Abstract
Adult T‐cell leukemia/lymphoma (ATLL) is a human malignancy associated with human T‐cell leukemia virus type 1 (HTLV‐1). The pathological features of the lymph nodes of ATLL change from those of lymphadenitis to Hodgkin's‐like features and those of lymphoma. Chemokines and their receptors are closely associated with T‐cell subgroups and immune responses. To clarify the relationship between chemokines and their receptor expression, as well as the development of ATLL, 17 cases with ATLL were analyzed using DNA chips of chemokines and their receptors. All cases showed a varied and mixed pattern of upregulated and downregulated gene expression of Th1, Th2, naïve, and cytotoxic cell‐associated chemokine genes. As CC chemokine ligand 18 (CCL18) accounted for the most upregulated gene and CX3C chemokine receptor 1 (CX3CR1) for the most downregulated gene, they were selected for immunohistochemical analysis. Immunohistochemical staining showed expression of the two genes in immunological cells, with a positive expression for reticulum cells, but not for ATLL cells. HTLV‐1‐associated lymphadenitis type (n = 13) and Hodgkin's‐like type (n = 12) cases showed significantly higher CCL18 expression than the non‐specific lymphadenitis cases (n = 10) (P < 0.05). However, all HTLV‐1‐associated cases showed significantly lower CX3CR1 expression than the non‐specific lymphadenitis cases (P < 0.05). These results suggest that upregulation of CCL18 expression and downregulation of CX3CR1 expression play a role in immune responses against the ATLL cells. (Cancer Sci 2007; 98: 1875–1880)
Abbreviations:
- ATLL
adult T‐cell leukemia/lymphoma
- CCL18
CC chemokine ligand 18
- CCR4
CC chemokine receptor 4
- CX3CR1
CX3C chemokine receptor 1
- DLBCL
diffuse large B‐cell lymphoma
- FoxP3
forkhead/winged helix transcription factor
- HPF
high‐power field
- HTLV‐1
human T‐cell leukemia virus type 1
- Treg
regulatory T cells
ATLL is a human malignancy associated with a retrovirus, HTLV‐1.( 1 ) Histologically, ATLL usually has the characteristics of peripheral T‐cell lymphoma, with a diffuse proliferation of tumor cells varying in size and form as well as the appearance of cerebriform giant cells.( 2 ) In ATLL lymph nodes, the pathological features change from those of lymphadenitis to Hodgkin's‐like features and those of lymphoma.( 2 , 3 ) HTLV‐1‐associated lymphadenitis is considered to be a non‐neoplastic HTLV‐1‐associated lymph node lesion,( 4 ) whereas Hodgkin's‐like histological features are associated with the initial stages of ATLL.( 5 , 6 )
Chemokines are members of a family of small secreted proteins and some of them have received considerable attention because they display selectivity of cell targets and receptors, and are closely associated with T‐cell subgroups. For example, the thymus and activation‐regulated chemokine is known to bind the CCR4, which is expressed on activated Th2 lymphocytes. Yoshie et al.( 7 ) reported a high frequency of CCR4 expression in ATLL and HTLV‐1 immortalized T cells. Moreover, CCR4 might facilitate ATLL cell invasion of the lymph nodes. Hasegawa et al.( 8 ) reported that increased surface expression of CCR7 correlated well with lymphoid organ involvement in ATLL. In addition, a recent study reported high‐level expression of CC chemokine ligand 5/regulated upon activation, normal T expressed and secreted in ATLL cells.( 9 )
Recent advances in technical and analytical methods, especially complementary DNA analysis, have facilitated the parallel quantitation of the expression of thousands of genes.( 10 ) Various pathologic conditions are also being scrutinized with these methods, from inflammatory diseases( 11 ) to neoplasia.( 12 ) Array technology is particularly well‐suited to studying changes in gene expression in normal tissues and their malignant counterparts. For example, Alizadeh et al.( 13 ) used DNA microarrays to identify two molecularly distinct forms of DLBCL, with one type expressing genes characteristic of germinal center B cells (germinal center B‐like DLBCL), and the other type expressing genes normally induced during in vitro activation of peripheral blood B cells (activated B‐like DLBCL). Patients with germinal center B‐like DLBCL had a significantly better overall clinical survival than those with activated B‐like DLBCL.
The aim of this study was to clarify the relationship between the expression of chemokines and chemokine receptors on the one hand, and the development of ATLL on the other. To make the DNA array method suitable for the identification of T‐cell subtypes and immune responses in ATLL, we designed a special microarray, the ‘chemokine, chemokine receptor and cytokine chip’, by selecting genes that are preferentially expressed in lymphoid cells. In addition, genes which are specifically associated with the ATLL subgroup were selected for analysis of DNA chips, and their specific antibodies were used for the immunological staining of protein expressions.
