Abstract
Glutathione S‐transferase P1 (GSTP1) participates in detoxification of potentially genotoxic compounds that may alter the efficacy and toxicity of platinum‐based chemotherapy. We analyzed the influence of I105V polymorphism of GSTP1 on clinico‐pathological features and outcomes in 166 Chinese patients with metastatic colorectal carcinoma who had been treated with first‐line FOLFOX‐4. Combined analysis of GSTP1 I105V, ERCC1‐118, and XPD‐751 polymorphisms was also conducted. The results showed that, in comparison with Caucasian populations, a remarkably lower prevalence of Val105 allele variants was noted (24.7%). Patients with Val105 allele variants had a higher response to FOLFOX‐4 (56.1%vs 37.6%, P = 0.04), and a longer progression‐free (P < 0.01) as well as overall (P < 0.01) survival. By adjusted analysis, this polymorphism was identified as an independent prognostic factor (P = 0.01). In combined analysis, patients without any risk genotype, including GSTP1‐105 Ile/Ile, ERCC1‐118 C/T or T/T, and XPD‐751 Lys/Gln, had significantly longer progression‐free and overall survivals (P < 0.01). In addition, patients with Val105 allele variants had a higher incidence of grade 3/4 cumulative neuropathy after different cycles of treatment. These data suggest that Asian populations have a lower prevalence of I105V polymorphism in GSTP1. I105V polymorphism in GSTP1, by reducing its enzymatic activity and consequential detoxification to oxaliplatin, could be a key determinant for a better outcome, but more neurotoxicity, to FOLFOX‐4 treatment. (Cancer Sci 2009)
Colorectal carcinoma (CRC) is one of the leading causes of cancer‐related mortality in Taiwan and its incidence has increased steadily over the past few decades. Oxaliplatin is very effective in treating metastatic CRC patients,( 1 , 2 ) and improves the disease‐free survival of patients with stage II/III CRC.( 3 ) Neuropathy is the major toxicity of oxaliplatin, and the incidence of oxaliplatin‐induced severe neuropathy has varied from 12% to 18%.( 3 , 4 , 5 ) Oxaliplatin‐induced neuropathy includes an acute, transient peripheral nerve hyperexcitability,( 6 ) and a chronic, dose‐related, peripheral sensory neuropathy with symptoms similar to those caused by cisplatin.( 7 ) Development of chronic neuropathy results in severe disturbance of neurologic functions. Therefore, identification of factors predictive of neurotoxicity to oxaliplatin treatment, including genes involved in the nucleotide excision repair (NER)( 8 , 9 ) and detoxification pathways,( 10 ) is of extreme interest.
Glutathione S‐transferases (GST) are a multigene family of enzymes which catalyze the conjugation of glutathione to electrophilic xenobiotics to inactivate them, and thus prevent DNA damage and adduct formation.( 11 ) At least five major classes of the GST superfamily have been identified,( 10 ) among them, the GSTP1, GSTT1, and GSTM1 genotypes have been studies extensively for their influences in the inter‐individual variations of outcome to chemotherapeutic agents.( 12 , 13 ) The isoenzyme GSTP1 is highly expressed in human CRC tissues,( 14 ) and participates in the detoxification of platinum drugs that may mediate the resistance to platinum‐based chemotherapy. A single nucleotide polymorphism (A313G) in exon 5 of the GSTP1 gene causing isoleucine to valine substitution in the 105th amino acid (I105V) significantly decreases GSTP1 activity,( 11 ) and has a profound impact on chemotherapy for CRC patients.( 12 , 13 ) GSTP1 I105V polymorphism also predicts cumulative neuropathy in patients treated with docetaxel‐ as well as oxaliplatin‐based chemotherapy,( 15 , 16 ) but controversies exist. In addition, this polymorphism is associated with the susceptibility to several cancers.( 17 )
In addition to detoxification pathways, genetic polymorphisms in genes involved in DNA repair also attribute to resistance to oxaliplatin.( 8 , 9 ) From a genetic viewpoint, in a multi‐factorial disease, it is sometimes difficult with single polymorphisms in single genes to alter the extent of a physiologic or pathologic phenotype. Therefore, combined analysis of genes involved in the NER pathways, including ERCC1, XPD, and detoxification pathways, may more actually identify patients with maximal benefit, or toxicity, from oxaliplatin‐based chemotherapy.
