Abstract
Cancer/testis (CT) antigens are protein antigens with normal expression restricted to adult testicular germ cells, and yet are aberrantly activated and expressed in a proportion of various types of human cancer. At least a subset of this group of antigens has been found to elicit spontaneous humoral and cell‐mediated immune responses in cancer patients, raising the possibility that these antigens could be cancer vaccine targets. More than 100 CT antigen genes have been reported in the literature, with approximately 30 being members of multigene families on the X chromosome, so‐called CT‐X genes. Most CT‐X genes are expressed at the spermatogonia stage of spermatogenesis, and their functions are mostly unknown. In cancer, the frequency of CT antigen expression is highly variable among different tumor types, but is more often expressed in high‐grade late‐stage cases in general. Cancer vaccine trials based on CT antigens MAGE‐A3 and NY‐ESO‐1 are currently ongoing, and these antigens may also play a role in antigen‐specific adoptive T‐cell transfer and in the immunomodulation approach of cancer therapy. (Cancer Sci 2009)
The search for human tumor antigens as potential immunotherapeutic targets, either for antibody‐based therapy or for cancer vaccines, has been a continuous task in the field of tumor immunology for several decades. For tumor antigens to be potential immunotherapeutic targets, the antigen must have no or highly restricted expression in normal tissues so that autoimmunity can be prevented. Over the decades, several categories of antigens were found to fulfill this requirement, including uniquely mutated antigens (e.g. p53), viral antigens (e.g. human papillomavirus antigens in cervical cancer), and differentiation antigens (e.g. CD20 in B‐cell lymphoma). More recently, a new category of antigen, namely the cancer/testis (CT) antigen, has emerged to be a unique group of antigen that could potentially be important antigen targets for antigen‐specific cancer immunotherapy.
Identification of CT Antigens
A major breakthrough in identifying tumor antigens recognized by host cytotoxic T‐lymphocytes (CTL) was the molecular cloning of MAGE‐1 by van der Bruggen et al. in 1991.( 1 ) Using the melanoma cell line MZ2‐MEL and autologous CTL clones cytolytic to this line, MAGE‐1 (subsequently re‐named as MAGE‐A1, melanoma antigen A1) was identified as the target antigen for one of the CTL clones, and this represented the first immunogenic tumor antigen shown to have elicited autologous cytotoxic T‐lymphocyte responses in a cancer patient. Pursuing the same strategy, Boon et al. successfully identified MAGE‐A3, another member of the MAGE‐A family, as well as two additional families of antigens, namely the BAGE and GAGE gene families.( 2 , 3 , 4 )
Analysis of the mRNA expression found the intriguing feature that MAGE‐A, BAGE, and GAGE genes are expressed in testis, but not in any other normal somatic tissues or cells, including melanocytes. In contrast, these genes are expressed in a proportion of many different types of cancers, including breast cancer, lung cancer, and ovarian cancer, etc. This expression pattern, in conjunction with the lack of MHC class I antigen in testicular germ cells, imply that these gene products are tumor‐specific antigens from the cancer vaccine perspective, and these antigens were referred to as ‘shared tumor‐specific antigens’ by Boon et al. ( 5 ) Shortly after the discovery of tumor antigens by this transfection‐based assay, Pfreundschuh et al. developed a serological approach to molecularly clone immunogenic tumor antigens that had elicited high‐titer IgG immune responses in autologous cancer patients. This methodology, termed SEREX (serological analysis of recombinant cDNA expression libraries), was based on the immunoscreening of tumor cDNA expression libraries with sera from the autologous patients.( 6 ) In their first experiments, Sahin et al. analyzed melanoma, renal cell carcinoma, astrocytoma, and Hodgkin lymphoma, and a large number of genes were isolated, including MAGE‐A1 and tyrosinase, two antigens previously shown to be targets for cytotoxic T‐lymphocytes, indicating that protein antigens that elicit antibody responses in cancer patients are likely to have elicited simultaneous T‐cell responses.( 7 ) This prompted a large‐scale SEREX screening of various cancer types, spearheaded by the Ludwig Institute for Cancer Research, and the screening strategy was broadened to include the screening of cDNA libraries derived from allogeneic tumors, tumor cell lines, and testis.( 8 , 9 , 10 , 11 , 12 , 13 , 14 ) This effort and similar efforts by other researchers led to the identification of more than 1000 SEREX‐defined antigens in several years (http://ludwig‐sun5.unil.ch/CancerImmunomeDB/; http://www.cancerimmunity.org/SEREX/). Intriguing, several of these newly defined tumor antigens, for example SSX2, NY‐ESO‐1, SCP1, and CT7, similarly had normal mRNA expression restricted to testis, with abnormal expression detected in various cancers. Recognizing this characteristic expression pattern, the term cancer/testis (CT) antigen was coined by Old and Chen( 9 , 15 ) to encompass this expanding category of tumor antigens, and CT antigens identified by SEREX to date include MAGE‐A,( 11 ) SSX2,( 16 ) SSX4, NY‐ESO‐1,( 9 ) SCP1,( 17 ) CT7,( 11 )NY‐SAR‐35,( 10 ) OY‐TES‐1,( 18 ) SLCO6A1,( 19 ) PASD1,( 20 ) CAGE‐1,( 21 ) and KK‐LC‐1,( 22 ) etc.
Following the recognition of this restricted mRNA expression pattern, multiple studies were launched to identify new CT genes based on their preferential expression in testis and cancer. By representational difference analysis and comparing melanoma versus normal skin, Gure et al. cloned a MAGE‐A related CT gene, CT10,( 23 ) and anti‐CT10 antibody was found in a melanoma patient, establishing its immunogenicity. Using a similar approach, Lucas et al. ( 24 ) independently isolated the same gene and CT7, and these two genes were later named MAGE‐C1 (CT7) and MAGE‐C2 (CT10). A second gene of the NY‐ESO‐1 family, LAGE1,( 25 ) as well as other new CT genes, for example SAGE1,( 26 ) were similarly identified. More recently, massively parallel signature sequencing (MPSS) was utilized to compare the mRNA expression profiles between testis, melanoma cell lines, and other somatic tissues.( 27 ) This resulted in the identification of >20 CT or CT‐like genes, including CT45. In addition to these experimental approaches, in silico analysis, for example by analyzing the EST (expressed sequence tags) databases for genes with cancer‐testis restricted expression, also resulted in the identification of CT antigens, including BRDT,( 28 ) CT46,( 29 ) XAGE1,( 30 , 31 ) and PLAC1.( 32 )
To comprehensively analyze the mRNA expression data at the genomic level and identify all potentially CT genes, Hofmann et al. recently analyzed all available data using a combination of four platforms: MPSS, ESTs, CAGE, and RT‐PCR.( 33 ) This thorough analysis resulted in the cataloguing of a total of 153 genes with mRNA expression in normal tissues restricted to, or at least preferentially in, testis, with evidence of tumor expression.
With this expansion of the CT genes, it became evident that a CT database would be highly desirable, and such a database has recently been established by the Ludwig Institute for Cancer Research (http://www.cta.lncc.br/).( 34 ) A total of 110 CT genes or gene families have been entered, reflecting all antigens that were published as CT antigens in the literature (Table S1). The CT database also includes the results of standardized RT‐PCR analysis of each CT antigen in a panel of 22 normal tissues and 32 cancer cell lines.