Materials and Methods
Patients. Patients with ATLL were identified in the lymph node registry files of the Department of Pathology, Fukuoka University (Fukuoka, Japan) and the Department of Pathology, Kurume University (Kurume, Japan). In all cases, Southern blot analysis was used to examine clonal integration of HTLV‐1 proviral DNA. The lymph nodes were fixed with buffered formalin, embedded in paraffin, and stained with hematoxylin–eosin. The patients were classified into four groups: (i) HTLV‐1‐associated lymphadenitis type; (ii) Hodgkin's‐like type; (iii) pleomorphic type; and (iv) anaplastic large cell type. Almost all of these cases were included in previous reports.( 14 , 15 )
Immunohistochemistry. The 60 patients enrolled in this study included 10 with non‐specific lymphadenitis, 13 with HTLV‐1‐associated lymphadenitis, 12 with Hodgkin's‐like type, 13 with pleomorphic type, and 12 with anaplastic large cell type lymph node lesions.
The paraffin‐embedded lymph nodes were used for immunohistochemical analysis of L26 (CD20) for B cells (DakoCytomation, Glostrup, Denmark), UCHL‐1 (CD45RO) and CD3 for T cells (DakoCytomation), and CD4, CD8, CD15 and CD30 (DakoCytomation), as reported previously.( 14 , 15 ) In addition, antibodies to CX3CR1 (Chemicon International, Temecula, CA) and CCL18/ pulmonary and activation‐regulated CC chemokine (R&D Systems, Minneapolis, MN) were used in the paraffin‐embedded samples. Heat‐mediated antigen retrieval was carried out, as was determination of the presence or absence of Epstein–Barr virus small RNA by in situ hybridization using Epstein–Barr virus‐encoded small RNA oligonucleotides (Dakopatts, Glostrup, Denmark).
Immunohistological scoring. For the determination of CX3CR1 and CCL18 expression, representative areas were used with a maximum density of positive cells per 400× HPF. Negative and positive staining controls for CX3CR1 and CCL18 were obtained using the paraffin‐embedded lymph nodes that were histologically diagnosed with non‐specific lymphadenitis.
DNA analysis. Other parts of the frozen materials were used for DNA isolation and gene analysis with methods that have been reported previously.( 16 ) After the frozen materials were confirmed to consist of lymphoma cells, proviral DNA of HTLV‐1 (full‐length probe, including gag, pol, env, pX, and LTR) was also examined. The monoclonal integration of HTLV‐1 DNA was examined by digestion with EcoRI, as previously reported.( 16 )
Microarray procedures. Seventeen cases of ATLL, four with HTLV‐1‐associated lymphadenitis, five with Hodgkin's‐like type, four with pleomorphic type, and four with anaplastic large cell type, were analyzed using DNA chips. Total RNA was prepared from frozen materials with the guanidinium thiocyanate–phenol–chloroform method using the Total RNA Separator Kit (Clontech, Paolo Alto, CA). DNA microarray analysis of gene expression was carried out essentially as described previously.( 17 ) The cDNA clones on the chemokine, chemokine receptor and cytokine DNA chips were modified versions of Chemokine Chip version 1.0 (Biomedical Science, Tokyo, Japan). Fluorescent images of hybridized DNA chips were obtained with a Scan Array 4000 scanner (GSI Lumonics, Boston, MA). Images were analyzed with QuantArray (GSI Lumonics), and fluorescence ratios (along with numerous quality control parameters) were stored in a customized database. Single spots or areas of the array with obvious blemishes were flagged and excluded from subsequent analysis. Fluorescence ratios were calibrated independently for each array by applying a single scaling factor to all fluorescent ratios from each array; this scaling factor was computed so that the median fluorescence ratio of well‐measured spots on each array was 1.0.
All cDNA microarray analyses were carried out using total RNA and the 3DNATM Submicro Expression Array Detection Kit (Genisphere, Montvale, NJ). For each experiment, fluorescent cDNA probes were prepared from an experimental total RNA sample (Cy5‐labeled) and a control total RNA sample (Cy3‐labeled) mixed from a pool of total RNA from the non‐specific lymphadenitis of 20 patients.
Data analysis. All non‐flagged array elements for which the fluorescence intensity in each channel was greater than 1.4 times the local background were considered well measured. Each cDNA probe had three spots on the DNA chips, and we used the median result from the three spots for data analysis. We calculated the mean of each cDNA probe in four cases of non‐specific lymphadenitis. The mean was used as the control ratio.
Hierarchical clustering was applied to both axes using the weighted pair‐group method with centroid average as implemented in the J‐Express software (http://www.molmine.com/). The distance matrices used were Pearson's correlation for clustering the arrays and the inner product of vectors normalized to magnitude 1 for the genes.
