnet et al. presented a model in which membrane‐bound plasmin generated by plasminogen and uPA derived from stromal cells induces selective proteolysis of IGFBP‐4 in the ECM and promotes autocrine IGF‐II bioavailability in human colon cancer cells.( 10 ) Manes et al. reported that metalloproteinase (MMP)‐9 derived from cancer cells triggered an IGF‐I autocrine response by degrading the membrane‐bound IGF‐I/IGFBP‐3 complex in human androgen‐independent prostate cancer cells.( 11 )
Unlike matrix MMPs synthesized by stromal cells, MMP‐7 is produced exclusively by cancer cells and participates directly in the process of invasion and metastasis through broad proteolytic activity against a variety of ECM substrates.( 12 , 13 , 14 , 15 ) In addition to this common role of MMPs, MMP‐7 has been reported to act as an activator of growth factors and cytokines by degrading their precursors or inhibitors.( 16 , 17 , 18 , 19 , 20 ) Our recent in vitro data demonstrated that MMP‐7 possesses pan‐IGFBP protease activity,( 21 ) but the biological implications of proteolysis by MMP‐7 have not been characterized fully for each IGFBP. In the present study, we first demonstrated that MMP‐7 derived from cancer cells triggered an autocrine response of IGF‐II by degrading the ECM‐bound IGF‐II/IGFBP‐2 complex. Some ECM‐associated growth factors can enhance their ability to bind to their receptors, which is called ‘matricrine’ action.( 22 ) Recently, Helmers et al. have reported a similar possibility that MMP7 can release IGF‐II from ECM through the IGFBP‐5 cleavage.( 23 ) These findings suggest that MMP‐7 contributes to tumor growth and survival by regulating the bioavailability of IGF in the surrounding tissue in a paracrine and a matricrine manner.
Materials and Methods
Proteins and reagents. Recombinant human IGF‐II and recombinant human IGFBPs (IGFBP‐1 to IGFBP‐6) were obtained from R & D Systems (Minneapolis, MN, USA). Recombinant human active MMP‐7 was obtained from Chemicon International (Temecula, CA, USA). Tissue inhibitor of matrix metalloproteinase‐1 (TIMP‐1) was obtained from Daiichi Fine Chemical (Toyama, Japan). The commercially available antibodies used in this study were anti IGFBP‐1 mouse monoclonal antibody (Ab) (R & D Systems), anti IGFBP‐2 goat polyclonal Ab (C‐18, Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti IGFBP‐3 goat polyclonal Ab (C‐19, Santa Cruz Biotechnology), anti IGFBP‐4 mouse monoclonal Ab (R & D Systems) anti IGFBP‐5 goat polyclonal antibody (R & D Systems), anti IGFBP‐6 goat polyclonal Ab (C‐20, Santa Cruz Biotechnology), anti‐IGF‐1R rabbit polyclonal Ab (C‐20, Santa Cruz Biotechnology), antiphospho‐IGF‐1R rabbit polyclonal Ab (44–804, BioSource International, Camarillo, CA, USA), anti‐Akt rabbit monoclonal Ab (Cell Signaling Technology, Beverly, MA, USA), antiphospho‐Akt rabbit monoclonal Ab (Cell Signaling Technology) and peroxidase labeled antimouse/rabbit/goat Abs (Zymed Laboratories, San Francisco, CA, USA). KM1468 (antihuman‐IGF‐II rat monoclonal Ab) and KM1762 (antiavermectin rat monoclonal Ab as a control for IGF) were developed at Kyowa Hakko Kogyo (Tokyo, Japan).( 24 , 25 ) Protease‐inhibitor cocktail tablets (Complete) were purchased from Roche Diagnostic (Mannheim, Germany). Heparinase III was purchased from Sigma (St Louis, MO, USA). Agarose and trichloroacetic acid were purchased from Wako (Osaka, Japan). Silver‐staining kit was purchased from Daiichi Pure Chemicals (Tokyo, Japan).
Cell culture. HT29 cells (ATCC HTB‐38, American Type Culture Collection, Manassas, VA, USA) and SW1116 cells (ATCC CCL‐233) were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Sigma) supplemented with 10% fetal bovine serum (FBS) in humidified incubators at 37°C in 5% CO2. Anchorage‐independent culture was carried out in 0.9% soft‐agar coated dishes. Tissue‐culture plastic ware was obtained from Corning Glass Works (Corning, NY, USA).
Western blotting. Cell lysates were prepared as described previously.( 8 ) Lysis buffer consisted of 20 mM Tris‐HCl (pH 7.6), 150 mM NaCl, 1 mM MgCl2, 1% Nonidet P‐40, 10% glycerol, 8 µL complete protease‐inhibitor cocktail (1 tablet per ml H2O), 1 mM sodium orthovanadate and 10 mM NaF. Equal volumes of lysate were separated by sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) and immunoblotted onto a polyvinylidene difluoride membrane (Millipore, Bedford, MA, USA). Nonspecific binding was blocked by incubation for 1 h at room temperature with phosphate‐buffered saline (PBS) (pH 7.4) containing 5% non‐fat dry milk, 1% bovine serum albumin (BSA) and 0.1% Tween 20. Analysis was carried out using the indicated antibodies (first antibodies, 4°C overnight; peroxidase‐labeled second antibodies, room temperature for 1 h). Bands were visualized with enhanced chemiluminescence (ECL) (Amersham Biosciences, Piscataway, NJ, USA) or ECL‐plus (Roche Diagnostic, Indianapolis, IN, USA) chemiluminescent reagents.
