Abstract
No ideal serum markers for screening colorectal cancer (CRC) have been identified. The aim of this study was to determine the usefulness of endothelial cell‐specific molecule‐1 (ESM‐1) as a serum marker for CRC. Illumina microarray was carried out to search CRC‐related biomarkers. cDNA microarray detected that ESM‐1 was one of the overexpressed genes in CRC. Overexpression of ESM‐1 mRNA was confirmed in tissues of CRC by RT‐PCR and real‐time PCR. Immunohistochemical staining showed strong expression of ESM‐1 in the cytoplasm of tumor cells. Overexpression of ESM‐1 in human serum with CRC was found by Western blot analysis. For quantitative analysis of ESM‐1 in serum, we determined the ESM‐1 levels in serum specimens using an ELISA kit. We showed that the ESM‐1 levels in the serum of patients with CRC were significantly elevated (70.1 ± 29.7 pg/mL) compared to healthy subjects (29.7 ± 14.9 pg/mL). The accuracy, sensitivity, and specificity of ESM‐1 for CRC were 0.94, 99%, and 73%, respectively, by receiver operating characteristics curve analysis. The positive predictive value and negative predictive value were 63% and 95%, respectively. The likelihood ratios of a positive or negative test result were 73 and 0.27, respectively. When analyzed with a Cox regression model, a higher serum ESM‐1 level (≥76.0 pg/mL) was correlated with poor prognosis. This study suggests that expression of ESM‐1 is increased in tissue and serum of CRC patients and that ESM‐1 can be used as a potential serum marker for the early detection of CRC. (Cancer Sci 2010); 101: xxx–xxx
Colorectal cancer is one of the most common cancers in the world. It is most effectively treated when detected at an early stage. Various methods, including the fecal occult blood test and colonoscopy, are currently used for screening CRC, and increase the rates of detection of early stage cancer. Regular colonoscopic examinations are recommended, but the high cost and invasiveness of the procedure is an obstacle to its application as a screening test for CRC. Furthermore, the sensitivity and specificity of fecal occult blood tests are low, although inexpensive and non‐invasive.( 1 , 2 , 3 ) Therefore, more accurate and acceptable tumor markers for the early detection of CRC are needed.
Recent studies have shown that CRC can be detected by non‐invasive markers, such as specific changes in serum proteins.( 4 , 5 , 6 , 7 ) Serum markers have the potential to greatly increase the effectiveness of CRC screening programs, as they can be analyzed relatively non‐invasively, conveniently, and economically.( 4 ) Although CEA is well‐known and commonly used as a serum tumor marker of CRC, it has no role in the early detection of CRC. Currently, no ideal serum markers for screening CRC have been identified. Therefore, we attempted to identify a potential serum marker for the early detection of CRC. First, we carried out cDNA microarray to screen the genes, the expression of which was increased in CRC tissue compared to adjacent normal mucosa, and selected candidate genes encoding a secretory protein among the list of genes overexpressed in CRC tissue. We then validated the overexpression in CRC tissue using RT‐PCR and IHC staining. Finally, we analyzed the candidate protein in serum by ELISA. In this study, we show that endothelial cell‐specific molecule‐1 (ESM‐1) has the potential as a serum marker for early detection of CRC.
Materials and Methods
Tissue specimens. For a cDNA microarray assay, 66 pairs of CRC tissue and adjacent normal mucosa samples were obtained from Samsung Medical Center (Seoul, Korea). Tissue specimens were frozen in liquid nitrogen and stored until use. Total RNA was extracted with an RNeasy midi‐kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions.
Serum specimens. Serum specimens were collected at Dankook Medical School (Chunan, Korea) and Samsung Medical Center. We obtained 10 mL of blood from patients with CRC before surgery in 2004 (n = 14/37/39/10, TNM stage I/II/III/IV, respectively)( 8 ) or advanced colorectal adenoma before polypectomy in 2009 (n = 10). An advanced colorectal adenoma was defined as >1 cm in diameter, ≥25% villous features, and/or high grade dysplasia. We centrifuged the blood within 8 h of collection and stored serum at −70°C. We also obtained serum in identical fashion from healthy subjects who visited our hospitals in 2008 and 2009 for routine health checks. They underwent medical examinations such as blood test, radiological test, and endoscopic exam. It was very unlikely that CRC patients would be included in controls, although we could not get any result of the examinations because of ethical issues. All protocols were carried out in accordance with the guidelines approved by the Ethics Committee of the Dankook Medical School and Samsung Medical Center. Clinical data with demographics were collected using hospital intranet resources. Overall survival data was obtained using the national registry of medical insurance. The technicians were blinded to clinical information of the serum samples.
