Abstract
Pirh2 (p53‐induced RING‐H2) is an E3 ubiquitin ligase that can target p53 for degradation and thereby repress a diverse group of biological activities regulated by p53. Notably, Pirh2, rather than MDM2, is the primary degrader of active p53 under conditions of DNA damage. Moreover, Pirh2 is highly expressed in multiple cancer cell lines regardless of p53 status. Recent research has shown that Pirh2 is involved in many signalling pathways related to the genesis and evolution of cancer. This review aims to summarize a comprehensive picture of the role of Pirh2 in cellular processes and its significance to tumorigenesis. Furthermore, this review focuses on its potential role as a cancer therapeutic target. (Cancer Sci 2011; 102: 909–917)
The tumor suppressor p53, known as “the guardian of the genome”, plays a key role in eliciting cellular responses to many signals of cell stress. Upon activation, p53 can affect cellular functions including transcription, DNA synthesis and repair, cell cycle arrest, senescence and apoptosis. Recent studies have demonstrated that p53 also influences metabolism and angiogenesis, regulates cell motility and immune responses and mediates cell–cell communication.( 1 ) By promoting cell cycle arrest, apoptosis, senescence and DNA repair, p53 helps prevent cancer development.( 2 , 3 ) The importance of p53 in tumor suppression is highlighted by the abundant, inactivating, somatic p53 mutations that are found in more than 50% of human cancer cells.( 4 ) p53 is subjected to a variety of post‐translational modifications, including phosphorylation, acetylation, ubiquitylation and methylation.( 5 , 6 ) Ubiquitylation is an important mechanism for controlling p53 activity.( 7 )
Pirh2, first identified as an androgen receptor N‐terminal‐interacting protein (ARNIP)( 8 ) and also known as ring finger and CHY zinc finger domain‐containing 1 (Rchy1), is a member of the RING finger family of E3 ubiquitin ligases, which can facilitate protein degradation via the ubiquitin‐proteasome pathway.( 9 ) The RING domain of Pirh2 contains a RING‐H2 (Cys3His2Cys3) motif, which is critical for maintaining the functions of several important E3 ubiquitin ligases including c‐Cbl, anaphase‐promoting complex (APC) and SCF complexes.( 10 , 11 , 12 ) The N‐terminal lobe of the Pirh2 N‐terminal domain is a member of the CHY zinc finger family (Fig. 1).( 13 ) Currently, the best understood function of Pirh2 is its role in the Pirh2–p53 feedback loop that is independent of MDM2. Pirh2 is transcriptionally activated by p53, and Pirh2, in turn, inhibits p53 activity in several ways. Notably, Pirh2 has been shown to degrade active p53 under conditions of DNA damage when MDM2 dissociates from and fails to degrade p53.( 14 ) In addition, the disruption of Pirh2 homeostasis is closely related to the formation of various human tumors (Table 1).( 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 ) Rchy1 is one of the key genes related to the locoregional recurrence control of breast cancer,( 23 , 24 ) and Pirh2 was elevated in approximately 93% of all murine lung tumors.( 19 , 25 ) Mouse models of Pirh2 functions have provided data suggesting that the overexpression of Pirh2 promotes tumorigenicity.( 26 , 27 ) Moreover, Pirh2 is highly expressed in multiple cancer cell lines regardless of p53 status, in a manner distinct from MDM2.( 28 ) Therefore, Pirh2 not only promotes degradation of p53 but also interacts with other proteins to function as an oncoprotein. Recent studies showed that Pirh2 is involved in a wide array of cellular signalling pathways; most of these are related to DNA damage signalling, which is closely linked to the genesis and evolution of cancer. These data suggest that Pirh2 could be a novel oncoprotein and a promising target of cancer therapy. In this review, we highlight the recent findings on the functions of Pirh2 in cells and focus on its role as a novel oncoprotein.
Figure 1.
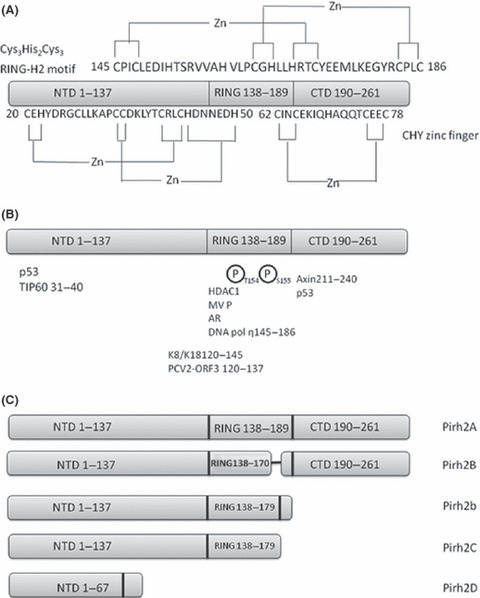
Schematic models depict Pirh2. The numbers in the figure represent the amino acid residues corresponding to full‐length Pirh2. (A) The CHY zinc finger domain and RING‐H2 domain structure. (B) Part of the Pirh2‐interacting proteins are indicated. (C) Schematic illustration of full‐length Pirh2 (Pirh2A) and Pirh2 isoforms. The Pirh2B isoform lacks amino acids 171–179, whereas Pirh2C misses C‐terminal amino acids 180–261. Pirh2b is a 188‐residue protein with 179 amino acids corresponding to Pirh2C and nine unique amino acids at the carboxyl‐terminal end. Pirh2D harbors 67 amino‐terminal amino acids identical to those in Pirh2A and has eight additional unique amino acids at the carboxyl‐terminal end.
Table 1.
Overview of the overexpression of pirh2 in human tumors investigated
Pirh2: rising from the shadow of MDM2
The RING‐finger‐containing oncoprotein MDM2 is the first and best characterised ubiquitin ligase that antagonises p53.( 29 , 30 ) Since 1992, numerous scientific studies have characterised this E3 ubiquitin ligase. This extensive focus on MDM2 is partly due to its function as an antagonist of the tumor suppressor protein p53. MDM2 can both degrade p53 and quench its activity by concealing its transactivation domain.( 31 ) Several lines of evidence support the role of MDM2 as an important regulator of a variety of fundamental cellular processes; the best characterised and understood is the MDM2–p53 feedback loop.( 32 ) Although 13 additional ubiquitin ligases have been shown to mediate p53 ubiquitylation, MDM2 is considered to be the most efficient protein to antagonise p53 activity.( 7 ) However, MDM2 dissociates from p53 after DNA damage.( 14 , 33 ) Furthermore, previous reports showed that MDM2 mainly regulates the basal levels of p53, and MDM2 is the main regulator of p53 in unstressed conditions.( 34 , 35 , 36 ) The phosphorylation of serine 15 on p53 is sufficient to prevent its degradation by MDM2, and the overexpression of MDM2 does not prevent the activation of p53.( 14 )
Tai showed that Pirh2 can target the serine 15 phosphorylated form of p53 for degradation even though this form does not appear to be regulated by MDM2.( 14 ) Serine 15 is one of two serines in p53 that is phosphorylated following DNA damage (induced by ionising or ultraviolet irradiation) in multiple cell types.( 37 ) Serine 15 is a key phosphorylation target during the p53 activation process and is critical for p53‐dependent transactivation.( 38 ) Mouse models showed that the phosphorylation of serine 18 (human serine 15) was required for robust p53‐mediated apoptosis.( 39 ) Pirh2 knockout mice are viable, develop normally and show no difference in the basal level of p53 compared with the wild type in unstressed conditions; however, these mice display elevated levels of phosphoserine 15 p53 after DNA damage.( 14 ) Among the ubiquitin ligases that target p53 for ubiquitin‐mediated proteolysis, only Pirh2 and CARP are reported to target p53 for degradation after DNA damage. CARP1/2 targets phosphoserine 20 of p53 for degradation. This form of p53 is active, is only present under conditions of double strand breaks and can only be induced by checkpoint kinase 2 (Chk2).( 40 , 41 ) Based on these data, Tai proposed that the role of Pirh2 in regulating p53 might be to fine‐tune the DNA damage response (Fig. 2). This could explain why Pirh2 promoted limited degradation of p53 in some cell lines( 42 , 43 ) and p53 did not accumulate in R1 ES cell lines, even though a significant reduction of Pirh2 abundance was detected after the transfection of Pirh2‐siRNA.( 44 )
Figure 2.