Genomic organization of CT antigen genes. Among the first several CT antigens identified, most were encoded by multigene families on chromosome X, particularly on the telomeric end between Xq24 to Xq28. These included MAGE‐A, NY‐ESO‐1, CT7/MAGE‐C1, CT10, and SAGE. In addition, SSX and GAGE were located at a more centromeric position of X chromosome, Xp11.2‐11.4. This unusual clustering of CT genes on the X chromosome was noticed repeatedly as additional CT genes were identified, leading to the classification of CT genes into CT‐X and non‐X CT genes by Simpson et al. in their review.( 35 ) Of the 110 CT genes listed in the current CT database, 30 were CT‐X genes, with Xq24‐q28 bearing the highest density of these genes (Fig. 1). One characteristic of the CT‐X genes is that they are often multicopy genes that resulted from recent gene duplications. These repeats can be inverted repeats, for example CT45, or direct repeats, for example CT47. Combining all multicopy CT‐X genes, it has been estimated that CT genes comprise ∼10% of DNA sequence on the X chromosome.( 36 ) Similar findings have been reported in mouse in which 36 multicopy genes were defined on chromosome X, with between two to 28 gene copies.( 37 ) When tested for their expression by RT‐PCR, 33 of the 36 genes were found to be exclusively or preferentially expressed in testis, including the homologs of human CT genes, for example MAGE, NXF2, and SSX genes. Eight of the 28 genes analyzed were shown to be expressed in the self‐renewing spermatogonia. On the other hand, the mRNA expression of the remaining 20 genes coincided with the appearance of post‐meiotic germ cells, that is secondary spermatocytes and spermatids, suggesting that they are only expressed in the haploid germ cells.( 37 )
Figure 1.
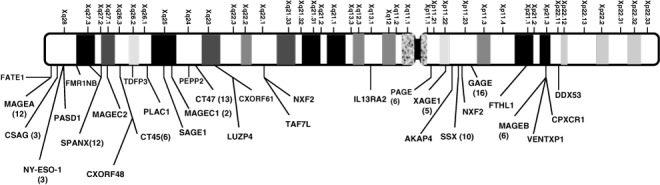
Distribution of cancer/testis (CT) families on the X‐chromosome. The number of CT genes in each family is indicated in parenthesis.
In contrast to the CT‐X genes, most of the non‐X CTs genes are single copy genes, and no additional chromosomal clustering is found. This drastic difference indicates that CT‐X and non‐X CT genes are evolutionarily distinctive. Such differences are also reflected in their expression patterns and functional characteristics (see below), and CT‐X antigens are currently considered to be more promising cancer vaccine targets.
Expression of CT antigen mRNA in normal tissues. In addition to testicular expression, a subset of CT antigens has been found to be expressed in placenta, including MAGE‐A3, MAGE‐A10, MAGE‐A8, XAGE2, and XAGE3. Conversely, placenta‐specific genes, for example PLAC1, have been shown to be expressed in testis, but at a low abundance level.( 32 ) Besides placental expression, it also became clear that many CT genes showed low‐level mRNA expression in a limited number of somatic tissues. However, based on quantitative RT‐PCR data, the mRNA expression levels of these genes in non‐testicular tissues are usually at <1% of their expression levels in testis. This low‐level CT expression has never been confirmed at the protein level by immunohistochemical analysis with anti‐CT antibodies, and whether such ‘leaky’ RNA expression translates to a biologically significant level of protein is debatable. On the other hand, some of the so‐called CT genes were subsequently shown to have broader mRNA expression than was initially recognized. These genes probably should not be considered CT genes, and examples include JARID1B and SPA17. Recognizing this mRNA expression spectrum in normal tissues, Hofmann et al. classified the 153 CT genes that they analyzed into ‘testis‐restricted’ and ‘testis‐selective’ categories, with the latter being genes that are predominantly, but not exclusively expressed in testis. (A third category, ‘testis/brain‐selective’, was also described that encompassed a small number of genes that showed expression limited to the testis and brain.) Significantly, 35 of the 39 testis‐restricted genes were CT‐X genes and only four were non‐CT‐X genes. This disproportional enrichment of testis‐restricted genes on the X chromosome, in conjunction to the fact that most immunogenic CT antigens, for example MAGE‐A, NY‐ESO‐1, and SSX, are all within this testis‐restricted subgroup of CT‐X genes, strongly imply that the CT‐X genes are likely also the most interesting genes from the immunotherapeutic standpoint.
mRNA expression of CT antigens in cancer. Aberrant activation and expression of mRNA transcripts in various human cancers in a lineage‐independent fashion is the defining criterion of CT antigens. The following expression characteristics, pertinent to the consideration of CT antigens as immunotherapeutic targets, have been observed: (a) different cancer types are significantly different in their frequency of CT mRNA expression; (b) for a given cancer type, tumors of higher histological grade and later clinical stage often show higher frequency of CT expression; and (c) CT antigens tend to be coordinated expressed; that is, tumors that are positive for CT antigens often show simultaneous expression of more than one CT antigen.
Of the different types of cancers, melanoma, ovarian cancer, and lung cancer, particularly the squamous cell type, have been found to have the highest frequency of CT expression, sometimes referred to as ‘CT‐rich’ tumor types. In contrast, hematopoietic malignancies, including lymphomas and leukemia as well as renal, colon, and pancreatic cancers, have notably low frequency of CT‐antigen expression. Other epithelial cancer types, for example breast cancer, bladder cancer, and prostate cancer, appear to be intermediate in their CT‐expression frequency. For instance, NY‐ESO‐1 mRNA expression has been observed in 52% of melanoma,( 38 ) 27% of non‐small‐cell lung carcinoma,( 39 ) and 35% of bladder cancer,( 40 ) but only in 10% of the colon cancer,( 41 ) and none of the renal cell carcinoma and lymphoma tested.( 9 ) Exceptions to this observation do occur, most notably the high frequency expression of NY‐ESO‐1 in synovial sarcoma,( 42 ) CT7/MAGE‐C1 in multiple myeloma,( 43 ) and CT45 in classical Hodgkin lymphoma.( 44 ) It is possible that these CT antigens might have specific biological roles in these specific tumor types, but such roles, if they do exist, remain to be elucidated.
For a given cancer type, higher frequency of CT expression is often correlated with worse outcome (Table 1). Higher grade and metastatic tumors have also been found to have more frequent CT expression than the primary tumors. For example, NY‐ESO‐1 has been found to be expressed in 40% of grade 3 bladder tumors and 23% of grade 2 tumors, but none of the grade 1 tumors.( 45 ) Similarly, MAGE‐A1 expression has been found in 48% of metastatic melanoma versus 16% of primary melanoma.( 46 )
Table 1.
