Abstract
Transcriptional inactivation of breast cancer gene 1 (BRCA1) by DNA methylation is a frequent event in sporadic breast cancers. To investigate whether BRCA1 methylation is associated with survival in Chinese patients with sporadic breast cancer, BRCA1 methylation was determined using methylation specific PCR in 536 sporadic breast cancers. Survival curves for patients with methylated and unmethylated BRCA1 were compared using the log‐rank tests. Twenty‐six percent (139/536) of patients exhibited BRCA1 methylation in their tumors. The degree of BRCA1 methylation was correlated with clinical stages of breast cancer, but was not significant. Patients with BRCA1 methylated tumors had a significantly worse 5‐year disease‐free survival (DFS) and 5‐year disease‐specific survival (DSS) than did patients with unmethylated tumors (DFS: 73.2%vs 82.6%, P = 0.045; DSS 80.5%vs 87%, P = 0.038, two‐sided). In conclusions, BRCA1 methylation is a frequent event in breast cancer and is associated with poor clinical outcome in Chinese women with breast cancer. (Cancer Sci 2009; 100: 1663–1667)
Breast cancer gene 1 (BRCA1) encodes a multifunctional protein involved in DNA repair, cell cycle control, protein ubiquitinylation, and chromatin remodeling.( 1 , 2 ) It is well known that germline mutations of BRCA1 lead to familial breast cancer. The reported BRCA1 mutation rate was up to 45% in familial breast cancer, but only 1% in sporadic breast cancer.( 3 ) However, downregulation of BRCA1 is a very frequent event in sporadic breast cancer and correlates with its progression.( 4 )
Most housekeeping genes and 40% of tissue‐specific genes contain CpG islands around the transcription starting sites. Methylation of these CpG islands silences gene transcription epigenetically. Aberrant methylation of tumor suppressor genes including BRCA1 is a frequent event that may play an important role in carcinogenesis.( 5 , 6 , 7 , 8 ) It was reported that DNA methylation was the major cause of transcriptional silence of BRCA1, ranging from 13–40% in sporadic breast cancer.( 9 , 10 , 11 ) Recently, Xu et al. reported that BRCA1 methylation was correlated with the prognosis of Caucasian patients with breast cancers.( 12 ) However, the role of BRCA1 methylation is not well characterized in prognosis of sporadic breast cancers among other populations. In the present study, we determined the methylation status of BRCA1 in 536 Chinese patients with sporadic breast cancer and investigated whether the BRCA1 methylation was associated with clinical outcomes.
Materials and Methods
Study patients. A total of 611 patients with operable primary breast cancer (stage I–III) who had enough tumor DNA samples were selected from a pool of 857 consecutive breast cancer patients treated at the Breast Center, Peking University School of Oncology, from December 1994 to September 1999. A PCR product could not be obtained due to poor‐quality DNA in 75 of 611 available DNA samples. Thus, 536 patients with sporadic primary breast cancer were analyzed in the present study. Pathological diagnosis was performed for all patients. The patient ages ranged from 25 to 86 years, with a median of 49 years. A total of 290 patients were premenopausal, and 246 patients were postmenopausal. The stage of the tumors was classified according to the tumor‐node‐metastasis classification of the International Union Against Cancer (UICC). Patients received a radical or modified radical mastectomy, and the axillary lymph nodes were routinely dissected to at least level I and II. The status of lymph node metastasis was determined based on the histological examination. The majority of patients received adjuvant treatment, including chemotherapy, endocrine therapy, or combined treatment, as summarized in Table 1.
Table 1.
