Abstract
Optimization of intracellular concentrations of dNTPs is critical for the fidelity of DNA synthesis during DNA replication and repair because levels that are too high or too low can easily lead to increased rates of mutagenesis. Recent advances in the analysis of intracellular concentrations of dNTPs have suggested that eukaryotes use diverse mechanisms in supplying dNTPs for DNA synthesis during DNA replication and repair. The enzyme ribonucleotide reductase (RNR) is a key enzyme involved in the synthesis of dNTPs. We found that Tip60‐dependent recruitment of RNR at sites of DNA damage is essential for supplying a sufficient amount of dNTPs for mammalian DNA repair. In this review, we focus on recent findings related to RNR regulation in eukaryotes of the dNTPs supplied for DNA synthesis. We also discuss the effect of this regulation on mutagenesis and tumorigenesis. (Cancer Sci 2010; 101: 2505–2509)
Ribonucleotide reductase (RNR) catalyzes the substitution of the 2′hydroxyl group of a ribonucleoside diphosphate with hydrogen, resulting in a deoxyribonucleoside diphosphate (dNDP) (Fig. 1A).( 1 ) The minimal form of this enzyme is an a2b2 heterotetramer, but its higher order forms (a6b6 or a6b2) also exist as active enzymes.( 2 , 3 ) In mammalian cells, the a‐protein and b‐protein are referred to as the R1 protein and R2 protein, respectively. The R1 protein (90 kDa) carries the active site as well as binding sites for allosteric effectors. The smaller R2 protein (45 kDa) contains an Fe–O–Fe center, which generates a tyrosyl‐free radical. During catalysis, this radical is continuously shuttled to a cysteine residue in the active site.( 4 ) Mammalian cells possess a single large subunit, R1 and two distinct small subunits, R2 and p53R2, the latter being induced by DNA damage in a manner dependent on p53 (Fig. 1A).( 5 ) Similarly, the RNR of budding yeast contains two large subunits of Rnr1 and one each of the small subunits Rnr2 and Rnr4.( 6 ) The expression of Rnr3, another large subunit, is highly elevated after DNA damage, suggesting its role in DNA repair,( 7 ) although the level of Rnr3 protein even after DNA damage never reaches more than one‐tenth of the Rnr1 protein level and a complex between Rnr3 and the Rnr2/Rnr4 heterodimer has a very low catalytic activity.( 8 ) Intriguingly, the Rnr3‐catalyzed CDP reduction is significantly stimulated by dATP, indicating that Rnr3 lacks a functional allosteric activity site that allows generation of higher levels of dNTPs for DNA repair( 8 ) (see below).
Figure 1.
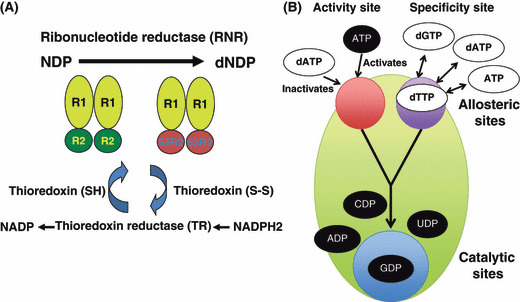
Ribonucleotide reductase (RNR) and its allosteric regulation. (A) The enzyme RNR catalyzes the de novo synthesis of dNTPs. Catalysis of ribonucleoside 5′‐diphosphate (NDP) involves a reduction at the 2′‐carbon of ribose 5‐phosphate to form the 2′‐deoxy derivative‐reduced 2′‐deoxyribonucleoside 5′‐diphosphate (dNDP). This reduction is initiated with the generation of a free radical. Following a single reduction, RNR requires electrons donated from the dithiol groups of the protein thioredoxin. Regeneration of thioredoxin occurs when nicotinamide adenine dinucleotide phosphate (NADPH) provides two hydrogen atoms that are used to reduce the disulfide groups of the thioredoxin protein. (B) Allosteric regulation of RNR. RNR is activated by binding ATP or inactivated by binding dATP to the activity site located on the large subunit Rnr1 (R1). When the enzyme is activated, substrates are reduced if the corresponding effectors bind to the allosteric substrate specificity site as follows. When dATP or ATP is bound at the allosteric site, the enzyme accepts UDP and CDP into the catalytic site. When dGTP is bound, ADP enters the catalytic site. When dTTP is bound, GDP enters the catalytic site. The substrates (UDP, CDP, ADP, and GDP) are converted to dNTPs. R2, small subunit Rnr2‐Rnr4.
