Abstract
PI3Ks (phosphatidylinositol 3‐kinases) are lipid kinases that regulate signalling pathways involved in cell proliferation, motility, and adhesion. Somatic mutations and amplification of the PIK3CA gene have been reported in various types of human cancers. However, little is known about the frequency and prognosis role of PIK3CA activation in nasopharyngeal carcinoma (NPC). This study was conducted with the aim to screen for PIK3CA mutations in the two hot spot regions (exons 9 and 20) and to investigate for the PIK3CA gene amplification combined with the expression analysis of the phosphorylated Akt (pAkt). We showed that among 88 specimens, none had mutation in the helical domain (exon 9) and only one (1.13%) had mutation in the kinase domain (exon 20). On the other hand, PIK3CA gene amplification was found in 21.6% of cases and was strongly associated with distant metastasis (P = 0.002), lymph node involvement (P = 0.032), and advanced tumor stage (P < 0.001). Moreover, patients with PIK3CA copy number gain have a significant reduced overall survival time (P log rank = 0.02). We concluded that PIK3CA gene amplification is frequent in NPC and occurs in the advanced stage of NPC. Moreover, our finding emphasizes the association of PIK3CA gene amplification with worse prognosis in nasopharyngeal carcinoma. (Cancer Sci 2009); 00: 000–000)
The phosphatidylinositol 3‐kinase (PI3K)/Akt signaling pathway plays an important role in cell growth and proliferation initiated by activation of receptor tyrosine kinases and in tumor genesis and progression.( 1 , 2 ) Activation of cell‐surface receptors recruits PI3K, which phosphorylates the phosphatidylinositol 4,5‐bisphosphate substrate to generate phosphatidylinositol 3,4,5‐trisphosphate, recognized by the protein kinase Akt and its regulator PDK1. In the cell membrane, Akt is activated through phosphorylation at serine and threonine residues and thus modulates the expression of several genes involved in suppression of apoptosis and cell cycle progression.( 3 ) PIK3CA, the gene encoding the 110‐kDa subunit of PI3K, was mapped to 3q26, an area amplified in various human cancers including ovarian, head and neck, breast, urinary tract, and cervical cancers.( 3 , 4 , 5 ) The PIK3CA copy number gains are associated with increased PIK3CA transcription, P110‐alpha protein expression, and PI3K activity in ovarian cancer.( 6 ) Beside gene amplification, high frequency of PIK3CA mutations have been reported in colorectal, ovarian, lung and breast cancers.( 7 , 8 ) A majority of somatic mutations in PIK3CA are missense mutations clustering in exons 9 and 20, which encode a part of the helical and kinase domains, respectively.( 7 , 8 , 9 ) In vitro study has demonstrated that one of the most frequently observed PIK3CA mutations in human cancers, the H1047R (exon 20), is associated with increased kinase activity, indicating that the PIK3CA mutations actually activate the PI3K pathway.( 10 , 11 )
In nasopharyngeal carcinoma only few reports that studied PIK3CA have been available. In one small previous study of 40 patients with primary NPC, no mutations were detected in exons 9 and 20 of the PIK3CA gene, whereas copy number gains were found in 75% of these tumors without PIK3CA mutations.( 12 ) In a second recent study, PIK3CA mutations were detected in 9.6% of primary NPC tumors, and no association was found with clinico‐pathological features and prognosis.( 13 ) Thus, the relation of PIK3CA amplification to major clinico‐pathological parameters and patient outcome still remains unknown in NPC.
In Tunisia, NPC represents the most frequent head and neck cancer with a bimodal pattern of age distribution.( 14 , 15 ) Indeed, in the endemic regions of the South‐East Asia, there is only one major peak of incidence at about the age of 50 while in North Africa including Tunisia, an additional minor peak of incidence occurs between the ages of 10–20 years, and represents about 25% of all NPC patients.( 15 , 16 , 17 )
In this study, we aimed to investigate the frequency of PIK3CA mutations and amplification in Tunisian patients with NPC and their effect on patient survival and major clinical parameters.
Materials and Methods
Tumor specimens and DNA extraction. Primary NPC biopsies were collected from 88 patients who underwent resection before any treatment at the Sfax University Hospital in the south of Tunisia. The median age is 40 years (range, 10 to 80 years) at the time of diagnosis. The clinical stage was determined according to the tumor, node, and metastasis (TNM) classification of the American Joint Committee on Cancer (AJCC)/International Union Against Cancer (UICC).( 18 ) Histological type was determined on tissues sections according to the World Health Organization criteria. Our patients were grouped as followed: 41cases (47%) of undifferentiated type (UCNT) and 47 cases (53%) of non‐keratinizing carcinoma. The majority of patients (83%) had palpable neck lymph nodes (N+). In addition, 10 histological normal hyperplasia tissues were collected from patients with clinical symptoms indicative of NPC but nasopharyngeal biopsies did not show tumoral cells. These specimens were used as controls.