Association between methylation statuses of BRCA1 CpG island and clinicopathological characteristics in 536 patients with breast cancer
| Characteristics | n | BRCA1 methylation (%) | |
|---|---|---|---|
| Age | <50 years | 273 | 26.0 |
| ≥50 years | 263 | 25.9 | |
| Tumor size | <2 cm | 246 | 25.2 |
| ≥2 cm | 288 | 26.7 | |
| (Unknown) | 2 | 0 | |
| Histological type | Invasive ductal | 397 | 25.7 |
| Medullary | 61 | 26.2 | |
| Invasive lobular | 27 | 29.6 | |
| Mucinous | 20 | 35.0 | |
| Others* | 29 | 20.7 | |
| (Unknown) | 2 | 0 | |
| Clinical stage | I | 232 | 22.8 |
| II | 189 | 26.5 | |
| III | 112 | 32.1** | |
| (Unknown) | 3 | 0 | |
| Lymph node status | Positive | 209 | 29.2 |
| Negative | 325 | 24.0 | |
| (Unknown) | 2 | 0 | |
| ER status | Positive | 339 | 24.2 |
| Negative | 182 | 30.2 | |
| (Unknown) | 15 | 13.3 | |
| PR status | Positive | 248 | 22.6 |
| Negative | 273 | 29.3 | |
| (Unknown) | 15 | 20.0 | |
| HER2 status | Positive | 117 | 31.6 |
| Negative | 396 | 24.5 | |
| (Unknown) | 23 | 21.7 | |
| Adjuvant therapy | C | 225 | 24.4 |
| C + TAM | 215 | 24.2 | |
| TAM alone | 52 | 26.9 | |
| No treatment | 17 | 47.1 | |
| (Unknown) | 27 | 37.0 | |
| Total | 536 | 25.9 | |
Including five lipid‐rich carcinomas, two Paget's disease, nine scirrhous adenocarcinomas;
stage I versus stage II versus stage III, χ2 trend test, P = 0.066. BRCA1, breast cancer gene 1; C, chemotherapy; ER, estrogen receptor; HER2, neuroblastoma/glioblastoma derived oncogene homolog; M, methylated; PR, progesterone receptor; TAM, tamoxifen; U, unmethylated.
The levels of ER and progesterone receptor (PR) were measured by the dextran‐coated charcoal method. ER or PR was considered positive when the samples contained at least 10‐fmol/mg protein. HER2 expression was determined by immunohistochemistry as described elsewhere.( 13 ) BRCA1 expression was also determined by immunohistochemistry using a BRCA1 polyclone antibody (clone Ab‐1423; dilution 1:100; Signalway, Pearland, TX, USA). BRCA1 immunostaining was considered positive when more than 10% of tumor cells showed positive nuclear staining. The follow‐up data was available for all patients, with a median follow‐up of 8 years (range, 0.4 to 11.6 years). During the follow‐up period, 133 patients had developed distant metastases or local recurrences. Among them, 96 died of breast cancer. This study was approved by the Research and Ethical Committee of Peking University School of Oncology.
DNA extraction and bisulfite modification. Tumor DNA was extracted from the paraffin‐embedded breast tumor specimens by a phenol‐chloroform method. Briefly, three to four sections, 10 µm thick, were cut from each sample. After xylene deparaffination and washing with absolute ethanol, the sections were digested overnight with proteinase K (0.1 mg/mL) in 200 mL of DNA extraction buffer at 56°C.
Tumor DNA (1–2 µg) and controls were treated with sodium bisulfite and purified by the Wizards DNA Clean‐Up System Kit (Promega, Madison, WI, USA). During the modification, the unmethylated cytosines of the genomic DNA were converted to uridines, but the methylated cytosines remained unchanged.( 14 )
Methylation‐specific PCR (MSP). The methylation status of the BRCA1 CpG islands (NC_000017) were determined by MSP as described.( 15 ) The primers for detection of the unmethylated BRCA1 CpG islands were 5′‐ttggt ttttg tggta atgga aaagt gt‐3′ (sense) and 5′‐caaaa aatct caaca aactc acacc a‐3′ (antisense). The primers for detecting methylated BRCA1 CpG islands were 5′‐tcgtg gtaac ggaaa agcgc‐3′ (sense) and 5′‐aaatc tcaac gaact cacgc cg‐3′ (antisense).( 16 ) Approximately 60 ng of the bisulfite‐modified DNA was used as a template for MSP. The PCR conditions were as follows: initial denaturation at 95°C for 15 min to activate HotStar Taq DNA polymerase followed by amplification at 95°C for 30 s, 58°C for 30 s, and 72°C for 45 s for 38 cycles, and a final extension at 72°C for 10 min. PCR products were loaded onto 8% PAGE gel and visualized under UV illumination. The unmethylated and methylated PCR products were 86 bp and 75 bp, respectively. Genomic DNA of the human breast cancer cell line MCF7 was used as the unmethylated BRCA1 negative control. The M.Sss I‐modified genomic DNA of blood from healthy persons was used as the methylated BRCA1 positive control.
Clone sequencing. Fresh MSP products of BRCA1 were cloned with the AT clone kit (Ao Ke Company, Beijing, China), and sequenced on the ABI Prism 3730 DNA Analyzer (Foster City, CA, USA).