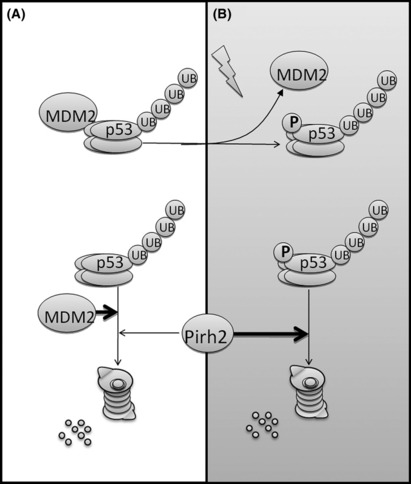
Schematic diagram illustrating the regulation of p53 by murine double minute 2 protein (MDM2) and Pirh2. (A) Under unstressed conditions, MDM2 is the main regulator of p53. (B) MDM2 and p53 are phosphorylated after DNA damage. MDM2 dissociates from p53, and Pirh2 becomes a main regulator of active p53. Adapted from Tai,( 14 ) with permission. UB, ubiquitin, p, phosphorylation.
Duan et al. ( 28 ) also showed that Pirh2 levels were not affected by the presence of wild‐type p53 in the detected cancer cells and were elevated in p53(−/−) cells under conditions of DNA damage. In contrast, MDM2 was upregulated by wild‐type p53 in detected cancer cells and was absent from the p53(−/−) cells.( 28 ) As mentioned above, Pirh2 is highly expressed in multiple cancer cell lines regardless of p53 status, and previous studies have shown that the MDM2 amplification and p53 mutation were mutually exclusive.( 30 ) The fact that Pirh2 overexpression is decoupled from the p53 mutational status is another important distinction between Pirh2 and MDM2. Therefore, in addition to targeting activated p53 for degradation, Pirh2 is perhaps a “key player” of another p53‐independent pathway to promote the genesis of cancer. Here, we describe the functions of Pirh2 in both a p53‐dependent and p53‐independent manner.
Functions of Pirh2
p53‐dependent functions of Pirh2. Leng et al. ( 9 ) were the first to show that Pirh2 can directly bind to p53 in vitro and in vivo. Pirh2 catalyses the ubiquitylation of tetrameric p53 primarily through the interaction between the C‐terminal domain residues 249–256 of Pirh2 and the tetramerisation domain of p53. The disruption of this interaction could represent a potential target for some cancer therapies. Additionally, a much weaker interaction was detected between the DNA binding domain of p53 and the N‐terminal domain of Pirh2.( 13 ) Intron 3 of Pirh2 has been shown to contain a p53 binding site. Pirh2 can repress p53‐dependent transactivation, and p53 transactivates Pirh2, which can target p53 for ubiquitin‐proteasome degradation; therefore, a feedback loop exists (Fig. S1).( 9 ) The interaction inhibits apoptosis and the growth‐inhibiting ability of p53, and it ultimately contributes to tumorigenicity, caused by the accumulation of genomic mutations that disrupt p53 homeostasis.( 9 , 27 ) Tight regulation of p53 levels is crucial for maintaining normal cell growth and preventing tumorigenesis. These data might explain why Pirh2 is overexpressed in many types of cancer cells. Sheng et al. ( 13 ) observed Pirh2‐mediated p53 ubiquitylation in vitro. However, Li et al. ( 26 ) showed that Pirh2 did not exhibit any E3 ubiquitin ligase activity towards p53 in vitro.
In response to sublethal DNA damage, Pirh2 also inhibits Axin‐HIPK2‐induced p53 phosphorylation of serine 46 by competing with HIPK2 for binding to Axin in a manner independent of the RING domain.( 26 ) An in vitro reconstitution assay showed that high levels of Pirh2 correlated with higher dissociation rates of HIPK2 from Axin. The competition results in a decreased level of activated p53, thus inhibiting p53‐induced apoptosis. The interaction was abolished by Tat‐interactive protein of 60 kDa (Tip60) because an Axin‐Tip60‐HIPK2‐p53 complex was formed when a lethal treatment was administered to reverse the Pirh2‐Axin‐induced p53 inactivation; this allowed for the maximal activation of p53, which triggered apoptosis (Fig. S1). Because Pirh2 and Tip60 can bind to the same sites of Axin, it was inferred that the two are in competition. ATM and ATR are also involved in this process by promoting the assembly of the Axin–Tip60 complex.( 26 ) DNA polymerase η, a Y‐family DNA polymerase that plays an important role in translesion DNA synthesis (TLS) through UV‐induced cyclobutane pyrimidine dimers and photoproducts, can be targeted for proteasomal degradation by Pirh2.( 45 , 46 , 47 ) DNA polymerase η might activate p53 in an ATM‐dependent manner, therefore activating p53 via the phosphorylation of p53 serine 15.( 48 ) DNA polymerase η is also involved in the regulation of ATM activity towards Chk2, which can phosphorylate p53 on serine 20 and promote its stability; this leads to cell cycle arrest and provides the cell with extra time to repair its DNA.( 48 ) Cells deficient in DNA polymerase η are hypersensitive to UV‐induced cell death. Pirh2‐mediated proteasomal degradation of DNA polymerase η might play a role in UV‐induced cancer formation.( 45 )
In addition to directly ubiquitylating p53 and targeting it for degradation, Pirh2 can also act synergistically with three additional major negative regulators of p53, MDM2, MDMX and COP1, to inhibit p53‐mediated transcriptional activity (Fig. S1).( 49 , 50 , 51 , 52 , 53 ) MDM2 and COP1 can both dramatically increase Pirh2 protein levels, and the overexpression of Pirh2 leads to a substantial increase in MDM2 and MDMX levels and a milder increase in COP1 levels.( 50 ) Pirh2 can also increase MDM2 levels indirectly by targeting the SCY1‐like 1 binding protein 1 (SCYL1‐BP1) for proteasomal degradation, which can accelerate MDM2 self‐ubiquitylation.( 54 )
Notably, Pirh2 can also degrade histone deacetylases 1 (HDAC1), which can inactivate p53 transcriptional activity. Recent biochemical studies revealed that Pirh2 can ubiquitylate HDAC1 and reduce HDAC1 levels, thus reducing the repressive activity of HDAC1 on transcription (Fig. S1).( 18 ) Previous reports have shown that acetylation controls p53 stability by potentially interfering with MDM2‐mediated ubiquitylation.( 55 , 56 ) The recruitment of HDAC1 by MDM2 promotes p53 degradation by removing these acetyl groups.( 57 ) HDAC1 also repressed the transcriptional activation of p53 and p21.( 58 , 59 ) Moreover, HDAC1 is involved in the repression of the E2F1 transcription factor that determines the timely expression of many genes that are required for entry into and progression through S phase of the cell cycle.( 60 ) E2F1 also indirectly regulates the levels and activity of p53.( 61 ) Based on these data, Pirh2 has a dual role as a tumor suppressor and as an oncoprotein. Additionally, the ubiquitin modification has been shown to promote the transcriptional activity of some transcription factors,( 62 , 63 ) so the ubiquitylation of HDAC1 might also promote its transcriptional repression of p53, p21 and E2F1. It is unclear whether HDAC1‐mediated deacetylation of p53 is essential for Pirh2 to facilitate p53 degradation.
p53‐independent functions of Pirh2. Under the stress of DNA damage, cells first initiate cell cycle arrest to make time for DNA repair.( 64 ) p27Kip1 can inhibit the activation of cyclinE‐CDK2 and cyclinA‐CDK2, which can activate the transcription of genes that are required for the G1‐S transition.( 65 , 66 ) Pirh2 is a key regulator of p27Kip1 (Fig. S1) and degrades p27Kip1 late in the G1 phase in a ubiquitin‐proteasome‐dependent manner.( 67 ) These data are in agreement with the fact that the Pirh2 expression level is low in G0 and early G1 phases and gradually increases toward the S phase in a cell cycle‐dependent manner. Skp2 and the KPC are able to ubiquitylate p27Kip1 for degradation. However, neither protein can degrade p27Kip1 at the G1‐S transition.( 68 , 69 ) High expression levels of Pirh2 are associated with low expression levels of p27 and a poor prognosis in head and neck cancers( 17 ) and lung cancer.( 20 ) Pirh2 might also affect the function of p21, another important cell cycle‐dependent kinase inhibitor. p21 is targeted by p53 and can be transactivated by p53. Transfection studies have shown that p21 accumulated in Pirh2 knockdown H1299 (p53−/−) cells.( 45 ) p21 can be targeted by several E3 ubiquitin ligases for degradation,( 70 ) but whether Pirh2 can directly regulate p21 warrants further investigation.