Microarray analysis. cDNA microarrays (48K, Human‐6 V2) were purchased from Illumina (San Diego, CA, USA). The microarray slides collectively contained a total of 2230 unique sequence‐verified clones and housekeeping genes. The human reference RNA, which contained the total RNA samples from adjacent normal colon mucosa, was used as a reference for microarray analysis. The purified and concentrated fluorescence‐labeled cDNA from the reference and experimental samples was processed. The signals from each immobilized cDNA target on a microarray slide were localized, and the expression ratio between the experimental and reference samples (Cy5/Cy3 ratio) was determined and analyzed using BeadStudio version 3 (Illumina).
RT‐PCR. Oligonucleotide sequences corresponding to the ESM‐1 gene were designed using Primer3 software (http://frodo.wi.mit.edu/primer3/). The cDNA mixture contained 5 μg total RNA as a template for RT‐PCR (or real‐time RT‐PCR) analysis and premix Ex Taq (or SYBR premix Ex Taq) (TaKaRa Bio, Otsu, Japan) were used for RT‐PCR (or real‐time RT‐PCR). The gene‐specific primers used for PCR were: 5′‐CTTGCTACCGCACAGTCTCA‐3′ (sense) and 5′‐GCGTGGATTTAACCATTTCC‐3′ (antisense). Optimized PCR was carried out as follows: 1 cycle of 94°C for 5 min; 35 cycles of 94°C for 40 s, 56°C for 40 s, and 72°C for 30 s; and a final extension at 72°C for 7 min. The primers used for ESM‐1 real‐time RT‐PCR were 5′‐TTGCTACCGCACAGTCTCAG‐3′ (sense) and 5′‐AGGGGAATTTCAGGCATTTT‐3′ (antisense). Optimized PCR was carried out as follows: 1 cycle of 95°C for 10 s; 40 cycles of 95°C for 5 s, 60°C for 30 s, and 95°C for 15 s; and a final extension at 60°C for 15 s. The relative levels of gene expression were normalized to GAPDH expression.
Western blot analysis. We carried out SDS‐PAGE using a Phast PAGE System (Pharmacia, Piscataway, NJ, USA) and a 12% gel according to the manufacturer’s instructions. Proteins were then transferred to a PVDF membrane and probed with an anti‐ESM‐1 antibody (R&D Systems, Minneapolis, MN, USA) followed by an HRP‐conjugated secondary antibody (Sigma‐Aldrich, St. Louis, MO, USA). Immunolabelled proteins were detected by incubation with ECL substrate, followed by exposure of the membrane to auto‐radiographic film. The band intensity was quantitated using a Gel Doc system (Bio‐Rad, Hercules, CA, USA).
Immunohistochemical staining. Tissue specimens were fixed in neutral buffered formalin (10% v/v formalin in water [pH 7.4]) and embedded in paraffin wax. Serial sections of 4 μm thickness were cut and mounted on charged glass slides (Superfrost Plus; Fisher Scientific, Rochester, NY, USA). Immunohistochemical conditions for ESM‐1 were optimized and evaluated. In brief, the tissue sections were deparaffinized, and antigen retrieval was carried out in citrate buffer (pH 6.0) for 10 min. The sections were then treated with 3% hydrogen peroxide in methanol to quench the endogenous tissue peroxidase activity, followed by incubation with 1% BSA to block the non‐specific binding. A mouse monoclonal antibody against ESM‐1 (Abnova, Taipei, Taiwan) was used at a dilution of 1:200. The sections were incubated with antibody overnight at 4°C in a wet chamber. The sections were stained using a standard EnVision‐HRP kit (Dako, Glostrup, Denmark) and developed with diaminobenzidine. The sections were counterstained with 10% Mayer’s hematoxylin. An irrelevant mouse IgG of the same isotype or antibody dilution solution served as a negative control.
Determination of ESM‐1 in human serum using a sandwich ELISA. The ESM‐1 ELISA kits were obtained from Atila Biosystems (Palo Alto, CA, USA) and used to detect ESM‐1 according to the manufacturer’s instructions. All samples were coded using a numbering system and were analyzed by technicians in our laboratory who were not aware of the disease state of the patients. A microwell plate was coated with 100 μL capture antibody (1 μg/mL) against ESM‐1, incubated overnight at 4°C, then coated again with 1% BSA. Then 100 μL serially diluted ESM‐1 standard solution and samples diluted fivefold in dilution buffer (2% BSA in PBST) were applied to wells in duplicate and incubated with 0.25 μg/mL detection antibody for 1 h at room temperature. After washing the wells with PBST solution, 100 μL diluted HRP‐conjugated strepatvidin was applied to each well for 30 min. Subsequently, tetramethylbenzidine (TMB) solution was added to the wells, and the reaction was stopped with 1N H2SO4 solution. The absorbance was then measured at 450 nm on a spectrophotometer (Molecular Devices, Sunnyvale, CA, USA).