Ribonucleotide reductase is controlled by various regulatory mechanisms, such as allosteric regulation, transcriptional regulation, post‐transcriptional modification, and regulation of subcellular localization.( 1 , 4 ) The eukaryotic large subunit of RNR contains two allosteric sites each consisting of a polypeptide capable of binding to effectors (dNTP or NTP). One of these is the specificity site that monitors balance among the four dNTPs and adjusts its activity to maintain their ratio, and the other is the activity site that regulates the total dNTP pool size by monitoring the dATP/ATP ratio. Both sites are required for maintaining all dNTPs at the correct overall levels (Fig. 1B).( 9 ) In the case of DNA damage, in order to produce higher levels of dNTPs, it might be possible that either the ATP level may also increase or negative feedback regulations may be suppressed through unknown mechanisms. A failure of dATP feedback inhibition in budding yeast Rnr1 mutants results in a drastic increase in the dNTP pool during DNA damage.( 10 )
In addition to RNR, thymidylate synthase (TS) is a key enzyme in the synthesis of 2′‐deoxythymidine‐5′‐monophosphate (dTMP) by catalyzing the methylation of 2′‐deoxyuridine‐5′‐monophosphate (dUMP). Thymidylate synthase provides the sole intracellular de novo source of dTMP. The hydrolysis of dUTP to dUMP is catalyzed by dUTPase and the resultant pyrophosphate simultaneously provides substrate to TS. In addition, 2′‐deoxycytidine‐5′‐monophosphate (dCMP) deaminase may produce dUMP by the deamination of dCMP. Nucleotide diphosphate kinases (NDPK) are also required for dNTP synthesis through phosphorylation of dNDPs. Therefore, these enzymes should also play an important role in the balanced supply of dNTPs for DNA synthesis. In this review, we summarize recent findings on the regulation of the dNTPs supplied for DNA synthesis. An insufficient or unbalanced supply of dNTPs into DNA strands can result in genetic abnormalities and cell death.( 11 ) Implications of dNTP supply related to carcinogenesis are also discussed.
Mechanism of dNTP supply during DNA replication and repair in yeasts
In budding yeast, dNTP levels during DNA replication are approximately threefold higher than in G1 phase.( 10 ) Ribonucleotide reduction in budding yeast shows maximum activity in S phase, which is explained by the cell cycle‐dependent expression of the Rnr1 gene.( 12 ) There is more than a 10‐fold fluctuation in Rnr1 mRNA concentrations in coordination with the induction of the POL1 gene during the cell cycle, whereas the Rnr2 transcript shows only a twofold fluctuation. Similarly, cdc22 R1 and suc22 R2 are also induced transcriptionally at the G1/S transition by the transcription factor MBF in fission yeast.( 13 ) Importantly, a constitutively high dNTP concentration due to a mutation in the allosteric activity site of Rnr1 arrests the cell cycle at late G1 phase and has an effect on origin firings,( 14 ) suggesting that fluctuation in the dNTP concentration controls the initiation of DNA replication.
The dNTP levels during the DNA damage response are approximately four‐fold higher than the levels at S phase.( 10 ) In response to DNA replication stress or DNA damage, the transcription of RNR genes increases rapidly through activation of the DNA damage response pathway, which includes the Mec1‐Rad53‐Dun1 kinase cascade, and the Rad3‐Cds1 and/or Rad3‐Chk1 pathways in budding and fission yeasts, respectively.( 15 , 16 ) Upon DNA damage, Mec1 and Rad53 activate Dun1, which in turn leads to hyperphosphorylation of the Crt1 repressor.( 17 ) This repressor encodes a DNA‐binding protein that recruits and forms a complex with the general repressors Ssn6 and Tup1 to the promoters of RNR genes. Hyperphosphorylation of Crt1 prevents its DNA binding, providing the mechanism for transcriptional induction of RNR genes in response to DNA damage or DNA replication stress.( 17 ) The FSH3 gene is also a target of Crt1.( 18 ) Although the exact function of family of serine hydrolases (FSH) remains unknown, it shares significant homo‐logy with dihydrofolate reductase, suggesting the interesting scenario that the activation of the Mec1‐Rad53‐Dun1‐Crt1 pathway upon DNA damage results in derepression of the FSH3 gene. The induced FSH activity then leads to increased tetrahydrofolate and N 10‐methylene tetrahydrofolate pools that enhance dNTP synthesis. In addition to Crt1, Nrm1, a corepressor of MBF, is also involved in transcriptional induction of cdc22 R1 after DNA replication stalling in fission yeast.( 19 ) In response to DNA replication block, Cds1, activated by Rad3, phosphorylates Nrm1 leading to its dissociation from MBF, which in turn results in induction of MBF‐dependent transcripts, such as cdc22 R1.