Preparation of protein samples in the conditioned medium. Ten micrograms of protein from conditioned medium after 24 h serum starvation was precipitated by the addition of trichloroacetic acid to a 10% final concentration, incubation of the samples at 4°C for 1 h and centrifugation for 14 000 g for 30 min. Pellets were washed three times with 50% ether and 50% ethanol and redissolved in Laemmli sample buffer.
Preparation of protein samples from extracellular matrix. When the cultures reached confluency, the medium was aspirated and each dish was rinsed twice with ice‐cold PBS. Cells were incubated for 30 min at 37°C in 1 mL 10 mM EDTA and harvested from the dishes by thorough pipetting. This procedure was repeated until all cells had been removed. The fraction that remained on the dish was used as the ECM (Fig. 1a). Dishes were washed twice with ice‐cold PBS, and ECM proteins were extracted with Laemmli sample buffer. To release the IGF‐II/IGFBP‐2 complex from the ECM, the ECM fraction was incubated with MMP‐7 (200 ng/mL) or Heparinase III (0.2 U/mL) at 37°C in fresh serum‐free medium for 30 min. When these supernatants were used as ligands for IGF‐1R, they were preincubated with TIMP‐1 at 37°C for 30 min to neutralize residual MMP‐7 activity (Fig. 2a).
Figure 1.
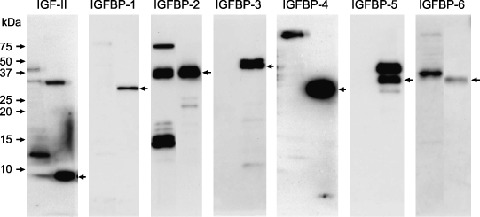
Expression of insulin‐like growth factor (IGF)‐II and insulin‐like growth factor binding proteins (IGFBPs) in HT29 cells. Ten micrograms of trichloroacetic acid‐precipitable protein from conditioned medium of HT29 cells were fractionated on a 15% sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) and analyzed by Western blotting for the expression levels of IGF‐II and all IGFBPs (IGFBP‐1 to IGFBP‐6). Arrow of each left lane indicates positive control (100 ng of recombinant protein, respectively).
Figure 2.
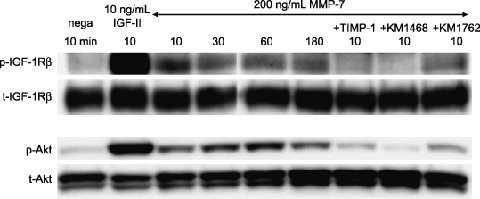
Matrix metalloproteinase (MMP)‐7 induced insulin‐like growth factor (IGF)‐II autocrine response. The IGF type1 receptor (IGF‐1R)/Akt signaling pathway was rapidly activated and sustained for at least 180 min by exogenous active MMP‐7 in HT29 cells. This IGF‐1R phosphorylation was inhibited by TIMP‐1(lane7) and IGF‐II neutralizing antibody (KM1468, lane8). Cell lysates from HT29 cells (10 µg of protein per lane) were separated by 7.5% (IGF‐1R) and 10% (Akt) sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE). The blotted proteins were detected by Western blotting. Bands of phosphorylated IGF‐1R (1st antibody, ×500; 2nd antibody, ×6000) were visualized with enhanced chemiluminescence (ECL)‐plus chemiluminescent reagent. Bands of total IGF‐1R/phosphorylated‐Akt/total‐Akt (1st antibody, ×500, ×1000, ×1000; 2nd antibody, ×3000) were visualized with ECL reagent. p‐, phosphorylated; t‐, total.
Enzyme cleavage assays and determination of cleavage sites. A total of 500 ng recombinant human IGFBP‐2 was cleaved by exposure to 14 ng of active MMP‐7 (MMP‐7: IGFBP‐2 molar ratio, 1:20) in a cleavage buffer (150 mM NaCl; 10 mM HEPES, pH 7.4; 5 mM CaCl2) in a final volume of 20 µL at 37°C for periods that varied from 5 min to 18 h. Reactions were terminated by the addition of sample buffer containing the reducing agent, 2‐mercaptoethanol. The reaction solution was boiled and then resolved by SDS‐PAGE using a 10–20% gradient gel (Bio‐Rad, Hercules, CA, USA). The proteolytic fragment patterns were evaluated using a silver‐staining kit (Daiichi Pure Chemicals, Tokyo, Japan). One‐hundred ng of IGFBP‐2 protein was applied to each lane. To determine the cleavage site, the proteolytic fragments (a mixture of samples obtained at 8 h, 12 h and 18 h) were separated by 15% SDS‐PAGE followed by blotting on a ProBlott membrane (Applied Biosystems, Foster City, CA, USA). Proteins were visualized by staining with the SYPRO Ruby protein blot stain (Molecular Probes, Eugene, OR, USA) according to the manufacturer's protocol. Amino acid sequences were determined by automated Edman degradation with a Procise cLC protein sequencer (Applied Biosystems).
In vitro assay of IGF‐1R phosphorylation after degradation of the IGF‐II/IGFBP‐2 complex by MMP‐7. Subconfluent HT29 cells were cultured and switched to 3 mL serum‐free medium. IGF‐II (15 ng) plus IGFBP‐2 (540 ng) was preincubated in vitro for 30 min at 37°C (IGF‐II/IGFBP‐2 complex). The IGF‐II/IGFBP‐2/MMP‐7 mixture was made by exposing the IGF‐II/IGFBP‐2 complex to 600 ng of MMP‐7 (MMP‐7: IGFBP‐2 molar ratio, 2:1) in a cleavage buffer in a final volume of 20 µL for 30 min at 37°C. IGF‐II (10 ng/mL), IGFBP‐2 (180 ng/mL), MMP‐7 (200 ng/mL), IGF‐II/IGFBP‐2 complex and the IGF‐II/IGFBP‐2/MMP‐7 mixture were added to serum‐free medium for 30 min at 37°C. Ten‐microgram aliquots of cell lysates were fractionated by 7.5% SDS‐PAGE under reducing conditions. The phospho‐IGF‐1R and total‐IGF‐1R protein levels were estimated using antiphospho‐IGF‐1R or anti‐IGF‐1R polyclonal antibody (both × 500).