Ethnic differences have a profound influence on the response and toxicity to chemotherapeutic agents. Due to a higher prevalence of epidermal growth factor receptor mutations, gefitinib becomes very effective in Asian patients with non‐small‐cell lung cancer.( 18 ) The UGT1A1*28 polymorphism is rare in Asian populations,( 19 ) leading to decreased risk of severe neutropenia after being treated with irinotecan. Furthermore, Asian populations have a remarkably lower prevalence of ERCC1 codon 118 C→T and XPD K751Q polymorphisms, which may affect platinum‐based chemotherapy.( 8 , 9 ) However, little is known about ethnic difference in GSTP1 I105V polymorphism and its impact on Asian CRC patients treated with oxaliplatin.
Based on these earlier findings, we propose that the GSTP1 I105V polymorphism may account for altered susceptibility and neuropathy to oxaliplatin‐based chemotherapy in CRC patients. Ethnic difference of this polymorphism in Asian populations was analyzed, and combined analysis of the GSTP1 I105V, ERCC1‐118, and XPD‐751 polymorphisms was conducted.
Materials and Methods
Patient characteristics. To understand the impact of the GSTP1 I105V polymorphism on response and neurotoxicity to FOLFOX‐4 treatment, we examined 217 Chinese patients with unresectable metastatic CRC, who had received FOLFOX‐4 as a first‐line treatment, from June 2003 to December 2007. Among them, 166 patients were enrolled and analyzed (patients’ characteristics are shown in Table 1). The remaining patients were excluded. They either lacked measurable lesions (n = 9), or did not have primary tumor removed to know the accurate T and N stages (n = 7), or died before blood sampling (n = 6), or were unwilling to participate (n = 4), or were lost in follow‐up (n = 3). Patients with diabetes mellitus (n = 10), central nervous system metastasis (n = 7), pre‐existing neuropathy (n = 3), and alcoholic disease (n = 2), were also excluded from this study. The FOLFOX‐4 regimen consisted of oxaliplatin (Sanofi‐Aventis, Paris, France) 85 mg/m2 (1‐h infusion, day 1) and folinic acid (FA) (200 mg/m2, 2‐h infusion, days 1 and 2) before bolus 5‐fluorouracil (5‐FU) (400 mg/m2, days 1 and 2) and infusional 5‐FU (600 mg/m2, 22‐h infusion immediately after bolus 5‐FU, days 1 and 2) was administered every 2 weeks. Patients with or without the GSTP1 I105V polymorphism were followed up at a similar intensity with a median duration of 18.0 months. The responses of treatment were evaluated on the basis of standard RECIST criteria. Patients with complete response (CR), partial response (PR), or stable disease remained in the protocol until progressive disease or unacceptable toxicity was documented. Common toxicities were assessed according to the National Cancer Institute Common Toxicity Criteria (NCI‐CTC).
Table 1.