Dun1 also upregulates RNR activity through regulating Sml1. Sml1 is an inhibitor of the RNR complex which binds to the large subunit of RNR through its C‐terminal portion.( 20 ) Sml1 is phosphorylated by Dun1 and degraded after DNA damage in a Mec1‐Rad53‐Dun1‐dependent manner.( 21 ) The degradation of Sml1 causes it to lose its ability for RNR inhibition. This strongly suggests that Dun1 kinase functions as the last step in the Mec1/Rad53 cascade to remove Sml1 during S phase and after DNA damage.
Another level of RNR regulation has been reported in which subcellular localization of RNR subunits plays a pivotal role in the formation of an active RNR complex in budding yeast.( 22 , 23 ) The small subunit Rnr2‐Rnr4 localizes in the nucleus, whereas the large subunit Rnr1 is cytoplasmic throughout the cell cycle. After DNA damage or during S phase, the Rnr2‐Rnr4 subunit enters into the cytoplasm enabling its binding to Rnr1, to form an active complex. Under unperturbed conditions or outside of S phase, Dif1 directly binds to the Rnr2‐Rnr4 complex through a Hug domain,( 24 , 25 ) which is a conserved domain present among Rnr‐inhibitor homologs, such as Spd1,( 26 ) Sml1,( 27 ) and Hug1. Dif1 binding to Rnr2‐Rnr4 promotes its nuclear import. Imported Rnr2‐Rnr4 then forms a complex with Wtm1, which anchors the complex in the nucleus (Fig. 2).( 28 , 29 ) In the presence of DNA damage, the association of Rnr2‐Rnr4 with Wtm1 is disrupted. The Dif1 protein level is significantly lowered during S phase( 24 ) and in response to DNA damage, as Dun1 directly phosphorylates Dif1, which leads to Dif1 inactivates and triggers its degradation. The reduction in Dif1 after DNA damage or during S phase allows Rnr2‐Rnr4 to move into the cytoplasm.
Figure 2.
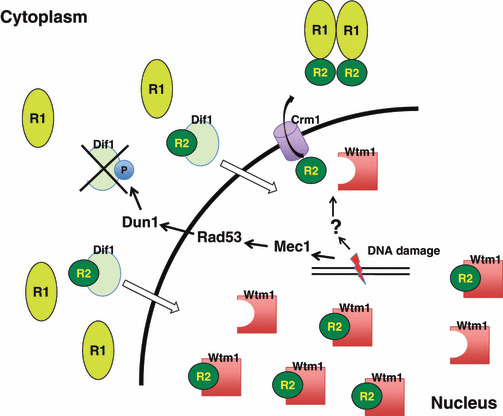
Regulation of subcellular localization of ribonucleotide reductase (RNR) in budding yeast. The small subunit Rnr2‐Rnr4 (R2) localizes at the nucleus, whereas the large subunit Rnr1 (R1) is cytoplasmic outside of S phase. After DNA damage or during S phase, the Rnr2‐Rnr4 subunit enters the cytoplasm enabling it to bind to Rnr1, forming an active complex. Dif1 directly binds to the R2 complex which promotes the import of R2 into the nucleus. The imported R2 then forms a complex with Wtm1, which anchors the complex in the nucleus. In the presence of DNA damage, the association of R2 with Wtm1 is disrupted. Furthermore, DNA damage‐induced activation of the Mec1‐Rad53‐Dun1 axis directly phosphorylates (P) Dif1, which inactivates and triggers its degradation. A reduction in Dif1, together with the dissociation of R2 from Wtm1 after DNA damage, allows R2 to enter the cytoplasm.
Very recently, Nestoras et al. ( 30 ) reported that although, in fission yeast, Spd1 functions to promote nuclear import of Suc22R2 as Dif1 does in budding yeast, it also regulates RNR activity at multiple levels, such as direct inhibition of RNR activity and modulation of RNR complex architecture. Intriguingly, using several lines of mutant Spd1, they clearly demonstrated that the major function of Spd1 in regulating dNTP synthesis is unrelated to its role in nuclear sequestration of Suc22R2. Therefore, the molecular system in which subcellular localization of RNR subunits enables the generation of an active RNR complex, and thereby regulates dNTP synthesis, may be specifically used in budding yeast.