Correlation of cancer/testis (CT) protein expression with clinicopathologic parameters and prognosis
| Tumor type | Antigen | Association | Reference |
|---|---|---|---|
| Melanoma | MAGE‐A1,MAGE‐A2, MAGE‐A3, MAGE‐A4 | Tumor thickness and metastasis | (46) |
| Non‐small‐cell lung cancer | MAGE‐A1,MAGE‐A3, MAGE‐A4,MAGE‐A10, MAGE‐C1 | Advanced tumor type, nodal and pathologic stages as well as pleural invasion | (39) |
| Pancreatic cancer | MAGE‐A3 | Poor survival | (88) |
| Hepatocellular carcinoma | MAGE‐C1 | Reduced overall survival | (89) |
| Multiple myeloma | MAGE‐A1,MAGE‐A3, MAGE‐A4, MAGE‐C1 | Stage and risk status of disease | (90, 91, 92) |
| Serous ovarian carcinomas | MAGE‐A4 | Inverse correlation between expression and patient survival | (93) |
| Melanoma | NY‐ESO‐1 | Thicker primary lesions and a higher frequency of metastatic disease | (94) |
Another expression characteristic that has been observed in multiple tumor types is the tendency for CT antigens to be coordinately expressed. In the analysis of expression of nine CT‐X genes in lung cancer, Gure et al. found that expression of one CT antigen by a tumor greatly enhanced the likelihood that it would also simultaneously express a second CT antigen.( 39 ) On the other hand, a subset of tumors showed no expression of any of the CT antigens tested. This phenomenon of coordinated expression (or non‐expression) has been observed in tumor samples as well as cell lines; SK‐MEL‐37, for example, expresses almost all the CT antigens tested.( 11 )
Protein expression of CT antigens in testis and in cancer. Using polyclonal and monoclonal antibodies, the expression of CT proteins in normal and tumor tissues have been analyzed using immunohistochemical techniques, and at least three common patterns of CT protein expression have been observed in testis (Fig. 2): (a) predominant expression in spermatogonia – the self‐renewing stem cell population of germ cell in adult testis, mostly as nuclear protein; (b) predominant expression in primary and/or secondary spermatocytes, again as nuclear antigens; and (c) restricted expression to the mature sperm cells, mostly as cytoplasmic protein. Most of the CT‐X antigens, including NY‐ESO‐1, MAGE‐A, CT7/MAGE‐C1, CT10/MAGE‐C2, GAGE, CT47, SAGE1, and NXF2, etc., belong to the first group, with strongest expression seen in the spermatogonia. Most of these are mainly nuclear antigens, but not infrequently also present in the cytoplasmic compartment. Predominant cytoplasmic expression is rare, with CT47 being an example. In comparison to this group, some CT antigens are expressed mainly or exclusively in the spermatocyte stage. This group is comprised mostly of nuclear proteins and includes meiosis‐related proteins, for example SCP1 and CT46/HORMAD1, as well as rare CT antigens in the CT‐X group, for example CT45. The third group consists of genes that are only expressed in the more mature, post‐meiotic sperm cells. COX6B2, a testis‐specific isoform of the cytochrome c oxidase subunit VIb, is an example of this group. The only CT‐X antigen analyzed so far that belongs to this group is the SPANX family, a family on Xq27 with at least five members.
Figure 2.
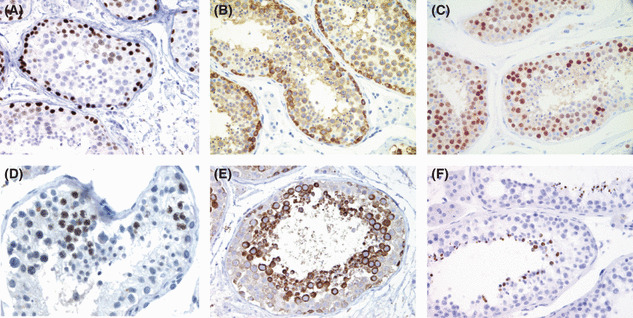
Different patterns of cancer/testis (CT) antigen expression in adult testis. Many CT‐X antigens are expressed as nuclear antigens in spermatogonia, including NXF2 (A), SAGE, and most melanoma antigen (MAGE‐A) antigens. An exception is CT47 (B), expressed as cytoplasmic antigens in spermatogonia. The third pattern is represented by CT45 (C), which shows strongest expression in the pre‐meiotic spermatocytes as nuclear antigens. The fourth pattern is observed in meiosis‐related CT antigens, which are expressed as nuclear antigens in cells undergoing meiosis, an example being CT46/HORMAD1 (D). THEG is the only antigen that shows the fifth pattern, being expressed as a cytoplasmic antigen in the post‐meiotic spermatids as a cytoplasmic antigen (E). The last pattern, expression in the most mature sperm cells, is seen in SPANX (F), COX6B2, etc.
Expression of CT proteins in tumor has only been analyzed for a few CT antigens, that is NY‐ESO‐1, MAGE‐A, GAGE, CT7/MAGE‐C1, and CT10/MAGE‐C2, and most recently CT45,( 47 ) examples of which are depicted in Figure 3. From these analyses, the following characteristics have been observed: (a) most CT genes evaluated have demonstrable protein expression in cancer, but with important exceptions; (b) CT protein expression correlates to the mRNA expression level, and tumors with higher CT mRNA levels, in general >1–10% of testicular mRNA expression level, usually have demonstrable protein expression; and (c) CT protein expression in tumor is often heterogeneous, and strong expression in a very small subset of tumor cells are not infrequently observed.
Figure 3.
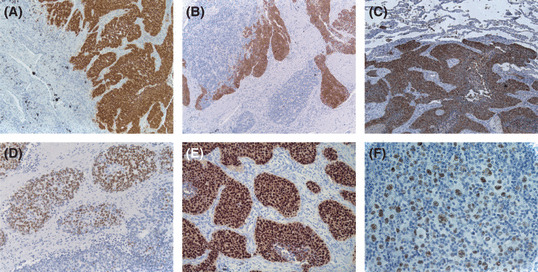
Protein expression of cancer/testis (CT) antigen in cancer. (A) Melanoma antigen (MAGE‐A) antigens are expressed as both nuclear and cytoplasmic proteins, showing diffuse expression in this lung cancer. (B) Similar nuclear and cytoplasmic expression is also seen in NY‐ESO‐1, which shows heterogeneous expression in this lung cancer. (C) Similar nuclear and cytoplasmic staining is observed for the GAGE gene family, as well as other CT antigens, including CT7 (not shown). In comparison, pure nuclear staining is seen for CT10 (D) and CT45 (E). In Hodgkin lymphoma, CT45 often showed diffuse expression in the neoplastic Reed‐Sternberg cells (F).
It has generally been assumed that CT mRNA expression correlates to protein expression. Studies to correlate mRNA and protein levels of NY‐ESO‐1, CT45, and GAGE have supported this concept of transcriptional regulation, and tumor samples with CT mRNA level at >10% of the testicular expression almost always have detectable CT protein expression. In comparison, tumors with <1% testicular expression have usually shown no detectable protein expression. In contrast to this mRNA‐protein correlation, however, expression of some CT antigens may also be regulated post‐transcriptionally, and the presence of CT mRNA may not guarantee protein expression. For instance, despite the detection of substantial SCP1 and HORMAD1/CT46 mRNA levels in some tumors and immunohistochemical detection of the protein expression in spermatocytes, we have not been able to detect expression of these proteins in mRNA‐positive tumors. It is possible that the expression of these genes, given their specific functions in meiosis, is tightly regulated physiologically, probably at both the transcriptional and translational levels.
Two different spatial distribution patterns of CT protein expression have been observed in cancer. In some cases, CT antigens are diffusely and homogenously expressed in almost all tumor cells, suggesting that CT gene activation is a clonal event. On the other hand, CT protein expression is heterogeneous in many tumor specimens, and sometimes a small cluster of tumor cells with strong expression is seen amongst a background of >99% of CT‐negative tumor cells. This heterogeneous staining pattern suggests that the activation might be epigenetic, for example due to changes in DNA methylation. Alternatively, it has also been proposed that the CT‐positive cells might represent the cancer stem cells in these cases.( 48 ) From the immunotherapeutic standpoint, the heterogeneous staining pattern raises the concern of immunoselection of CT‐negative cells following vaccination. However, the observation of ‘antigen‐spreading’ following the killing of a subset of tumor cells would argue against this concern.( 49 )
Regulation of CT Antigen Expression
One common future of CT antigen gene expression, particularly for the CT‐X genes, is the induction by the DNA methyl‐transferase 1 inhibitors, 5‐aza‐2‐deoxycytidine (5DC), and/or by histone deacetylase (HDAC) inhibitors.( 50 ) This has been shown for MAGE‐A, NY‐ESO‐1, and SSX, etc.( 50 , 51 , 52 ) This finding, together with the inclination of global hypomethylation in cancer, suggests CpG island hypomethylation at the promoter regions as the likely mechanism for transcriptional activation of CT genes in cancer. However, it has been observed that 5DC cannot induce CT expression in primary fibroblasts as it readily does in most tumor cell lines,( 53 ) and addition of HDAC also does not lead to significant expression of the CT genes. Although it is possible that CpG islands in the promoter regions of these genes are so densely methylated in normal cells that 5DC and HDAC were ineffective, the possibility that other mechanisms of transcriptional, and even post‐transcriptional, control of gene expression might in effect exist is highly likely.