Clinico‐pathological features of metastatic colorectal cancer patients with or without glutathione S‐transferase P1 (GSTP1) codon 105 Ile→Val polymorphism (n = 166)
| Characteristics | Ile/Ile (wild‐type) (%) | Ile/Val or Val/Val (%) | P‐values |
|---|---|---|---|
| All patients | 125 (100) | 41 (100) | |
| Age (years) | |||
| <50 | 43 (34.4) | 13 (31.7) | 0.75 |
| ≥50 | 82 (65.6) | 28 (68.3) | |
| Gender | |||
| Male | 77 (61.6) | 27 (68.9) | 0.63 |
| Female | 48 (38.4) | 14 (34.1) | |
| Performance status | |||
| 0 | 94 (75.2) | 29 (70.7) | 0.57 |
| 1, 2 | 31 (24.8) | 12 (29.3) | |
| Primary tumor | |||
| Colon | 85 (68.0) | 27 (65.9) | 0.80 |
| Rectum | 40 (32.0) | 14 (34.1) | |
| Histological differentiation | |||
| Well/moderate | 111 (88.8) | 36 (87.8) | 0.86 |
| Poorly/unknown | 14 (11.2) | 5 (12.2) | |
| Extent of invasion | |||
| T1‐2 | 44 (35.2) | 16 (39.0) | 0.66 |
| T3‐4 | 81 (64.8) | 25 (61.0) | |
| Lymph node involvement | |||
| N0 | 26 (20.8) | 11 (26.8) | 0.42 |
| N1‐3 | 99 (79.2) | 30 (73.2) | |
| Serum CEA level (ng/mL) | |||
| ≤6 | 13 (10.4) | 5 (29.3) | 0.75 |
| >6 | 112 (89.6) | 36 (87.8) | |
| TSER 28‐bp polymorphism | |||
| 2R/2R or 2R/3R | 42 (33.6) | 14 (34.1) | 0.95 |
| 3R/3R | 83 (66.4) | 27 (65.9) | |
| ERCC1 codon 118 genotype | |||
| C/C (wild type) | 59 (47.2) | 19 (46.3) | 0.92 |
| C/T or T/T | 66 (52.8) | 22 (53.7) | |
| XPD codon 751 status | |||
| Lys/Lys (wild type) | 105 (84.0) | 34 (82.9) | 0.87 |
| Lys/Gln | 20 (16.0) | 7 (17.1) | |
CEA, carcinoembryonic antigen; TSER, 5′‐enhancer region of the thymidylate synthase gene.
In this study, we primarily focused on oxaliplatin‐induced ‘chronic cumulative neuropathy’ because this neuropathy may result in a severe disturbance of neurologic function, and have a significant impact on oxaliplatin treatment. A detailed neurological history was obtained including possible risk factors for the development of peripheral neuropathy (e.g. diabetes mellitus, alcohol abuse, central nervous system diseases, or prior history of neurotoxic chemotherapy or neuropathy). Symptoms (paresthesias, dysesthesias, numbness, etc.) as well as whether symptoms interfered with function were assessed separately and were graded according to the NCI‐CTC criteria. Complete neurological examinations were performed at baseline and after four, eight, and 12 cycles of treatment. Treatment was delayed until recovery if grade 3–4 toxicity occurred, and the doses of oxaliplatin and 5‐FU were reduced by 20% in subsequent cycles. In the case of intolerable toxicity or failure on front‐line FOLFOX‐4, treatment was discontinued, and irinotecan‐based or fluoropyrimidine‐only regimens were subsequently administered according to physicians’ decision. During treatments, chest X‐ray, ultrasonography of the abdomen, or computed tomography scan was conducted every 2 months. An institutional review board approved this study and an informed consent was given by all patients before blood testing for genotyping.
Examination of the GSTP1 I105V polymorphism. Genomic DNA, extracted from patients’ leukocytes obtained via 0.5‐mL whole blood using standard phenol‐chloroform procedures, was subjected to GSTP1 codon 105 testing. The GSTP1 I105V polymorphism was examined by the PCR–restriction fragment length polymorphism (PCR‐RFLP) method as previously described.( 17 ) 0.1 μg of genomic DNA, forward primer 5′‐ACC CCA GGG CTC TAT GGG AA‐3′ and reverse primer 5′‐TGA GGG CAC AAG AAG CCC CT‐3′, were used for PCR amplification. Initial denaturation was carried out at 95°C for 5 min. Cycling conditions were: primer annealing at 55°C for 30 s, polymerization at 72°C for 30 s, and strand separation at 94°C for 30 s. Thirty cycles were carried out. A final polymerization step of 72°C for 5 min was carried out to complete the elongation processes. PCR products, after being digested by BsmAI restriction enzyme (New England Biolabs, Beverly, MA, USA) at 37°C for 2 h, were separated on 2% Nusieve ethidium bromide‐stained agarose gels to visualize the bands.