Reverse transcription–polymerase chain reaction (RT‐PCR). cDNA was reverse transcribed from approximately 2.5 µg total RNA using Ready‐To‐Go You‐Prime First‐Strand Beads (GE Healthcare, Buckinghamshire, UK) and was primed by oligo(dT) oligonucleotide (GE Healthcare). Three microliters of cDNA was subjected to PCR reaction using AmpliTaq Gold DNA Polymerase (Applied Biosystems, Foster City, CA). The oligonucleotide primers were as follows: for CCL18, sense, 5′‐CTT GCA GCT GCC CTC CTT GTC‐3′ and antisense, 5′‐CAC TTC TTA TTG GGG TCA GCA‐3′; for CX3CR1, sense, 5′‐TGG CTG ACT GGC AGA TCC AGA GGT T‐3′ and antisense, 5′‐TGA GTC CAG AAG GGC AAA GTG GCT A‐3′; for β‐actin, sense, 5′‐CAA GAG ATG GCC ACG GCT GCT‐3′ and antisense, 5′‐TCC TTC TGC ATC CTG TCG GCA‐3′. Product sizes were 221 bp for CCL18, 320 bp for CX3CR1, and 275 bp for β‐actin. Amplification conditions were denaturation at 95°C for 30 s (10 min for the first cycle), annealing at 60°C for 30 s and extension at 72°C for 1 min (10 min for the last cycle) for 25 cycles for CCL18, 35 cycles for CX3CR1, and 30 cycles for β‐actin. The amplified products were evaluated in 2% agarose gel and visualized by ethidium bromide staining under ultraviolet light. The stained bands were subjected to densitometric analysis using ImageJ software (http://rsb.info.nih.gov/ij). The quality of cDNA was monitored using RT‐PCR with β‐actin primers. cDNAs yielding a 275‐bp product for β‐actin mRNA without contamination with the 370‐bp genomic amplification product were used for experiment amplification.
Statistical analysis. Expression of each chemokine receptor was compared among the subgroups of HTLV‐1‐associated lymph node lesions and non‐specific lymphadenitis, using Student's t‐test. Differences were considered significant if the P‐value was less than 0.05.
Results
Histological classification. Histological examination of the lymph nodes of HTLV‐1‐associated lymphadenitis showed a preserved nodal architecture with small lymph follicles, enlargement of the paracortex, and diffuse infiltration of small or intermediate‐sized lymphocytes. None of the cases showed rearrangement of the T‐cell receptor genes, or of the immunoglobulin heavy chain gene. In Hodgkin's‐like type lymph nodes, small aggregations of foci or clusters of a few giant cells with irregularly lobulated, highly convoluted, Reed–Sternberg or Hodgkin's cell‐like nuclei were scattered throughout the expanded paracortex. Giant cells reacted with CD30 and/or CD15, whereas the background lymphocytes reacted with CD3 and CD4. Receptor gene analysis showed a rearrangement and/or deletion of the TCR gene. Proviral HTLV‐1 DNA bands were found in all cases, although the bands were weaker than those usually seen in typical ATLL. In pleomorphic type lymph nodes, lymphoma cells showed irregular or convoluted nuclear contours, and/or a few scattered giant cells with cerebriform or Reed–Sternberg type nuclei. The lymphoma cells reacted with CD3 and CD4. The lymph nodes of the pleomorphic type showed typical ATLL characteristics. In the anaplastic large cell type, tumor cells were much larger than those of large cell lymphoma and a uniform pattern of cell proliferation was observed. The lymphoma cells reacted with CD3, CD4 and CD30. Proviral HTLV‐1 DNA bands were detected in all pleomorphic type and anaplastic large cell type cases.
Expression of chemokines, chemokine receptors and cytokines by disease type (DNA chips). Compared with non‐specific lymphadenitis, all cases showed a varied mixed pattern of upregulated and downregulated gene expression of Th1, Th2, naïve, and cytotoxic cell‐associated genes. We calculated the mean ratio of each gene expression in ATLL subgroups versus that in non‐specific lymphadenitis (Table 1). In the pleomorphic type, only three genes were upregulated (ratio, >1.20): CCL18 (1.33); MIB‐1α (1.31); and MIB‐1β (1.22). In the anaplastic large cell type, all genes showed a downregulated pattern (ratio, <0.8). In the Hodgkin's‐like type, half of all genes (16 genes) were upregulated, for example, CCL18 (2.25), MCP‐2 (1.85), and MCP‐3 (1.83). In HTLV‐1‐associated lymphadenitis, six genes were upregulated, for example, CCL18 (1.72) and MCP‐2 (1.56). In all subtypes, the CX3CR‐1 gene was downregulated.
Table 1.