Results
Phosphorylation of IGF‐1R by MMP‐7 is inhibited by IGF‐II neutralizing antibody. We confirmed that HT29 cells did not express IGF‐I mRNA (data not shown) and that they secreted IGF‐II, IGFBP‐2 and IGFBP‐6 proteins into the conditioned medium (Fig. 3). The IGF‐1R was rapidly phosphorylated by MMP‐7; the level of phosphorylation was sustained for at least 3 h (Fig. 1, lane 3 to lane 6) and inhibited by TIMP‐1 (Fig. 1, lane 7) and KM1468 (human‐IGF‐II neutralizing antibody, Fig. 1, lane 8). A similar phosphoprotein banding pattern was obtained for Akt. These results demonstrate that MMP‐7 triggers autocrine IGF‐II‐induced signal transduction.
Figure 3.
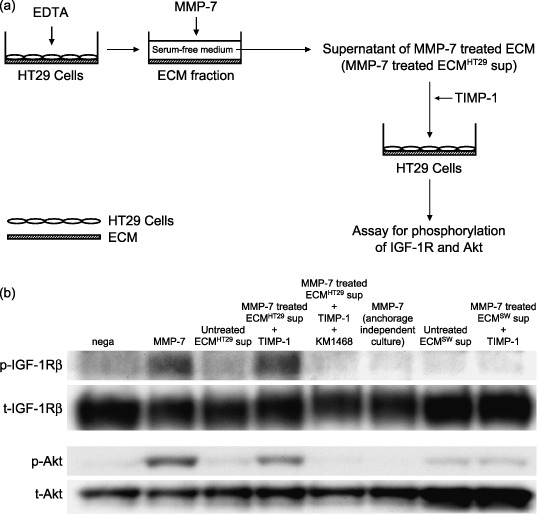
Matrix metalloproteinase (MMP)‐7 acts as a trigger for the matricrine action of insulin‐like growth factor (IGF)‐II. (a) The supernatant fraction of extracellular matrix (ECM) from HT29 cells after MMP‐7 treatment (200 ng/mL at 37°C for 30 min) was used to determine whether MMP‐7 activates IGF‐II in the ECM. After residual MMP‐7 activity was neutralized by addition of TIMP‐1, the supernatant fraction was added to serum‐starved HT29 cells. (b) Cell lysates from HT29 cells (10 µg of protein per lane) were separated by 7.5% (IGF‐1R) and 10% (Akt) sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE). The blotted proteins were detected by Western blotting. Bands of phosphorylated IGF‐1R (1st antibody, ×500; 2nd antibody, ×6000) were visualized with enhanced chemiluminescence (ECL)‐plus chemiluminescent reagent. Bands of total IGF‐1R/phosphorylated‐Akt/total‐Akt (1st antibody, ×500, ×1000, ×1000; 2nd antibody, ×3000) were visualized with ECL chemiluminescent reagent. p‐, phosphorylated; t‐, total; sup, supernatant; SW, SW1116 cells.
Metalloproteinase‐7 acts as a trigger for the matricrine action of IGF‐II. The supernatant from the MMP‐7‐treated ECM fraction phosphorylated the IGF‐1R and Akt (Fig. 2b, lane 4). This signal transduction was completely inhibited by KM1468 (Fig. 2b, lane 5). To establish that the IGF‐II was derived from the cancer cells and not from the FBS, SW1116 cells were used as a source of ECM. This cell line does not express IGF‐I or IGF‐II mRNA. As expected, no IGF‐1R phosphorylation was observed when ECM derived from SW1116 cells was used (Fig. 2b, lane 8). These results demonstrate that MMP‐7 activates cancer‐producing IGF‐II in the ECM. Interestingly, MMP‐7 only induced phosphorylation of IGF‐1R in anchorage‐dependent culture (Fig. 2b, lane 2), not in anchorage‐independent culture (Fig. 2b, lane 6).
Metalloproteinase‐7 releases IGF‐II/IGFBP‐2 complex binding to heparan sulfate proteoglycan in the ECM and generates bioactive IGF‐II. We speculated that IGF‐II formed a complex with one of the IGFBPs in the ECM and confirmed that IGF‐II and IGFBP‐2 coexisted in the ECM fraction (data not shown). The concentration of IGFBP‐2 in the ECM fraction was rapidly decreased by MMP‐7, and gradually decreased by heparinase III (Fig. 4a). Phosphorylation of IGF‐1R was induced by supernatant from MMP‐7‐treated ECM (Fig. 4b, lane 2), but not by supernatant from heparinase‐III‐treated ECM (Fig. 4b, lane 3). In other words, MMP‐7 released IGF‐II/IGFBP‐2 complex from ECM and activated IGF‐II; Heparinase III released IGF‐II/IGFBP‐2 complex but did not activate IGF‐II. These results suggest that MMP‐7 releases the IGF‐II/IGFBP‐2 complex from binding to heparan sulfate proteoglycan in the ECM, generating bioactive IGF‐II.
Figure 4.