Mechanisms of dNTP supply during DNA replication and repair in mammals
During DNA replication, in mammalian cells the whole dNTP pool is increased to a level 20‐fold higher than the level in G1 phase cells.( 31 ) This increase is mainly regulated at transcriptional and post‐transcriptional levels. The transcription of R1 and R2 genes is cell cycle‐regulated, with undetectable mRNA levels in G0/G1 phase and maximal levels in S phase when cells are arrested at a quiescent state by serum starvation and then released into G1 phase by serum addition.( 32 ) Promoter–reporter analysis revealed that the mouse R1 gene promoter is TATA‐less and its S phase‐specific activation is partly regulated by YY1.( 33 ) The mouse R2 gene promoter possesses a typical TATA box and its S phase‐specific activation is regulated by transcriptional activation through the upstream activating region.( 34 ) Another R2 gene, p53R2, was first identified as a p53‐inducible gene using differential display.( 5 , 35 ) Unlike R2, p53R2 is constitutively expressed at a low level during the cell cycle, but is significantly induced when p53 is activated.( 36 ) Therefore, p53R2 has been proposed to play a role in dNTP synthesis in resting cells in which R2 is undetectable. More recently, various mutations in p53R2 have been identified in several patients with mitochondrial DNA (mtDNA) depletion syndrome.( 37 ) Severe mtDNA depletion was also found in various tissues of p53R2‐deficient mice that died from severe renal failure through enhancement of p53‐induced apoptosis.( 38 ) Taken together, p53R2 has a non‐redundant role in supplying dNTP for mtDNA synthesis in resting cells.
R2 protein degradation at mitosis also appears to be important for S phase‐specific expression of R2 protein. The mouse R2 protein specifically binds to the Cdh1‐anaphase‐promoting complex through its KEN box.( 39 ) This interaction leads to polyubiquitination and degradation of R2 protein by way of proteasomes. p53R2 protein, however, lacks a KEN box.
Other enzymes involved in dNTP synthesis are also upregulated during DNA replication. Expression of NDPK and TS is induced during G1 to S phase.( 40 , 41 ) The activity of the dCMP deaminase shows periodic regulation during the cell cycle( 42 ) and the expression of the nuclear isoform of dUTPase is induced during S phase,( 43 ) suggesting the existence of a regulatory network of the enzymatic activities toward dTTP supplied for DNA replication.
Recent advances in the methodology of measuring dNTP have revealed that whole dNTP pools in mammalian cells are almost unchanged after DNA damage,( 31 ) suggesting the existence of a unique mechanism in mammals by which dNTPs are compartmentalized close to the damage site during the DNA repair process, thereby providing a high local dNTP concentration (Fig. 3). Although the exact reason why mammalian cells possess this unique mechanism is not known, compartmentalization of dNTPs may be less wasteful for providing sufficient amounts of dNTPs. Alternatively, high concentrations of dNTPs outside S phase might be deleterious in mammals. We detected traces but significant signals of both R1 and R2 proteins in a chromatin fraction,( 44 ) although both R1 and R2 predominantly localized in the cytoplasm as reported.( 45 ) We also found that both R1 and R2 proteins accumulated very rapidly at sites of DNA damage on an evaluation using ionizing irradiation treatment and UVA microirradiation. R1 constitutively forms a complex with Tip60 histone acetyltransferase, which is rapidly recruited to DNA damage sites( 46 ) through its chromo‐domain.( 47 ) Hence, the accumulation of R1 and R2 proteins requires an R1 interaction with Tip60 (Fig. 4). In this regard, it might be possible that Tip60 enhances the formation of active RNR complexes as Spd1 does in fission yeast.( 30 ) Tip60‐dependent recruitment of RNR to these damage sites is essential for efficient DNA repair because an active R1 mutant lacking Tip60 binding shows an impaired DNA repair. Intriguingly, recruitment of RNR is specifically required for effective DNA repair in cells with low levels of dNTPs, such as G1 phase cells. Although Tip60‐dependent recruitment of RNR is not involved in the dTTP supply to DNA damage sites, it should be noted that γ‐radiation in mice significantly induced the enzymatic activity of TS as well as that of dihydrofolate reductase,( 48 ) which may produce a sufficient amount of dTMP for DNA repair. Importantly, DNA damage has been reported to induce nuclear localization of NDPK,( 49 ) suggesting its role in dNDPs to dNTPs conversion in DNA repair.
Figure 3.