Statistical analysis. The correlation between BRCA1 methylation status, clinicopathologic characteristics, and adjuvant treatment was determined using Pearson's χ2‐test. DFS was defined as the time from the date of diagnosis to first recurrence (local or distant) or death from breast cancer without a recorded relapse. DSS was defined as the time from the date of diagnosis to death from breast cancer. Survival curves were assessed using the Kaplan–Meier method with log‐rank tests. A Cox regression model was applied to determine whether a factor was an independent predictor of survival in multivariate analysis. All statistical tests were two‐sided, and P‐values less than 0.05 were considered statistically significant. The statistical analyses were performed using SPSS 15.0 software (SPSS, Chicago, IL, USA).
Results
Prevalence of BRCA1 methylation in sporadic breast cancer. In this study of 611 tested patients, information on BRCA1 methylation status in primary breast carcinoma tissues was obtained for 536 patients (87.7%). Twenty‐six percent (139/536) exhibited BRCA1 methylation in tumors by MSP (Fig. 1). To confirm the results of MSP, six representatives of MSP products were further analyzed by sequencing. The sequencing data showed that all cytosines in the CpG sites in the methylated samples remained as cytosines and those cytosines not in the CpG sites were converted to thymidines, indicating that all CpG sites were indeed methylated (Suppl. Fig.). The degree of BRCA1 methylation was correlated with the clinical stages, but was not significant (P = 0.066) (Table 1). In addition, prevalence of BRCA1 methylation in 95 basal‐like related breast cancers (ER‐, PR‐, and HER2‐concomitant triple negative) was slightly higher than that in 423 cancers without the concomitant negative (32.6%vs 24.8%, P = 0.118).
Figure 1.
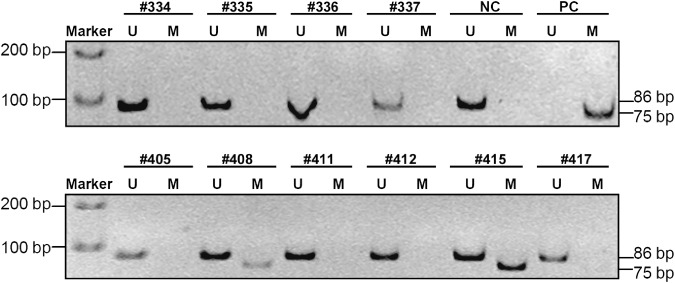
Detection of methylated and unmethylated breast cancer gene 1 (BRCA1) by methylation‐specific PCR. TE buffer was used as reagent control. Genomic DNA of human breast cancer cell line MCF7 was used as the unmethylated BRCA1 negative control (NC). The M.Sss I‐modified genomic DNA of blood from healthy persons was used as the methylated BRCA1 positive control (PC). Methylation‐specific PCR products (M, methylated, 75 bp; U, unmethylated, 86 bp) were run on an 8% PAGE gel.
Breast cancer gene 1 (BRCA1) protein expression was analyzed by immunohistochemistry in 41 representative samples of breast tumors with or without BRCA1 methylation in this cohort. The clinicopatholigical characteristics were not significantly different between the BRCA1‐methylated and ‐unmethylated tumors in these 41 patients (data not shown). There was a trend association that BRCA1‐methylated tumors exhibited a lower level (29%, 6/21) of BRCA1 expression as compared with BRCA1‐unmethylated tumors (55%, 11/20) (P = 0.086).
Association between BRCA1 methylation and survival. BRCA1 methylation was significantly associated with poorer DFS (P = 0.045, Fig. 2a) and DSS (P = 0.038, Fig. 2b). The 5‐year DFS rate was 73.2% in patients with BRCA1 methylation as compared with 82.6% in patients without BRCA1 methylation; and the 5‐year DSS rate was 80.5% in patients with BRCA1 methylated tumors compared with 87% in patients with unmethylated tumors.
Figure 2.