Statistical analysis. Statistical analysis was carried out using PASW Statistics 17.0 (SPSS, Chicago, IL, USA). Serum ESM‐1 levels were shown as the mean ± SD. The means among groups were compared using an unpaired Student’s t‐test. Receiver operating characteristic curves were calculated to assess the diagnostic value of ESM‐1. The sensitivity, specificity, PPV, NPV, LR+, and LR− for the diagnosis of CRC were calculated.( 9 , 10 ) Survival curves by the Kaplan–Meier method were plotted to display overall survival distributions and the log–rank test was used for comparison. A Cox regression analysis was carried out to assess independent prognostic factors for overall survival in CRC, including stage, serum ESM‐1 level, and CEA level. P < 0.05 was considered to be statistically significant.
Results
Expression of ESM‐1 in CRC tissues. To identify the genes associated with the development of human CRC, we analyzed the gene expression profiles of CRC and adjacent non‐tumor tissues from 66 patients using Illumina microarray chips. The expression of 281 genes was upregulated more than twofold in at least 60% of the CRC tissues compared to normal colorectal mucosa. We carried out cellular component ontology analysis to identify the genes of which products are located in the extracellular region. It was hypothesized that these products could be elevated in the serum of CRC patients. Among these genes, we focused on ESM‐1 encoding a secretory protein. The expression of ESM‐1 was upregulated more than twofold in 65 of 66 cases (P = 8.75e‐17). We determined the level of mRNA of 30 paired samples of CRC by RT‐PCR. We confirmed the results of DNA chip analysis as the levels of ESM‐1 expression were significantly higher in all of the CRC tissues compared to the normal colon tissues (Fig. 1a). A quantitative real‐time RT‐PCR analysis using 20 pairs also showed that ESM‐1 mRNA levels were much higher in CRC than the corresponding normal colon tissue (Fig. 1b). The relative level of expression of ESM‐1 was determined by the ratio of ESM‐1 to GAPDH. The value of tumor‐originated ESM‐1 was a relative value against ESM‐1/GAPDH (value = 1.0) of matched normal controls. The ESM‐1 RNA levels were much higher (123.1 ± 239.7) in most of the CRC tissues compared to the corresponding non‐tumor tissue.
Figure 1.

Expression of the endothelial cell‐specific molecule‐1 (ESM‐1) gene in human colorectal tissues. (a) RT‐PCR analysis was done with 30 pairs of non‐tumor colon (N) and colorectal cancer (CRC) tumor (C) tissues for ESM‐1, with GAPDH used as the loading control. (b) Real‐time RT‐PCR analysis. The level of relative expression of ESM‐1 was determined by the ratio of ESM‐1 to GAPDH. The value of tumor‐originated ESM‐1 was a relative value against ESM‐1/GAPDH (value = 1.0) of matched normal controls (n = 20).
Expression of ESM‐1 in cytoplasm of tumor cells and serum with CRC. We carried out IHC for ESM‐1 protein of colon tissues with a monoclonal antibody against human ESM‐1. The results showed a strong expression of ESM‐1 in the cytoplasm of tumor cells in CRC tissues compared to normal colonic mucosa. The ESM‐1 protein was also intensely expressed on the luminal side of the glandular structures in CRC (Fig. 2).
Figure 2.
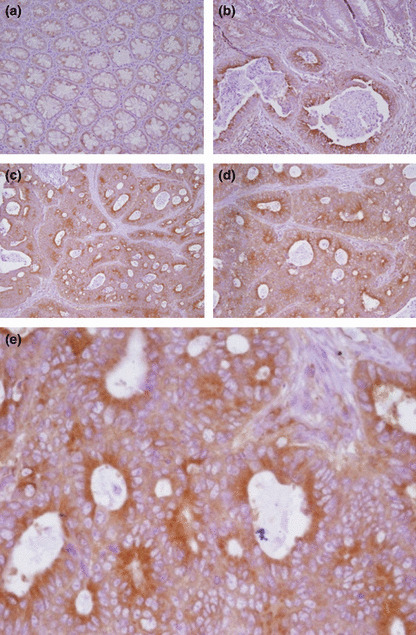
Expression of endothelial cell‐specific molecule‐1 (ESM‐1) in normal and colorectal cancer (CRC) tissues. Expression was high in the cytoplasm of tumor cells of CRC tissues. (a) Expression of ESM‐1 in normal tissue (magnification, ×200). (b) Expression of ESM‐1 in normal tumor tissues from CRC patients (magnification, ×200); ESM‐1 was not expressed in normal colonic epithelial cells. (c–e) Expression of ESM‐1 in tumor tissues from CRC patients (magnification, ×200 and ×400). The expression of ESM‐1 was increased in 19 of 31 cancer tissues.