Genomic DNA was extracted from frozen sections using the Wizard SV Genomic DNA purification system according to the manufacturer’s protocol (Promega, France).
Informed consent for use of all human specimens in this study was obtained under a protocol approved by CHU Habib Bourguiba (Sfax‐Tunisia) institutional review board.
PCR‐SSCP. PCR amplification targeting the PIK3CA gene areas of interest were performed using two primer pairs based on the PIK3CA sequence obtained from Gene Bank NM_006218.
To optimize the size of the PCR fragments for polymerase chain reaction (PCR)‐single strand conformation polymorphism (SSCP) analysis and avoid co‐amplification of the pseudogene, primer pairs were chosen to individually amplify exon 9 of PIK3CA using two‐step PCR according to the protocol described previously( 19 ) Primary PCR was performed using the following primers: forward 5′‐AACTT CAGCAGTTACTATTCTGTGAC‐3′ and reverse: 5′‐GATTTTCCACA AATATCA ATT TA CAA‐3′ generating 575 bp. PCR amplification was performed in 25 μL containing 2 nM of dNTPs, 1.5 mm MgCl2, 50 mm KCl, 0.2 mm of each primer, and 1 U of Taq DNA polymerase (Fermentas, France). PCRs were done with the touchdown conditions: an initial 3 min denaturation at 95°C followed by 25 cycles of 1 min each at 94°C, 61°C, and 72°C, and a single final extension step for 5 min at 72°C. The nested PCR was done using the following primers: forward 5′‐CTGTGAATCCAGAGGGGAAA‐3′ and reverse: 5′‐ACAG AGAATCTCCATTTTAGCA‐3′ to generate 217 bp products. PCR were done with an initial 3 min denaturation at 95°C followed by 25 cycles of 1 min each at 94°C, 60°C, 72°C, and a final extension step for 5 min at 72°C. After amplification, PCR products were diluted 5‐fold in loading buffer containing 20 mm/L EDTA, 94% formamide and 0.05% of bromophenol blue and xylene cyanol. After denaturation at 94°C for 10 min, each sample was immediately chilled on ice for 5 min and applied on 6% non denatured polyacrylamide gel containing 10% glycerol. Electrophoresis was performed at 6°C with constant voltage (100 V) in 1X TBE buffer for 20 h. DNA was visualized using the DNA Silver Staining Kit according to the manufacture’s recommendations (Amersham‐Biosciences, Athens, Greece).
PCR‐DHPLC analysis. PIK3CA exon 20 was screened for mutations by denaturating high performance liquid chromatography (DHPLC) (Wave Nucleic Acid Fragment Analysis System; Transgenomic, Omaha, USA). Briefly, PCR was performed using primers: forward 5′‐TTT GCTCCAAACTGACCAAAC‐3′ and reverse: 5′‐CAGTGCAGTGTGGAATCCAG‐3′ giving a 360 bp DNA fragment. The cycling conditions consisted of an initial 5 min denaturation at 95°C followed by 35 cycles of 45 s each at 94°C, 60°C, 72°C, and a single final extension for 5 min at 72°C. PCR products displaying different profiles compared to the wild type were sequenced using the Big Dye Cycle Sequencing Kit with the ABI Prism 3100 DNA sequencer (Applied Biosystems, Foster City, CA, USA).
PIK3CA copy number quantification. To determine the level of PIK3CA gene amplification, copy number quantification was carried out using quantitative real‐time PCR (iCycler iQ System; Bio‐Rad, France) with the GAPDH gene (glyceraldehyde‐3‐phosphate dehydrogenase) as the internal control. The PCR reaction was carried out with the QTM Syber Green Supermix (Bio‐Rad) using 50ng of DNA in a 25 μL final reaction volume. The thermal cycling conditions were as follows: 5 min at 95°C and 50 cycles of 15 s at 95°C, 15 s at 58°C, and 15 s at 72°C. The GAPDH primers were: 5′‐ACCCACTCCTCCACCTTTG‐3′ (forward) and 5′‐ACCCACTCCTCCACCTTTG‐3′ (reverse) to generate a 188 bp fragment. The PIK3CA primers were: 5′AAATGAAAGCTCACTCTGGATT‐3′ (forward) and 5′‐TGTGCAATTCCTATGCAATCG‐3′ (reverse) to generate a fragment of 81 bp. Each sample was run in duplicate for both target and endogenous genes. Each case was repeated twice and the mean value was calculated.