Cells are targeted for apoptosis if DNA damage is too severe for recovery. Pirh2 can inhibit the apoptosis pathway by binding to Kertain8/18 (K8/18); the phosphorylation of either target leads to apoptosis.( 71 ) Duan et al. showed that the Pirh2‐K8/18 association is significant for cells to maintain the K8/18 filament and that the mitochondria in these cells are normal (Fig. S1).( 71 ) Mitochondria play a key role in controlling cell life and death by releasing cytochrome c into the cytoplasm; cytochrome c is a primary activator of the caspase cascade and its release activates the apoptotic process.( 72 ) K8/18 is preferentially bound to unphosphorylated Pirh2 in the cytoplasm. The disruption of this association sensitises the cells to UV‐induced apoptosis. This process is caused, in part, by enhancing the release of pro‐apoptotic proteins, such as cytochrome c and Smac/DIABLO, from the mitochondria to the cytoplasm.( 71 ) This partially explains why Pirh2 mainly exists in its unphosphorylated form in most tumor cell lines. Pancreatic tumor epithelial cells contained an increased level of phosphorylated K8/18,( 73 ) and another group showed that K8/18 was hyperphosphorylated after an apoptotic challenge.( 74 ) These data suggest that the phosphorylation of K8/18 by JNK or p38 leads to morphological changes in the mitochondria, caused by the dissociation of Pirh2‐K8/18.( 71 , 73 , 74 ) Studies also revealed that K8/18 can modulate the cellular response to specific pro‐apoptotic signals. Moreover, it is involved in resisting TNF‐ and Fas‐induced cytotoxicity or apoptosis.( 75 , 76 ) Until now, it was unclear whether the Pirh2‐K8/18 interaction was involved in these processes.
Although it was determined long ago that Pirh2 and p73 interacted( 9 ) and Pirh2 could not directly degrade p73,( 43 ) no new information about this interaction has emerged. p73 can induce cell apoptosis by functioning with many cofactors in both a p53‐dependent and p53‐independent manner.( 77 ) Therefore, it is worthwhile to further investigate the relationship between them.
It has been shown that Pirh2 inhibits the androgen‐dependent secretion of prostate‐specific antigen (PSA); this implies that Pirh2 can negatively regulate protein secretion.( 78 ) Previous studies have shown that some cellular proteins can utilise ubiquitin modification as their targeting signal.( 79 ) Pirh2 ubiquitylates the signal recognition particle receptor β (SR β), which is a subunit of the signal recognition particle receptor (SR) heterodimer composed of SR α and SR β.( 80 ) SR β contains a transmembrane segment, providing the membrane anchor for SR α. SR β also plays an important role in both the assembly and disassociation of SR subunits, which are essential for protein transportation.( 81 , 82 , 83 ) The SR is located on the endoplasmic reticulum membrane and can associate with a signal recognition particle that recognises the N‐terminal hydrophobic signal sequence of the secretory protein; this process targets the ribosome‐nascent chain complex to the endoplasmic reticulum (ER). Once a protein has passed the ER quality control process it is transported to the Golgi. The continuous loss of material at the trans‐face of the ER is antagonised by retrograde cargo retrieval to the ER, which is predominantly mediated by the coatomer complex. Pirh2 can promote the ubiquitylation of ε‐COP, a subunit of COPI, and target it for degradation.( 78 ) Because ε‐COP plays an important role in the assembly and disassembly of COPI, the ubiquitylation and degradation of ε‐COP can block the normal transportation of secretory proteins (Fig. S1).( 78 ) Notably, the overexpression of Pirh2 causes a morphological change in the trans‐Golgi network.( 78 ) Previous studies have shown that K8/18, another Pirh2‐interacting protein, helps to orchestrate the positioning and function of Golgi and protein secretion.( 84 ) Further studies will help elucidate this process. Finally, it is worth noting that knockdown of Rchy1 specifically downregulated epidermal growth factor (EGF) internalisation.( 85 )
Pirh2, which was first identified as an ARNIP,( 8 ) was able to enhance androgen receptor (AR) signalling by inhibiting HDAC1’s repressive activity towards AR.( 18 ) Pirh2 can inhibit the AR N‐C terminal interaction, which helps determine its transcriptional activity by regulating the androgen dissociation rate and the efficient recruitment of coactivators.( 86 , 87 , 88 ) Although two groups showed that Pirh2 upregulates AR‐mediated transcription of the PSA target gene,( 18 , 78 ) Beitel et al. ( 8 ) reported that Pirh2 does not significantly affect AR transcriptional activation. Because AR signalling plays an important role in prostate cancer and Pirh2 overexpression was detected in prostate cancer cells, Pirh2 might contribute to prostate cancer formation via the AR signalling pathway.( 89 )
Regulation of Pirh2 activity
Phosphorylation and ubiquitylation are the best understood post‐translational modifications of Pirh2. When Calmodulin‐dependent kinase II (CaMKII) is at its maximal activity level, it hyperphosphorylates Pirh2 in the G2/M phase of the cell cycle at residues threonine 154 and serine 155.( 27 ) The phosphorylation of Pirh2 enhances its self‐ubiquitylation, which causes the phosphorylated form of Pirh2 to be more unstable than the unphosphorylated form. Phosphorylated Pirh2 mainly exists in the cytoplasm, and the Pirh2–p53 interaction putatively occurs in the nucleus. Because p53 preferentially interacts with the unphosphorylated form of Pirh2, the phosphorylation of Pirh2 by CaMKII impairs its E3 ubiquitin ligase activity towards p53. In normal cells, Pirh2 primarily exists in the phosphorylated form, but in tumor cells and tissues, Pirh2 mainly exists in the unphosphorylated form.( 27 ) These data could explain why the levels of CaMKII are decreased in tumor cells.( 90 )
Pirh2 is a short‐lived protein and is rapidly degraded by ubiquitin‐dependent proteolysis.( 91 ) Pirh2 can act as an E3 ubiquitin ligase on itself, resulting in autoubiquitylation and proteasomal degradation; some Pirh2‐interacting proteins have been shown to influence the autoubiquitylation activity of Pirh2.( 92 ) The mutation of a single cysteine residue (Cys145 or Cys164) in the RING‐H2 domain abolished this E3 ubiquitin ligase activity.( 8 , 25 , 26 , 78 ) The fact that Pirh2 containing a mutant RING domain can still be ubiquitylated indicates that Pirh2 can be both autoubiquitylated and ubiquitylated by an unidentified E3 ubiquitin ligase.( 91 ) Both Pleomorphic Adenoma Gene like 2 (PLAGL2) and Tip60 can promote Pirh2 stability by inhibiting its ubiquitin‐dependent degradation.( 91 , 93 ) PLAGL2 is an oncoprotein involved in many malignancies.( 94 , 95 ) Tip60 can also target p53 into the nucleus, where Pirh2 mainly interacts with p53 and stabilises Pirh2.( 91 ) EGR1 can induce the expression of Pirh2, thus decreasing the p53 levels.( 96 , 97 ) Measles virus phosphoprotein and porcine circovirus type 2 open reading frame 3 are two pathogenic proteins that affect the stability of Pirh2.( 98 , 99 , 100 )
For proteins targeted for ubiquitin‐proteasomal destruction, cellular localisation is an important mechanism that can regulate protein stability. This is the case for p53, for which nuclear export is essential for its degradation by MDM2.( 101 ) Pirh2 displays a diffuse nuclear and cytoplasmic subcellular localisation.( 26 , 27 , 71 , 91 , 92 , 97 , 98 ) The subcellular localisation of Pirh2 is critical for its regulation and stability. Pirh2 primarily meditates the polyubiquitylation of p53 in the nucleus; as a result, p53 is exported to the cytoplasm and subsequently removed via proteasomal degradation.( 27 ) Measles virus phosphoprotein, porcine circovirus type 2 open reading frame 3 and the presence of androgens can recruit Pirh2 into the cytoplasm.( 98 , 99 , 100 ) In addition, NTKL‐BP1 and ARF4 can interact with and colocalise with Pirh2 in the cytoplasm surrounding the nucleus.( 102 , 103 ) Tip60 can induce the migration of Pirh2 to the nucleus,( 91 ) whereas phosphorylation of Pirh2 leads to its migration to the cytoplasm.( 27 )
Pirh2 as a promising target for cancer therapy
Pirh2 was overexpressed in several kinds of cancers (Table 1).( 15 , 16 , 17 , 18 , 19 , 20 ) Increasing levels of Pirh2 were correlated with poor survival in patients with hepatocellular carcinoma.( 16 ) Furthermore, high expression of Pirh2 was associated with a poor prognosis in head and neck cancers.( 17 ) Increased Pirh2 expression affects lung tumorigenesis by reducing p53 activity.( 19 ) Another group showed that Pirh2 shRNA mediated by the psiRNA‐hH1 vector plasmid effectively inhibits the proliferation of lung carcinoma cells.( 104 ) Pirh2 is also involved in the measles virus’ association with lung cancer and the human papillomavirus’ association with cervical cancer.( 105 , 106 ) Moreover, Pirh2 can regulate AR activity and PSA secretion, which are tightly associated with prostate cancer.( 18 , 78 ) Importantly, the overexpression of Pirh2 is also detected in prostate cancer cells.( 18 ) Pirh2 can also account for the correlation of the overexpression of PRL‐1 and PRL‐3 with tumorgenesis in various cancer cells.( 96 , 97 , 107 , 108 ) Because abundant inactivating, somatic mutations of p53 are found in more than 50% of human cancer cells, Pirh2 might be an important regulator of p53 during DNA damage. The association between Pirh2 and p53 is well characterised at the structural and biological levels; inhibiting the Pirh2–p53 interaction is a promising approach for activating p53.( 13 , 109 , 110 ) In summary, several lines of evidence strongly suggest that Pirh2 positively regulates cell cycle progression and tumor growth. These findings also indicate that Pirh2 might be a novel prognostic marker for some types of cancer.