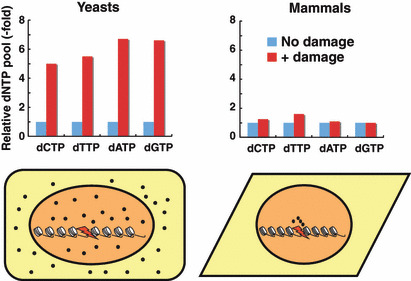
Sharp contrasts between yeasts and mammals in the regulation of dNTP pools after DNA damage. After DNA damage, whole dNTP pools in mammalian cells are almost unchanged, but those in yeasts are drastically increased.( 10 ) These observations suggest that there is a unique mechanism in mammals by which dNTPs are compartmentalized close to the damage sites during the DNA repair process, thereby providing a high local concentration. Black dots represent dNTPs (lower panels).
Figure 4.
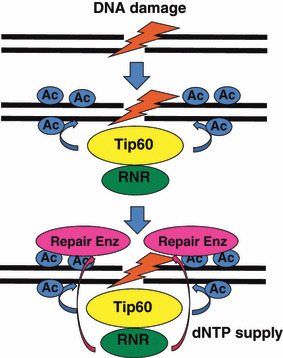
Mechanism of dNTP supply to DNA damage sites in mammals After DNA damage, the Tip60/ribonucleotide reductase (RNR) complex is rapidly recruited to the damage sites. Tip60 acetylates (Ac) histone H4 surrounding these sites and this acetylation enhances recruitment of repair enzymes and several checkpoint proteins. Recruited RNR can supply sufficient amounts of dNTPs for proper DNA repair.
Implications of dNTP supply in tumorigenesis
Budding yeast RNR mutants with increased dNTP pools show increased survival following DNA damage even with higher mutation rates,( 10 ) possibly due to reduced fidelity of the replicative polymerases and/or activation of error‐prone translesion polymerases during DNA synthesis. The features of RNR underlying its essential function in dNTPs supply strongly suggest that it is involved in a tumorigenic environment and is a potential biomarker for the clinical outcome and the chemotherapeutic efficacy of anticancer drugs. Ectopic expression of R1 in ras‐transformed mouse fibroblasts results in reduced anchorage‐independent growth and tumor formation.( 50 ) Increased expression of R1 in human lung cancer cells also results in reduced cellular migration, invasion, and metastasis.( 51 ) Finally, transgenic mice overexpressing R1 show a strong suppression of carcinogen‐induced lung tumor formation.( 52 ) Thus, the high level of dNTP supply for DNA synthesis might protect mammalian cells from progressive malignant transformation.
In contrast, an insufficient level of dNTPs can lead to malignant transformation. Fission yeast mutants with reduced dNTP pools show an increased rate in spontaneous mutations, including base substitution and small insertion/deletion,( 53 ) presumably due to recruitment of error‐prone translesion polymerases to extend stalled replication forks. Tip60 is reported to be a haplo‐insufficient tumor suppressor required for an oncogene‐induced DNA damage response.( 54 ) Although Tip60 is required for the tumor suppressor function of ATM, and there is still no direct evidence implicating Tip60‐dependent recruitment of RNR to DNA damage sites in tumorigenesis, an insufficient dNTP supply to these sites in Tip60 heterozygote mice might explain their predisposition to cancer.
Gemcitabine (27deoxy‐2′/2′‐difluorocytidine monohydrochloride), one of the principal chemotherapeutic agents in the treatment of non‐small‐cell lung cancer (NSCLC), is a potent and specific pyrimidine nucleoside antimetabolite with structural relatedness to deoxycytidine. Low R1 mRNA expression was reported to be associated with a significantly longer overall survival in NSCLC patients treated with gemcitabine/cisplatin,( 55 , 56 , 57 ) although the molecular basis underlying this association remains elusive. R1 polymorphisms have also been reported to be associated with the clinical outcome in NSCLC patients treated with this drug.( 58 )
In summary, accumulating evidence for the physiological importance of RNR regulation in maintaining genome integrity strongly suggests that components of the pathways regulating RNR function may be potent candidates as molecular targets in novel anticancer strategies. Although the exact function of each protein in mammalian ATM‐Chk2‐p53‐RNR signaling upon DNA damage is somewhat different from that found in yeast (Mec1‐Rad53‐Dun1‐Crt1‐RNR), conservation of these pathways is intriguing as both pathways determine sensitivity of cell survival to DNA damage agents. More importantly, p53 and ATM are two of the 10 most frequently mutated genes in lung adenocarcinoma, but their mutations are mutually exclusive,( 59 ) suggesting that mutations in ATM and p53 may be independently sufficient for the loss of RNR regulation. Therefore, prospective, epidemiologic, and clinical studies are needed to delineate the effects of factors regulating RNR in carcinogenesis and during ongoing therapy in humans.