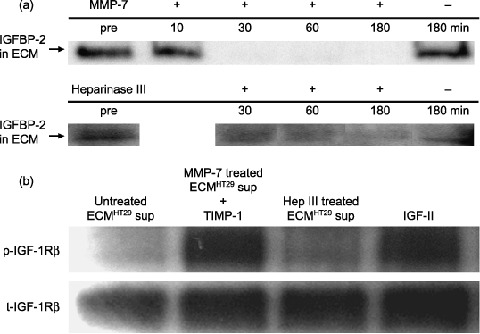
Matrix metalloproteinase (MMP)‐7 decreased binding of the insulin‐like growth factor (IGF)‐II/insulin‐like growth factor binding protein (IGFBP)‐2 complex to heparan sulfate proteoglycan in the extracellular matrix (ECM) and released bioactive IGF‐II. (a) MMP‐7 (200 ng/mL) and heparinase III (0.2 U/mL) released IGFBP‐2 from the ECM. ECM protein derived from one 6‐cm dish was used in each lane. Samples were separated by 15% sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) and analyzed by Western blotting. Bands of IGFBP‐2 (1st antibody, ×500; 2nd antibody, ×3000) were visualized with enhanced chemiluminescence (ECL) reagent. (b) Supernatant from MMP‐7‐treated (200 ng/mL at 37°C for 30 min) ECM induced phosphorylation of IGF type‐1 receptor (IGF‐1R), but supernatant from heparinase‐III‐treated (0.2 U/mL at 37°C for 3 h) ECM did not. 3T3‐IGF‐1R cells were used in the assay. Cell lysates from HT29 cells (10 µg of protein per lane) were separated by 7.5% SDS‐PAGE and analyzed by Western blotting. Bands of phosphorylated IGF‐1R (1st antibody, ×500; 2nd antibody, ×6000) were visualized with ECL‐plus chemiluminescent reagent. Bands of total IGF‐1R (1st antibody, ×500; 2nd antibody, ×3000) were visualized with ECL chemiluminescent reagent. p‐, phosphorylated; sup, supernatant; t‐, total.
Metalloproteinase‐7 degrades IGFBP‐2 at three novel cleavage sites. To characterize the proteolytic cleavage of IGFBP‐2 by MMP‐7, we carried out a cleavage assay with recombinant human IGFBP‐2 and analyzed the products using silver staining (Fig. 5). MMP‐7 degraded IGFBP‐2 into five distinct fragments with apparent molecular masses of 18.3 kDa (Fragment A), 16.6 kDa (Fragment B), 14.7 kDa (Fragment C), 13.9 kDa (Fragment D) and 11.5 kDa (Fragment E). To identify the cleavage site of MMP‐7, the IGFBP‐2 proteolysis fragments were N‐terminal sequenced directly from bands electrotransferred to nylon membranes. The N‐terminal sequences of each of the fragments are shown in Table 1. Western blotting showed that all but Fragment A contained the carboxy‐terminal of IGFBP‐2 (data not shown). The amino acid sequence of IGFBP‐2 and cleavage sites for MMP‐7 are shown in Fig. 6.
Figure 5.
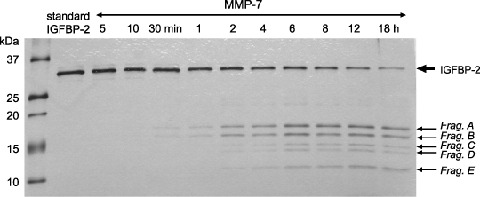
Identification of matrix metalloproteinase (MMP)‐7 cleavage fragments using recombinant insulin‐like growth factor binding protein (IGFBP)‐2. Time‐course analysis was carried out. Recombinant IGFBP‐2 (500 ng) was incubated with active MMP‐7 (14 ng) in a final volume of 20 µL as described under ‘Materials and Methods’. The reaction products were separated by 20% to 10% gradient sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE). The proteolytic fragment patterns were evaluated with a silver‐staining kit according to the manufacture's protocol. 100 ng of IGFBP‐2 protein was used in each lane. Frag., fragment.
Table 1.
Cleavage sites in insulin‐like growth factor binding protein (IGFBP)‐2 produced by matrix metalloproteinase (MMP)‐7. Cleavage sites in IGFBP‐2 produced by MMP‐7 were determined by N‐terminal sequence analysis as described in Materials and Methods. The N‐terminal sequence of each fragment is shown with its apparent molecular weight. Each fragment was analyzed for the presence of the carboxy‐terminal of IGFBP‐2 by Western blotting with the anticarboxy‐terminal peptides of IGFBP‐2 monoclonal antibody (1st antibody, ×500; 2nd antibody, ×3000)
| Molecular weight (apparent) | NH2‐terminal sequence | † COOH‐ terminal | |
|---|---|---|---|
| ‡ Frag. A | 18.3 kDa | E1VLFRXPP | – |
| ‡ Frag. B | 16.6 kDa | L152AVFREKVTE | + |
| ‡ Frag. C | 14.7 kDa | L176EEPKKLRPP | + |
| ‡ Frag. D | 13.9 kDa | L182RPPPARTP | + |
| ‡ Frag. E | 11.5 kDa | ND | + |
COOH‐terminal, presence (+) or absence (–) of 000H terminal of IGFBP‐2;
the letters correspond to the cleavage products represented in Fig. 5. ND, not determined.
Figure 6.

Amino acid sequence of human insulin‐like growth factor binding protein (IGFBP)‐2. Arrowheads indicate the cleavage sites for MMP‐7. The underlined sequence is located in the short heparin‐binding domain.
Co‐incubation of the IGF‐II/IGFBP‐2 complex with MMP‐7 Restored IGF‐II‐induced IGF‐1R phosphorylation in vitro. The consequence of IGFBP‐2 proteolysis by MMP‐7 for IGF‐induced signal transduction was analyzed in vitro. Fig. 7 shows that IGF‐II markedly phosphorylated the IGF‐1R (lane 2), but that IGFBP‐2 had no effect on IGF‐1R phosphorylation (lane 3). MMP‐7 alone had a slight stimulatory effect on IGF‐1R (lane 4). Fig. 7 also shows that IGFBP‐2 completely inhibited IGF‐II stimulation to the baseline level (lane 5). However, adding MMP‐7 to IGF‐II and IGFBP‐2 completely restored the stimulatory effect on the IGF‐1R (lane 6).