Inflammation plays a crucial role in tumorigenesis, and some of the underlying molecular mechanisms have been elucidated.( 111 , 112 ) Pirh2 might also impact the immune system to promote tumor growth. Pirh2 can target DNA polymerase η for ubiquitin‐proteasome degradation; this is involved in the diversification of immunoglobulin genes at different stages of the B cell differentiation pathway, known as somatic hypermutation (SHM).( 113 ) Pirh2 may regulate RhoA activation via p27Kip1.( 114 ) The inhibition of RhoA decreases B cell proliferation and T cell differentiation.( 115 ) Pirh2 could also promote tumorigenicity by interfering with the normal immune responses and may be involved in the genesis of lymphoma. Therefore, Pirh2 could be a promising candidate as a molecular target for cancer therapy. The use of molecule inhibitors to block the action of Pirh2 has not been previously reported.
Future perspectives
Previous data showed that Pirh2 serves as an important regulator in tumorigenicity (Table 2). Pirh2 is functionally connected to a broad range of cellular signalling pathways with important functions in cell proliferation, apoptosis, AR signalling and protein secretion (Fig. 3). In response to DNA damage in cells, Pirh2 operates in a manner distinct from MDM2. Under unstressed conditions, MDM2 is the primary regulator of p53. MDM2 and p53 are phosphorylated after DNA damage, and MDM2 dissociates from p53, whereas Pirh2 becomes a primary regulator of active p53. Another important difference between Pirh2 and MDM2 is that Pirh2 overexpression is decoupled from the mutational status of p53. The involvement of Pirh2 in tumor genesis is extensive. Numerous activities of Pirh2 affect tumor growth, both in a p53‐dependent and p53‐independent manner.
Table 2.
Pirh2‐interacting proteins with a proven or putative role in tumorigenesis
| Protein | Function | References |
|---|---|---|
| AR | Mediates androgen action | 122, 123 |
| ARF4 | Cooperates with other ADP‐ribosylation factors | 124 |
| Axin | A major scaffold for many signalling pathways, including mTOR, JNK MAPK, parathyroid hormone, Wnt and p53 signalling | 26, 125, 126, 127, 128 |
| CaMKII | Regulates neurite outgrowth and adipogenesis and cell cycle | 90 |
| COP1 | Mediates the ubiquitylation and degradation of c‐Jun and p53 | 51, 129, 130 |
| DNA polymerase η | A Y‐family DNA polymerase that plays an important role in Translesion DNA synthesis and somatic hypermutation | 131 |
| ε‐COP | Participates in assembly and disassembly of COPI | 132, 133, 134 |
| HDAC1 | A histone deacetyltransferase that actively reduces the level of histone acetylation | 135 |
| K8/18 | A major component of intermediate filaments, modulating cellular response to apoptotic stimuli | 75, 76 |
| MDM2 | An E3 ubiquitin ligase for p53 | 49, 52 |
| MDMX | Inhibits p53‐mediated transcriptional activity | 53 |
| p27 | Suppresses cell cycle progression from G1‐S | 65, 66 |
| p53 | Transcription factor that is activated by multiple stress signals, including DNA damage, hypoxia and oncogene activation | 2, 3 |
| p73 | Member of the p53‐family of transcription factors that have tumor‐suppressive activity | 77, 136 |
| PKCδ | Regulates cell growth, differentiation and apoptosis. | 137 |
| PLAGL2 | A transcription factor with a DNA‐binding and transactivation domain | 138 |
| SCYL1‐BP1 | Accelerates MDM2 self‐ubiquitylation | 54 |
| SRβ | A subunit of SR | 81, 82, 83 |
| Tip60 | A histone acetyltransferase that has a role in gene activation | 139, 140 |
ARF4, ADP‐ribosylation factor 4; COP1, constitutive photomorphogenesis protein 1; COPI, coatomer complex I; HDAC1, histone deacetylases 1; MDM2, murine double minute 2 protein; MDMX, MDM4 p53 binding protein homolog; PKCδ, protein kinase Cδ; PLAGL2, Pleomorphic Adenoma Gene like 2; SCYL1‐BP1, SCY1‐like 1 binding protein 1; SR β, signal recognition particle receptor β; Tip60, Tat‐interactive protein of 60 kDa.
Figure 3.
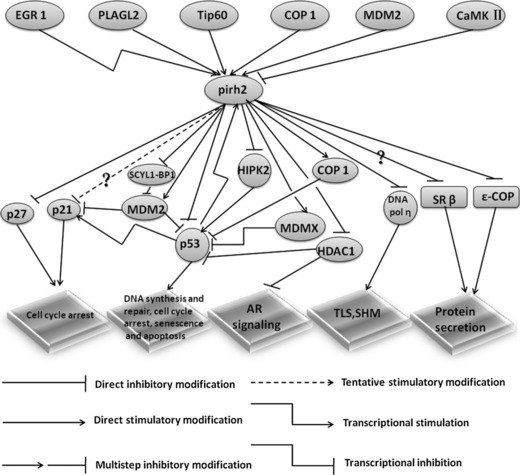
A non‐comprehensive list of recently described substrates and regulators of Pirh2. This model depicts the roles of Pirh2 in cellular signalling pathways and its key regulators. The Pirh2 levels and stability are tightly regulated in cells. AR, androgen receptor; COP1, constitutive photomorphogenesis protein 1; EGR1, early growth response 1; HDAC1, histone deacetylases 1; HIPK2, homeodomain‐interacting protein kinase 2; MDM2, murine double minute 2 protein; MDMX, MDM4 p53 binding protein homolog; PLAGL2, Pleomorphic Adenoma Gene like 2; SCYL1‐BP1, SCY1‐like 1 binding protein 1; SHM, somatic hypermutation; SR β, signal recognition particle receptor β; Tip60, Tat‐interactive protein of 60 kDa; TLS, translesion DNA synthesis.
Currently, the best understood function of Pirh2 is its role in a Pirh2‐p53 feedback loop. However, several questions concerning the function of Pirh2 in this pathway remain. Pirh2 can decrease the level of HDAC1, the activity of which is required for the degradation of p53 by MDM2.( 57 ) In this respect, Pirh2 plays dual roles. Pirh2 can enhance AR signalling through the inhibition of the AR corepressor HDAC1. Tip60 is an AR coactivator( 56 ) that can be ubiquitylated and degraded by both MDM2 and a p300‐associated E4 ubiquitin ligase, and possibly by Pirh2.( 116 , 117 ) Pirh2 might also target Tip60 for degradation.