References
- 1. Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem 2006; 75: 681–706. [DOI] [PubMed] [Google Scholar]
- 2. Kashlan OB, Scott CP, Lear JD, Cooperman BS. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP‐ and dATP‐induced oligomerization of the large subunit. Biochemistry 2002; 41: 462–74. [DOI] [PubMed] [Google Scholar]
- 3. Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J Biol Chem 2006; 281: 27705–11. [DOI] [PubMed] [Google Scholar]
- 4. Kolberg M, Strand KR, Graff P, Andersson KK. Structure, function, and mechanism of ribonucleotide reductases. Biochim Biophys Acta 2004; 1699: 1–34. [DOI] [PubMed] [Google Scholar]
- 5. Tanaka H, Arakawa H, Yamaguchi T et al. A ribonucleotide reductase gene involved in a p53‐dependent cell‐cycle checkpoint for DNA damage. Nature 2000; 404: 42–9. [DOI] [PubMed] [Google Scholar]
- 6. Chabes A, Domkin V, Larsson G et al. Yeast ribonucleotide reductase has a heterodimeric iron‐radical‐containing subunit. Proc Natl Acad Sci USA 2000; 97: 2474–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Elledge SJ, Davis RW. Two genes differentially regulated in the cell cycle and by DNA‐damaging agents encode alternative regulatory subunits of ribonucleotide reductase. Genes Dev 1990; 4: 740–51. [DOI] [PubMed] [Google Scholar]
- 8. Domkin V, Thelander L, Chabes A. Yeast DNA damage‐inducible Rnr3 has a very low catalytic activity strongly stimulated after the formation of a cross‐talking Rnr1/Rnr3 complex. J Biol Chem 2002; 277: 18574–8. [DOI] [PubMed] [Google Scholar]
- 9. Reichard P, Eliasson R, Ingemarson R, Thelander L. Cross‐talk between the allosteric effector‐binding sites in mouse ribonucleotide reductase. J Biol Chem 2000; 275: 33021–6. [DOI] [PubMed] [Google Scholar]
- 10. Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell 2003; 112: 391–401. [DOI] [PubMed] [Google Scholar]
- 11. Kunz BA, Kohalmi SE, Kunkel TA, Mathews CK, McIntosh EM, Reidy JA. International Commission for Protection Against Environmental Mutagens and Carcinogens. Deoxyribonucleoside triphosphate levels: a critical factor in the maintenance of genetic stability. Mutat Res 1994; 318: 1–64. [DOI] [PubMed] [Google Scholar]
- 12. Elledge SJ, Zhou Z, Allen JB, Navas TA. DNA damage and cell cycle regulation of ribonucleotide reductase. Bioessays 1993; 15: 333–9. [DOI] [PubMed] [Google Scholar]
- 13. Fernandez Sarabia MJ, McInerny C, Harris P, Gordon C, Fantes P. The cell cycle genes cdc22+ and suc22+ of the fission yeast Schizosaccharomyces pombe encode the large and small subunits of ribonucleotide reductase. Mol Gen Genet 1993; 238: 241–51. [DOI] [PubMed] [Google Scholar]
- 14. Chabes A, Stillman B. Constitutively high dNTP concentration inhibits cell cycle progression and the DNA damage checkpoint in yeast Saccharomyces cerevisiae . Proc Natl Acad Sci USA 2007; 104: 1183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Osborn AJ, Elledge SJ, Zou L. Checking on the fork: the DNA‐replication stress‐response pathway. Trends Cell Biol 2002; 12: 509–16. [DOI] [PubMed] [Google Scholar]
- 16. Lambert S, Carr AM. Checkpoint responses to replication fork barriers. Biochimie 2005; 87: 591–602. [DOI] [PubMed] [Google Scholar]
- 17. Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell 1998; 94: 595–605. [DOI] [PubMed] [Google Scholar]
- 18. Zaim J, Speina E, Kierzek AM. Identification of new genes regulated by the Crt1 transcription factor, an effector of the DNA damage checkpoint pathway in Saccharomyces cerevisiae . J Biol Chem 2005; 280: 28–37. [DOI] [PubMed] [Google Scholar]