Examinations of genetic polymorphisms involved in the NER pathway and expression of the thymidylate synthase gene. An enhanced repair of DNA‐platinum lesions through the NER pathway results in poor response to platinum‐based chemotherapy. Because polymorphisms of genes involved in the NER pathway, including ERCC1 and XPD, have contributed to resistance to FOLFOX‐4 treatment in Asian populations,( 8 , 9 ) the influence of these polymorphisms between different GSTP1 codon 105 genotypes warrants further analysis. ERCC1 codon 118 C→T and XPD K751Q polymorphisms were examined by PCR‐RFLP as previously described.( 8 , 9 ) Primers 5′‐GCA GAG CTC ACC TGA GGA AC‐3′ (forward) and 5′‐GAG GTG CAA GAA GAG GTG GA‐3′ (reverse) were used for PCR amplification for examining ERCC1‐118 C→T, whereas 5′‐CCT CTC CCT TTC CTC TGT TC‐3′ (forward) and 5′‐CAG GTG AGG GGG ACA TCT‐3′ (reverse) were used for examining XPD K751Q. PCR products, after being digested with BsrD1 (for ERCC1) or MboII (for XPD) restriction enzymes (New England Biolabs), were separated on Nusieve ethidium bromide‐stained agarose gels.
Since 5‐FU was used in combination with oxaliplatin for treating these patients, and germ‐line polymorphisms of a 28‐bp tandemly repeated sequence in the 5′‐enhancer region of the thymidylate synthase gene (TSER) remarkably affect the response and survival of CRC patients receiving 5‐FU,( 20 ) the influence of this polymorphism on patients with different GSTP1 codon 105 genotypes was also analyzed. The primers used for TSER analysis were 5′‐GTG GCT CCT GCG TTT CCC CC‐3′ (forward) and 5′‐CCA AGC TTG GCT CCG AGC CGG CCA CAG GCA TGG CGC GG‐3′ (reverse).( 21 ) Then the amplified DNA fragments were analyzed by electrophoresis on a 4% agarose gel to determine the number of a 28‐bp tandemly repeated sequence in TSER.
Statistical analysis and survival curve plotting. All statistical analyses were performed using the SPSS software system (SPSS for Windows, version 14.0; SPSS, Chicago, IL, USA). The progression‐free and overall survival curves were plotted using the Kaplan–Meier product limit method, and the statistical differences in survival among subgroups were compared by log‐rank test. The correlations between different GSTP1‐105 genotypes and clinico‐pathological characteristics, response, and neurotoxicity to FOLFOX‐4 treatment were analyzed with the chi‐squared test. To assess the independent prognostic values of different polymorphisms, we used Cox’s proportional hazards regression analysis (multivariate) including GSTP1‐105, ERCC1‐118, and XPD‐751 genotypes and other clinico‐pathologic parameters. Cox hazard ratio and its 95% confidence interval (CI) were analyzed accordingly. Two‐sided P‐values less than 0.05 were considered statistically significant.
Results
Significantly lower prevalence of GSTP1 I105V polymorphism is identified in Asian populations, but no difference in this polymorphism between patients with or without CRC. Representative PCR‐RFLP patterns of different GSTP1‐105 genotypes are shown in Figure 1. The prevalence of Ile/Ile, Ile/Val, and Val/Val genotypes was 75.3% (n = 125), 22.9% (n = 38), and 1.8% (n = 3), respectively, in Chinese populations. It has been reported that in Caucasian populations, the prevalence of Ile/Ile, Ile/Val, and Val/Val genotypes was 45% (n = 34), 46% (n = 35), and 9% (n = 7), respectively.( 12 ) In another study, the prevalence of Ile/Ile, Ile/Val, and Val/Val genotypes was 61% (n = 39), 31% (n = 20), and 8% (n = 5), respectively.( 16 ) In comparison with Caucasian populations, a significantly lower prevalence of Val105 allele variants was clearly demonstrated in our cases (24.7%), indicating the existence of ethnic difference.