We also measured ESM‐1 expression in human serum from patients with CRC by Western blot analysis. When the levels of expression of ESM‐1 in human serum were compared, ESM‐1 was highly expressed in serum from patients with CRC (Fig. 3).
Figure 3.
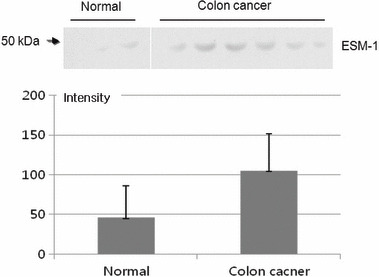
Expression of serum endothelial cell‐specific molecule‐1 (ESM‐1) protein by Western blot analysis. We carried out SDS‐PAGE under a reducing condition (12% gels, 0.5% mercaptoethanol). Proteins were transferred onto a membrane and probed with mouse anti‐ESM‐1 antibody. The expression of serum ESM‐1 was higher in cancer patients (n = 6) than in normal subjects (n = 3) (108.6 ± 40.6 vs. 43.5 ± 43.1; P = 0.0306).
Serum ESM‐1 concentration in patients with CRC or advanced colorectal adenoma. The ESM‐1 concentration in serum was detected using an ELISA kit. The relationship between colorimetric intensity and ESM‐1 concentration was in the range of 0–25.0 pg/mL. The minimal concentration of ESM‐1 protein that could be detected was 0.3 pg/mL when serum was diluted fivefold. Serum specimens from 100 patients with CRC and 78 healthy subjects were evaluated in this study. As per the manual instructions, the serum ESM‐1 concentration was determined after a fivefold dilution and in duplicate using the ESM‐1 ELISA kit. The results were analyzed, and are summarized in Table 1 and Figure 4(a,b). The serum ESM‐1 concentrations of patients with CRC were significantly elevated when compared with those of normal individuals (70.1 ± 29.7 and 29.7 ± 14.9 pg/mL, respectively; P = 8.43e‐35). The serum level of ESM‐1 was higher in patients with late stage CRC (stages III and IV) compared to patients with early stage CRC (stages I and II) (74.3 ± 17.9 pg/mL vs. 66.2 ± 20.8 pg/mL; P = 0.039). The level of ESM‐1 was lower in 5‐year survivors than in patients who died within 5 years of diagnosis for CRC (67.8 ± 19.5 pg/mL vs. 80.3 ± 18.2 pg/mL; P = 0.013). The results of serum CEA levels are shown in Figure S1(a,b). The serum CEA concentrations of patients with CRC were significantly elevated when compared with those of normal individuals (4.32 ± 7.17 and 1.37 ± 0.62 ng/mL, respectively; P = 0.001). However, the serum level of CEA was not significantly higher in patients with late stage CRC compared to patients with early stage CRC (5.35 ± 9.25 ng/mL vs. 3.28 ± 4.09 ng/mL; P = 0.148). The serum CEA level was not significantly lower in 5‐year survivors than in patients who died within 5 years after diagnosis for CRC (3.25 ± 3.97 ng/mL vs. 8.85 ± 13.57 ng/mL; P = 0.091). There was no correlation between the serum level of ESM‐1 and CEA (R 2 = 0.01).
Table 1.
Serum endothelial cell‐specific molecule‐1 (ESM‐1) con‐centration as measured by ELISA
| Serum ESM‐1 (pg/mL) (mean ± SD) | P‐value | |
|---|---|---|
| Gender | ||
| Male (n = 61) | 69.3 ± 21.2 | 0.563 |
| Female (n = 39) | 71.5 ± 17.5 | |
| Age | ||
| <50 (n = 18) | 66.8 ± 15.0 | 0.343 |
| ≥50 (n = 82) | 70.97 ± 20.79 | |
| Location | ||
| Proximal (n = 28) | 70.4 ± 18.79 | 0.946 |
| Distal (n = 72) | 70.1 ± 20.3 | |
| Differentiation | ||
| Well/moderate (n = 88) | 70.2 ± 20.2 | 0.739 |
| Poor/mucinous (n = 12) | 68.3 ± 16.9 | |
| Stage | ||
| Early (I, II) (n = 51) | 66.2 ± 20.8 | 0.039 |
| Late (III, IV) (n = 49) | 74.3 ± 17.9 | |
| Five‐year survival | ||
| Yes (n = 81) | 67.8 ± 19.5 | 0.013 |
| No (n = 19) | 80.3 ± 18.2 | |
Figure 4.