An interesting theory that has been proposed to explain the activation of CT antigens in cancer is that the activation may be the consequence of induction of a gametogenic program in cancer.( 35 , 54 ) According to this hypothesis, the different CT expression profiles seen in cancer may correspond to the profiles of CT antigens normally expressed at various stages of gametogenesis or trophoblastic development, and the triggering event for this activation could be the switch‐on of a master gene in germ cell development, for example by a mutational event. This hypothesis, although interesting, remains to be proven.
Functions of CT Antigens
The biological role of CT‐X in both germ line tissues and tumors remains poorly understood. Recent studies have provided some evidence that CT antigens may play a role in human tumorigenesis. Yeast two‐hybrid studies using cancer‐related genes as bait have twice pulled out MAGE proteins: MAGE‐A11 and MAGE‐A4.( 55 , 56 ) MAGE‐A11 was found to have a role in the regulation of androgen‐receptor function,( 55 ) and MAGE‐A4 was identified in a search for binding partners of the oncoprotein gankyrin.( 56 ) Overexpression of MAGE‐A4 in human embryonic kidney cells (293 cells) was found to increase apoptosis while MAGE‐A4 mRNA silencing decreased caspase‐3 activity in a squamous cell lung cancer and in 293/MAGE‐A4 cells.( 57 ) Mage‐A2 protein was shown to strongly down‐regulate p53 transactivation function, and association between MAGE‐A expression levels and resistance to etoposide (ET) treatment was shown in short‐term cell lines obtained from melanoma biopsies harboring wild‐type‐p53.( 58 ) Multiple MAGE proteins including human MAGE‐A3, MAGE‐C2, and murine mage‐b1 (mMage‐b) proteins were shown to form complexes with Kap‐1, a known co‐repressor of p53, and siRNA suppression of these MAGE genes induces apoptosis and causes increased p53 expression in vitro. Thus, MAGE gene expression may protect cells from programmed cell death and contribute to the development of malignancies by promoting survival.( 59 ) In pituitary tumors, a reciprocal profile of FGFR2‐IIIb and MAGE‐A3 expression was identified.( 60 ) While FGFR2‐IIIb plays a growth‐inhibitory tumor‐suppressive role, down‐regulation of MAGE‐A3 resulted in p53 transcriptional induction and p21 accumulation, suggesting that MAGE‐A3 might be oncogenic.( 60 )
Similar to MAGE, antiapoptotic properties of GAGE‐7 have also been reported, as GAGE‐7C was shown to render cells resistant to apoptosis mediated by IFN‐γ or by Fas.( 61 ) Significantly higher expression of MAGE and GAGE were observed in paclitaxel‐ and doxorubicin‐resistant cells lines than in the parental cell lines,( 62 ) and GAGE appeared to render cells resistant to Taxol and γ‐irradiation. This anti‐apoptotic activity and the resistance to the clinically relevant agents may explain the reported correlation between GAGE expression and poor prognosis in some cancers.
We used the yeast two‐hybrid system to identify putative novel MAGE‐homology domain (MHD)‐interacting proteins.( 63 ) The MHD of MAGE‐C1/CT7 was used as a bait to screen a human testis cDNA library, and NY‐ESO‐1 was found to be a MAGE‐C1/CT7 binding partner. This was the first report of a direct interaction between two CT antigens and may be pertinent to the frequently coordinated expression of these proteins.
While few clues have emerged so far in relation to the function of the CT‐X, most of the non‐X CTs are conserved during evolution and have known roles in spermatogenesis and fertilization. OY‐TES‐1 (ACRBP) is similar to the proacrosin binding protein sp32 precursor found in mice, guinea pigs, and pigs. Located in the sperm acrosome, this protein is thought to function as a binding protein to proacrosin for packaging and condensation of the acrosin zymogen in the acrosomal matrix.( 18 ) Three proteins involved in germ cell meiosis were identified as CT antigens: SCP1, SYCE, and HORMAD1/CT46. SCP1 and SYCE are part of the synaptonemal complex lateral and central elements, respectively.( 64 , 65 )ADAM2 (fertilin beta) and PRM2 were found to contribute to successful fertilization and also may have an important impact in development of preimplantation embryos.( 66 ) Protamines are small sperm nuclear‐specific proteins that replace somatic histones during early spermiogenesis.( 67 )SEMG1 is the predominant protein in semen and it is involved in the formation of a gel matrix that encases ejaculated spermatozoa.( 68 ) SEMG1 and/or its proteolytic fragments were also found be involved in regulating spermatozoon motility and capacitation, and also in presenting antibacterial activity.( 69 )
Immunogenicity of CT Antigens
As potential cancer vaccine targets, the demonstration of immunogenicity in the human host is considered crucial for CT antigens. To date, however, only several CT antigens have been shown to elicit coordinated humoral and cell mediated responses, including MAGE‐A1 and MAGE‐A3, initially identified by cytotoxic T‐cell cloning, and NY‐ESO‐1 and SSX, initially identified by SEREX.
The T‐cell responses to CT antigens are typically investigated by the screening of overlapping peptide panels with CD8+ or CD4+ T‐cells from peripheral blood. Many HLA‐restricted T‐cell epitopes have been identified this way, particularly for the MAGE‐A, NY‐ESO‐1, and SSX genes, forming the basis for peptide‐based CT cancer vaccine trials and for the monitoring of post‐vaccination T‐cell responses (see below). These data on T‐cell epitopes have been compiled into the peptide database of T‐cell‐defined tumor antigens (http://www.cancerimmunity.org/peptidedatabase/Tcellepitopes.htm). In contrast to this abundance of data on MAGE‐A, NY‐ESO‐1, and SSX, studies to investigate T‐cell recognition of other CT antigens have been very limited to date.( 22 , 70 , 71 , 72 )
In comparison, humoral immune responses have been investigated more broadly, usually by ELISA testing against recombinant CT proteins. NY‐ESO‐1 is the prototype example, and anti‐NY‐ESO‐1 antibody has been detected in many cancer types, including lung cancer, ovarian cancer, breast cancer, bladder cancer, and melanoma, etc. The frequency of anti‐NY‐ESO‐1 antibody response in patients with advanced NY‐ESO‐1 positive tumors has been estimated to be at the range of 25–50%, and the titer of the antibody appears to increase with progressive disease and decrease upon removal of the tumor or upon tumor regression.( 73 ) Investigation of anti‐NY‐ESO‐1 T‐cell responses has demonstrated NY‐ESO‐1‐specific CD8+ T‐cells in the majority of patients with positive anti‐NY‐ESO‐1 antibodies, whereas T‐cell response in the absence of a concurrent NY‐ESO‐1 antibody response is very rare. This high frequency of coordinated humoral and cell‐mediated responses indicates that NY‐ESO‐1 is among the most immunogenic of CT antigens known to date, making it one of the most attractive targets for cancer vaccines.