Figure 1.
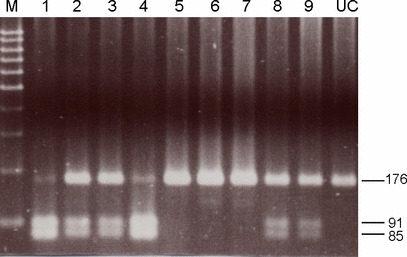
Representative PCR‐restriction fragment length polymorphism (RFLP) patterns of different glutathione S‐transferase P1 (GSTP1) codon 105 genotypes examined by patients’ blood samples. Genomic DNA obtained from patients’ leukocytes was subjected to PCR amplification using 5′‐ACC CCA GGG CTC TAT GGG AA‐3′ and 5′‐TGA GGG CAC AAG AAG CCC CT‐3′ as forward and reverse primers, respectively. PCR products after being digested by BsmAI were separated by agarose gel electrophoresis. Lanes 1 and 4 represent Val/Val; lanes 2, 3, 8, and 9 represent Ile/Val; lanes 5, 6, and 7 represent Ile/Ile. Lane UC indicates PCR product that has not been digested.
Regarding clinico‐pathologic characteristics, there were no between‐group differences in patients with or without GSTP1 I105V polymorphism (Table 1). Because I105V polymorphism of GSTP1 is associated with reduced enzymatic activity and detoxification efficiency,( 11 ) and is associated with susceptibility to bladder and testicular cancers,( 17 ) we wondered whether a higher percentage of 105V allele variants may present, and thus contribute to the malignant progression, in CRC patients. A total of 63 patients free of malignant diseases were analyzed, including hypertension (n = 23), diabetes mellitus (n = 22), chronic obstructive pulmonary diseases (n = 10), upper respiratory tract infection (n = 6), and urinary tract infection (n = 2). The percentages of Ile/Ile, Ile/Val, and Val/Val genotypes were 74.6% (n = 47), 23.8% (n = 15), and 1.6% (n = 1), respectively (P = 0.88), indicating no between‐group difference in patients with or without CRC.
GSTP1 I105V polymorphism leads to a higher response to and favorable prognosis for FOLFOX‐4 treatment. In comparison with Ile/Ile (wild type), patients with Ile/Val or Val/Val genotypes in GSTP1 have a significantly higher response to FOLFOX‐4 treatment (56.1%vs 37.6%, P = 0.04) (Table 2). Accordingly, longer progression‐free (12.0 months vs 6.0 months, P < 0.01) and overall (25.0 months vs 16.0 months, P < 0.01) survivals were observed in those with Val105 allele variants (Fig. 2), which is consistent with previous findings.( 12 , 13 ) By adjusted analysis, this polymorphism was further identified as an independent prognostic factor (P = 0.01; Table 3).
Table 2.