The amount of PIK3CA‐specific template was estimated in each carcinoma with respect to the control gene. The relative representation of the PIK3CA copy number (PIK3CA gain) in each tumor with respect to non‐tumor tissue is given by the formula 2−ΔΔCt, which is a simplified calculation tool derived from the ΔΔCt method for gene expression analysis.( 20 )
The ΔΔCt = (Ct PIK3CA−Ct GAPDH)samples−(Ct PIK3CA−Ct GAPDH) calibrator. Normal nasopharyngeal mucosa (10 cases) were used to normalize the data. The differences between the cycle threshold value (Ct: the cycle at which the fluorescence rises appreciably above the background fluorescence) of each tumor and the mean cycle threshold value of the normal nasopharyngeal mucosa were designated as ΔCt. PIK3CA copy number gain or amplification ≥3 were considered significant. For statistical evaluation, the cases were sub‐divided into three groups with respect to the PIK3CA gene copies number (0–3 no amplification, 3–4 moderate amplification, >4 high amplification).
PAkt protein expression. To evaluate Akt phosphorylation status, we performed western blot analysis on available NPC samples that allowed us to perform protein extraction. Frozen tumors were collected in sodium dodecyl sulfate lysis buffer and proteins were extracted by Trizol Reagent (Invitrogen, France). Approximately 50 μg of total proteins was denatured in loading buffer for 10 min, electrophoresed on 10% SDS‐PAGE, and electroblotted to Hybond‐P (Amersham Biosciences). The membrane was incubated overnight with primary antibody anti‐Akt Thr308 (rabbit polyclonal IgG, 1:200; Santa Cruz Biotechnology, Hildelberg, Germany), then with anti‐β‐actin (antimouse monoclonal antibody, 1:500; Santa Cruz Biotechnology) at 4°C. The membrane was washed three times in PBS with 0.1% Tween 20 at room temperature and incubated with horseradish peroxidase–labeled secondary antibody (goat antirabbit IgG or goat antimouse IgG; Sigma‐Aldrich, France) for 1 h at room temperature. Signal detection was performed by horseradish peroxidase chemiluminescent reaction (ECL plus; Amersham Biosciences).
Statistical analysis. Statistical analyses were performed using SPSS (SPSS, Inc., Chicago, IL, USA) 13.0 statistical software for Windows. The χ2 test was used to determine associations between the amplification of PIK3CA gene and various clinico‐pathological features of NPC. Analyses of disease free survival (end‐point = loco‐regional recurrence or distant metastases) and cancer‐specific survival (end‐point = death from NPC) were performed using Kaplan–Meier survival plots, and the significance was tested using the log‐rank test. P‐values of < 0.05 were considered significant.
Results
Mutations analysis. Because more than 80% of mutations in the PIK3CA gene occur in helical (exon 9) and kinase (exon 20) domains, we screened these exons in 88 primary NPC tumors using PCR‐SSCP (for exon 9) and PCR‐DHPLC (for exon 20). For SSCP analysis, we used nested PCR with a primer design that avoids confounding PIK3CA pseudogene sequences because PIK3CA shows a high homology (98%) with a genomic fragment in the Cat Eye Syndrome region. As positive control, we used two DNA samples with G1633A (E545K) or G1624A (E542K) mutations. A representative example of PCR‐SSCP is shown in Figure 1. No aberrant shifted bands were found in the exon 9 of the 88 NPC specimens tested compared with DNA from non‐tumoral nasopharyngeal mucosa used as negative control (Fig. 1).
Figure 1.
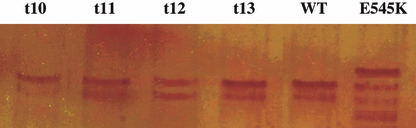
Single strand conformation performance liquid (SSCP) analysis of PIK3CA exon 9 (wild type [WT]). WT DNA sample from non‐tumoral tissues. t10, t11, t12, and t13, DNA from nasopharyngeal carcinoma (NPC) tumors. E545K is a DNA with mutation E>K in position 545 of PIK3CA exon 9.
For exon 20 (360 bp) we used the PCR‐DHPLC as prescreening approach for primary NPC specimens using as a control one DNA with a wild‐type sequence. Only one synonymous point mutation (C3231T) leading to the replacement of the codon “cct” with “tct” (both encoding leucin 2710) was found in exon 20 in one NPC case (Fig. 2A,B). No other mutations were detected in exons 9 and 20.
Figure 2.