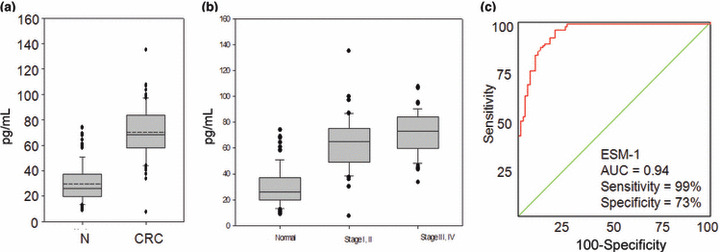
Serum concentration and receiver operating characteristics (ROC) curve of endothelial cell‐specific molecule‐1 (ESM‐1). (a) Serum ESM‐1 concentrations in normal subjects (N) and in patients with colorectal cancer (CRC). (b) Serum concentrations of ESM‐1 in normal samples (normal), patients with early stage CRC (stages I and II), and those with late stage CRC (stages III and IV). The horizontal line inside the box is the mean, and the box represents the lower and upper quartiles. The whiskers indicate the 95% confidence interval of the values. The values outside the whiskers represent individual outliers. (c) The ROC curves for ESM‐1 in serum samples collected form healthy subjects and CRC patients. In a ROC curve the true positive rate (sensitivity) is plotted in function of the false rate (1‐specificity) for different cut‐off points. Each point on the ROC plot represents a sensitivity/specificity pair corresponding to a particular decision threshold. A test with perfect discrimination has a ROC plot that passes through the upper left corner (100% sensitivity, 100‐specificity). Therefore the closer the ROC plot is to the upper left corner, the higher the overall accuracy of the test.
We determined the serum ESM‐1 level in 10 patients with advanced colorectal adenoma. The serum ESM‐1 level was not significantly different from those of normal individuals (24.4 ± 9.0 pg/mL vs. 29.7 ± 14.9 pg/mL; Table S1). The serum ESM‐1 level is not thought to be useful for detection of colorectal adenoma.
Accuracy, specificity, sensitivity, LR+, and LR− of serum ESM‐1 in patients with CRC. The ROC curve of the patient samples with CRC was used to evaluate ESM‐1 as a potential serum maker for CRC.( 11 ) As shown in Figure 4(c), the area under the curve for ESM‐1 was 0.94 in patients with CRC. The ROC curve also provides a cut‐off point (33.3 pg/mL) to show the effects on sensitivity and specificity. As shown in the curve, the sensitivity and specificity were 99% and 73%, respectively. The PPV and NPV were 63% and 95%, respectively, and the LR+ and LR− were 73 and 0.27, respectively. The ROC curve of serum CEA of the patient and control samples is shown in Figure S1(c). The sensitivity and specificity was 48% and 99%, respectively (cut‐off point; 2.59 ng/mL).
Comparison of prognoses between CRC with low or high serum ESM‐1 levels. Figure 5(a) shows the Kaplan–Meier curves for the effect of serum ESM‐1 levels. The cut‐off point for survival analysis (76.0 pg/mL) was calculated using a ROC curve. In CRC, a high serum ESM‐1 level was significantly associated with a poor prognosis. The 5‐year overall survival rates were 90.5% (57/63) and 64.9% (24/37) for the patients with low serum ESM‐1 levels and those with high serum ESM‐1 levels, respectively. Univariate analysis initially included gender, age at diagnosis, location, tumor differentiation, TNM stage, serum ESM‐1 level, and serum CEA level for survival analysis. The TNM stage, serum ESM‐1 level, and serum CEA level were associated with survival and were introduced into the multivariate analysis. The multivariate analysis showed that late stage, high serum ESM‐1 level (≥76.0 pg/mL) and high CEA level (≥5 ng/mL) were shown to have a statistically independent prognostic value with respect to survival (Table 2). In addition, we carried out a subgroup analysis with stratification by tumor stage. For early stage CRC cases, 5‐year overall survival rates were 100% (36/36) and 80.0% (12/15) for the patients with low serum ESM‐1 level and those with high serum ESM‐1 levels, respectively, and there was a significant difference between the two groups (P = 0.0054; Fig. 5b). For late stage CRC, the 5‐year overall survival rates were 77.8% (21/27) and 54.5% (12/22) for cases with low serum ESM‐1 levels and those with high serum ESM‐1 levels, respectively, but it did not reach statistical significance (P = 0.076; Fig. 5c).
Figure 5.