DNA chip data of human T‐cell leukemia virus type 1 (HTLV‐1)‐associated diseases
| Pleomorphic ATLL | Anaplastic ATLL | Hodgkin's‐like ATLL | HTLV‐1 lymphadenitis | ||||
|---|---|---|---|---|---|---|---|
| CCL18 | 1.33 | — | — | CCL18 | 2.25 | CCL18 | 1.72 |
| MIP‐1α | 1.31 | — | — | MCP‐2 | 1.85 | MCP‐2 | 1.56 |
| MIP‐1β | 1.22 | — | — | MCP‐3 | 1.83 | MIP‐1α | 1.46 |
| — | — | — | — | SCYA 14/15 | 1.55 | SCYA 14/15 | 1.42 |
| — | — | — | — | MCP‐4 | 1.54 | IL‐5 | 1.26 |
| — | — | — | — | IL‐5 | 1.52 | MCP‐4 | 1.21 |
| — | — | — | — | MIG | 1.50 | — | — |
| — | — | — | — | I‐Tac | 1.46 | — | — |
| — | — | — | — | IFN‐γ | 1.40 | — | — |
| — | — | — | — | IP‐10 | 1.35 | — | — |
| — | — | — | — | Other factors | 6 | — | — |
| CX3CR‐1 | 0.20 | SLC | 0.25 | CX3CR‐1 | 0.35 | CX3CR‐1 | 0.41 |
| CCR‐3 | 0.28 | CX3CR‐1 | 0.32 | CCR‐9 | 0.49 | BLC | 0.49 |
| SDF‐1α | 0.32 | CCR‐3 | 0.34 | CCR‐2 | 0.54 | FTN | 0.49 |
| CCR‐2 | 0.33 | SDF‐1α | 0.34 | SDF‐1α | 0.57 | CCR‐3 | 0.51 |
| BLC | 0.34 | FTN | 0.34 | XCR‐1 | 0.58 | CXCR‐1 | 0.51 |
| CCR‐9 | 0.36 | BLC | 0.35 | CCR‐3 | 0.58 | SDF‐1α | 0.53 |
| FTN | 0.37 | SDF‐1β | 0.38 | FTN | 0.62 | XCR‐1 | 0.54 |
| TARC | 0.38 | BRAK/CXCL14 | 0.38 | IL‐10 | 0.65 | IL‐13 | 0.58 |
| SDF‐1β | 0.39 | XCR‐1 | 0.40 | SDF‐1β | 0.68 | TARC | 0.58 |
| CXCR‐1 | 0.39 | IL‐13 | 0.40 | IL‐13 | 0.68 | SDF‐1β | 0.60 |
| Other factors | 41 | Other factors | 44 | Other factors | 6 | Other factors | 14 |
The mean ratio of each gene expression; subgroups of adult T‐cell leukemia/lymphoma (ATLL) versus non‐specific lymphadenitis. BLC, B lymphocyte chemoattractant; BRAK/CXCL14, breast and kidney/CXC chemokine ligand 14; CCL18, CC chemokine ligand 18; CCR, CC chemokine receptor; CX3CR1, CX3C chemokine receptor 1; CXCR‐1, CXC chemokine receptor 1; FTN, fractalkine; IFN, interferon; IL, interleukin; IP‐10, interferon‐inducible protein‐10; I‐Tac, interferon‐inducible T‐cell α chemoattractant; MCP, monocyte chemtactic protein; MIG, monokine induced by γ‐interferon; MIP, macrophage inflammatory protein; SCYA, small cytokine group A; SDF, stromal cell‐derived factor; SLC, secondary lymphoid‐tissue chemokine; TARC, thymus and activation‐regulated chemokine; XCR‐1, chemokine (C motif) receptor 1; —, no data produced.
DNA chip analysis and gene expression patterns. In the tree views of DNA chips using all cases, none of the HTLV‐1‐associated diseases showed a specific tendency to develop distinct cluster patterns (Fig. 1a). However, the tree views of HTLV‐1‐associated lymphadenitis and Hodgkin's‐like type showed a tendency to make small clusters in each group, but were not clearly separated (Fig. 1b). In the clustering analysis of HTLV‐1‐associated lymphadenitis and ATLL, HTLV‐1‐associated lymphadenitis also showed a tendency to develop a small cluster, but ATLL did not (Fig. 1c). The analysis of Hodgkin's‐like type and ATLL did not show clustering patterns in any group (Fig. 1d). Some cases of HTLV‐1‐associated lymphadenitis showed a tendency to develop a small cluster, but Hodgkin's‐like type and ATLL did not.
Figure 1.
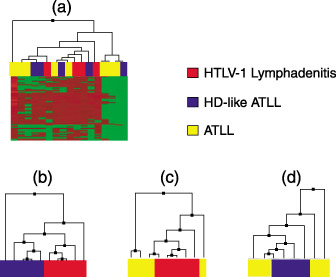
Tree view of DNA chips using all 57 cases of adult T‐cell leukemia/lymphoma (ATLL) did not show any tendency to develop cluster patterns (a). In the tree view of human T‐cell leukemia virus type 1 (HTLV‐1)‐associated lymphadenitis versus Hodgkin's‐like type (b), HTLV‐1‐associated lymphadenitis versus ATLL (c), and Hodgkin's‐like type versus ATLL (d), some cases of HTLV‐1‐associated lymphadenitis showed a tendency to develop small clusters.
Expression of mRNA for CCL18 and CX3CR1. In order to study the regulation of expression of CCL18 and CX3CR1, we carried out RT‐PCR on cDNA delivered from the frozen materials. RT‐PCR products for CCL18 were found in HTLV‐1‐associated disease, but mRNA in non‐specific lymphadenitis (control cases) was rarely detected [control versus HTLV‐1‐associated diseases; mean (CCL18/β‐actin) 0.04 versus 0.15]. In contrast, CX3CR1 expression was relatively clearly detected in all cases. The density of the RT‐PCR bands in HTLV‐1‐associated lymphadenitis and Hodgkin's‐like type were the same as those in the control cases [control versus HTLV‐1‐associated lymphadenitis and Hodgkin's‐like type; mean (CX3CR1/β‐actin) 0.38 versus 0.42]. But the density in pleomorphic type and anaplastic large cell type were weak compared with other type [control versus pleomorphic type and anaplastic large cell type; mean (CX3CR1/β‐actin) 0.38 versus 0.23] (Fig. 2).
Figure 2.