Response to FOLFOX‐4 treatment in patients with different genetic polymorphisms (n = 166)
| Genotypes | Responders (%) | Non‐responders (%) | P‐values |
|---|---|---|---|
| Overall | 70 (42.2) | 96 (57.8) | |
| GSTP1‐105 status | |||
| Ile/Ile (n = 125) | 47 (37.6) | 78 (62.4) | 0.04 |
| Ile/Val or Val/Val (n = 41) | 23 (56.1) | 18 (43.9) | |
| ERCC1‐118 genotype | |||
| C/C (n = 78) | 41 (52.6) | 7 (47.4) | 0.01 |
| C/T or T/T (n = 88) | 29 (33.0) | 59 (67.0) | |
| XPD‐751 status | |||
| Lys/Lys (n = 139) | 64 (46.0) | 75 (54.0) | 0.02 |
| Lys/Gln (n = 27) | 6 (22.2) | 21 (77.8) | |
| TSER 28‐bp polymorphism | |||
| 2R/2R or 2R/3R (n = 56) | 26 (46.4) | 30 (53.6) | 0.43 |
| 3R/3R (n = 110) | 44 (40.0) | 66 (60.0) | |
| Any risk genotype | |||
| Absence (n = 26) | 18 (69.2) | 8 (30.8) | <0.01 |
| Presence (n = 140) | 52 (37.1) | 88 (62.9) | |
Risk genotypes: GSTP1‐105 Ile/Ile, ERCC1‐118 C/T or T/T, and XPD‐751 Lys/Gln. Non‐responders, patients with stable or progressive diseases; responders, patients with complete or partial remission after treatment with FOLFOX‐4.
Figure 2.
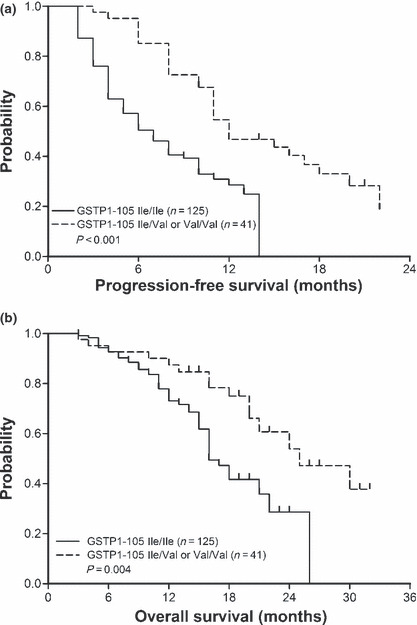
Patients with glutathione S‐transferase P1 (GSTP1) codon 105 Ile→Val polymorphism have significantly longer progression‐free and overall survivals after being treated with FOLFOX‐4. (a) Progression‐free survival curves of 166 patients with GSTP1‐105 Ile/Ile (wild type) or Ile/Val, Val/Val genotypes are plotted by Kaplan–Meier method (P < 0.01, log‐rank test). (b) A similar method has been used to plot overall survival curves of patients with different GSTP1 codon 105 genotypes (P < 0.01).
Table 3.
Analysis of factors that may affect the survival of patients with metastatic colorectal carcinoma receiving FOLFOX‐4 (n = 166)
| Characteristics | P‐values (univariate) | P‐values (multivariate) | HR | 95% CI |
|---|---|---|---|---|
| TSER 28‐bp polymorphism | ||||
| 2R/3R + 3R/3R vs 3R/3R | 0.06 | NA | NA | NA |
| GSTP1‐105 status | ||||
| Ile/Ile vs Ile/Val + Val/Val | 0.04 | 0.01 | 2.45 | 1.30–4.62 |
| ERCC1‐118 genotype | ||||
| T/T+T/C vs C/C | 0.03 | <0.01 | 3.15 | 1.89–5.23 |
| XPD‐751 status | ||||
| Lys/Gln vs Lys/Lys | 0.02 | <0.01 | 4.41 | 2.51–7.75 |
| Any risk genotype | ||||
| Presence vs absence | <0.01 | <0.01 | 5.73 | 2.22–14.78 |
Risk genotypes: GSTP1‐105 Ile/Ile, ERCC1‐118 C/T or T/T, and XPD‐751 Lys/Gln. 95% CI, 95% confidence interval for Cox hazard ratio; HR, Cox hazard ratio; NA, not analyzed.