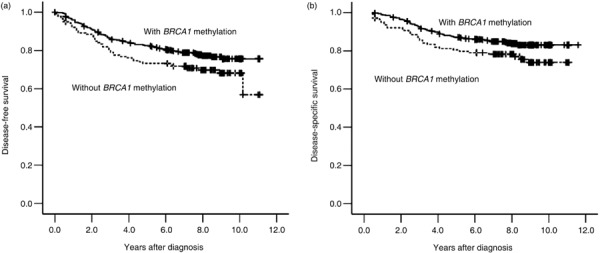
Kaplan–Meier survival curves for disease‐free survival (a) and disease‐specific survival (b) for patients with breast cancer gene 1 (BRCA1)‐methylated and ‐unmethylated sporadic breast cancer. Patients with BRCA1‐methylated breast cancer had a shorter disease‐free survival (P = 0.045, two‐sided) and shorter disease‐specific survival (P = 0.038, two‐sided) in univariate analysis.
Using univariate analysis, the clinical stage, lymph node status, HER2 status, and tumor size were also significantly linked to both DFS and DSS. ER status was significantly associated with DFS but not with DSS, whereas PR status and age of diagnosis were not associated with survival. Thus, multivariate analysis was used further to evaluate whether BRCA1 methylation was an independent factor of survival in this study. In multivariate analysis, clinical stage, lymph node involvement, and HER2 expression were significant independent factors, but BRCA1 methylation was not (DFS: HR = 1.23, 95% CI: 0.84–1.80, P = 0.295; DSS: HR = 1.27, 95% CI: 0.81–1.99, P = 0.290; Table 2).
Table 2.
Univariate and multivariate analysis of prognostic factors in Chinese patients with breast cancer *
| Factor | DFS | DSS | ||
|---|---|---|---|---|
| HR (95% CI) | P‐values** | HR (95% CI) | P‐values | |
| Univariate Analysis | ||||
| BRCA1 methylation | 1.45 (1.01–2.09) | 0.045 | 1.56 (1.02–2.37) | 0.038 |
| Clinical stage (I&II vs III) | 5.67 (4.02–8.01) | <0.001 | 5.35 (3.58–7.99) | <0.001 |
| Positive lymph node status | 4.71 (3.24–6.86) | <0.001 | 5.55 (3.50–8.81) | <0.001 |
| Tumor size (≥2 cm) | 1.52 (1.07–2.16) | 0.020 | 1.87 (1.22–2.86) | 0.004 |
| Positive HER2 status | 1.46 (0.99–2.15) | 0.055 | 2.07 (1.35–3.16) | 0.001 |
| Negative ER status | 1.73 (1.22–2.44) | 0.002 | 1.36 (0.90–2.04) | 0.142 |
| Multivariate analysis | ||||
| BRCA1 methylation | 1.23 (0.84–1.80) | 0.295 | 1.27 (0.81–1.99) | 0.290 |
| Clinical stage (I&II vs III) | 3.30 (2.04–5.33) | <0.001 | 2.69 (1.57–4.64) | <0.001 |
| Positive lymph node status | 2.13 (1.26–3.58) | 0.005 | 2.85 (1.53–5.32) | 0.001 |
| Tumor size (≥2 cm) | 0.98 (0.67–1.45) | 0.932 | 1.20 (0.75–1.94) | 0.444 |
| Positive HER2 status | 1.42 (0.95–2.10) | 0.086 | 2.06 (1.33–3.20) | 0.001 |
| Negative ER status | 1.60 (1.12–2.29) | 0.009 | 1.13 (0.71–1.80) | 0.604 |
Including treatment adjustment;
P‐value, two‐sided. BRCA1, breast cancer gene 1; CI, confidence interval; DFS, disease‐free survival; DSS, disease‐specific survival; ER, estrogen receptor; HER2, neuroblastoma/glioblastoma derived oncogene homolog; HR, hazard ratio.
Discussion
Previous studies showed that the methylation rate for BRCA1 varies from 12% to 40%.( 9 , 10 , 11 ) In the present study, we investigated the methylation status of BRCA1 in 536 Chinese patients with sporadic breast cancer. Twenty‐six percent of the patients exhibited BRCA1 methylation in their tumors. Our study extends previous research and is consistent with the observations that BRCA1 methylation is a relatively frequent event in sporadic breast cancers and that methylation of BRCA1 CpG islands might play an important role in breast carcinogenesis.