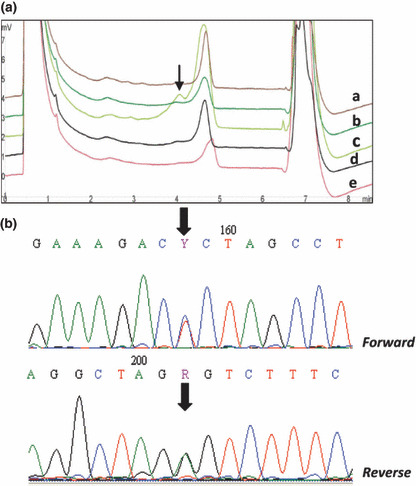
(a) Denaturating high performance liquid chromatography (DHPLC) profiles of PIK3CA exon 20 (a) wild type DNA sample (parts b, c, d, and e) correspond to DNA from NPC biopsies: note the small peak (shown by the black arrows) at 4.3 min that was not observed in the control sample which indicates the existence of heterozygous change. (b) Sequence analysis of exon 20 (NPC126) showing an additional peak on the DHPLC profile. The arrows indicate the heterozygous mutation Y 3231R.
PIK3CA gene amplification in NPC. To determine the PIK3CA gene copy number, we performed quantitative PCR on 88 primary tumors, and 10 normal nasopharyngeal mucosa were used as calibrator to normalize the data. Standard curves for PIK3CA and GAPDH amplification were generated and showed linearity over the range used. Most samples showed no difference in GAPDH amplification between tumor and normal samples. A representative example of GAPDH and PIK3CA amplification in tumor and normal samples is shown in Figure 3.
Figure 3.
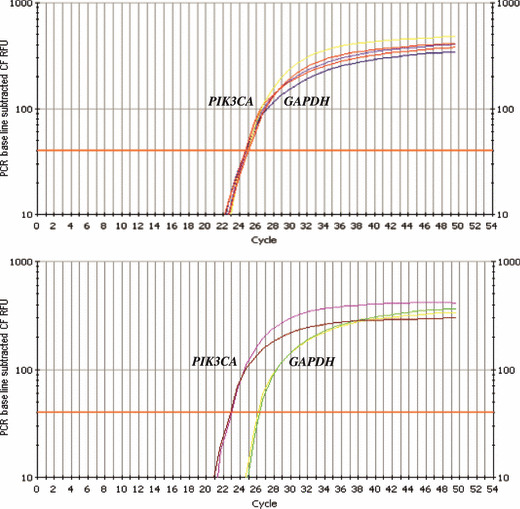
Determination of PIK3CA gene copy number by quantitative PCR. Relative fluorescence of PIK3CA and GAPDH reactions are plotted against cycle number. The horizontal line marks the threshold used for assessment of C t value. Upper, normal nasopharyngeal epithelia used as a negative control. Bottom, example of nasopharyngeal carcinoma (NPC) case showing genomic amplification of the PIK3CA gene.
Among the 88 cases, 19 (21.6%) showed a gain of gene copy number; among them, 16 cases (84%) showed more than four copies and three (16%) had a moderate amplification with three to four copies (Table 1).
Table 1.
Clinical features of nasopharyngeal carcinoma (NPC) patients with PIK3CA gene amplification and pAkt expression
| Cases | Age (year) | Histology (WHO type)‡ | Tumor stage | Lymphe node | Distant metastasis | pAKT expression | PIK3CA copy number | |
|---|---|---|---|---|---|---|---|---|
| 3–4 | >4 | |||||||
| T1† | 64 | Non‐keratinizing carcinoma | T3 | N+ | M1 | + | ND | + |
| T2 | 17 | Non‐keratinizing carcinoma | T4 | N+ | M1 | ND | + | ND |
| T3 | 41 | Undifferentiated (UCNT) | T4 | N+ | M0 | ND | ND | + |
| T4 | 46 | Undifferentiated (UCNT) | T4 | N+ | M1 | ND | ND | + |
| T5 | 28 | Undifferentiated (UCNT) | T4 | N+ | M1 | ND | ND | + |
| T6 | 20 | Non‐keratinizing carcinoma | T4 | N+ | M1 | ND | + | ND |
| T7† | 13 | Non‐keratinizing carcinoma | T4 | N+ | M1 | + | ND | + |
| T8† | 64 | Non‐keratinizing carcinoma | T4 | N+ | M0 | + | ND | + |
| T9 | 77 | Undifferentiated (UCNT) | T4 | N+ | M1 | ND | ND | + |
| T10† | 74 | Undifferentiated (UCNT) | T4 | N+ | M0 | + | ND | + |
| T11 | 64 | Non‐keratinizing carcinoma | T4 | N+ | M0 | ND | + | ND |
| T12† | 66 | Undifferentiated (UCNT) | T4 | N+ | M0 | + | ND | + |
| T13† | 40 | Non‐keratinizing carcinoma | T4 | N+ | M1 | + | ND | + |
| T14† | 54 | Non‐keratinizing carcinoma | T4 | N+ | M0 | + | ND | + |
| T15† | 40 | Non‐keratinizing carcinoma | T4 | N+ | M0 | + | ND | + |
| T16 | 48 | Undifferentiated (UCNT) | T4 | N+ | M0 | ND | ND | + |
| T17† | 43 | Non‐keratinizing carcinoma | T3 | N+ | M1 | + | ND | + |
| T18 | 55 | Non‐keratinizing carcinoma | T4 | N+ | M1 | ND | ND | + |
| T19 | 42 | Non‐keratinizing carcinoma | T4 | N+ | M1 | ND | ND | + |
†The nine cases tested for pAkt expression. ‡American Joint Committee on Cancer (AJCC, 5th edition), N+, lymph node involvement. Metastatic status defined as M0 in the absence of clinical or radiological evidence of distant metastasis at the initial workup, M1 in the other cases (synchronous metastasis).