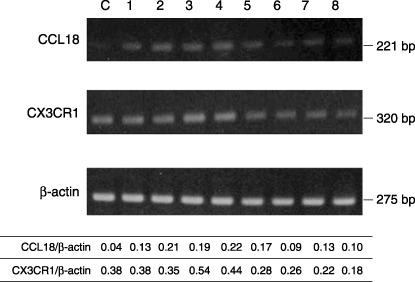
Expression of mRNA for CC chemokine ligand 18 (CCL18) and CX3C chemokine receptor 1 (CX3CR1). Lane C, non‐specific lymphadenitis; lanes 1 and 2, human T‐cell leukemia virus type 1 (HTLV‐1)‐associated lymphadenitis; lanes 3 and 4, Hodgkin's‐like type lymph node lesion; lanes 5 and 6, pleomorphic type lymph node lesion; lanes 7 and 8, anaplastic large cell type lymph node lesion. Reverse transcription–polymerase chain reaction (RT‐PCR) products for CCL18 were found in HTLV‐1‐associated diseases, but was rarely detected in the control case. The density of RT‐PCR bands for CX3CR1 in HTLV‐1‐associated lymphadenitis and Hodgkin's‐like type were the same as in the control case, but the density in pleomorphic type and anaplastic large cell type were weak. β‐Actin was used an internal standard. Clear β‐actin product can be seen for all cases used in this experiment.
Immunostaining of CX3CR‐1 and CCL18. In non‐specific lymphadenitis cases, many follicular lymphocytes and a few interfollicular lymphocytes were found to be positive for CX3CR‐1 (Fig. 3g). These positive cells were medium in size, with slight nuclear irregularities, indistinct nucleoli and abundant cytoplasm. A few follicular dendritic cells were positive for CCL18, and interfollicular lymphocytes only rarely (Fig. 3a). These cells were large to medium in size with abundant cytoplasm, and looked like interdigitated reticulum cells (Fig. 3b). Immunostaining for CX3CR‐1 and CCL18 was observed in cytoplasm. In HTLV‐1‐associated diseases, cells positive for CCL18 and CX3CR1 were the same as those in non‐specific lymphadenitis cases, whereas lymphoma cells were all negative (Fig. 3c–f,h,i). In addition, CCL18 expression in the Hodgkin's‐like type, Hodgkin's‐like cells were also positive (Fig. 3d). In the pleomorphic and anaplastic large cell types, however, only interdigitated but not large‐sized tumor cells were positive (Fig. 3e,f).
Figure 3.
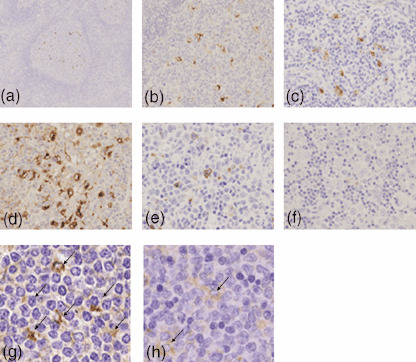
Immunohistochemical staining for CC chemokine ligand 18 (CCL18) (a–f). In non‐specific lymphadenitis, a few follicular dendritic cells were positive (a, low magnification; b, high magnification). In human T‐cell leukemia virus type 1‐associated lymphadenitis (c), Hodgkin's‐like type lymph node lesion (d), pleomorphic type lymph node lesion (e), and anaplastic large cell type lymph node lesion (f), dendritic cells but not tumor cells were positive, and Hodgkin's‐like giant cells were also positive (d). Immunohistochemical staining for CX3C chemokine receptor 1 (CX3CR1) (g, h). In non‐specific lymphadenitis, many follicular lymphocytes and a few interfollicular lymphocytes were positive (g). In pleomorphic type (h), the same cells were positive for CX3CR1 and were fewer than those in non‐specific lymphadenitis.
The median number of cells showing positive CCL18 expression in the non‐specific lymphadenitis type was 5.30/HPF, in the HTLV‐1‐associated lymphadenitis type it was 10.53/HPF, in the Hodgkin's‐like type 30.25/HPF, in the pleomorphic type 14.61/HPF and in the anaplastic large cell type 8.33/HPF (Fig. 4). HTLV‐1‐associated lymphadenitis type and Hodgkin's‐like type showed significantly higher expression than the control cases (control versus HTLV‐1‐associated lymphadenitis type, P = 0.0046; control versus Hodgkin's‐like type, P = 0.0018).
Figure 4.
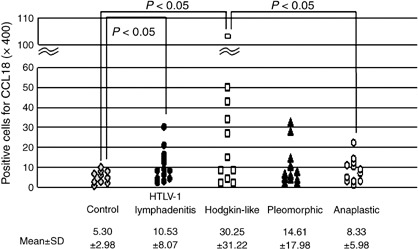
Comparison of CC chemokine ligand 18 expression in control cases (non‐specific lymphadenitis) and human T‐cell leukemia virus type 1 (HTLV‐1)‐associated diseases. HTLV‐1‐associated lymphadenitis and Hodgkin's‐like type lymph node lesion showed significantly higher expression than control cases (control versus HTLV‐1‐associated lymphadenitis, P = 0.0046; control versus Hodgkin's‐like type, P = 0.0018).