With regard to genes involved in the NER pathway, patients with ERCC1‐118 C/T or T/T genotypes had a lower response to FOLFOX‐4 than those with C/C genotype (33.0%vs 52.6%, P = 0.01). And patients with XPD‐751 Lys/Gln genotype also had a lower response rate (22.2%vs 46.0%, P = 0.02) (Table 2). In combined analysis with three risk genotypes, including GSTP1‐105 Ile/Ile, ERCC1‐118 C/T or T/T, and XPD‐751 Lys/Gln, patients without the risk genotypes had a remarkably higher response rate than those with any one of the risk genotypes (69.2%vs 37.1%, P < 0.01) (Table 2). Accordingly, remarkably longer progression‐free (17.0 months vs 6.0 months, P < 0.01) and overall (30.0 months vs 16.0 months; P < 0.01) survivals were observed in patients without risk genotypes (Fig. 3). Furthermore, a better effect in predicting survival was found in combined analysis with three risk genotypes (HR = 5.73, Table 3). Although polymorphisms in TSER affect the response to 5‐FU in CRC patients;( 20 ) in the current study, patients with different TSER genotypes had a similar response to FOLFOX‐4 treatment (Table 2).
Figure 3.
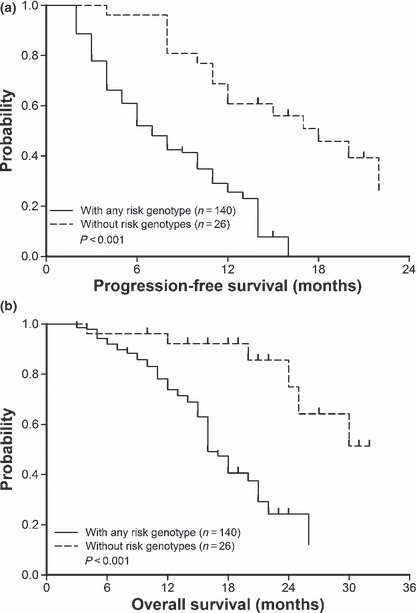
Patients without risk genotypes (GSTP1‐105 Ile/Ile, ERCC1‐118 C/T or T/T, and XPD‐751 Lys/Gln) have a significantly longer progression‐free as well as overall survival time after treatment with FOLFOX‐4. (a) Progression‐free survival curves of 166 patients with or without any risk genotype plotted by Kaplan–Meier method (P < 0.01, log‐rank test). (b) A similar method has been used to plot overall survival curves of patients with or without any risk genotype (P < 0.01).
GSTP1 I105V polymorphism is associated with higher incidence of severe oxaliplatin‐related cumulative neuropathy. Later, we proposed that the GSTP1 I105V polymorphism, by reducing enzymatic activity of GSTP1 and thus decreased metabolism of potentially cytotoxic compounds, might account for a higher incidence of severe neuropathy after treatment with oxaliplatin. As shown in Table 4, there were significantly more neurotoxicities in patients with Val105 allele variants than those without them. After eight cycles of treatment, the percentage of grade 3–4 peripheral sensory neuropathy was higher in patients with Val105 allele variants (19.5%vs 7.2%, P = 0.02). After 12 cycles, the percentage of grade 3–4 neuropathy remained higher in patients with Val105 allele variants (36.6%vs 14.4%, P < 0.01). In the current study, patients with different ERCC1‐118 and XPD‐751 genotypes had a similar incidence of grade 3–4 neurotoxicity after treatment with FOLFOX‐4 (P = 0.56 and 0.85, respectively).
Table 4.
Incidence of oxaliplatin‐induced neuropathy after different cycles of treatment of patients with different glutathione S‐transferase P1 (GSTP1) codon 105 status (n = 166)
| Neurotoxicity | Ile/Ile (wild type) (%) | Ile/Val or Val/Val (%) | P‐values |
|---|---|---|---|
| All patients enrolled | 125 (100) | 41 (100) | |
| After 4 cycles | |||
| Grade 0–2 | 124 (99.2) | 40 (97.6) | 0.40 |
| Grade 3–4 | 1 (0.8) | 1 (2.4) | |
| After 8 cycles | |||
| Grade 0–2 | 116 (92.8) | 33 (80.5) | 0.02 |
| Grade 3–4 | 9 (7.2) | 8 (19.5) | |
| After 12 cycles | |||
| Grade 0–2 | 107 (85.6) | 26 (63.4) | <0.01 |
| Grade 3–4 | 18 (14.4) | 15 (36.6) | |
Neurological toxicity defined by the National Cancer Institute Common Toxicity Criteria (NCI‐CTC).