Figure 7.
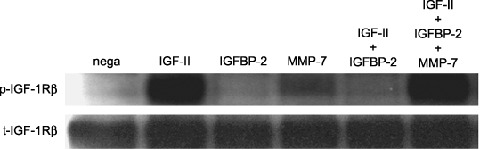
Recovery of insulin‐like growth factor (IGF)‐II mediated phosphorylation of IGF type1 receptor (IGF‐1R) through Insulin‐like growth factor binding protein (IGFBP)‐2 degradation by matrix metalloproteinase (MMP)‐7. Cell lysates from HT29 cells (10 µg of protein per lane) exposed to IGF system component molecules (IGF‐II/IGFBP‐2/MMP‐7, preincubation conditions; see ‘Materials and Methods’) were separated by 7.5% sodium dodecylsulfate–polyacrylamide gel electrophoresis (SDS‐PAGE) and analyzed by Western blotting. Bands of phosphorylated IGF‐1R (1st antibody, ×500; 2nd antibody, ×6000) were visualized with enhanced chemiluminescence (ECL)‐plus chemiluminescent reagent. Bands of total IGF‐1R (1st antibody, ×500; 2nd antibody, ×3000) were visualized with ECL chemiluminescent reagent. p‐, phosphorylated; t‐, total.
Discussion
The evidence suggests that MMPs contribute to tumor growth and survival by regulating access to growth factors in the ECM surrounding the tumor through a proteolytic cascade.( 26 , 27 , 28 ) Until now, there were no reports about the protease‐induced activation of IGF associated with ECM. In the present study, we first demonstrated that MMP‐7 facilitates ECM‐bound IGF‐II bioavailability by degrading the IGF‐II/IGFBP‐2 complex. We propose that this mechanism be designated ‘protease‐triggered matricrine’ action. Secretion of IGFBP‐2 and IGFBP‐6 into the conditioned medium in which the HT29 cells were incubated was confirmed (Fig. 3). We focused our attention on IGFBP‐2 for two reasons: (i) Although MMP‐7 degrades all six IGFBPs in vitro, IGFBP‐2 is more susceptible to MMP‐7 than IGFBP‐6;( 21 ) and (ii) the level of mRNA expression in HT29 cells is greater for IGFBP‐2 than for IGFBP‐6 (data not shown).
IGFBP‐3 and IGFBP‐5 interact with the ECM through their putative long heparan binding domains (HBD) of the form B‐B‐B‐X‐X‐B (where B is a basic residue) in the conserved carboxy‐terminal domain (K220KKQCR225 in IGFBP‐3, K206RKQCK210 in IGFBP‐5).( 7 , 29 ) Likewise, it has been shown that non‐glycosylated IGFBP‐6 can bind to heparan sulfate proteoglycan (HSPG) through a consensus long HBD at residues R173KRQCR178 in the conserved carboxyl‐terminal domain.( 7 , 30 ) Therefore, IGF‐II may be stored in the ECM of HT29 cells in the form of the IGF‐II/IGFBP‐6 complex. On the other hand, IGFBP‐2 is predicted to associate with the ECM through the central domain, a consensus short HBD of the form B‐B‐X‐B (K180 K L R183).( 7 , 29 , 31 , 32 ) Interestingly, IGFBP‐2 binding to ECM was markedly enhanced in the presence of IGFs (especially IGF‐II), possibly because of a conformational change of IGFBP‐2.( 33 , 34 , 35 ) Moreover, Yu et al. reported that HSPG was the extracellular docking molecule for pro‐MMP‐7 and that it potentiated the enzymatic activity of pro‐MMP‐7.( 36 ) In other words, the inactive form of the growth factor (the IGF‐II/IGFBP‐2 complex) and the latent form of the growth factor activator (pro‐MMP‐7) coexisted in the ECM because of HSPG. This extracellular molecular system is similar to that described in an earlier report in which CD44 anchors the assembly of MMP‐7 with the heparin‐binding epidermal growth factor (HB‐EGF) precursor and ErbB4.( 18 ) In some cell lines, once MMP‐7 is activated, the IGF and EGF signaling pathway are simultaneously switched on in a matricrine manner. These mechanisms are very advantageous for the survival of tumors in a tissue microenvironment that is on the verge of growth‐factor starvation. A hypothetical representation of the interaction between HSPG‐bound pro‐MMP‐7, the IGF‐II/IGFBP‐2 complex and the IGF‐1R‐mediated signaling pathway is shown in Fig. 8.
Figure 8.
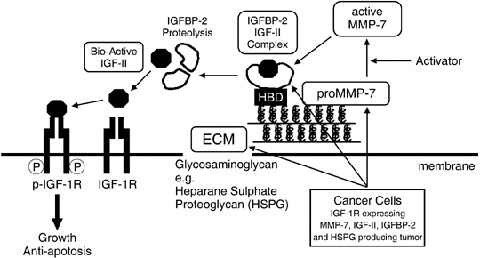
Matrix metalloproteinase (MMP)‐7 triggers the matricrine action of insulin‐like growth factor (IGF)‐II via proteinase activity on insulin‐like growth factor binding protein (IGFBP)‐2 in the ECM. MMP‐7, IGF‐II, IGFBP‐2 and heparan sulfate proteoglycan (HSPG) were produced by cancer cells (HT29 cells). HSPG anchors the IGF‐II/IGFBP‐2 complex to the ECM through the heparin‐binding domain of IGFBP‐2. MMP‐7, once activated, degrades the IGF‐II/IGFBP‐2 complex and releases bioactive IGF‐II followed by IGF‐II‐induced signal transduction. HBD, heparin‐binding domain; p‐, phosphorylated.