- 19. De Bruin RA, Kalashnikova TI, Aslanian A et al. DNA replication checkpoint promotes G1‐S transcription by inactivating the MBF repressor Nrm1. Proc Natl Acad Sci USA 2008; 105: 11230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhao X, Georgieva B, Chabes A et al. Mutational and structural analyses of the ribonucleotide reductase inhibitor Sml1 define its Rnr1 interaction domain whose inactivation allows suppression of mec1 and rad53 lethality. Mol Cell Biol 2000; 20: 9076–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA 2002; 99: 3746–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yao R, Zhang Z, An X et al. Subcellular localization of yeast ribonucleotide reductase regulated by the DNA replication and damage checkpoint pathways. Proc Natl Acad Sci USA 2003; 100: 6628–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Liu C, Powell KA, Mundt K, Wu L, Carr AM, Caspari T. Cop9/signalosome subunits and Pcu4 regulate ribonucleotide reductase by both checkpoint‐dependent and ‐independent mechanisms. Genes Dev 2003; 17: 1130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA‐damage‐regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell 2008; 32: 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wu X, Huang M. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol 2008; 28: 7156–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hakansson P, Dahl L, Chilkova O, Domkin V, Thelander L. The Schizosaccharomyces pombe replication inhibitor Spd1 regulates ribon‐ucleotide reductase activity and dNTPs by binding to the large Cdc22 subunit. J Biol Chem 2006; 281: 1778–83. [DOI] [PubMed] [Google Scholar]
- 27. Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell 1998; 2: 329–40. [DOI] [PubMed] [Google Scholar]
- 28. Lee YD, Elledge SJ. Control of ribonucleotide reductase localization through an anchoring mechanism involving Wtm1. Genes Dev 2006; 20: 334–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang Z, An X, Yang K et al. Nuclear localization of the Saccharomyces cerevisiae ribonucleotide reductase small subunit requires a karyopherin and a WD40 repeat protein. Proc Natl Acad Sci USA 2006; 103: 1422–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nestoras K, Mohammed AH, Schreurs AS et al. Regulation of ribonucleotide reductase by Spd1 involves multiple mechanisms. Genes Dev 2010; 24: 1145–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hakansson P, Hofer A, Thelander L. Regulation of mammalian ribonucleotide reduction and dNTP pools after DNA damage and in resting cells. J Biol Chem 2006; 281: 7834–41. [DOI] [PubMed] [Google Scholar]
- 32. Bjorklund S, Skog S, Tribukait B, Thelander L. S‐phase‐specific expression of mammalian ribonucleotide reductase R1 and R2 subunit mRNAs. Biochemistry 1990; 29: 5452–8. [DOI] [PubMed] [Google Scholar]
- 33. Johansson E, Hjortsberg K, Thelander L. Two YY‐1‐binding proximal elements regulate the promoter strength of the TATA‐less mouse ribonucleotide reductase R1 gene. J Biol Chem 1998; 273: 29816–21. [DOI] [PubMed] [Google Scholar]
- 34. Chabes AL, Bjorklund S, Thelander L. S Phase‐specific transcription of the mouse ribonucleotide reductase R2 gene requires both a proximal repressive E2F‐binding site and an upstream promoter activating region. J Biol Chem 2004; 279: 10796–807. [DOI] [PubMed] [Google Scholar]
- 35. Nakano K, Balint E, Ashcroft M, Vousden KH. A ribonucleotide reductase gene is a transcriptional target of p53 and p73. Oncogene 2000; 19: 4283–9. [DOI] [PubMed] [Google Scholar]
- 36. Guittet O, Hakansson P, Voevodskaya N et al. Mammalian p53R2 protein forms an active ribonucleotide reductase in vitro with the R1 protein, which is expressed both in resting cells in response to DNA damage and in proliferating cells. J Biol Chem 2001; 276: 40647–51. [DOI] [PubMed] [Google Scholar]
- 37. Bourdon A, Minai L, Serre V et al. Mutation of RRM2B, encoding p53‐controlled ribonucleotide reductase (p53R2), causes severe mitochondrial DNA depletion. Nat Genet 2007; 39: 776–80. [DOI] [PubMed] [Google Scholar]
- 38. Kimura T, Takeda S, Sagiya Y, Gotoh M, Nakamura Y, Arakawa H. Impaired function of p53R2 in Rrm2b‐null mice causes severe renal failure through attenuation of dNTP pools. Nat Genet 2003; 34: 440–5. [DOI] [PubMed] [Google Scholar]