In addition to NY‐ESO‐1, coordinated antibody and T‐cell responses have also been observed for MAGE‐A and SSX antigens, but at a much lower frequency. For instance, MAGE‐A genes are highly expressed in melanoma, and yet spontaneous MAGE‐A1 and MAGE‐A3 antibodies have only been found in <3% of these patients in our hands. After immunization with MAGE‐A3 recombinant protein, many patients do develop MAGE‐A3‐specific antibodies, but not accompanied by detectable CD8+ T‐cell responses in most cases.( 74 )
Antibody responses to other CT antigens have been reported, some of them at very high frequencies, including SCP1 in 50% breast cancer patients,( 75 ) cTAGE in 33% of cutaneous T‐cell lymphoma patients,( 76 ) and SSX2 in 18% of melanoma patients,( 6 ) etc. However, it would be prudent to interpret these reports of exceptional high antibody frequency with caution, as much lower frequency of antibody response has also been reported.
Cancer Vaccine Trials Targeting CT Antigens
To evaluate CT antigens as therapeutic cancer vaccine targets, multiple clinical trials have been carried out, targeting MAGE‐A3 or NY‐ESO‐1. For both antigens, the trials have tested peptide‐based vaccines and recombinant protein vaccines. In melanoma patients, both peptide vaccines have led to regression of individual tumor nodules, including occasional complete regression. Immunological responses have also been documented, significantly more frequent in NY‐ESO‐1 than in MAGE‐A3‐vaccinated patients.( 77 , 78 )
Unlike peptide vaccines, recombinant protein vaccines are expected to induce a broader spectrum of CD8+ and CD4+ immune responses, with the additional advantage of being unrestricted by the HLA types of the patients, thus suitable for a larger patient population. For the first MAGE‐A3 recombinant protein trial, a His‐tag MAGE‐A3 protein with protein D at N‐terminus, produced by GlaxoSmithKline (gsk; Brentford, UK), was administered to stage III/IV melanoma patients, and clinical responses were seen in five of 26 patients, including one partial response and four mixed responses. This was followed by a phase II trial in 182 non‐small‐cell lung cancer patients, for which an improvement of disease‐free survival (hazard ratio = 0.666, P = 0.12) was observed at the interim analysis. Based on this promising result, a phase III trial that involves 2270 lung cancer patients has been launched and is ongoing. Additional phase II melanoma trials on MAGE‐A3 proteins have also been conducted to evaluate different immunological adjuvants (AS15 vs AS02B), and the results are being analyzed.( 79 )
For NY‐ESO‐1, the first recombinant protein trial was in melanoma patients after complete tumor resection, using a His‐tagged recombinant protein either with or without ISCOMATRIX adjuvant.( 80 ) The results showed anti‐NY‐ESO‐1 antibody responses in almost all patients receiving NY‐ESO‐1 with ISCOMATRIX, and integrated CD4+ and CD8+ T‐cell responses were also induced in a subset of patients, reacting to a broad range of NY‐ESO‐1 epitopes, most of them previously undefined. Clinically, it was found that NY‐ESO‐1 vaccination might reduce the risk of melanoma recurrence, as only two of 19 patients in the group of NY‐ESO‐1 with ISCOMATRIX showed tumor recurrence, in comparison to 14 of 23 in other groups (placebo or NY‐ESO‐1 alone). However, a more recent study using the same vaccine on stage III/IV melanoma patients showed objective response in only one of 27 patients (in the form of stable disease), and T‐cell responses in these patients appeared to be inferior to those seen in the prior group of patients with minimal residual disease.( 81 ) The reason for this inferior T‐cell response was attributed to immunosuppression by regulatory T‐cells, and the authors proposed that vaccine‐based treatment might be more beneficial at the setting of early or minimal residual disease, when the tumor load and the extent of immunosuppression are both minimized.
In addition to the ISCOMATRIX adjuvants, other forms of NY‐ESO‐1 protein vaccine constructs are also being evaluated, including the fusion of NY‐ESO‐1 with cholesterol‐bearing hydrophobized pullulan (CHP‐NY‐ESO‐1),( 82 ) or the use of other adjuvants, for example CpG, Montanide ISA‐51, imiquimod, etc.( 83 , 84 ) The possibility of producing NY‐ESO‐1 protein in vivo using DNA vaccine constructs has also been examined, either using naked plasmid DNA,( 85 ) vaccinia/fowlpox viral constructs,( 77 ) or bacterial vectors such as Salmonella typhimurium.( 86 ) Most of these phase I/II trials showed safety of the vaccine preparations, with variable capability of inducing NY‐ESO‐1‐specific immune responses. Whether the observed immune responses will correlate to beneficial clinical outcomes remains to be proven.
Aside from its potential as an antigen‐specific cancer vaccine, NY‐ESO‐1 has been found to be useful in adoptive immunotherapy. In the study of Hunder et al.,( 49 ) a CD4+ T‐cell clone specifically targeting a HLA‐APB1*0401‐restricted NY‐ESO‐1 epitope was isolated from a melanoma patient, expanded in vitro, and infused back to the patient. Complete resolution of pulmonary and nodal disease was observed in this patient, who remains disease‐free 2 years after treatment. In vitro testing showed that this patient has also generated previously undetected anti‐MAGE‐A3 and anti‐Melan A T‐cell responses following the adoptive transfer, supporting the notion of ‘antigen spreading’.
Most recently, NY‐ESO‐1 has also been found to be potentially useful if combined with non‐specific immunotherapeutic approaches such as CTLA‐4 blockade.( 87 ) In this study, 15 melanoma patients were treated with anti‐CTLA4 monoclonal antibody (ipilimumab), and five of eight patients with evidence of clinical benefit were found to be NY‐ESO‐1 antibody positive, whereas none of seven clinical non‐responders had NY‐ESO‐1 antibody in serum. This finding suggests that anti‐CTLA‐4 therapy following induction of anti‐NY‐ESO‐1 immune responses by vaccination could have a synergistic effect and this possibility should be explored.
Concluding Remarks
Identification of appropriate target antigens is the first and most crucial step in the successful development of antigen‐specific immunotherapy, and the discovery and characterization of CT antigens has provided the first group of target antigens that can be used in various common epithelial cancers. As none of the CT antigens appear to be cell surface antigens, they are currently considered cancer vaccine targets rather than targets for antibody‐based therapy. However, recent studies have also shown them to be potentially useful in adoptive T‐cell transfer approaches and in a non‐specific immunotherapeutic approach that aims at CTLA‐4 checkpoint blockade. This broadened role of CT antigens is exciting and will likely be further explored in the coming years.
Supporting information
Table S1. The cancer/testis antigens.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgments
The authors would like to thank Drs. Lloyd J. Old and Andy J.G. Simpson of the Ludwig Institute for Cancer Research for their unwavering support over the years.