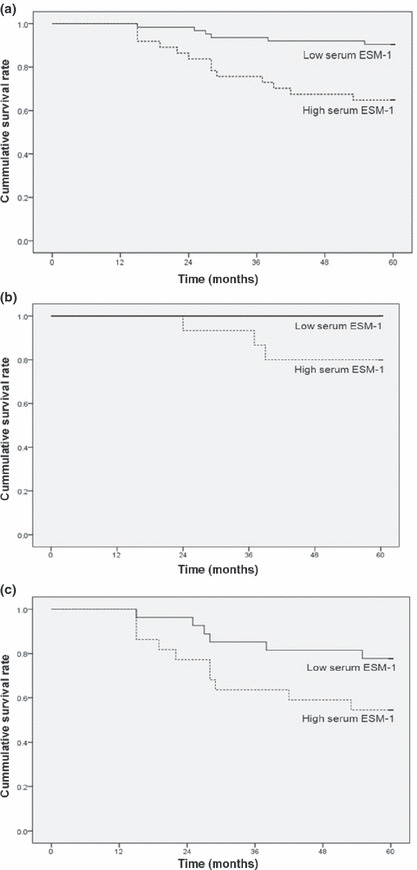
Kaplan–Meier survival analyses for high serum endothelial cell‐specific molecule‐1 (ESM‐1) level (≥76 pg/mL) versus low serum ESM‐1 level (<76 pg/mL) in (a) total CRC grade, (b) early stage CRC (TNM stage I, II), and (c) late stage CRC (stage III, IV).
Table 2.
Independent predictors for overall survival in colorectal cancer according to Cox regression model
| Variable | Hazard ratio (95% CI) | P‐value |
|---|---|---|
| TNM stage | ||
| Early (I, II) | 1 | 0.007 |
| Late (III, IV) | 5.516 (1.600–19.011) | |
| Serum ESM‐1 level | ||
| Low (<76 pg/mL) | 1 | 0.014 |
| High (≥76 pg/mL) | 3.394 (1.285–8.963) | |
| Serum CEA level | ||
| Low (<5 ng/mL) | 1 | 0.037 |
| High (≥5 ng/mL) | 2.714 (1.065–6.920) | |
CEA, carcinoembryonic antigen; CI, confidence interval; ESM‐1, endothelial cell‐specific molecule‐1.
Discussion
Detection and treatment at an earlier stage can reduce the risk of death from CRC. Therefore, one of the most important requirements of screening markers for CRC is that disease can be detected at an earlier stage. However, fecal occult blood tests and colonoscopy have limitations. Therefore, the development of non‐invasive markers can enhance the acceptance of a screening program and the detection of early CRC.
In the current study we showed the efficacy of a systematic approach to identify new markers in serum. Candidate genes were selected by consideration of the function of overexpressed genes after comparison of mRNA expression between tumor tissues and normal mucosa. Candidate proteins were then validated in serum.
In our studies, the AUC value, sensitivity, and specificity in our ROC analysis were 0.94, 99%, and 73%, respectively. The PPV, NPV, LR+, and LR− were 63%, 95%, 73, and 0.27, respectively. Interestingly, the serum ESM‐1 level was not only elevated in advanced stage, but also in early stage CRC, which is the most important requirement for serum markers in terms of early diagnosis. As the sensitivity was 99%, the serum ESM‐1 test may be very useful for ruling out CRC if a person tests negative.( 9 ) Thus, serum ESM‐1 can reduce the burden of colonoscopy in screening for CRC. As sensitivity and specificity cannot be used to estimate the probability of disease in a patient, and PPV and NPV are affected by prevalence of the disease, we calculated LR. Likelihood ratio provides a summary of how many times more (or less) likely patients with the disease are to have that particular result than patients without the disease.
This study also shows that higher serum ESM‐1 levels appear to be a predictor of bad prognosis in CRC, independent of stage. However, further studies are mandatory because this study was carried out retrospectively and all the prognostic factors for CRC could not be considered.
The origin of serum ESM‐1 is obscure. However, IHC showed strong expression of ESM‐1 in CRC cells on the luminal side of the glandular structures, which is consistent with the notion that the increased ESM‐1 in serum of CRC patients is derived from tumor cells.
Also known as endocan, ESM‐1 is a 50‐kDa secretory proteoglycan comprised of a mature polypeptide of 165 amino acids and a single dermatan sulfate chain covalently linked to the serine residue at position 137.( 12 ) It is a key player in the regulation of cell adhesion, inflammatory disorders, and tumor progression. Endothelial cell‐specific molecule‐1 specifically associates with insulin‐like growth factor to promote cell growth and proliferation, and its expression is induced by tumor necrosis factor‐α and interleukin‐1β.( 12 , 13 ) It is expressed by the vascular endothelium and freely circulates in the bloodstream and has only one glycosaminoglycan chain.( 12 ) Recently, it was reported that ESM‐1 is expressed in tumor endothelium and induces tumor formation.( 14 , 15 ) Levels of ESM‐1 are increased in vascular endothelial growth factor‐A treated endothelial cells and in human renal clear cell carcinoma.( 16 ) In human cancer tissues, high expression of ESM‐1 has been shown to be correlated with poor prognosis and metastasis in several types of cancers, including breast, renal, lung, liver, and brain cancer.( 12 , 17 , 18 ) Also, serum ESM‐1 level was correlated with survival and time to tumor progression in lung cancer.( 19 ) To our knowledge, this is the first study to show the utility of serum ESM‐1 for early detection of CRC, and the relationship between serum ESM‐1 levels and prognosis in CRC. In contrast, Zuo et al. ( 20 ) reported that ESM‐1 was downregulated in CRC tissue and was positively correlated with tissue differentiation. We cannot explain the difference between the results of Zuo et al. and our own. However, we confirmed the consistent results of cDNA microarray, RT‐PCR, and IHC using tissue and ELISA using serum. In addition, the expression of ESM‐1 was increased in tumor tissues according to most other studies using different malignant tumors.