Correlation between PIK3CA amplification and clinico‐pathological parameters. The relationship of PIK3CA amplification with clinico‐pathologic variables is summarized in Table 2. Overall, tumors with PIK3CA amplification show more aggressive behavior. In univariate analysis, PIK3CA amplification is significantly higher in tumors with advanced tumor stage (T4, 45%vs T1‐3, 4%; P = 0.001), with lymph node (N+,100%vs N0, 0%; P = 0.032), and with distant metastasis (M+, 60%vs M0, 19.6%; P = 0.009). Multivariate analysis using the logistic regression model showed a strong association with tumor stage (P = 0.003, odds ratio [OR] = 0.084, 95% confidence interval [CI] = 0.017–0.42) and distant metastasis (P = 0.009, OR = 0.147, 95% CI = 0.035–0.617) whereas the association with lymph node involvement was lost.
Table 2.
Correlation between PIK3CA amplification and clinical features
| Number | PIK3CA amplification (%) | |
|---|---|---|
| Cases | 88 | 19 (21.6) |
| Age (years) | ||
| ≤30 | 23 | 4 (21) |
| >30 | 65 | 15 (79) |
| Histology (WHO type)† | ||
| Undifferentiated (UCNT) | 41 | 7 (37) |
| Non‐keratinizing carcinoma | 47 | 12 (63) |
| Tumor stage | ||
| T1–3 | 48 | 2 (11) |
| T4 | 40 | 17 (89) |
| P‐value (χ2 test) | 0.001 | |
| Lymph node | ||
| N0 | 15 | 0 (0) |
| N+ | 73 | 19 (100) |
| P‐value (χ2 test) | 0.032 | |
| Metastasis | ||
| M0 | 72 | 8 (42) |
| M1 | 16 | 11 (58) |
| P‐value (χ2 test) | 0.002 | |
| Clinical stage | ||
| Stage I–III | 18 | 0 (0) |
| Stage IV | 70 | 19 (100) |
| P‐value (χ2 test) | 0.013 | |
†American Joint Committee on Cancer (AJCC, 5th edition), N+, lymph node involvement. Metastatic status defined as M0 in the absence of clinical or radiological evidence of distant metastasis at the initial workup, M1 in the other cases (synchronous metastasis). WHO, World Health Organization.
Relationship of PIK3CA amplification and patient survival. Survival data was available for only 53 patients, among them 27 patients died of their disease. The Kaplan–Meier method for overall survival revealed significant association in the survival times of patients with respect to PIK3CA amplification (log‐rank test, P = 0.028, Fig. 4a). Moreover in the group of patients with advanced NPC (TNM stage IV), PIK3CA amplification was associated with a poor survival (Fig. 4b) suggesting once again that activation of PIK3CA via copy number gain is associated with worse prognosis in NPC. No significant association between PIK3CA amplification and disease free survival was noted in our series.
Figure 4.