References
- 1. Van Der Bruggen P, Traversari C, Chomez P et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 1991; 254: 1643–7. [DOI] [PubMed] [Google Scholar]
- 2. Boel P, Wildmann C, Sensi ML et al. BAGE: a new gene encoding an antigen recognized on human melanomas by cytolytic T lymphocytes. Immunity 1995; 2: 167–75. [DOI] [PubMed] [Google Scholar]
- 3. Van den Eynde B, Peeters O, De Backer O, Gaugler B, Lucas S, Boon T. A new family of genes coding for an antigen recognized by autologous cytolytic T lymphocytes on a human melanoma. J Exp Med 1995; 182: 689–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gaugler B, Van den Eynde B, Van Der Bruggen P et al. Human gene MAGE‐3 codes for an antigen recognized on a melanoma by autologous cytolytic T lymphocytes. J Exp Med 1994; 179: 921–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boon T, Coulie PG, Van den Eynde B. Tumor antigens recognized by T cells. Immunol Today 1997; 18: 267–8. [DOI] [PubMed] [Google Scholar]
- 6. Sahin U, Tureci O, Pfreundschuh M. Serological identification of human tumor antigens. Curr Opin Immunol 1997; 9: 709–16. [DOI] [PubMed] [Google Scholar]
- 7. Sahin U, Tureci O, Schmitt H et al. Human neoplasms elicit multiple specific immune responses in the autologous host. Proc Natl Acad Sci U S A 1995; 92: 11810–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Obata Y, T TA, Tamaki H et al. Identification of cancer antigens in breast cancer by the SEREX expression cloning method. Breast Cancer 1999; 6: 305–11. [DOI] [PubMed] [Google Scholar]
- 9. Chen YT, Scanlan MJ, Sahin U et al. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc Natl Acad Sci U S A 1997; 94: 1914–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee SY, Obata Y, Yoshida M et al. Immunomic analysis of human sarcoma. Proc Natl Acad Sci U S A 2003; 100: 2651–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen YT, Gure AO, Tsang S et al. Identification of multiple cancer/testis antigens by allogeneic antibody screening of a melanoma cell line library. Proc Natl Acad Sci U S A 1998; 95: 6919–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jager D, Stockert E, Gure AO et al. Identification of a tissue‐specific putative transcription factor in breast tissue by serological screening of a breast cancer library. Cancer Res 2001; 61: 2055–61. [PubMed] [Google Scholar]
- 13. Obata Y, Takahashi T, Sakamoto J et al. SEREX analysis of gastric cancer antigens. Cancer Chemother Pharmacol 2000; 46 (Suppl): S37–42. [DOI] [PubMed] [Google Scholar]
- 14. Scanlan MJ, Gordan JD, Williamson B et al. Antigens recognized by autologous antibody in patients with renal‐cell carcinoma. Int J Cancer 1999; 83: 456–64. [DOI] [PubMed] [Google Scholar]
- 15. Old LJ, Chen YT. New paths in human cancer serology. J Exp Med 1998; 187: 1163–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Tureci O, Sahin U, Schobert I et al. The SSX‐2 gene, which is involved in the t(X;18) translocation of synovial sarcomas, codes for the human tumor antigen HOM‐MEL‐40. Cancer Res 1996; 56: 4766–72. [PubMed] [Google Scholar]
- 17. Tureci O, Sahin U, Zwick C, Koslowski M, Seitz G, Pfreundschuh M. Identification of a meiosis‐specific protein as a member of the class of cancer/testis antigens. Proc Natl Acad Sci U S A 1998; 95: 5211–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ono T, Kurashige T, Harada N et al. Identification of proacrosin binding protein sp32 precursor as a human cancer/testis antigen. Proc Natl Acad Sci U S A 2001; 98: 3282–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lee SY, Williamson B, Caballero OL et al. Identification of the gonad‐specific anion transporter SLCO6A1 as a cancer/testis (CT) antigen expressed in human lung cancer. Cancer Immun 2004; 4: 13. [PubMed] [Google Scholar]
- 20. Liggins AP, Brown PJ, Asker K, Pulford K, Banham AH. A novel diffuse large B‐cell lymphoma‐associated cancer testis antigen encoding a PAS domain protein. Br J Cancer 2004; 91: 141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Park S, Lim Y, Lee D et al. Identification and characterization of a novel cancer/testis antigen gene CAGE‐1. Biochim Biophys Acta 2003; 1625: 173–82. [DOI] [PubMed] [Google Scholar]
- 22. Fukuyama T, Hanagiri T, Takenoyama M et al. Identification of a new cancer/germline gene, KK‐LC‐1, encoding an antigen recognized by autologous CTL induced on human lung adenocarcinoma. Cancer Res 2006; 66: 4922–8. [DOI] [PubMed] [Google Scholar]
- 23. Gure AO, Stockert E, Arden KC et al. CT10: a new cancer‐testis (CT) antigen homologous to CT7 and the MAGE family, identified by representational‐difference analysis. Int J Cancer 2000; 85: 726–32. [DOI] [PubMed] [Google Scholar]
- 24. Lucas S, De Smet C, Arden KC et al. Identification of a new MAGE gene with tumor‐specific expression by representational difference analysis. Cancer Res 1998; 58: 743–52. [PubMed] [Google Scholar]
- 25. Lethe B, Lucas S, Michaux L et al. LAGE‐1, a new gene with tumor specificity. Int J Cancer 1998; 76: 903–8. [DOI] [PubMed] [Google Scholar]
- 26. Martelange V, De Smet C, De Plaen E, Lurquin C, Boon T. Identification on a human sarcoma of two new genes with tumor‐specific expression. Cancer Res 2000; 60: 3848–55. [PubMed] [Google Scholar]
- 27. Chen YT, Scanlan MJ, Venditti CA et al. Identification of cancer/testis‐antigen genes by massively parallel signature sequencing. Proc Natl Acad Sci U S A 2005; 102: 7940–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scanlan MJ, Altorki NK, Gure AO et al. Expression of cancer‐testis antigens in lung cancer: definition of bromodomain testis‐specific gene (BRDT) as a new CT gene, CT9. Cancer Lett 2000; 150: 155–64. [DOI] [PubMed] [Google Scholar]
- 29. Chen YT, Venditti CA, Theiler G et al. Identification of CT46/HORMAD1, an immunogenic cancer/testis antigen encoding a putative meiosis‐related protein. Cancer Immun 2005; 5: 9. [PubMed] [Google Scholar]
- 30. Brinkmann U, Vasmatzis G, Lee B, Pastan I. Novel genes in the PAGE and GAGE family of tumor antigens found by homology walking in the dbEST database. Cancer Res 1999; 59: 1445–8. [PubMed] [Google Scholar]
- 31. Sato S, Noguchi Y, Ohara N et al. Identification of XAGE‐1 isoforms: predominant expression of XAGE‐1b in testis and tumors. Cancer Immun 2007; 7: 5. [PMC free article] [PubMed] [Google Scholar]
- 32. Silva WA Jr, Gnjatic S, Ritter E et al. PLAC1, a trophoblast‐specific cell surface protein, is expressed in a range of human tumors and elicits spontaneous antibody responses. Cancer Immun 2007; 7: 18. [PMC free article] [PubMed] [Google Scholar]
- 33. Hofmann O, Caballero OL, Stevenson BJ et al. Genome‐wide analysis of cancer/testis gene expression. Proc Natl Acad Sci U S A 2008; 105: 20422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Almeida LG, Sakabe NJ, DeOliveira AR et al. CTdatabase: a knowledge‐base of high‐throughput and curated data on cancer‐testis antigens. Nucleic Acids Res 2009; 37: D816–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Simpson AJ, Caballero OL, Jungbluth A, Chen YT, Old LJ. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer 2005; 5: 615–25. [DOI] [PubMed] [Google Scholar]