Phosphorylation plays an important role in regulating protein activity; COP1 and MDM2 can be phosphorylated by ATM, which impedes the assembly of the Axin–Pirh2 complex.( 36 , 118 ) It is not clear whether Pirh2 can be phosphorylated by ATM. The monoubiquitylation of p53 can contribute to mitochondrial p53 migration;( 119 ) it is worth investigating whether Pirh2 can monoubiquitylate p53. Pirh2 can also interact with protein kinase Cδ (PKCδ), but the nature of the interaction is currently unclear.( 14 ) Pirh2 has also been shown to interact with Gfi1b and Atrophin 1 in the yeast‐two‐hybrid assay.( 120 ) Logan et al. ( 18 ) showed that Pirh2 can be recruited to androgen response elements in the PSA gene in response to androgens. Just as zinc‐finger proteins can bind to DNA, Pirh2 might also be involved in blocking the DNA damage response by directing its binding to DNA to act as a transcriptional factor.( 121 ) Furthermore, identification of the Pirh2 degron will facilitate the identification of more Pirh2 interactors.
An ultimate goal of these studies is to translate them into clinical applications. As a novel oncoprotein involved in a diverse group of biological activities, Pirh2 might be a novel prognostic cancer marker and a new target of cancer therapeutics. The development of pharmaceutics can enable the discovery of small molecules that can inhibit the Pirh2 ligase activity.
Disclosure Statement
The authors have no conflict of interest.
Abbreviations
- ARF4
ADP‐ribosylation factor 4
- ATM
ataxia telangiectasia mutated
- ATR
ATM‐ and rad3‐related
- CARP
caspase 8/10‐associated RING proteins
- Cbl
casitas B‐lineage lymphoma
- COP1
constitutive photomorphogenesis protein 1
- COPI
coatomer complex I
- EGR1
early growth response 1
- Gfi1b
growth factor independent 1B transcription repressor
- HIPK2
homeodomain‐interacting protein kinase 2
- KPC
kip1 ubiquitylation‐promoting complex
- MDM2
murine double minute 2 protein
- MDMX
MDM4 p53 binding protein homolog
- NTKL‐BP1
N‐terminal kinase‐like protein–binding protein 1
- PRL
phosphatase of regenerating liver
- RhoA
Ras homologue gene family member A
- SCF
Skp1–Cul1–F‐box protein
- Skp2
S‐phase kinase‐associated protein 2
- Smac/DIABLO
second mitochondria‐derived activator of caspase/direct inhibitor of apoptosis (IAP) binding protein with low protein isoelectric point (PI)
Supporting information
Fig. S1. A comprehensive picture of the overall Pirh2‐involved cellular processes.
Supporting info item
Acknowledgments
This work was supported by the National Natural Sciences Foundation of China (No. 30870856 and 30970145), the Program for New Century Excellent Talents in Wuhan University by the Ministry of Education of China (No. NCET‐07‐0630), the Scientific Research Foundation for the Returned Overseas Scholars by the Ministry of Education of China, the Research Fund for the Doctoral Program of Higher Education of China (No. 20090141110010), National Program on Key Basic Research Project (973 Program, No. 2010CB529803) and the Fundamental Research Funds for the Central Universities of China (No. 3081002).
References
- 1. Menendez D, Inga A, Resnick MA. The expanding universe of p53 targets. Nat Rev Cancer 2009; 9: 724–37. [DOI] [PubMed] [Google Scholar]
- 2. Junttila MR, Evan GI. p53 – a Jack of all trades but master of none. Nat Rev Cancer 2009; 9: 821–9. [DOI] [PubMed] [Google Scholar]
- 3. Zuckerman V, Wolyniec K, Sionov RV, Haupt S, Haupt Y. Tumour suppression by p53: the importance of apoptosis and cellular senescence. J Pathol 2009; 219: 3–15. [DOI] [PubMed] [Google Scholar]
- 4. Whibley C, Pharoah PD, Hollstein M. p53 polymorphisms: cancer implications. Nat Rev Cancer 2009; 9: 95–107. [DOI] [PubMed] [Google Scholar]
- 5. Appella E, Anderson CW. Post‐translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem 2001; 268: 2764–72. [DOI] [PubMed] [Google Scholar]
- 6. Kruse JP, Gu W. SnapShot: p53 posttranslational modifications. Cell 2008; 133: 930–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lee JT, Gu W. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ 2010; 17: 86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beitel LK, Elhaji YA, Lumbroso R et al. Cloning and characterization of an androgen receptor N‐terminal‐interacting protein with ubiquitin‐protein ligase activity. J Mol Endocrinol 2002; 29: 41–60. [DOI] [PubMed] [Google Scholar]
- 9. Leng RP, Lin Y, Ma W et al. Pirh2, a p53‐induced ubiquitin‐protein ligase, promotes p53 degradation. Cell 2003; 112: 779–91. [DOI] [PubMed] [Google Scholar]
- 10. Joazeiro CA, Wing SS, Huang H, Leverson JD, Hunter T, Liu YC. The tyrosine kinase negative regulator c‐Cbl as a RING‐type, E2‐dependent ubiquitin‐protein ligase. Science 1999; 286: 309–12. [DOI] [PubMed] [Google Scholar]
- 11. Seol JH, Feldman RM, Zachariae W et al. Cdc53/cullin and the essential Hrt1 RING‐H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev 1999; 13: 1614–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zachariae W, Shevchenko A, Andrews PD et al. Mass spectrometric analysis of the anaphase‐promoting complex from yeast: identification of a subunit related to cullins. Science 1998; 279: 1216–9. [DOI] [PubMed] [Google Scholar]
- 13. Sheng Y, Laister RC, Lemak A et al. Molecular basis of Pirh2‐mediated p53 ubiquitylation. Nat Struct Mol Biol 2008; 15: 1334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tai E. Characterization of the E3 Ubiquitin Ligase Pirh2. Medical Biophysics. Toronto: University of Toronto, 2010, 124. [Cited 1 Sep 2010.] Available from URL: http://hdl.handle.net/1807/24889. [Google Scholar]
- 15. Zhang C, Tang L, Ouyang H, Ren L, Wu Y, G L. The significance Of Pirh2 mRNA expression in primary breast cancer. Chin J Gen Surg 2008; 17: 444–8. [Google Scholar]
- 16. Wang XM, Yang LY, Guo L, Fan C, Wu F. p53‐induced RING‐H2 protein, a novel marker for poor survival in hepatocellular carcinoma after hepatic resection. Cancer 2009; 115: 4554–63. [DOI] [PubMed] [Google Scholar]
- 17. Shimada M, Kitagawa K, Dobashi Y et al. High expression of Pirh2, an E3 ligase for p27, is associated with low expression of p27 and poor prognosis in head and neck cancers. Cancer Sci 2009; 100: 866–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Logan IR, Gaughan L, McCracken SR, Sapountzi V, Leung HY, Robson CN. Human PIRH2 enhances androgen receptor signaling through inhibition of histone deacetylase 1 and is overexpressed in prostate cancer. Mol Cell Biol 2006; 26: 6502–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duan W, Gao L, Druhan LJ et al. Expression of Pirh2, a newly identified ubiquitin protein ligase, in lung cancer. J Natl Cancer Inst 2004; 96: 1718–21. [DOI] [PubMed] [Google Scholar]
- 20. Su Y, Bai P, L Z. Expression and significance of Pirh2 and p27Kip1 in lung cancer. Chin J Histochem Cytochem 2007; 16: 336–9. [Google Scholar]
- 21. Peng D, Sheta EA, Powell SM et al. Alterations in Barrett’s‐related adenocarcinomas: a proteomic approach. Int J Cancer 2008; 122: 1303–10. [DOI] [PubMed] [Google Scholar]
- 22. Fang M, Toher J, Morgan M, Davison J, Tannenbaum S, Claffey K. Genomic differences between estrogen receptor (ER)‐positive and ER‐negative human breast carcinoma identified by single nucleotide polymorphism array comparative genome hybridization analysis. Cancer 2010; DOI: 10.1002/cncr.25770 [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cheng SH, Horng CF, West M et al. Genomic prediction of locoregional recurrence after mastectomy in breast cancer. J Clin Oncol 2006; 24: 4594–602. [DOI] [PubMed] [Google Scholar]