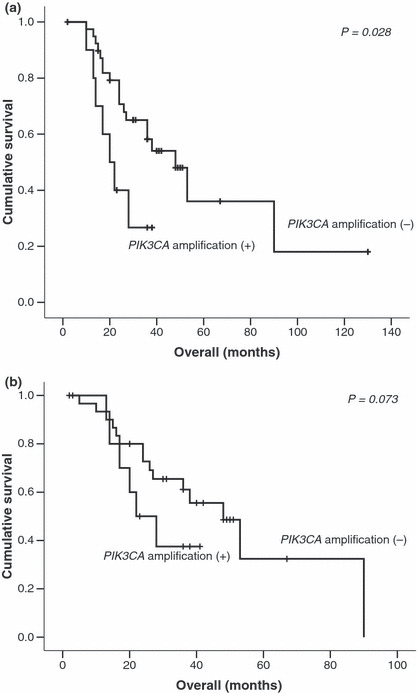
Kaplan–Meier curves for (a) overall survival and (b) overall survival in patients with stage IV nasopharyngeal carcinoma (NPC) according to PIK3CA amplification status.
pAkt‐protein expression in NPC tissues. To further evaluate the functional significance of the PIK3CA gene amplification, we studied the expression of active Akt (pAkt) on 19 available NPC samples, 10 of them without PIK3CA amplification and nine with high amplification level (Table 1). We found a perfect correlation between pAkt expression and PIK3CA amplification since all tested specimens with more than four copies of the PIK3CA gene showed pAkt expression, whereas none of the 10 cases without PIK3CA amplification exhibited the activated Akt (Fig. 5).
Figure 5.
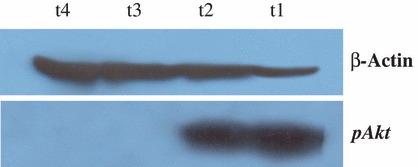
Western blot analysis of Akt phosphorylation in nasopharyngeal carcinoma (NPC) samples. Anti‐pAkt was used as a primary antibody to visualize pAkt (52 kDa), and β‐actin levels were examined to ensure the integrity of the protein preparations. Note that t1 and t2 samples harbor more than four copies of the PIK3CA gene, while t3 and t4 do not carry PIK3CA amplification as determined by quantitative PCR.
Furthermore, and as shown in Table 1, all patients with tumors exhibiting PIK3CA gene amplification and pAkt expression had lymph nodes and advanced T‐stage, and 11 out of 19 patients developed distant metastasis. Our result suggests that PIK3CA gene amplification and pAkt expression occur in advanced NPC stage.
Discussion
The PI3K/Akt pathway plays a central role in many physiological processes and is implicated in malignant transformation by promoting cell cycle progression, and invasion.( 21 ) Many studies have already reported PIK3CA somatic mutation in a wide variety of human cancers, such as lung, colorectal, ovarian, gastric, and breast cancer.( 22 , 23 , 24 , 25 ) Functional significance of these missense mutations was suggested because they are clustered in the helical (exon 9) and kinase domains (exon 20); also the mutant protein showed increased kinase activity( 26 ) and oncogenic potential.( 22 )
In NPC, there are only a few published reports on PIK3CA mutations and whether it affects the clinical behavior and patient prognosis; however, to the best of our knowledge, the effect of PIK3CA amplification on patient survival is not documented in NPC. In this study, we confirmed that PIK3CA mutations are rare in NPC (1.13%) compared to other cancers such as colon( 27 ) and breast cancer.( 28 ) Our results are concordant with those of two independent studies conducted on Asian patients, showing the absence or the low frequency (9%) of PIK3CA mutations in the two hotspot regions.( 12 , 13 )
Beside mutation, PIK3CA amplification has been reported in many human cancers. In this study, we investigated by quantitative PCR (qPCR) the status of PIK3CA in 88 NPC. Nineteen out of 88 cases (21.6%) harbored amplification. A previous study employing array‐based comparative genomic hybridization and FISH analysis showed that PIK3CA amplification was observed in 24 out of 32 cases (75%). Discrepancies between qPCR and FISH or CGH methods were previously observed in a study assessing the PIK3CA gene in ovarian carcinoma.( 26 ) This could be due to the fact that FISH or CGH methods are not able to detect changes restricted only to a small genomic region and the qPCR approach seems to be more appropriate to assess genetic aberrations limited to a single gene.