- 36. Ross MT, Grafham DV, Coffey AJ et al. The DNA sequence of the human X chromosome. Nature 2005; 434: 325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 2008; 40: 794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Goydos JS, Patel M, Shih W. NY‐ESO‐1 and CTp11 expression may correlate with stage of progression in melanoma. J Surg Res 2001; 98: 76–80. [DOI] [PubMed] [Google Scholar]
- 39. Gure AO, Chua R, Williamson B et al. Cancer‐testis genes are coordinately expressed and are markers of poor outcome in non‐small cell lung cancer. Clin Cancer Res 2005; 11: 8055–62. [DOI] [PubMed] [Google Scholar]
- 40. Sharma P, Gnjatic S, Jungbluth AA et al. Frequency of NY‐ESO‐1 and LAGE‐1 expression in bladder cancer and evidence of a new NY‐ESO‐1 T‐cell epitope in a patient with bladder cancer. Cancer Immun 2003; 3: 19. [PubMed] [Google Scholar]
- 41. Li M, Yuan YH, Han Y et al. Expression profile of cancer‐testis genes in 121 human colorectal cancer tissue and adjacent normal tissue. Clin Cancer Res 2005; 11: 1809–14. [DOI] [PubMed] [Google Scholar]
- 42. Jungbluth AA, Antonescu CR, Busam KJ et al. Monophasic and biphasic synovial sarcomas abundantly express cancer/testis antigen NY‐ESO‐1 but not MAGE‐A1 or CT7. Int J Cancer 2001; 94: 252–6. [DOI] [PubMed] [Google Scholar]
- 43. Jungbluth AA, Ely S, DiLiberto M et al. The cancer‐testis antigens CT7 (MAGE‐C1) and MAGE‐A3/6 are commonly expressed in multiple myeloma and correlate with plasma‐cell proliferation. Blood 2005; 106: 167–74. [DOI] [PubMed] [Google Scholar]
- 44. Heidebrecht HJ, Claviez A, Kruse ML et al. Characterization and expression of CT45 in Hodgkin’s lymphoma. Clin Cancer Res 2006; 12: 4804–11. [DOI] [PubMed] [Google Scholar]
- 45. Kurashige T, Noguchi Y, Saika T et al. Ny‐ESO‐1 expression and immunogenicity associated with transitional cell carcinoma: correlation with tumor grade. Cancer Res 2001; 61: 4671–4. [PubMed] [Google Scholar]
- 46. Brasseur F, Rimoldi D, Lienard D et al. Expression of MAGE genes in primary and metastatic cutaneous melanoma. Int J Cancer 1995; 63: 375–80. [DOI] [PubMed] [Google Scholar]
- 47. Chen YT, Hsu M, Lee P et al. Cancer/testis antigen CT45: analysis of mRNA and protein expression in human cancer. Int J Cancer 2009; 124: 2893–8. [DOI] [PubMed] [Google Scholar]
- 48. Gedye C, Quirk J, Browning J et al. Cancer/testis antigens can be immunological targets in clonogenic CD133(+) melanoma cells. Cancer Immunol Immunother 2009; 58: 1635–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hunder NN, Wallen H, Cao J et al. Treatment of metastatic melanoma with autologous CD4+ T cells against NY‐ESO‐1. N Engl J Med 2008; 358: 2698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. De Smet C, Lurquin C, Lethe B, Martelange V, Boon T. DNA methylation is the primary silencing mechanism for a set of germ line‐ and tumor‐specific genes with a CpG‐rich promoter. Mol Cell Biol 1999; 19: 7327–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Oi S, Natsume A, Ito M et al. Synergistic induction of NY‐ESO‐1 antigen expression by a novel histone deacetylase inhibitor, valproic acid, with 5‐aza‐2′‐deoxycytidine in glioma cells. J Neurooncol 2009; 92: 15–22. [DOI] [PubMed] [Google Scholar]
- 52. Sigalotti L, Fratta E, Coral S et al. Intratumor heterogeneity of cancer/testis antigens expression in human cutaneous melanoma is methylation‐regulated and functionally reverted by 5‐aza‐2′‐deoxycytidine. Cancer Res 2004; 64: 9167–71. [DOI] [PubMed] [Google Scholar]
- 53. Weber J, Salgaller M, Samid D et al. Expression of the MAGE‐1 tumor antigen is up‐regulated by the demethylating agent 5‐aza‐2′‐deoxycytidine. Cancer Res 1994; 54: 1766–71. [PubMed] [Google Scholar]
- 54. Old LJ. Cancer/testis (CT) antigens – a new link between gametogenesis and cancer. Cancer Immun 2001; 1: 1. [PubMed] [Google Scholar]
- 55. Bai S, He B, Wilson EM. Melanoma antigen gene protein MAGE‐11 regulates androgen receptor function by modulating the interdomain interaction. Mol Cell Biol 2005; 25: 1238–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nagao T, Higashitsuji H, Nonoguchi K et al. MAGE‐A4 interacts with the liver oncoprotein gankyrin and suppresses its tumorigenic activity. J Biol Chem 2003; 278: 10668–74. [DOI] [PubMed] [Google Scholar]
- 57. Peikert T, Specks U, Farver C, Erzurum SC, Comhair SA. Melanoma antigen A4 is expressed in non‐small cell lung cancers and promotes apoptosis. Cancer Res 2006; 66: 4693–700. [DOI] [PubMed] [Google Scholar]
- 58. Monte M, Simonatto M, Peche LY et al. MAGE‐A tumor antigens target p53 transactivation function through histone deacetylase recruitment and confer resistance to chemotherapeutic agents. Proc Natl Acad Sci U S A 2006; 103: 11160–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yang B, O’Herrin SM, Wu J et al. MAGE‐A, mMage‐b, and MAGE‐C proteins form complexes with KAP1 and suppress p53‐dependent apoptosis in MAGE‐positive cell lines. Cancer Res 2007; 67: 9954–62. [DOI] [PubMed] [Google Scholar]
- 60. Zhu X, Asa SL, Ezzat S. Fibroblast growth factor 2 and estrogen control the balance of histone 3 modifications targeting MAGE‐A3 in pituitary neoplasia. Clin Cancer Res 2008; 14: 1984–96. [DOI] [PubMed] [Google Scholar]
- 61. Cilensek ZM, Yehiely F, Kular RK, Deiss LP. A member of the GAGE family of tumor antigens is an anti‐apoptotic gene that confers resistance to Fas/CD95/APO‐1, Interferon‐gamma, taxol and gamma‐irradiation. Cancer Biol Ther 2002; 1: 380–7. [PubMed] [Google Scholar]
- 62. Duan Z, Duan Y, Lamendola DE et al. Overexpression of MAGE/GAGE genes in paclitaxel/doxorubicin‐resistant human cancer cell lines. Clin Cancer Res 2003; 9: 2778–85. [PubMed] [Google Scholar]
- 63. Cho HJ, Caballero OL, Gnjatic S et al. Physical interaction of two cancer‐testis antigens, MAGE‐C1 (CT7) and NY‐ESO‐1 (CT6). Cancer Immun 2006; 6: 12. [PubMed] [Google Scholar]
- 64. Meuwissen RL, Meerts I, Hoovers JM, Leschot NJ, Heyting C. Human synaptonemal complex protein 1 (SCP1): isolation and characterization of the cDNA and chromosomal localization of the gene. Genomics 1997; 39: 377–84. [DOI] [PubMed] [Google Scholar]
- 65. Hamer G, Gell K, Kouznetsova A, Novak I, Benavente R, Hoog C. Characterization of a novel meiosis‐specific protein within the central element of the synaptonemal complex. J Cell Sci 2006; 119: 4025–32. [DOI] [PubMed] [Google Scholar]
- 66. Depa‐Martynow M, Kempisty B, Lianeri M, Jagodzinski PP, Jedrzejczak P. Association between fertilin beta, protamines 1 and 2 and spermatid‐specific linker histone H1‐like protein mRNA levels, fertilization ability of human spermatozoa, and quality of preimplantation embryos. Folia Histochem Cytobiol 2007; 45 (Suppl 1): S79–85. [PubMed] [Google Scholar]