The median number of cells showing positive CX3CR‐1 expression in non‐specific lymphadenitis cases was 18.0/HPF, in the HTLV‐1‐associated lymphadenitis type it was 12.23/HPF, in the Hodgkin's‐like type 6.91/HPF, in the pleomorphic type 5.38/HPF and in the anaplastic large cell type 7.16/HPF (Fig. 5). All types showed significantly lower expression than the control cases (control versus HTLV‐1‐associated diseases, P < 0.05).
Figure 5.
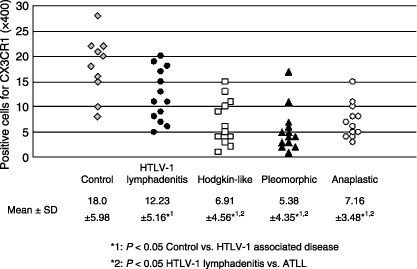
Comparison of CX3C chemokine receptor 1 expression in control cases (non‐specific lymphadenitis) and human T‐cell leukemia virus type 1 (HTLV‐1)‐associated diseases. All types of HTLV‐1‐associated diseases showed significantly lower expression than control cases.
Discussion
ATLL is a human malignancy associated with HTLV‐1 and features monoclonal integration of the HTLV‐1 provirus. HTLV‐1 is transmitted in a cell‐to‐cell fashion, and, after infection, promotes clonal proliferation of infected cells by pleiotropic actions of Tax and other viral proteins. A model of HTLV‐1 proviral DNA integration displayed change from undetectable to polyclonal, and then to monoclonal malignant transformation.( 18 ) After a long period of latency, ATLL develops in approximately 5% of asymptomatic carriers. In vivo, during the carrier state, infected cells are selected by the host's immune system, the genetic and epigenetic environment of proviral integration sites, and other factors.( 19 )
Previous studies( 3 ) have shown a clear correlation between the histological subtype of HTLV‐1 lymph node lesion and prognosis. The pleomorphic type was associated with a high mortality in the initial stages (2‐year survival rate was 31.3%), followed by a continuous and rapid decline in survival (5‐year survival rate was 11.0%). The anaplastic large cell type was also associated with a highly aggressive course (2‐year survival rate was 23.3%). In contrast, patients with HTLV‐1‐associated lymphadenitis were still alive at the end of that study (the duration of follow‐up was 2–20 years). Patients with Hodgkin's‐like type lymph node lesions showed a continuous decline in survival (2‐ and 5‐year survival rates were 62.1% and 25.9%, respectively). These findings indicate that the survival patterns of patients with lymph node lesions of the pleomorphic and anaplastic types are similar to those of ATLL, whereas the lymphadenitis type was confirmed to represent a non‐neoplastic state and the Hodgkin's‐like type a pre or early neoplastic state. A multistep process is thus thought to lead to the selection of neoplastic cells, although the exact mechanism remains to be elucidated.
Chemokines are key players in immune reaction by directing the migration and the activation of leukocytes throughout the body under various physiological and immunopathological conditions. Furthermore, they are involved in various other processes, including angiogenesis, hematopoiesis, tumor growth, and metastasis. CCR4, CC chemokine receptor 7 and CC chemokine ligand 5 have been identified as homing receptors and tumor markers in ATLL. In our study, CCL18 was found to be upregulated and CX3CR1 downregulated.
Fractalkine/CX3CL1 is a membrane‐bound chemokine that functions not only as a chemoattractant but also as an adhesion molecule, and is expressed on endothelial cells activated by proinflammatory cytokines. The fractalkine receptor, CX3CR1, is expressed on cytotoxic effector lymphocytes including NK cells and cytotoxic effector T cells, as well as on mature monocytes/macrophages, and mucosal dendritic cells, all of which play important roles in the elimination of pathogens and cancer cells.( 20 , 21 )
Proliferation of HTLV‐1 infected cells was shown to be controlled by cytotoxic T cells in vivo. Host cellular immune responses against HTLV‐1, especially the outgrowth of cytotoxic T cells, are frequently found in peripheral blood mononuclear cell culture of asymptomatic HTLV‐1 carriers but infrequently in ATLL patients.( 22 , 23 ) Treg are characterized by coexpression of CD4 and CD25. FoxP3 is expressed on Treg and is associated with Treg cell activity and phenotype. A previous study of the expression of FoxP3 on ATLL cells( 24 ) suggested that such expression might have a regulatory effect on cytotoxic T cells. In another study( 25 ) infection with HTLV‐1 was found to be significantly associated with poor survival in B‐cell lymphoma of HTLV‐1 carriers. Staining for TIA‐1, a marker of cytotoxic T cells, showed only a few T cells in HTLV‐1 carriers, compared to an abundance of T cells in B‐cell lymphoma without HTLV‐1. The HTLV‐1 carriers in our study, including those with HTLV‐1‐associated lymphadenitis, showed a low expression of CX3CR1 compared with non‐specific lymphadenitis cases. These findings suggest that HTLV‐1 infection correlates with host immunity through a reduction in cytotoxic T cells.