- 39. Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: a new target for anaphase‐promoting complex‐Cdh1‐mediated proteolysis. Proc Natl Acad Sci USA 2003; 100: 3925–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keim D, Hailat N, Melhem R et al. Proliferation‐related expression of p19/nm23 nucleoside diphosphate kinase. J Clin Invest 1992; 89: 919–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Le Francois BG, Maroun JA, Birnboim HC. Expression of thymidylate synthase in human cells is an early G(1) event regulated by CDK4 and p16INK4A but not E2F. Br J Cancer 2007; 97: 1242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gelbard AS, Kim JH, Perez AG. Fluctuations in deoxycytidine monophosphate deaminase activity during the cell cycle in synchronous populations of HeLa cells. Biochim Biophys Acta 1969; 182: 564–6. [DOI] [PubMed] [Google Scholar]
- 43. Ladner RD, Caradonna SJ. The human dUTPase gene encodes both nuclear and mitochondrial isoforms. Differential expression of the isoforms and characterization of a cDNA encoding the mitochondrial species. J Biol Chem 1997; 272: 19072–80. [DOI] [PubMed] [Google Scholar]
- 44. Niida H, Katsuno Y, Sengoku M et al. Essential role of Tip60‐dependent recruitment of ribonucleotide reductase at DNA damage sites in DNA repair during G1 phase. Genes Dev 2010; 24: 333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pontarin G, Fijolek A, Pizzo P et al. Ribonucleotide reduction is a cytosolic process in mammalian cells independently of DNA damage. Proc Natl Acad Sci USA 2008; 105: 17801–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Murr R, Loizou JI, Yang YG et al. Histone acetylation by Trrap‐Tip60 modulates loading of repair proteins and repair of DNA double‐strand breaks. Nat Cell Biol 2006; 8: 91–9. [DOI] [PubMed] [Google Scholar]
- 47. Sun Y, Jiang X, Xu Y et al. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol 2009; 11: 1376–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Batra V, Kesavan V, Mishra KP. Modulation of enzymes involved in folate dependent one‐carbon metabolism by gamma‐radiation stress in mice. J Radiat Res (Tokyo) 2004; 45: 527–33. [DOI] [PubMed] [Google Scholar]
- 49. Yoon JH, Singh P, Lee DH et al. Characterization of the 3′ ‐‐> 5′ exonuclease activity found in human nucleoside diphosphate kinase 1 (NDK1) and several of its homologues. Biochemistry 2005; 44: 15774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Fan H, Huang A, Villegas C, Wright JA. The R1 component of mammalian ribonucleotide reductase has malignancy‐suppressing activity as demonstrated by gene transfer experiments. Proc Natl Acad Sci USA 1997; 94: 13181–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gautam A, Li ZR, Bepler G. RRM1‐induced metastasis suppression through PTEN‐regulated pathways. Oncogene 2003; 22: 2135–42. [DOI] [PubMed] [Google Scholar]
- 52. Gautam A, Bepler G. Suppression of lung tumor formation by the regulatory subunit of ribonucleotide reductase. Cancer Res 2006; 66: 6497–502. [DOI] [PubMed] [Google Scholar]
- 53. Holmberg C, Fleck O, Hansen HA et al. Ddb1 controls genome stability and meiosis in fission yeast. Genes Dev 2005; 19: 853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gorrini C, Squatrito M, Luise C et al. Tip60 is a haplo‐insufficient tumour suppressor required for an oncogene‐induced DNA damage response. Nature 2007; 448: 1063–7. [DOI] [PubMed] [Google Scholar]
- 55. Rosell R, Danenberg KD, Alberola V et al. Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin‐treated advanced non‐small cell lung cancer patients. Clin Cancer Res 2004; 10: 1318–25. [DOI] [PubMed] [Google Scholar]
- 56. Rosell R, Felip E, Taron M et al. Gene expression as a predictive marker of outcome in stage IIB‐IIIA‐IIIB non‐small cell lung cancer after induction gemcitabine‐based chemotherapy followed by resectional surgery. Clin Cancer Res 2004; 10: 4215s–9s. [DOI] [PubMed] [Google Scholar]
- 57. Ceppi P, Volante M, Novello S et al. ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non‐small‐cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol 2006; 17: 1818–25. [DOI] [PubMed] [Google Scholar]
- 58. Bepler G, Zheng Z, Gautam A et al. Ribonucleotide reductase M1 gene promoter activity, polymorphisms, population frequencies, and clinical relevance. Lung Cancer 2005; 47: 183–92. [DOI] [PubMed] [Google Scholar]
- 59. Ding L, Getz G, Wheeler DA et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature 2008; 455: 1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]