- 67. Aoki VW, Liu L, Jones KP et al. Sperm protamine 1/protamine 2 ratios are related to in vitro fertilization pregnancy rates and predictive of fertilization ability. Fertil Steril 2006; 86: 1408–15. [DOI] [PubMed] [Google Scholar]
- 68. Robert M, Gagnon C. Semenogelin I: a coagulum forming, multifunctional seminal vesicle protein. Cell Mol Life Sci 1999; 55: 944–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Zhao H, Lee WH, Shen JH, Li H, Zhang Y. Identification of novel semenogelin I‐derived antimicrobial peptide from liquefied human seminal plasma. Peptides 2008; 29: 505–11. [DOI] [PubMed] [Google Scholar]
- 70. Ait‐Tahar K, Liggins AP, Collins GP et al. Cytolytic T‐cell response to the PASD1 cancer testis antigen in patients with diffuse large B‐cell lymphoma. Br J Haematol 2009; 146: 396–407. [DOI] [PubMed] [Google Scholar]
- 71. Xing Q, Pang XW, Peng JR et al. Identification of new cytotoxic T‐lymphocyte epitopes from cancer testis antigen HCA587. Biochem Biophys Res Commun 2008; 372: 331–5. [DOI] [PubMed] [Google Scholar]
- 72. Frank C, Hundemer M, Ho AD, Goldschmidt H, Witzens‐Harig M. Cellular immune responses against the cancer‐testis antigen SPAN‐XB in healthy donors and patients with multiple myeloma. Leuk Lymphoma 2008; 49: 779–85. [DOI] [PubMed] [Google Scholar]
- 73. Jager E, Stockert E, Zidianakis Z et al. Humoral immune responses of cancer patients against “Cancer‐Testis” antigen NY‐ESO‐1: correlation with clinical events. Int J Cancer 1999; 84: 506–10. [DOI] [PubMed] [Google Scholar]
- 74. Atanackovic D, Altorki NK, Cao Y et al. Booster vaccination of cancer patients with MAGE‐A3 protein reveals long‐term immunological memory or tolerance depending on priming. Proc Natl Acad Sci U S A 2008; 105: 1650–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mischo A, Kubuschok B, Ertan K et al. Prospective study on the expression of cancer testis genes and antibody responses in 100 consecutive patients with primary breast cancer. Int J Cancer 2006; 118: 696–703. [DOI] [PubMed] [Google Scholar]
- 76. Eichmuller S, Usener D, Thiel D, Schadendorf D. Tumor‐specific antigens in cutaneous T‐cell lymphoma: expression and sero‐reactivity. Int J Cancer 2003; 104: 482–7. [DOI] [PubMed] [Google Scholar]
- 77. Jager E, Karbach J, Gnjatic S et al. Recombinant vaccinia/fowlpox NY‐ESO‐1 vaccines induce both humoral and cellular NY‐ESO‐1‐specific immune responses in cancer patients. Proc Natl Acad Sci U S A 2006; 103: 14453–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Marchand M, Van Baren N, Weynants P et al. Tumor regressions observed in patients with metastatic melanoma treated with an antigenic peptide encoded by gene MAGE‐3 and presented by HLA‐A1. Int J Cancer 1999; 80: 219–30. [DOI] [PubMed] [Google Scholar]
- 79. Lehmann F LJ, Gaulis S, Gruselle O, Brichard V. Clinical Response to the MAGE‐A3 Immunotherapeutic in Metastatic Melanoma Patients is associated with a Specific Gene Profile Present Prior to Treatment. XVIth meeting in the Cancer Research Institute International Cancer Immunotherapy Symposium Series and the 2008 Meeting of the Cancer Vaccine Consortium. New York City, 2008; S25. [Google Scholar]
- 80. Davis ID, Chen W, Jackson H et al. Recombinant NY‐ESO‐1 protein with ISCOMATRIX adjuvant induces broad integrated antibody and CD4(+) and CD8(+) T cell responses in humans. Proc Natl Acad Sci U S A 2004; 101: 10697–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nicholaou T, Ebert LM, Davis ID et al. Regulatory T‐cell‐mediated attenuation of T‐cell responses to the NY‐ESO‐1 ISCOMATRIX vaccine in patients with advanced malignant melanoma. Clin Cancer Res 2009; 15: 2166–73. [DOI] [PubMed] [Google Scholar]
- 82. Harada N, Hoshiai K, Takahashi Y et al. Preclinical safety pharmacology study of a novel protein‐based cancer vaccine CHP‐NY‐ESO‐1. Kobe J Med Sci 2008; 54: E23–34. [PubMed] [Google Scholar]
- 83. Valmori D, Souleimanian NE, Tosello V et al. Vaccination with NY‐ESO‐1 protein and CpG in Montanide induces integrated antibody/Th1 responses and CD8 T cells through cross‐priming. Proc Natl Acad Sci U S A 2007; 104: 8947–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Odunsi K, Qian F, Matsuzaki J et al. Vaccination with an NY‐ESO‐1 peptide of HLA class I/II specificities induces integrated humoral and T cell responses in ovarian cancer. Proc Natl Acad Sci U S A 2007; 104: 12837–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gnjatic S, Altorki NK, Tang DN et al. NY‐ESO‐1 DNA vaccine induces T‐cell responses that are suppressed by regulatory T cells. Clin Cancer Res 2009; 15: 2130–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Nishikawa H, Sato E, Briones G et al. In vivo antigen delivery by a Salmonella typhimurium type III secretion system for therapeutic cancer vaccines. J Clin Invest 2006; 116: 1946–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yuan J, Gnjatic S, Li H et al. CTLA‐4 blockade enhances polyfunctional NY‐ESO‐1 specific T cell responses in metastatic melanoma patients with clinical benefit. Proc Natl Acad Sci U S A 2008; 105: 20410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kim J, Reber HA, Hines OJ et al. The clinical significance of MAGEA3 expression in pancreatic cancer. Int J Cancer 2006; 118: 2269–75. [DOI] [PubMed] [Google Scholar]
- 89. Riener MO, Wild PJ, Soll C et al. Frequent expression of the novel cancer testis antigen MAGE‐C2/CT‐10 in hepatocellular carcinoma. Int J Cancer 2009; 124: 352–7. [DOI] [PubMed] [Google Scholar]
- 90. Atanackovic D, Luetkens T, Hildebrandt Y et al. Longitudinal analysis and prognostic effect of cancer‐testis antigen expression in multiple myeloma. Clin Cancer Res 2009; 15: 1343–52. [DOI] [PubMed] [Google Scholar]
- 91. Condomines M, Quittet P, Lu ZY et al. Functional regulatory T cells are collected in stem cell autografts by mobilization with high‐dose cyclophosphamide and granulocyte colony‐stimulating factor. J Immunol 2006; 176: 6631–9. [DOI] [PubMed] [Google Scholar]
- 92. Dhodapkar MV, Osman K, Teruya‐Feldstein J et al. Expression of cancer/testis (CT) antigens MAGE‐A1, MAGE‐A3, MAGE‐A4, CT‐7, and NY‐ESO‐1 in malignant gammopathies is heterogeneous and correlates with site, stage and risk status of disease. Cancer Immun 2003; 3: 9. [PubMed] [Google Scholar]
- 93. Yakirevich E, Sabo E, Lavie O, Mazareb S, Spagnoli GC, Resnick MB. Expression of the MAGE‐A4 and NY‐ESO‐1 cancer‐testis antigens in serous ovarian neoplasms. Clin Cancer Res 2003; 9: 6453–60. [PubMed] [Google Scholar]
- 94. Velazquez EF, Jungbluth AA, Yancovitz M et al. Expression of the cancer/testis antigen NY‐ESO‐1 in primary and metastatic malignant melanoma (MM) – correlation with prognostic factors. Cancer Immun 2007; 7: 11. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The cancer/testis antigens.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