CCL18 is mainly expressed by a broad range of monocytes/macrophages and dendritic cells. Within a severe inflammatory context, the CCL18‐mediated attraction of naïve T cells toward the fully matured, interdigitating dendritic cells could assist in the mounting of a primary immune response. Enhanced CCL18 production has been established in several diseases, including various malignancies (acute lymphoblastic leukemia,( 26 ) gastric cancer,( 27 ) and ovarian carcinoma( 28 )) and inflammatory joint, lung, and skin disease.( 29 ) Leung et al.,( 27 ) in particular, using microarray and quantitative RT‐PCR analysis, found that high CCL18 levels were associated with prolonged survival of gastric cancer patients regardless of tumor stage. Immunohistochemical and in situ hybridization analysis showed that CCL18 was expressed by a subpopulation of tumor‐associated macrophages that were preferentially located at the tumor invasion front but not by neoplastic or non‐neoplastic gastric mucosal cells. Immunohistochemical staining used in our study showed the presence of CCL18 expression in dendritic cells but not ATLL cells. Hodgkin's‐like type cases showed higher levels of CCL18 expression than did those of non‐specific lymphadenitis, pleomorphic type and anaplastic large cell type. In a previous study( 3 ) we reported that the prognosis for pleomorphic type and anaplastic large cell type lymph node lesions was poor, whereas that for Hodgkin's‐like type was intermediate between HTLV‐1‐associated lymphadenitis type and other types. On the basis of these findings of improved patient outcome associated with CCL18 expression in Hodgkin's‐like type and HTLV‐1‐associated lymphadenitis type, it can be plausibly hypothesized that the expression of CCL18 in the ATLL cell environment might attract and activate immune cells, with the effect of targeting and augmenting immune responses against the ATLL cells. Our study's results also underscore a role for CCL18 in the generation of a more efficient antitumor response as a result of the attraction and activation of specific immune cells in ATLL. ATLL cells are reported to originate from CD4+ 25 + FoxP3 + Treg cells, and have regulatory T cell function.( 30 ) As ATLL cells are predominant in pleomorphic and anaplastic large cell types, it is possible that ATLL cells suppress CCL18 expression. However, in the present study we did not establish whether ATLL cells directly suppress CCL18 expression.
In conclusion, the expression of a panel of chemokines and/or their receptors showed that gene expression of CCL18 is upregulated and CX3CR1 downregulated compared with gene expression in non‐specific lymphadenitis. Immunohistochemical staining evidenced their expression in immunological but not in ATLL cells. These results suggest that an increase in CCL18 expression and a decrease in CX3CR1 expression play a role in immune responses against ATLL cells. As we could not carry out a functional analysis in this study, such an analysis of chemokines and their receptors seems to be necessary to further clarify the pathogenesis of ATLL.
Acknowledgments
This study was supported in part by a Grant‐in‐Aid from the Ministry of Education, Science and Culture, Japan. K.K. is a Research Fellow of the Japanese Society for the Promotion of Science (JSPS). We wish to thank Mrs Konomi Takasu for her technical assistance.
References
- 1. Takatsuki K. Overview of adult T‐cell leukemia/lymphoma. Jpn Soc Res 1985; 25: 97–103. [Google Scholar]
- 2. Kikuchi M, Mitsui T, Eimoto T, Toyooka R, Nishiuchi M. Biopsy of adult T cell leukemia. Gann Mono Can Res 1982; 28: 37–50. [Google Scholar]
- 3. Ohshima K, Suzumiya J, Sato K et al . Survival of patients with HTLV‐I‐associated lymph node lesions. J Pathol 1999; 189: 539–45. [DOI] [PubMed] [Google Scholar]
- 4. Ohshima K, Kikuchi M, Masuda Y et al . HTLV‐I associated lymphadenitis. Cancer 1992; 69: 239–48. [DOI] [PubMed] [Google Scholar]
- 5. Ohshima K, Kikuchi M, Yoshida T, Masuda Y, Kimura N. Lymph node in incipient adult T‐cell leukemia lymphoma with Hodgkin's disease‐like histologic features. Cancer 1991; 67: 1622–8. [DOI] [PubMed] [Google Scholar]
- 6. Ohshima K, Suzumiya J, Kato A, Tashiro K, Kikuchi M. Clonal HTLV‐I‐infected CD4+ T‐lymphocytes and non‐clonal non‐HTLV‐I‐infected giant cells in incipient ATLL with Hodgkin‐like histologic features. Int J Cancer 1997; 72: 592–8. [DOI] [PubMed] [Google Scholar]
- 7. Yoshie O, Fujisawa R, Nakayama T et al . Frequent expression of CCR4 in adult T‐cell leukemia and human T‐cell leukemia virus type 1‐transformed T cells. Blood 2002; 99: 1505–11. [DOI] [PubMed] [Google Scholar]
- 8. Hasegawa H, Nomura T, Kohno M et al . Increased chemokine receptor CCR7/EBI1 expression enhances the infiltration of lymphoid organs by adult T‐cell leukemia cells. Blood 2000; 95: 30–8. [PubMed] [Google Scholar]