Several studies investigated the association between the BRCA1 methylation and tumorigenesis.( 16 , 17 ) BRCA1 methylation can occur in early stages of breast and ovarian carcinogenesis.( 18 ) But few studies have been performed to investigate the prognostic role of BRCA1 methylation in breast cancer. In the present study, we found that Chinese patients with BRCA1 methylated tumors had a significantly poorer DFS and DSS than patients with BRCA1 unmethylated tumors in univariate analysis (Table 2). Xu et al. also observed that BRCA1 methylation was an independent risk factor for high mortality in 851 Caucasian women with sporadic breast cancer.( 12 ) It was reported that the clinicopathological characteristics of breast cancers from Caucasian women and Asian women were different.( 19 , 20 ) Whether these differences might result in different contribution of BRCA1 methylation to patient survival could not be excluded. Anyway, in view of the positive observation in both populations, it is highly likely that BRCA1 methylation correlates with poor prognosis in patients with breast cancer.
Breast cancer gene 1 (BRCA1) is a tumor suppressor gene; thus, mutations within this gene increase a person's susceptibility to breast cancer and ovarian carcinoma. In familial breast cancer, the mutation rate is as high as 45%.( 21 ) However, the somatic mutation rate for BRCA1 is only 1.23% in sporadic breast cancer.( 3 ) The prevalence of somatic mutations of BRCA1 in the present tested samples is under investigation. Aberrant methylation of BRCA1 CpG islands may occur in a substantial number of sporadic breast cancers and may be associated with poor survival. It is well known that BRCA1 methylation results in downregulation of BRCA1 expression.( 16 , 18 ) We also observed a low positive rate of BRCA1 protein in breast tumors with BRCA1 methylation as compared with tumors without BRCA1 unmethylation (P = 0.086). However, the exact underlying mechanism(s) of BRCA1 dysfunction /methylation and its association with clinical outcomes in sporadic breast cancer is not clear.
Breast cancer gene 1 (BRCA1) is a crucial component for double‐strand DNA break repair. Dysfunction of BRCA1 might promote loss of PTEN expression, thus promoting cell transformation, proliferation, migration, angiogenesis, and genomic instability; inhibiting apoptosis; maintaining stem cell compartments; and finally leading to poor prognosis in breast carcinoma through the PTEN–PI3K pathway.( 22 ) There are number of factors related to the prognosis of breast cancer;( 23 ) whether BRCA1 methylation promotes proliferation and metastasis of sporadic breast cancer should be studied further.
In conclusion, the present study shows that methylation of BRCA1 CpG islands is associated with poor survival in patients with sporadic breast cancer, suggesting that inactivation of the BRCA1 gene by CpG methylation may be involved in the progression of breast cancer and affect the clinical course and treatment of patients with this disease.
Abbreviations
| BRCA1 | breast cancer gene 1 |
| CI | confidence interval |
| DFS | disease‐free survival |
| DSS | disease‐specific survival |
| ER | estrogen receptor |
| HR | hazard ratio |
| HER2 | neuroblastoma/glioblastoma derived oncogene homolog |
| MSP | methylation‐specific polymerase chain reaction |
| PR | progesterone receptor |
| PTEN | phosphatase and tensin homolog |
Supporting information
Fig. S1. Sequencing chromatograms of the methylation‐specific PCR products of breast cancer gene 1 (BRCA1) CpG islands in a representative sample by clone sequencing. The sequence of the methylated BRCA1 CpG islands with bisulfite modification was listed above each corresponding chromatogram. Red Ts were converted from the unmethylated Cs. The highlighted Cs were the methylated CpG sites that were not converted. Underlined fragments were partial primer sequences.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Acknowledgment
The present work was supported by the National Basic Research Program (973) of China (no. 2005CB522403 to D.D.).