The expression of IGFBP genes and the secretion of IGFBPs have been shown to be common properties of tumor cells. In particular, IGFBP‐2 is overexpressed not only in many tumor cell lines( 37 ) but also in many human malignancies. Interestingly, this often correlates with an increasingly malignant tumor status,( 38 ) although it is generally thought to inhibit IGF (especially IGF‐II) action.( 7 ) This discrepancy may be resolved by the concept that IGFBP‐2‐producing tumors secrete IGFBP‐2 protease in the presence of IGF‐II. In fact, expression of IGF‐II and IGFBP‐2 in colon cancer tissues was greater than that of normal controls.( 39 , 40 ) Our results may partially explain the mechanisms by which IGFBP‐2 affects tumor growth and progression.
Proteolyzed IGFBP‐2 has been detected in serum,( 41 ) milk( 42 ) and cerebrospinal fluid,( 43 ) but little attention has been paid to the implications of IGFBP‐2 proteolysis in pathological states such as cancer. Michell et al. reported that proteolysis of IGFBP‐2 occurred specifically in colon cancer tissue and did not occur in normal colonic mucosa,( 44 ) which is concordant with the fact that MMP‐7 is exclusively synthesized by cancer cells. Cleavage by MMP‐7 shows a preference for hydrophobic residues such as Leu and Ile in the P1′ position.( 45 ) The three cleavage sites (E151–L152, G175–L176 and K181–L182) of IGFBP‐2 that we identified are consistent with this preference, as in the case of IGFBP‐3.( 8 ) To date, only three proteases, plasmin,( 46 ) cathepsin D( 47 ) and MMP‐1,( 48 , 49 ) have been reported to act as IGFBP‐2 proteases. A few reports are available on the cleavage site of IGFBP‐2( 42 , 50 ) but the three cleavage sites reported in this paper have not been described before. It is reasonable that one cleavage site (K181–L182) was located in the short HBD of IGFBP‐2.
In conclusion, MMP‐7 degrades IGFBP‐2 in the ECM and facilitates IGF‐II bioavailability in the tissue microenvironment. The term, ‘protease‐triggered matricrine’ may constitute a suitable description for this mechanism of action. Our findings emphasize the importance of the ECM as a reservoir of cancer‐producing IGF‐II/IGFBP‐2 complexes.
Acknowledgments
The technical assistance of Chie Okumura, Yoko Okuhara and Keiko Hiraishi and the secretarial support of Motoko Suzaki are gratefully acknowledged.
Grant support: Supported in part by a Grant‐in‐Aid for Cancer Research (11–12) from the Ministry of Health and Welfare of Japan and by a Grant‐in‐Aid for the Second Term Comprehensive 10‐year Strategy for Cancer Control from the Ministry of Health and Welfare of Japan. S.M., M.N., T.S., H.M and Z.S. are recipients of Research Resident Fellowships from the Foundation for Promotion of Cancer Research in Japan.
References
- 1. Foulstone E, Prince S, Zaccheo O et al. Insulin‐like growth factor ligands, receptors, and binding proteins in cancer. J Pathol 2005; 205: 145–53. [DOI] [PubMed] [Google Scholar]
- 2. Pollak MN, Schernhammer ES, Hankinson SE. Insulin‐like growth factors and neoplasia. Nat Rev Cancer 2004; 4: 505–18. [DOI] [PubMed] [Google Scholar]
- 3. LeRoith D, Roberts CT Jr. The insulin‐like growth factor system and cancer. Cancer Lett 2003; 195: 127–37. [DOI] [PubMed] [Google Scholar]
- 4. Furstenberger G, Senn HJ. Insulin‐like growth factors and cancer. Lancet Oncol 2002; 3: 298–302. [DOI] [PubMed] [Google Scholar]
- 5. Rajaram S, Baylink DJ, Mohan S. Insulin‐like growth factor binding proteins in serum and other biological fluids: regulations and functions. Endocr Rev 1997; 18: 801–31. [DOI] [PubMed] [Google Scholar]
- 6. Jones JI, Clemmons DR. Insulin‐like growth factors and their binding proteins: biological actions. Endocr Rev 1995; 16: 3–34. [DOI] [PubMed] [Google Scholar]
- 7. Firth SM, Baxter RC. Cellular actions of the insulin‐like growth factor binding proteins. Endocr Rev 2002; 23: 824–54. [DOI] [PubMed] [Google Scholar]
- 8. Jones JI, Gockerman A, Busby WH Jr, Camacho‐Hubner C, Clemmons DR. Extracellular matrix contains insulin like growth factor binding protein‐5: potentiation of the effects of IGF‐I. J Cell Biol 1993; 121: 679–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Reeve JG, Morgan J, Schwander J, Bleehen NM. Role for membrane and secreted insulin‐like growth factor‐binding protein‐2 in the regulation of insulin‐like growth factor action in lung tumors. Cancer Res 1993; 53: 4680–5. [PubMed] [Google Scholar]
- 10. Remacle‐Bonnet MM, Garrouste FL, Pommier GJ. Surface‐bound plasmin induces selective proteolysis of insulin‐like‐growth‐factor (IGF)‐binding protein‐4 (IGFBP‐4) and promotes autocrine IGF‐II bioavailability in human colon‐carcinoma cells. Int J Cancer 1997; 72: 835–43. [DOI] [PubMed] [Google Scholar]