- 24. Nuyten DS, van de Vijver MJ. Using microarray analysis as a prognostic and predictive tool in oncology: focus on breast cancer and normal tissue toxicity. Semin Radiat Oncol 2008; 18: 105–14. [DOI] [PubMed] [Google Scholar]
- 25. Duan W, Villanlona‐calero MA. RCHY1 (ring finger and CHY zinc finger domain containing 1). Atlas Genet Cytogenet Oncol Haematol 2006. [Cited 1 Sep 2010.] Available from URL: http://atlasgeneticsoncology.org/Genes/RCHY1ID43012ch04q21.html. [Google Scholar]
- 26. Li Q, Lin S, Wang X et al. Axin determines cell fate by controlling the p53 activation threshold after DNA damage. Nat Cell Biol 2009; 11: 1128–34. [DOI] [PubMed] [Google Scholar]
- 27. Duan S, Yao Z, Hou D, Wu Z, Zhu WG, Wu M. Phosphorylation of Pirh2 by calmodulin‐dependent kinase II impairs its ability to ubiquitinate p53. The EMBO J 2007; 26: 3062–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duan W, Gao L, Wu X, Zhang Y, Otterson GA, Villalona‐Calero MA. Differential response between the p53 ubiquitin‐protein ligases Pirh2 and MdM2 following DNA damage in human cancer cells. Exp Cell Res 2006; 312: 3370–8. [DOI] [PubMed] [Google Scholar]
- 29. Momand J, Zambetti GP, Olson DC, George D, Levine AJ. The mdm‐2 oncogene product forms a complex with the p53 protein and inhibits p53‐mediated transactivation. Cell 1992; 69: 1237–45. [DOI] [PubMed] [Google Scholar]
- 30. Oliner JD, Kinzler KW, Meltzer PS, George DL, Vogelstein B. Amplification of a gene encoding a p53‐associated protein in human sarcomas. Nature 1992; 358: 80–3. [DOI] [PubMed] [Google Scholar]
- 31. Thut CJ, Goodrich JA, Tjian R. Repression of p53‐mediated transcription by MDM2: a dual mechanism. Genes Dev 1997; 11: 1974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Marine JC, Lozano G. Mdm2‐mediated ubiquitylation: p53 and beyond. Cell Death Differ 2010; 17: 93–102. [DOI] [PubMed] [Google Scholar]
- 33. Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage‐induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997; 91: 325–34. [DOI] [PubMed] [Google Scholar]
- 34. O’Leary KA, Mendrysa SM, Vaccaro A, Perry ME. Mdm2 regulates p53 independently of p19(ARF) in homeostatic tissues. Mol Cell Biol 2004; 24: 186–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mendrysa SM, McElwee MK, Michalowski J, O’Leary KA, Young KM, Perry ME. mdm2 Is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol Cell Biol 2003; 23: 462–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Maya R, Balass M, Kim ST et al. ATM‐dependent phosphorylation of Mdm2 on serine 395: role in p53 activation by DNA damage. Genes Dev 2001; 15: 1067–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Siliciano JD, Canman CE, Taya Y, Sakaguchi K, Appella E, Kastan MB. DNA damage induces phosphorylation of the amino terminus of p53. Genes Dev 1997; 11: 3471–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. The EMBO J 1999; 18: 7002–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sluss HK, Armata H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol Cell Biol 2004; 24: 976–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kruse JP, Gu W. Modes of p53 regulation. Cell 2009; 137: 609–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang W, Rozan LM, McDonald ER III et al. CARPs are ubiquitin ligases that promote MDM2‐independent p53 and phospho‐p53ser20 degradation. J Biol Chem 2007; 282: 3273–81. [DOI] [PubMed] [Google Scholar]
- 42. Kaeser MD, Pebernard S, Iggo RD. Regulation of p53 stability and function in HCT116 colon cancer cells. J Biol Chem 2004; 279: 7598–605. [DOI] [PubMed] [Google Scholar]
- 43. Wu L, Zhu H, Nie L, Maki CG. A link between p73 transcriptional activity and p73 degradation. Oncogene 2004; 23: 4032–6. [DOI] [PubMed] [Google Scholar]
- 44. Solozobova V, Blattner C. Regulation of p53 in embryonic stem cells. Exp Cell Res 2010; 316: 2434–46. [DOI] [PubMed] [Google Scholar]
- 45. Jung YS, Liu G, Chen X. Pirh2 E3 ubiquitin ligase targets DNA polymerase eta for 20S proteasomal degradation. Mol Cell Biol 2010; 30: 1041–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Prakash S, Johnson RE, Prakash L. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu Rev Biochem 2005; 74: 317–53. [DOI] [PubMed] [Google Scholar]
- 47. Johnson RE, Prakash S, Prakash L. Efficient bypass of a thymine‐thymine dimer by yeast DNA polymerase, Poleta. Science 1999; 283: 1001–4. [DOI] [PubMed] [Google Scholar]
- 48. Liu G, Chen X. DNA polymerase eta, the product of the xeroderma pigmentosum variant gene and a target of p53, modulates the DNA damage checkpoint and p53 activation. Mol Cell Biol 2006; 26: 1398–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature 1997; 387: 296–9. [DOI] [PubMed] [Google Scholar]
- 50. Wang L, He G, Zhang P, Wang X, Jiang M, Yu L. Interplay between MDM2, MDMX, Pirh2 and COP1: the negative regulators of p53. Mol Biol Rep 2011; 38: 229–36. [DOI] [PubMed] [Google Scholar]
- 51. Dornan D, Wertz I, Shimizu H et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature 2004; 429: 86–92. [DOI] [PubMed] [Google Scholar]
- 52. Kubbutat MH, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature 1997; 387: 299–303. [DOI] [PubMed] [Google Scholar]
- 53. Shvarts A, Steegenga WT, Riteco N et al. MDMX: a novel p53‐binding protein with some functional properties of MDM2. EMBO J 1996; 15: 5349–57. [PMC free article] [PubMed] [Google Scholar]
- 54. Yan J, Zhang D, Di Y, Shi H, Rao H, Huo K. A newly identified Pirh2 substrate SCYL1‐BP1 can bind to MDM2 and accelerate MDM2 self‐ubiquitination. FEBS Lett 2010; 584: 3275–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Legube G, Linares LK, Tyteca S et al. Role of the histone acetyl transferase Tip60 in the p53 pathway. J Biol Chem 2004; 279: 44825–33. [DOI] [PubMed] [Google Scholar]
- 56. Legube G, Linares LK, Lemercier C, Scheffner M, Khochbin S, Trouche D. Tip60 is targeted to proteasome‐mediated degradation by Mdm2 and accumulates after UV irradiation. EMBO J 2002; 21: 1704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ito A, Kawaguchi Y, Lai CH et al. MDM2‐HDAC1‐mediated deacetylation of p53 is required for its degradation. EMBO J 2002; 21: 6236–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim MM, Yoon SO, Cho YS, Chung AS. Histone deacetylases, HDAC1 and HSIR2, act as a negative regulator of ageing through p53 in human gingival fibroblast. Mech Ageing Dev 2004; 125: 351–7. [DOI] [PubMed] [Google Scholar]
- 59. Lagger G, Doetzlhofer A, Schuettengruber B et al. The tumor suppressor p53 and histone deacetylase 1 are antagonistic regulators of the cyclin‐dependent kinase inhibitor p21/WAF1/CIP1 gene. Mol Cell Biol 2003; 23: 2669–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Timmermann S, Lehrmann H, Polesskaya A, Harel‐Bellan A. Histone acetylation and disease. Cell Mol Life Sci 2001; 58: 728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer 2009; 9: 738–48. [DOI] [PubMed] [Google Scholar]
- 62. Kim SY, Herbst A, Tworkowski KA, Salghetti SE, Tansey WP. Skp2 regulates Myc protein stability and activity. Mol Cell 2003; 11: 1177–88. [DOI] [PubMed] [Google Scholar]