In line with our result, recent studies based on qPCR detected PIK3CA amplification in 24.6% and 22% of ovarian cancers.( 26 , 29 ) Moreover, it was reported in thyroid tumors that PIK3CA gene mutation is not a common mechanism in the activation of PIK3CA, whereas gene amplification occurs with a relatively high frequency. This suggests that the genetic alteration may play an important role in the tumorigenesis of thyroid cancer.( 30 )
In the current study, the univariate and multivariate analysis revealed a significant association between PIK3CA amplification and tumors with aggressive behavior, i.e. advanced tumor stage and presence of distant metastasis. All cases with stage IV showed PIK3CA amplification compared to specimens in stage I–III. Recently, Chou et al. reported that PIK3CA mutation was slightly influenced by sex but had no significant relationship to other clinicopathological characteristics in NPC.( 13 )
On the other hand, little has been known regarding the impact of PIK3CA mutation in NPC on patient survival. One recent study has shown that disease‐specific survival was not significantly affected by PIK3CA mutations.( 13 ) However, in colon cancer, PIK3CA mutations were associated with a significant increase in colon cancer–specific mortality.( 27 , 31 )
To the best of our knowledge, this is the first study demonstrating the prognosis value of PIK3CA amplification in NPC. We showed that PIK3CA amplification is associated with a poor overall survival (P log rank = 0.02). In addition, patients with TNM stage IV have a worse survival in a background where PIK3CA is amplified than those in the same stage without PIK3CA amplification. This suggests once again that activation of PIK3CA is a marker of poor prognosis in NPC. Further, to investigate the functional implication of PIK3CA amplification, we evaluated the expression of active Akt (pAkt) on 19 available NPC samples. The pAkt was observed in all tumors carrying PIK3CA amplification (nine out of nine) while pAkt was not detected in tumors without PIK3CA amplification. This result is in line with previous reports and confirms the importance of the protein kinase Akt as a downstream effector of PIK3CA in NPC. In lung cancer, it was shown that PIK3CA mutations and gains were strongly correlated with expression of activated pAkt.( 32 )
In summary, our study shows that PIK3CA amplification is associated with aggressive tumor behavior and poor survival in patients with nasopharyngeal carcinoma. We also confirm that mutations in exons 9 and 20 of the PIK3CA gene are not a common event in NPC as in other epithelial cancers. Our findings suggest the implication of PIK3CA as an oncogene with a prognostic relevance in NPC. Specific inhibitors of the PI3K/Akt pathway could be a promising target for the treatment of patients with NPC.
Disclosure Statement
The authors guarantee that there are no financial or personal relationships with other people or organizations that might pose a conflict of interest in connection with this work.
Acknowledgments
We wish to thank Professor P. Busson (IGR Pans, France), Professor P. Soderkvist (Faculty of Health Science, Sweden), and Professor K.S. Al‐Kuraya (King Fahad National Center for Children’s Cancer and Research, Ryiad, Saudi Arabia) for providing us with β‐actin antibody, DNA samples carrying E545K and E542K mutations, and PIK3CA amplication to use as controls. We are also grateful to Professor Fattoum and Professor Ben Masseoud from the Children’s Hospital (Tunis, Tunisia) for DHPLC facilities. This work was supported by a grant from the Ministère de l’Enseignement Supérieur et de la Recherche Scientifique of Tunisia.
References
- 1. Manning BD, Cantley LC. AKT/PKB signalling: navigating downstream. Cell 2007; 129: 1261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fresno Vara JA, Casado E, Castro J et al. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev 2004; 30: 193–204. [DOI] [PubMed] [Google Scholar]
- 3. Knuutila S, Bjorkqvist AM, Autio K et al. DNA copy number amplifications in human neoplasms: review of comparative genomic hybridization studies. Am J Pathol 1998; 152: 1107–23. [PMC free article] [PubMed] [Google Scholar]
- 4. Rooney PH, Murray GI, Stevenson DA et al. Comparative genomic hybridization and chromosomal instability in solid tumours. Br J Cancer 1999; 80: 862–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Larramendy ML, Lushnikova T, Bjorkqvist AM et al. Comparative genomic hybridization reveals complex genetic changes in primary breast cancer tumors and their cell lines. Cancer Genet Cytogenet 2000; 119: 132–8. [DOI] [PubMed] [Google Scholar]
- 6. Shayesteh L, Lu Y, Kuo WL et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet 1999; 21: 99–102. [DOI] [PubMed] [Google Scholar]