- 11. Manes S, Llorente M, Lacalle RA et al. The matrix metalloproteinase‐9 regulates the insulin‐like growth factor‐triggered autocrine response in DU‐145 carcinoma cells. J Biol Chem 1999; 274: 6935–45. [DOI] [PubMed] [Google Scholar]
- 12. Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: an overview. Mol Cell Biochem 2003; 253: 269–85. [DOI] [PubMed] [Google Scholar]
- 13. Shiomi T, Okada Y. MT1‐MMP and MMP‐7 in invasion and metastasis of human cancers. Cancer Metastasis Rev 2003; 22: 145–52. [DOI] [PubMed] [Google Scholar]
- 14. Lynch CC, Matrisian LM. Matrix metalloproteinases in tumor‐host cell communication. Differentiation 2002; 70: 561–73. [DOI] [PubMed] [Google Scholar]
- 15. Stamenkovic I. Matrix metalloproteinases in tumor invasion and metastasis. Semin Cancer Biol 2000; 10: 415–33. [DOI] [PubMed] [Google Scholar]
- 16. Gearing AJ, Beckett P, Christodoulou M et al. Processing of tumour necrosis factor‐alpha precursor by metalloproteinases. Nature 1994; 370: 555–7. [DOI] [PubMed] [Google Scholar]
- 17. Marcotte PA, Kozan IM, Dorwin SA, Ryan JM. The matrix metalloproteinase pump‐1 catalyzes formation of low molecular weight (pro) urokinase in cultures of normal human kidney cells. J Biol Chem 1992; 267: 13 803–6. [PubMed] [Google Scholar]
- 18. Yu WH, Woessner JF Jr, McNeish JD, Stamenkovic I. CD44 anchors the assembly of matrilysin/MMP‐7 with heparin‐binding epidermal growth factor precursor and ErbB4 and regulates female reproductive organ remodeling. Genes Dev 2002; 16: 307–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyamoto S, Yano K, Sugimoto S et al. Matrix metalloproteinase‐7 facilitates insulin‐like growth factor bioavailability through its proteinase activity on insulin‐like growth factor binding protein 3. Cancer Res 2004; 64: 665–71. [DOI] [PubMed] [Google Scholar]
- 20. Hashimoto G, Inoki I, Fujii Y, Aoki T, Ikeda E, Okada Y. Matrix metalloproteinases cleave connective tissue growth factor and reactivate angiogenic activity of vascular endothelial growth factor 165. J Biol Chem 2002; 277: 36 288–95. [DOI] [PubMed] [Google Scholar]
- 21. Nakamura M, Miyamoto S, Maeda H et al. Matrix metalloproteinase‐7 degrades all insulin‐like growth factor binding proteins and facilitates insulin‐like growth factor bioavailability. Biochem Biophys Res Comm 2005; 333: 1011–16. [DOI] [PubMed] [Google Scholar]
- 22. Tanaka Y, Kimata K, Adams DH, Eto S. Modulation of cytokine function by heparan sulfate proteoglycans: sophisticated models for the regulation of cellular responses to cytokines. Proc Assoc Am Physicians 1998; 110: 118–25. [PubMed] [Google Scholar]
- 23. Hemers E, Duval C, McCaig C, Handley M, Dockray GJ, Varro A. Insulin‐like growth factor binding protein‐5 is a target of matrix metalloproteinase‐7: implications for epithelial‐mesenchymal signaling. Cancer Res 2005; 65: 7363–9. [DOI] [PubMed] [Google Scholar]
- 24. Goya M, Miyamoto S, Nagai K et al. Growth inhibition of human prostate cancer cells in human adult bone implanted into non‐obese diabetic/severe combined immunodeficient mice by a ligand‐specific antibody to human insulin‐like growth factors. Cancer Res 2004; 64: 6252–8. [DOI] [PubMed] [Google Scholar]
- 25. Miyamoto S, Nakamura M, Shitara K et al. Blockade of paracrine supply of insulin‐like growth factors using neutralizing antibodies suppresses the liver metastasis of human colorectal cancers. Clin Cancer Res 2005; 11: 3494–502. [DOI] [PubMed] [Google Scholar]
- 26. Whitelock JM, Murdoch AD, Iozzo RV, Underwood PA. The degradation of human endothelial cell‐derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin, and heparinases. J Biol Chem 1996; 271: 10 079–86. [DOI] [PubMed] [Google Scholar]
- 27. Imai K, Hiramatsu A, Fukushima D, Pierschbacher MD, Okada Y. Degradation of decorin by matrix metalloproteinases: identification of the cleavage sites, kinetic analyses and transforming growth factor‐beta1 release. Biochem J 1997; 322: 809–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li Q, Park PW, Wilson CL, Parks WC. Matrilysin shedding of syndecan‐1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell 2002; 111: 635–46. [DOI] [PubMed] [Google Scholar]
- 29. Cardin AD, Weintraub HJ. Molecular modeling of protein‐glycosaminoglycan interactions. Arteriosclerosis 1989; 9: 21–32. [DOI] [PubMed] [Google Scholar]
- 30. Marinaro JA, Neumann GM, Russo VC, Leeding KS, Bach LA. O‐glycosylation of insulin‐like growth factor (IGF) binding protein‐6 maintains high IGF‐II binding affinity by decreasing binding to glycosaminoglycans and susceptibility to proteolysis. Eur J Biochem 2000; 267: 5378–86. [DOI] [PubMed] [Google Scholar]
- 31. Russo VC, Bach LA, Fosang AJ, Baker NL, Werther GA. Insulin‐like growth factor binding protein‐2 binds to cell surface proteoglycans in the rat brain olfactory bulb. Endocrinology 1997; 138: 4858–67. [DOI] [PubMed] [Google Scholar]