References
- 1. Miki Y, Swensen J, Shattuck‐Eidens D et al . A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1 . Science 1994; 266: 66–71. [DOI] [PubMed] [Google Scholar]
- 2. Ralhan R, Kaur J, Kreienberg R, Wiesmüller L. Links between DNA double strand break repair and breast cancer. Accumulating evidence from both familial and nonfamilial cases. Cancer Lett 2007; 248: 1–17. [DOI] [PubMed] [Google Scholar]
- 3. Van Der Looij M, Cleton‐Jansen AM, Van Eijk R et al . A sporadic breast tumor with a somatically acquired complex genomic rearrangement in BRCA1 . Genes Chromosomes Cancer 2000; 27: 295–302. [DOI] [PubMed] [Google Scholar]
- 4. Thompson ME, Jensen RA, Obermiller PS, Page DL, Holt JT. Decreased expression of BRCA1 accelerates growth and is often present during sporadic breast cancer progression. Nat Genet 1995; 9: 444–50. [DOI] [PubMed] [Google Scholar]
- 5. Costello JF, Frühwald MC, Smiraglia DJ et al . Aberrant CpG‐island methylation has non‐random and tumour‐type–specific patterns. Nat Genet 2000; 24: 132–8. [DOI] [PubMed] [Google Scholar]
- 6. Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 2002; 21: 5427–40. [DOI] [PubMed] [Google Scholar]
- 7. Herman JG, Baylin SB. Mechanisms of disease: gene silencing in cancer in association with promoter hypermethylation. N Engl J Med 2003; 349: 2042–54. [DOI] [PubMed] [Google Scholar]
- 8. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet 2002; 3: 415–28. [DOI] [PubMed] [Google Scholar]
- 9. Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res 2001; 61: 3225–9. [PubMed] [Google Scholar]
- 10. Jeronimo C, Costa I, Martins MC et al . Detection of gene promoter hypermethylation in fine needle washings from breast lesions. Clin Cancer Res 2003; 9: 3413–7. [PubMed] [Google Scholar]
- 11. Jing F, Zhang J, Tao J et al . Hypermethylation of tumor suppressor genes BRCA1, p16 and 14–3–3sigma in serum of sporadic breast cancer patients. Onkologie 2007; 30: 14–9. [DOI] [PubMed] [Google Scholar]
- 12. Xu X, Gammon MD, Zhang Y et al . BRCA1 promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat 2009; 115: 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xu Y, Yao L, Li H et al . Presence of erbB2 mRNA in the plasma of breast cancer patients is associated with circulating tumor cells and negative estrogen and progesterone receptor status. Breast Cancer Res Treat 2006; 97: 49–55. [DOI] [PubMed] [Google Scholar]
- 14. Eads CA, Laird PW. Combined bisulfite restriction analysis (COBRA). In: Mills KI, Ramsahoye BH, eds. DNA Methylation Protocols (Methods in Molecular Biology, Vol. 200). Totowa, NJ: Humana Press, 2002; 53–70. [DOI] [PubMed] [Google Scholar]
- 15. Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation‐specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci USA 1996; 93: 9821–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Esteller M, Silva JM, Dominguez G et al . Promoter hypermethylation and BRCA1 inactivation in sporadic breast and ovarian tumors. J Natl Cancer Inst 2000; 92: 564–9. [DOI] [PubMed] [Google Scholar]
- 17. Gordon S, Kim A, Maureen I. Primary Ovarian Carcinomas Display Multiple Methylator Phenotypes Involving Known Tumor Suppressor Genes. Am J Pathol 2001; 158: 1121–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cuiju W, Akiko H, Tsutomu I. Expression of BRCA1 protein in benign, borderline, and malignant epithelial ovarian neoplasms and its relationship to methylation and allelic loss of the BRCA1 gene. J Pathol 2004; 202: 215–23. [DOI] [PubMed] [Google Scholar]
- 19. Kwong A, Cheung P, Chan S, Lau S. Breast cancer in Chinese women younger than age 40: are they different from their older counterparts? World J Surg 2008; 32: 2554–61. [DOI] [PubMed] [Google Scholar]
- 20. Li CI, Malone KE, Daling JR. Differences in breast cancer stage, treatment, and survival by race and ethnicity. Arch Intern Med 2003; 163: 49–56. [DOI] [PubMed] [Google Scholar]
- 21. Hutter RV. The role of the pathologist in breast cancer management. Cancer 1990; 66(6 Suppl): 1363–72. [DOI] [PubMed] [Google Scholar]
- 22. Saal LH, Gruvberger‐Saal SK, Persson C et al . Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet 2007; 40: 102–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Soerjomataram I, Louwman MWJ, Ribot JG, Roukema JA, Coebergh JWW. An overview of prognostic factors for long‐term survivors of breast cancer. Breast Cancer Res Treat 2008; 107: 309–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Sequencing chromatograms of the methylation‐specific PCR products of breast cancer gene 1 (BRCA1) CpG islands in a representative sample by clone sequencing. The sequence of the methylated BRCA1 CpG islands with bisulfite modification was listed above each corresponding chromatogram. Red Ts were converted from the unmethylated Cs. The highlighted Cs were the methylated CpG sites that were not converted. Underlined fragments were partial primer sequences.
Please note: Wiley‐Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item