- 9. Mori N, Krensky AM, Ohshima K et al . Elevated expression of CCL5/RANTES in adult T‐cell leukemia cells: possible transactivation of the CCL5 gene by human T‐cell leukemia virus type I Tax. Int J Cancer 2004; 111: 548–57. [DOI] [PubMed] [Google Scholar]
- 10. Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995; 270: 467–70. [DOI] [PubMed] [Google Scholar]
- 11. Heller RA, Schena M, Chai A et al . Discovery and analysis of inflammatory disease‐related genes using cDNA microarrays. Proc Natl Acad Sci USA 1997; 94: 2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Backert S, Gelos M, Kobalz U et al . Differential gene expression in colon carcinoma cells and tissues detected with a cDNA array. Int J Cancer 1999; 82: 868–74. [DOI] [PubMed] [Google Scholar]
- 13. Alizadeh AA, Eisen MB, Davis RE et al . Distinct types of diffuse large B‐cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–11. [DOI] [PubMed] [Google Scholar]
- 14. Ohshima K, Suzumiya J, Sato K et al . Nodal T‐cell lymphoma in an HTLV‐I‐endemic area: proviral HTLV‐I DNA, histological classification and clinical evaluation. Br J Haematol 1998; 101: 703–11. [DOI] [PubMed] [Google Scholar]
- 15. Tsuchiya T, Ohshima K, Karube K et al . Th1, Th2, and activated T‐cell marker and clinical prognosis in peripheral T‐cell lymphoma, unspecified: comparison with AILD, ALCL, lymphoblastic lymphoma, and ATLL. Blood 2004; 103: 236–41. [DOI] [PubMed] [Google Scholar]
- 16. Ohshima K, Kikuchi M, Masuda Y et al . Defective provirus form of human T‐cell leukemia virus type I in adult T‐cell leukemia/lymphoma: clinicopathological features. Cancer Res 1991; 51: 4639–42. [PubMed] [Google Scholar]
- 17. Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol 1999; 303: 179–205. [DOI] [PubMed] [Google Scholar]
- 18. Ohshima K, Suzumiya J, Izumo S, Mukai Y, Tashiro K, Kikuchi M. Detection of human T‐lymphotrophic virus type‐I DNA and mRNA in the lymph nodes; using polymerase chain reaction in situ hybridization (PCR/ISH) and reverse transcription (RT‐PCR/ISH). Int J Cancer 1996; 66: 18–23. [DOI] [PubMed] [Google Scholar]
- 19. Taylor GP, Matsuoka M. Natural history of adult T‐cell leukemia/lymphoma and approaches to therapy. Oncogene 2005; 24: 6047–57. [DOI] [PubMed] [Google Scholar]
- 20. Imai T, Nishimura M, Nanki T, Umehara H. Fractalkine and inflammatory disease. Jpn J Clin Immunol 2005; 28: 131–9. [DOI] [PubMed] [Google Scholar]
- 21. Nishimura M, Umehara H, Nakayama T et al . Dual functions of fractalkine/CX3C ligand 1 in trafficking of perforin+/granzyme B+ cytotoxic effector lymphocytes that are defined by CX3CR1 expression. J Immunol 2002; 168: 6173–80. [DOI] [PubMed] [Google Scholar]
- 22. Kannagi M, Matsushita S, Shida H, Harada S. Cytotoxic T cell response and expression of the target antigen in HTLV‐I infection. Leukemia 1994; 8 (Suppl 1): S54–9. [PubMed] [Google Scholar]
- 23. Kannagi M, Sugamura K, Kinoshita K, Uchino H, Hinuma Y. Specific cytolysis of fresh tumor cells by an autologous killer T cell line derived from an adult T cell leukemia/lymphoma patient. J Immunol 1984; 133: 1037–41. [PubMed] [Google Scholar]
- 24. Karube K, Ohshima K, Tsuchiya T et al . Expression of FoxP3, a key molecule in CD4+CD25+ regulatory T cells, in adult T cell leukemia/lymphoma cells. Br J Haematol 2004; 126: 81–4. [DOI] [PubMed] [Google Scholar]
- 25. Suefuji H, Ohshima K, Hayabuchi N, Nakamura K, Kikuchi M. HTLV‐1 carriers with B‐cell lymphoma. Prognosis, clinical and immunopathological features. Br J Haematol 2003; 123: 606–12. [DOI] [PubMed] [Google Scholar]
- 26. Struyf S, Schutyser E, Gouwy M et al . PARC/CCL18 is a plasma CC chemokine with increased levels in childhood acute lymphoblastic leukemia. Am J Pathol 2003; 163: 2065–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leung SY, Yuen ST, Chu KM et al . Expression profiling identifies chemokine (C–C motif) ligand 18 as an independent prognostic indicator in gastric cancer. Gastroenterology 2004; 127: 457–69. [DOI] [PubMed] [Google Scholar]
- 28. Schutyser E, Struyf S, Proost P et al . Identification of biologically active chemokine isoforms from ascetic fluid and elevated levels of CCL18/pulmonary and activation‐regulated chemokine in ovarian carcinoma. J Biol Chem 2002; 277: 24 584–93. [DOI] [PubMed] [Google Scholar]
- 29. Schutyser E, Richmond A, Damme JV. Involvement of CC chemokine ligand 18 (CCL18) in normal and pathological processes. J Leukoc Biol 2005; 78: 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yano H, Ishida T, Inagaki A et al . Regulatory T‐cell function of adult T‐cell leukemia/lymphoma cells. Int J Cancer 2007; 120: 2052–7. [DOI] [PubMed] [Google Scholar]