- 63. von der Lehr N, Johansson S, Wu S et al. The F‐box protein Skp2 participates in c‐Myc proteosomal degradation and acts as a cofactor for c‐Myc‐regulated transcription. Mol Cell 2003; 11: 1189–200. [DOI] [PubMed] [Google Scholar]
- 64. Zhou BB, Elledge SJ. The DNA damage response: putting checkpoints in perspective. Nature 2000; 408: 433–9. [DOI] [PubMed] [Google Scholar]
- 65. Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1‐phase progression. Genes Dev 1999; 13: 1501–12. [DOI] [PubMed] [Google Scholar]
- 66. Hengst L, Reed SI. Inhibitors of the Cip/Kip family. Curr Top Microbiol Immunol 1998; 227: 25–41. [DOI] [PubMed] [Google Scholar]
- 67. Hattori T, Isobe T, Abe K et al. Pirh2 promotes ubiquitin‐dependent degradation of the cyclin‐dependent kinase inhibitor p27Kip1. Cancer Res 2007; 67: 10789–95. [DOI] [PubMed] [Google Scholar]
- 68. Nakayama K, Nagahama H, Minamishima YA et al. Targeted disruption of Skp2 results in accumulation of cyclin E and p27(Kip1), polyploidy and centrosome overduplication. EMBO J 2000; 19: 2069–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kamura T, Hara T, Matsumoto M et al. Cytoplasmic ubiquitin ligase KPC regulates proteolysis of p27(Kip1) at G1 phase. Nat Cell Biol 2004; 6: 1229–35. [DOI] [PubMed] [Google Scholar]
- 70. Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer 2009; 9: 400–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Duan S, Yao Z, Zhu Y et al. The Pirh2‐keratin 8/18 interaction modulates the cellular distribution of mitochondria and UV‐induced apoptosis. Cell Death Differ 2009; 16: 826–37. [DOI] [PubMed] [Google Scholar]
- 72. Jemmerson R, LaPlante B, Treeful A. Release of intact, monomeric cytochrome c from apoptotic and necrotic cells. Cell Death Differ 2002; 9: 538–48. [DOI] [PubMed] [Google Scholar]
- 73. Coulombe PA, Wong P. Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat Cell Biol 2004; 6: 699–706. [DOI] [PubMed] [Google Scholar]
- 74. Oshima RG. Apoptosis and keratin intermediate filaments. Cell Death Differ 2002; 9: 486–92. [DOI] [PubMed] [Google Scholar]
- 75. Inada H, Izawa I, Nishizawa M et al. Keratin attenuates tumor necrosis factor‐induced cytotoxicity through association with TRADD. J Cell Biol 2001; 155: 415–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Caulin C, Ware CF, Magin TM, Oshima RG. Keratin‐dependent, epithelial resistance to tumor necrosis factor‐induced apoptosis. J Cell Biol 2000; 149: 17–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Deyoung MP, Ellisen LW. p63 and p73 in human cancer: defining the network. Oncogene 2007; 26: 5169–83. [DOI] [PubMed] [Google Scholar]
- 78. Maruyama S, Miyajima N, Bohgaki M et al. Ubiquitylation of epsilon‐COP by PIRH2 and regulation of the secretion of PSA. Mol Cell Biochem 2008; 307: 73–82. [DOI] [PubMed] [Google Scholar]
- 79. Hicke L. A new ticket for entry into budding vesicles‐ubiquitin. Cell 2001; 106: 527–30. [DOI] [PubMed] [Google Scholar]
- 80. Abe K, Hattori T, Isobe T et al. Pirh2 interacts with and ubiquitylates signal recognition particle receptor beta subunit. Biomed Res 2008; 29: 53–60. [DOI] [PubMed] [Google Scholar]
- 81. Fulga TA, Sinning I, Dobberstein B, Pool MR. SRbeta coordinates signal sequence release from SRP with ribosome binding to the translocon. EMBO J 2001; 20: 2338–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Halic M, Beckmann R. The signal recognition particle and its interactions during protein targeting. Curr Opin Struct Biol 2005; 15: 116–25. [DOI] [PubMed] [Google Scholar]
- 83. Schwartz T, Blobel G. Structural basis for the function of the beta subunit of the eukaryotic signal recognition particle receptor. Cell 2003; 112: 793–803. [DOI] [PubMed] [Google Scholar]
- 84. Toivola DM, Tao GZ, Habtezion A, Liao J, Omary MB. Cellular integrity plus: organelle‐related and protein‐targeting functions of intermediate filaments. Trends Cell Biol 2005; 15: 608–17. [DOI] [PubMed] [Google Scholar]
- 85. Collinet C, Stoter M, Bradshaw CR et al. Systems survey of endocytosis by multiparametric image analysis. Nature 2010; 464: 243–9. [DOI] [PubMed] [Google Scholar]
- 86. Alen P, Claessens F, Verhoeven G, Rombauts W, Peeters B. The androgen receptor amino‐terminal domain plays a key role in p160 coactivator‐stimulated gene transcription. Mol Cell Biol 1999; 19: 6085–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH(2)‐terminal domain. J Biol Chem 1999; 274: 37219–25. [DOI] [PubMed] [Google Scholar]
- 88. Ikonen T, Palvimo JJ, Janne OA. Interaction between the amino‐ and carboxyl‐terminal regions of the rat androgen receptor modulates transcriptional activity and is influenced by nuclear receptor coactivators. J Biol Chem 1997; 272: 29821–8. [DOI] [PubMed] [Google Scholar]
- 89. Abate‐Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev 2000; 14: 2410–34. [DOI] [PubMed] [Google Scholar]
- 90. Tombes RM, Mikkelsen RB, Jarvis WD, Grant S. Downregulation of delta CaM kinase II in human tumor cells. Biochim Biophys Acta 1999; 1452: 1–11. [DOI] [PubMed] [Google Scholar]
- 91. Logan IR, Sapountzi V, Gaughan L, Neal DE, Robson CN. Control of human PIRH2 protein stability: involvement of TIP60 and the proteosome. J Biol Chem 2004; 279: 11696–704. [DOI] [PubMed] [Google Scholar]
- 92. Corcoran CA, Montalbano J, Sun H, He Q, Huang Y, Sheikh MS. Identification and characterization of two novel isoforms of Pirh2 ubiquitin ligase that negatively regulate p53 independent of RING finger domains. J Biol Chem 2009; 284: 21955–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Zheng G, Ning J, Yang YC. PLAGL2 controls the stability of Pirh2, an E3 ubiquitin ligase for p53. Biochem Biophys Res Commun 2007; 364: 344–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Castilla LH, Perrat P, Martinez NJ et al. Identification of genes that synergize with Cbfb‐MYH11 in the pathogenesis of acute myeloid leukemia. Proc Natl Acad Sci U S A 2004; 101: 4924–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Landrette SF, Kuo YH, Hensen K et al. Plag1 and Plagl2 are oncogenes that induce acute myeloid leukemia in cooperation with Cbfb‐MYH11. Blood 2005; 105: 2900–7. [DOI] [PubMed] [Google Scholar]
- 96. Min SH, Kim DM, Heo YS et al. New p53 target, phosphatase of regenerating liver 1 (PRL‐1) downregulates p53. Oncogene 2009; 28: 545–54. [DOI] [PubMed] [Google Scholar]
- 97. Min SH, Kim DM, Heo YS, Kim HM, Kim IC, Yoo OJ. Downregulation of p53 by phosphatase of regenerating liver 3 is mediated by MDM2 and PIRH2. Life Sci 2010; 86: 66–72. [DOI] [PubMed] [Google Scholar]
- 98. Chen M, Cortay JC, Logan IR, Sapountzi V, Robson CN, Gerlier D. Inhibition of ubiquitination and stabilization of human ubiquitin E3 ligase PIRH2 by measles virus phosphoprotein. J Virol 2005; 79: 11824–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Karuppannan AK, Liu S, Jia Q, Selvaraj M, Kwang J. Porcine circovirus type 2 ORF3 protein competes with p53 in binding to Pirh2 and mediates the deregulation of p53 homeostasis. Virology 2010; 398: 1–11. [DOI] [PubMed] [Google Scholar]
- 100. Liu J, Zhu Y, Chen I et al. The ORF3 protein of porcine circovirus type 2 interacts with porcine ubiquitin E3 ligase Pirh2 and facilitates p53 expression in viral infection. J Virol 2007; 81: 9560–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. O’Keefe K, Li H, Zhang Y. Nucleocytoplasmic shuttling of p53 is essential for MDM2‐mediated cytoplasmic degradation but not ubiquitination. Mol Cell Biol 2003; 23: 6396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhang L, Li J, Wang C, Ma Y, Huo K. A new human gene hNTKL‐BP1 interacts with hPirh2. Biochem Biophys Res Commun 2005; 330: 293–7. [DOI] [PubMed] [Google Scholar]