- 7. Samuels Y, Wang Z, Bardelli A et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004; 304: 554. [DOI] [PubMed] [Google Scholar]
- 8. Racz A, Brass N, Heckel D et al. Expression analysis of genes at 3q26‐q27 involved in frequent amplification in squamous cell lung carcinoma. Eur J Cancer 1999; 35: 641–6. [DOI] [PubMed] [Google Scholar]
- 9. Wang Y, Helland R, Holm A et al. PIK3CA mutation in advanced ovarian carcinoma. Hum Mutat 2005; 25: 316–22. [DOI] [PubMed] [Google Scholar]
- 10. Ikenoue T, Kanai F, Hikiba Y et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res 2005; 65: 4562–7. [DOI] [PubMed] [Google Scholar]
- 11. Samuels Y, Diaz LA, Schmidt‐Kittler O et al. Mutant PIK3CA promotes cell growth and invasion of human cancer cells. Cancer Cell 2005; 7: 561–73. [DOI] [PubMed] [Google Scholar]
- 12. Or YY, Hui AB, To KF et al. PIK3CA mutation in nasopharyngeal carcinoma. Int J Cancer 2006; 118: 1065–7. [DOI] [PubMed] [Google Scholar]
- 13. Chou CC, Chou MJ, Tzen CY et al. PIK3CA mutation occurs in nasopharyngeal carcinoma but does not significantly influence the disease‐specific survival. Med Oncol 2008; 10: 9121–7. [DOI] [PubMed] [Google Scholar]
- 14. Busson P, Keryer C, Ooka T et al. EBV‐associated nasopharyngeal carcinomas: from epidemiology to virus‐targeting strategies. Trends Microbiol 2004; 12: 356–60. [DOI] [PubMed] [Google Scholar]
- 15. Kammoun M, Vogt‐Hoerner G, Mourali N. Tumors of the nasopharynx in Tunisia: an anatomic and clinical study based on 143 cases. Cancer 1974; 33: 184–91. [DOI] [PubMed] [Google Scholar]
- 16. Khabir A, Sellami A, Sakka M et al. Contrasted frequencies of p53 accumulation in the two age groups of North African nasopharyngeal carcinomas. Clin Cancer Res 2000; 6: 3932–6. [PubMed] [Google Scholar]
- 17. Daoud J, Toumi N, Bouaziz M et al. Nasopharyngeal carcinoma in childhood and adolescence: analysis of a series of 32 patients treated with combined chemotherapy and radiotherapy. Eur J Cancer 2003; 39: 2349–54. [DOI] [PubMed] [Google Scholar]
- 18. Sobin LH, Wittekind CH. International Union Against Cancer, TNM Classification of Malignant Tumours, 5th edn. New York: Wiley‐Liss, 1997: 25–32. [Google Scholar]
- 19. Hummerdal P, Andersson P, Willander K et al. Absence of hot spot mutations of the PIK3CA gene in acute myeloid leukaemia. Eur J Haematol 2006; 77: 581–5. [DOI] [PubMed] [Google Scholar]
- 20. Livak K J, Thomas D S. Analysis of relative gene expression data using real‐time quantitative pcr and the 2−ΔΔCT method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- 21. Osaki M, Oshimura M, Ito H. PI3K–Akt pathway: its functions and alterations in human cancer. Apoptosis 2000; 49: 667–76. [DOI] [PubMed] [Google Scholar]
- 22. Kang S, Bader AG, Vogt PK et al. Phosphatidylinositol 3‐kinase mutations identified in human cancer are oncogenic. Proc Natl Acad Sci 2005; 102: 802–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li VS, Wong CW, Chan TL et al. Mutations of PIK3CA in gastric adenocarcinoma. BMC Cancer 2005; 5: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee JW, Soung YH, Kim SY et al. PIK3CA gene is frequently mutated in breast carcinomas and hepatocellular carcinomas. Oncogene 2005; 24: 1477–80. [DOI] [PubMed] [Google Scholar]
- 25. Bachman KE, Argani P, Samuels Y et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther 2004; 3: 772–5. [DOI] [PubMed] [Google Scholar]
- 26. Woenckhaus J, Steger K, Sturm K et al. Prognostic value of PIK3CA and phosphorylated AKT expression in ovarian cancer. Virchows Arch 2007; 450: 387–95. [DOI] [PubMed] [Google Scholar]
- 27. Kato S, Iida S, Higuchi T et al. PIK3CA mutation is predictive of poor survival in patients with colorectal cancer. Int J Cancer 2007; 121: 1771–8. [DOI] [PubMed] [Google Scholar]
- 28. Maruyama N, Miyoshi Y, Taguchi T et al. Clinicopathologic analysis of breast cancers with PIK3CA mutations in Japanese women. Clin Cancer Res 2007; 13: 408–14. [DOI] [PubMed] [Google Scholar]
- 29. Campbell IG, Russell SE, Choong DC et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res 2004; 64: 7678–81. [DOI] [PubMed] [Google Scholar]
- 30. Trink B, Paul WL, Sidransky D et al. Uncommon mutation, but common amplifications, of the PIK3CA gene in thyroid tumors. J Clin Endocrinol Metab 2005; 90: 4688–93. [DOI] [PubMed] [Google Scholar]
- 31. Ogino S, Nosho K, Gregory J et al. PIK3CA mutation is associated with poor prognosis among patients with curatively resected colon cancer. J Clin Oncol 2009; 193: 110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Amamoto H, Shigematsu H, Nomura M et al. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res 2008; 68: 6913–23. [DOI] [PMC free article] [PubMed] [Google Scholar]