The precise function of ESM‐1 is not known, but it has been suggested to regulate inflammatory processes by inhibiting the interation between ICAM‐1 and the integrin CD11a/CD18 (LFA‐1) on lymphocytes and monocytes.( 21 ) Inflammatory cytokines such as tumor necrosis factor‐α and interleukin‐1β stimulate ESM‐1 production and increased plasma levels have been found in patients suffering from inflammatory syndrome/septic shock.( 13 , 22 ) Little is known about the molecular mechanism of ESM‐1 contributing to colorectal carcinogenesis. In our unpublished data (2010), we showed that ESM‐1 silencing by siRNA inhibited CRC cell migration and invasion and decreased cell survival through the inhibition of the Akt/NF‐κB survival pathway. In these results, we suggest that ESM‐1 is involved with colorectal cancer cell migration and invasion and regulated cell survival through the Akt/NF‐κB pathway.
It is uncertain whether serum levels of ESM‐1 change in other malignant or inflammatory diseases. Grigoriu et al. ( 19 ) reported that serum levels of ESM‐1 were significantly higher for a group of 30 lung cancer patients compared with that for a group of 20 healthy volunteers. We also observed elevated serum levels of ESM‐1 in 33 lung cancer patients (72.2 ± 61.3 pg/mL), but not in 31 breast cancer patients (26.43 ± 29.2 pg/mL), although the data (2010) were not shown in results. Therefore, we need to evaluate serum levels of ESM‐1 in various malignant diseases. However, the results of this study warrant further investigation in large cohorts. We also need to compare the usefulness of serum ESM‐1 levels with fecal occult blood test or colonoscopy as a screening method for CRC. If confirmed, a test for serum ESM‐1 levels can be used as a valuable screening method, which will contribute to the early detection and treatment of CRC.
In summary, we have reported the differential expression of ESM‐1 in CRC tissues and serum. Our results suggest that ESM‐1 could be a useful marker for the diagnosis and treatment of CRC. Further investigation of the molecular mechanism involved in ESM‐1 action could reveal a novel approach for the treatment of CRC.
Abbreviations
- CEA
carcinoembryonic antigen
- CRC
colorectal cancer
- ESM‐1
endothelial cell‐specific molecule‐1
- IHC
immunohistochemical staining
- LR−
likelihood ratio of a negative test result
- LR+
likelihood ratio of a positive test result
- NPV
negative predictive value
- PPV
positive predictive value
- ROC
receiver operating characteristics
Supporting information
Fig. S1. Serum concentration and receiver operating characteristics (ROC) curve of carcinoembryonic antigen (CEA).
Table S1. Clinical characteristics and serum endothelial cell‐specific molecule‐1 (ESM‐1) level of patients with advanced colorectal adenoma.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Acknowledgments
We gratefully acknowledge financial support from the 21C Frontier Function Human Genomic Project and the KRIBB Research Initiative Fund from the Ministry of Education, Science, and Technology, Korea. We also thank the staff of the Samsung Medical Center (Seoul, Korea) for supplying the serum and tissue samples.