- 32. Russo VC, Schutt BS, Andaloro E et al. IGFBP‐2 binding to extracellular matrix plays a critical role in neuroblastoma cell proliferation, migration and invasion. Endocrinology 2005; 146: 4445–55. [DOI] [PubMed] [Google Scholar]
- 33. Arai T, Busby W Jr, Clemmons DR. Binding of insulin‐like growth factor (IGF) I or II to IGF‐binding protein‐2 enables it to bind to heparin and extracellular matrix. Endocrinology 1996; 137: 4571–5. [DOI] [PubMed] [Google Scholar]
- 34. Khosla S, Hassoun AA, Baker BK et al. Insulin‐like growth factor system abnormalities in hepatitis C‐associated osteosclerosis. Potential insights into increasing bone mass in adults. J Clin Invest 1998; 101: 2165–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Conover CA, Khosla S. Role of extracellular matrix in insulin‐like growth factor (IGF) binding protein‐2 regulation of IGF‐II action in normal human osteoblasts. Growth Horm IGF Res 2003; 13: 328–35. [DOI] [PubMed] [Google Scholar]
- 36. Yu WH, Woessner JF Jr. Heparan sulfate proteoglycans as extracellular docking molecules for matrilysin (matrix metalloproteinase 7). J Biol Chem 2000; 275: 4183–91. [DOI] [PubMed] [Google Scholar]
- 37. Reeve JG, Kirby LB, Brinkman A, Hughes SA, Schwander J, Bleehen NM. Insulin‐like growth‐factor‐binding protein gene expression and protein production by human tumour cell lines. Int J Cancer 1992; 51: 818–21. [DOI] [PubMed] [Google Scholar]
- 38. Hoeflich A, Reisinger R, Lahm H et al. Insulin‐like growth factor‐binding protein 2 in tumorigenesis: protector or promoter? Cancer Res 2001; 61: 8601–10. [PubMed] [Google Scholar]
- 39. Nosho K, Yamamoto H, Taniguchi H et al. Interplay of insulin‐like growth factor‐II, insulin‐like growth factor‐I, insulin‐like growth factor‐I receptor, COX‐2, and matrix metalloproteinase‐7, play key roles in the early stage of colorectal carcinogenesis. Clin Cancer Res 2004; 10: 7950–7. [DOI] [PubMed] [Google Scholar]
- 40. Mishra L, Bass B, Ooi BS, Sidawy A, Korman L. Role of insulin‐like growth factor‐I (IGF‐I) receptor, IGF‐I, and IGF binding protein‐2 in human colorectal cancers. Growth Horm IGF Res 1998; 8: 473–9. [DOI] [PubMed] [Google Scholar]
- 41. McCusker RH, Cohick WS, Busby WH, Clemmons DR. Evaluation of the developmental and nutritional changes in porcine insulin‐like growth factor‐binding protein‐1 and ‐2 serum levels by immunoassay. Endocrinology 1991; 129: 2631–8. [DOI] [PubMed] [Google Scholar]
- 42. Ho PJ, Baxter RC. Characterization of truncated insulin‐like growth factor‐binding protein‐2 in human milk. Endocrinology 1997; 138: 3811–8. [DOI] [PubMed] [Google Scholar]
- 43. Roghani M, Hossenlopp P, Lepage P, Balland A, Binoux M. Isolation from human cerebrospinal fluid of a new insulin‐like growth factor‐binding protein with a selective affinity for IGF‐II. FEBS Lett 1989; 255: 253–8. [DOI] [PubMed] [Google Scholar]
- 44. Michell NP, Langman MJ, Eggo MC. Insulin‐like growth factors and their binding proteins in human colonocytes: preferential degradation of insulin‐like growth factor binding protein 2 in colonic cancers. Br J Cancer 1997; 76: 60–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Netzel‐Arnett S, Sang QX, Moore WG, Navre M, Birkedal‐Hansen H, Van Wart HE. Comparative sequence specificities of human 72‐ and 92‐kDa gelatinases (type IV collagenases) and PUMP (matrilysin). Biochemistry 1993; 32: 6427–32. [DOI] [PubMed] [Google Scholar]
- 46. Menouny M, Binoux M, Babajko S. Role of insulin‐like growth factor binding protein‐2 and its limited proteolysis in neuroblastoma cell proliferation. modulation by transforming growth factor‐beta and retinoic acid. Endocrinology 1997; 138: 683–90. [DOI] [PubMed] [Google Scholar]
- 47. Claussen M, Kubler B, Wendland M et al. Proteolysis of insulin‐like growth factors (IGF) and IGF binding proteins by cathepsin D. Endocrinology 1997; 138: 3797–803. [DOI] [PubMed] [Google Scholar]
- 48. Rajah R, Nachajon RV, Collins MH, Hakonarson H, Grunstein MM, Cohen P. Elevated levels of the IGF‐binding protein protease MMP‐1 in asthmatic airway smooth muscle. Am J Respir Cell Mol Biol 1999; 20: 199–208. [DOI] [PubMed] [Google Scholar]
- 49. Rajah R, Nunn SE, Herrick DJ, Grunstein MM, Cohen P. Leukotriene D4 induces MMP‐1, which functions as an IGFBP protease in human airway smooth muscle cells. Am J Physiol 1996; 271: L1014–22. [DOI] [PubMed] [Google Scholar]
- 50. Wang JF, Hampton B, Mehlman T, Burgess WH, Rechler MM. Isolation of a biologically active fragment from the carboxy terminus of the fetal rat binding protein for insulin‐like growth factors. Biochem Biophys Res Comm 1988; 157: 718–26. [DOI] [PubMed] [Google Scholar]