- 103. Zhang W, Wu G, Yan X, Shi H, K H. Preliminary study on the interaction between human PIRH2b and ARF4. J Med Mol Biol 2007; 4: 469–74. [Google Scholar]
- 104. Su Y, Bai M, Zhu LP, Jin Y, Zhang XJ, Zhou Q. [Pirh2 shRNA mediated by psiRNA‐hH1 vector plasmid effectively inhibits the proliferation of lung carcinoma cells: in vitro and in vivo experiments]. Zhonghua Yi Xue Za Zhi 2007; 87: 1199–203 (In Chinese). [PubMed] [Google Scholar]
- 105. Sion‐Vardy N, Lasarov I, Delgado B, Gopas J, Benharroch D, Ariad S. Measles virus: evidence for association with lung cancer. Exp Lung Res 2009; 35: 701–12. [DOI] [PubMed] [Google Scholar]
- 106. Koivusalo R, Mialon A, Pitkanen H, Westermarck J, Hietanen S. Activation of p53 in cervical cancer cells by human papillomavirus E6 RNA interference is transient, but can be sustained by inhibiting endogenous nuclear export‐dependent p53 antagonists. Cancer Res 2006; 66: 11817–24. [DOI] [PubMed] [Google Scholar]
- 107. Miskad UA, Semba S, Kato H, Yokozaki H. Expression of PRL‐3 phosphatase in human gastric carcinomas: close correlation with invasion and metastasis. Pathobiology 2004; 71: 176–84. [DOI] [PubMed] [Google Scholar]
- 108. Wang Q, Holmes DI, Powell SM, Lu QL, Waxman J. Analysis of stromal‐epithelial interactions in prostate cancer identifies PTPCAAX2 as a potential oncogene. Cancer Lett 2002; 175: 63–9. [DOI] [PubMed] [Google Scholar]
- 109. Friedler A, Veprintsev DB, Rutherford T, von Glos KI, Fersht AR. Binding of Rad51 and other peptide sequences to a promiscuous, highly electrostatic binding site in p53. J Biol Chem 2005; 280: 8051–9. [DOI] [PubMed] [Google Scholar]
- 110. Lee W, Zhang Y, Mukhyala K, Lazarus RA, Zhang Z. Bi‐directional SIFT predicts a subset of activating mutations. PLoS ONE 2009; 4: e8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Balkwill F, Mantovani A. Cancer and inflammation: implications for pharmacology and therapeutics. Clin Pharmacol Ther 2010; 87: 401–6. [DOI] [PubMed] [Google Scholar]
- 112. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010; 140: 883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Martomo SA, Saribasak H, Yokoi M, Hanaoka F, Gearhart PJ. Reevaluation of the role of DNA polymerase theta in somatic hypermutation of immunoglobulin genes. DNA Repair (Amst) 2008; 7: 1603–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Besson A, Gurian‐West M, Schmidt A, Hall A, Roberts JM. p27kip1 modulates cell migration through the regulation of RhoA activation. Genes Dev 2004; 18: 862–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Tybulewicz VL, Henderson RB. Rho family GTPases and their regulators in lymphocytes. Nat Rev Immunol 2009; 9: 630–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Sapountzi V, Logan IR, Robson CN. Cellular functions of TIP60. Int J Biochem Cell Biol 2006; 38: 1496–509. [DOI] [PubMed] [Google Scholar]
- 117. Col E, Caron C, Chable‐Bessia C et al. HIV‐1 Tat targets Tip60 to impair the apoptotic cell response to genotoxic stresses. The EMBO J 2005; 24: 2634–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Dornan D, Shimizu H, Mah A et al. ATM engages autodegradation of the E3 ubiquitin ligase COP1 after DNA damage. Science 2006; 313: 1122–6. [DOI] [PubMed] [Google Scholar]
- 119. Marchenko ND, Wolff S, Erster S, Becker K, Moll UM. Monoubiquitylation promotes mitochondrial p53 translocation. EMBO J 2007; 26: 923–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Lim J, Hao T, Shaw C et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell 2006; 125: 801–14. [DOI] [PubMed] [Google Scholar]
- 121. Laity JH, Lee BM, Wright PE. Zinc finger proteins: new insights into structural and functional diversity. Curr Opin Struct Biol 2001; 11: 39–46. [DOI] [PubMed] [Google Scholar]
- 122. Brinkmann AO, Blok LJ, de Ruiter PE et al. Mechanisms of androgen receptor activation and function. J Steroid Biochem Mol Biol 1999; 69: 307–13. [DOI] [PubMed] [Google Scholar]
- 123. Edwards DP. Coregulatory proteins in nuclear hormone receptor action. Vitam Horm 1999; 55: 165–218. [DOI] [PubMed] [Google Scholar]
- 124. Volpicelli‐Daley LA, Li Y, Zhang CJ, Kahn RA. Isoform‐selective effects of the depletion of ADP‐ribosylation factors 1–5 on membrane traffic. Mol Biol Cell 2005; 16: 4495–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Inoki K, Ouyang H, Zhu T et al. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 2006; 126: 955–68. [DOI] [PubMed] [Google Scholar]
- 126. Rui Y, Xu Z, Lin S et al. Axin stimulates p53 functions by activation of HIPK2 kinase through multimeric complex formation. EMBO J 2004; 23: 4583–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Rui Y, Xu Z, Xiong B et al. A beta‐catenin‐independent dorsalization pathway activated by Axin/JNK signaling and antagonized by aida. Dev Cell 2007; 13: 268–82. [DOI] [PubMed] [Google Scholar]
- 128. Peifer M, Polakis P. Wnt signaling in oncogenesis and embryogenesis – a look outside the nucleus. Science 2000; 287: 1606–9. [DOI] [PubMed] [Google Scholar]
- 129. Yi C, Deng XW. COP1 – from plant photomorphogenesis to mammalian tumorigenesis. Trends Cell Biol 2005; 15: 618–25. [DOI] [PubMed] [Google Scholar]
- 130. Wertz IE, O’Rourke KM, Zhang Z et al. Human De‐etiolated‐1 regulates c‐Jun by assembling a CUL4A ubiquitin ligase. Science 2004; 303: 1371–4. [DOI] [PubMed] [Google Scholar]
- 131. Guo C, Kosarek‐Stancel JN, Tang TS, Friedberg EC. Y‐family DNA polymerases in mammalian cells. Cell Mol Life Sci 2009; 66: 2363–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Bethune J, Wieland F, Moelleken J. COPI‐mediated transport. J Membr Biol 2006; 211: 65–79. [DOI] [PubMed] [Google Scholar]
- 133. Beck R, Rawet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett 2009; 583: 2701–9. [DOI] [PubMed] [Google Scholar]
- 134. Eugster A, Frigerio G, Dale M, Duden R. COP I domains required for coatomer integrity, and novel interactions with ARF and ARF‐GAP. EMBO J 2000; 19: 3905–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 2006; 6: 38–51. [DOI] [PubMed] [Google Scholar]
- 136. Collavin L, Lunardi A, Del Sal G. p53‐family proteins and their regulators: hubs and spokes in tumor suppression. Cell Death Differ 2010; 17: 901–11. [DOI] [PubMed] [Google Scholar]
- 137. Kikkawa U, Matsuzaki H, Yamamoto T. Protein kinase C delta (PKC delta): activation mechanisms and functions. J Biochem 2002; 132: 831–9. [DOI] [PubMed] [Google Scholar]
- 138. Hensen K, Van Valckenborgh IC, Kas K, Van de Ven WJ, Voz ML. The tumorigenic diversity of the three PLAG family members is associated with different DNA binding capacities. Cancer Res 2002; 62: 1510–7. [PubMed] [Google Scholar]
- 139. Sun Y, Jiang X, Chen S, Fernandes N, Price BD. A role for the Tip60 histone acetyltransferase in the acetylation and activation of ATM. Proc Natl Acad Sci U S A 2005; 102: 13182–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Squatrito M, Gorrini C, Amati B. Tip60 in DNA damage response and growth control: many tricks in one HAT. Trends Cell Biol 2006; 16: 433–42. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. A comprehensive picture of the overall Pirh2‐involved cellular processes.
Supporting info item