References
- 1. Ahlquist DA, Wieand HS, Moertel CG et al. Accuracy of fecal occult blood screening for colorectal neoplasia. A prospective study using Hemoccult and HemoQuant tests. JAMA 1993; 269: 1262–7. [PubMed] [Google Scholar]
- 2. Strul H, Arber N. Fecal occult blood test for colorectal cancer screening. Ann Oncol 2002; 13: 51–6. [DOI] [PubMed] [Google Scholar]
- 3. Collins JF, Lieberman DA, Durbin TE, Weiss DG. Accuracy of screening for fecal occult blood on a single stool sample obtained by digital rectal examination: a comparison with recommended sampling practice. Ann Intern Med 2005; 142: 81–5. [DOI] [PubMed] [Google Scholar]
- 4. Roessler M, Rollinger W, Palme S et al. Identification of nicotinamide N‐methyltransferase as a novel serum tumor marker for colorectal cancer. Clin Cancer Res 2005; 11: 6550–7. [DOI] [PubMed] [Google Scholar]
- 5. Melle C, Ernst G, Schimmel B et al. Discovery and identification of alpha‐defensins as low abundant, tumor‐derived serum markers in colorectal cancer. Gastroenterology 2005; 129: 66–73. [DOI] [PubMed] [Google Scholar]
- 6. Habermann JK, Roblick UJ, Luke BT et al. Increased serum levels of complement C3a anaphylatoxin indicate the presence of colorectal tumors. Gastroenterology 2006; 131: 1020–9. quiz 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kim HJ, Yu MH, Kim H, Byun J, Lee C. Noninvasive molecular biomarkers for the detection of colorectal cancer. BMB Rep 2008; 41: 685–92. [DOI] [PubMed] [Google Scholar]
- 8. Greene FL. TNM staging for malignancies of the digestive tract: 2003 changes and beyond. Semin Surg Oncol 2003; 21: 23–9. [DOI] [PubMed] [Google Scholar]
- 9. Akobeng AK. Understanding diagnostic tests 1: sensitivity, specificity and predictive values. Acta Paediatr 2007; 96: 338–41. [DOI] [PubMed] [Google Scholar]
- 10. Akobeng AK. Understanding diagnostic tests 2: likelihood ratios, pre‐ and post‐test probabilities and their use in clinical practice. Acta Paediatr 2007; 96: 487–91. [DOI] [PubMed] [Google Scholar]
- 11. Akobeng AK. Understanding diagnostic tests 3: receiver operating characteristic curves. Acta Paediatr 2007; 96: 644–7. [DOI] [PubMed] [Google Scholar]
- 12. Sarrazin S, Adam E, Lyon M et al. Endocan or endothelial cell specific molecule‐1 (ESM‐1): a potential novel endothelial cell marker and a new target for cancer therapy. Biochim Biophys Acta 2006; 1765: 25–37. [DOI] [PubMed] [Google Scholar]
- 13. Lassalle P, Molet S, Janin A et al. ESM‐1 is a novel human endothelial cell‐specific molecule expressed in lung and regulated by cytokines. J Biol Chem 1996; 271: 20458–64. [DOI] [PubMed] [Google Scholar]
- 14. Aitkenhead M, Wang SJ, Nakatsu MN, Mestas J, Heard C, Hughes CC. Identification of endothelial cell genes expressed in an in vitro model of angiogenesis: induction of ESM‐1, (beta)ig‐h3, and NrCAM. Microvasc Res 2002; 63: 159–71. [DOI] [PubMed] [Google Scholar]
- 15. Scherpereel A, Gentina T, Grigoriu B et al. Overexpression of endocan induces tumor formation. Cancer Res 2003; 63: 6084–9. [PubMed] [Google Scholar]
- 16. Rennel E, Mellberg S, Dimberg A et al. Endocan is a VEGF‐A and PI3K regulated gene with increased expression in human renal cancer. Exp Cell Res 2007; 313: 1285–94. [DOI] [PubMed] [Google Scholar]
- 17. Huang GW, Tao YM, Ding X. Endocan expression correlated with poor survival in human hepatocellular carcinoma. Dig Dis Sci 2009; 54: 389–94. [DOI] [PubMed] [Google Scholar]
- 18. Maurage CA, Adam E, Mineo JF et al. Endocan expression and localization in human glioblastomas. J Neuropathol Exp Neurol 2009; 68: 633–41. [DOI] [PubMed] [Google Scholar]
- 19. Grigoriu BD, Depontieu F, Scherpereel A et al. Endocan expression and relationship with survival in human non‐small cell lung cancer. Clin Cancer Res 2006; 12: 4575–82. [DOI] [PubMed] [Google Scholar]
- 20. Zuo L, Zhang SM, Hu RL et al. Correlation between expression and differentiation of endocan in colorectal cancer. World J Gastroenterol 2008; 14: 4562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bechard D, Scherpereel A, Hammad H et al. Human endothelial‐cell specific molecule‐1 binds directly to the integrin CD11a/CD18 (LFA‐1) and blocks binding to intercellular adhesion molecule‐1. J Immunol 2001; 167: 3099–106. [DOI] [PubMed] [Google Scholar]
- 22. Bechard D, Meignin V, Scherpereel A et al. Characterization of the secreted form of endothelial‐cell‐specific molecule 1 by specific monoclonal antibodies. J Vasc Res 2000; 37: 417–25. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Serum concentration and receiver operating characteristics (ROC) curve of carcinoembryonic antigen (CEA).
Table S1. Clinical characteristics and serum endothelial cell‐specific molecule‐1 (ESM‐1) level of patients with advanced colorectal adenoma.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