Discussion
In comparison with Caucasian populations,( 12 , 16 ) a remarkably lower prevalence of the GSTP1 I105V polymorphism was noted in our patients (24.7%). Ethnic differences have a profound influence on chemotherapy for CRC patients. For example, the prevalence of homozygous triple‐repeat polymorphism in TSER was twice as common in Chinese as in Caucasian subjects, which accounts for an impaired response to fluoropyrimidine regimens.( 20 ) With regard to genes involved in the NER pathway, a remarkably lower prevalence of ERCC1 codon 118 C/T or T/T genotypes and XPD Gln751 allele variants have been found in Asian populations, which may account for an increased susceptibility to platinum‐based chemotherapy.( 8 , 9 ) It has been shown that Asian populations seem to have a lower prevalence of GSTP1 Val105 allele variants,( 10 ) but the sample size was quite small (n = 11). In our study, a larger sample size (n = 166) was analyzed, and a remarkably lower percentage of Val105 allele variants (24.7%) was identified, compared to that in Caucasian populations.( 12 , 16 ) These results implicated that Asian populations might have a better detoxification pathway. However, to date, no randomized study has been conducted to compare the outcome of FOLFOX treatment in different racial populations. Whether these polymorphisms may account for altered treatment outcome in Asian patients warrants further studies.
Altered expression of GST genes has been associated with different susceptibility to cancer.( 22 , 23 , 24 ) For example, methylation of the regulatory sequences near the GSTP1, resulting in its inactivation, has been identified in the early stages of prostatic carcinogenesis.( 22 ) Polymorphisms in GSTM1 and GSTT1 are associated with the susceptibility to colon cancer and age at onset,( 23 ) and I105V polymorphism of the GSTP1 has been associated with susceptibility to testicular and bladder cancers.( 17 ) Because I105V polymorphism of the GSTP1 is associated with reduced detoxification efficiency in comparison with the wild‐type one,( 11 ) a higher prevalence of the 105V allele variants might contribute to susceptibility in CRC. However, in the current study, there was no statistically difference between patients with CRC and control groups in different GSTP1 codon 105 genotypes, which was similar to a previous report.( 24 )
It has been reported that the ERCC1 codon 118 C→T and XPD K751Q polymorphism results in a higher NER proficiency;( 8 , 9 ) we proposed that these polymorphisms may have an influence in the development of neuropathy after treatment with oxaliplatin. But in the current study, patients with different ERCC1 codon 118 and XPD codon 751 genotypes had a similar incidence of severe neurotoxicity after treatment with FOLFOX‐4 (P = 0.56 and 0.85, respectively). These results indicate that different NER proficiency and oxaliplatin metabolisms between nerve and tumor tissues might exist. Another explanation is that nerve cells are not in actively dividing status; therefore, oxaliplatin neurotoxicity may be mediated by a different mechanism than the platinum‐DNA lesions. For example, oxalate, an oxaliplatin metabolite, may alter the properties of voltage‐gated sodium channels or slow down the clearance of platinum compounds from the peripheral nervous system.( 25 )
We found that patients with GSTP1 Val105 allele variants had a significantly higher incidence of grade 3/4 cumulative neuropathy after treatment with FOLFOX‐4 (Table 4). To date, in spite of the biochemical evidence that GSTs mediate the inactivation of platinum drugs, the association between polymorphisms in the GST genes and platinum‐induced neurotoxicity remains controversial.( 16 , 26 , 27 ) It has been shown that the Val105 allele variants of the GSTP1 were an unfavorable factor for developing neurotoxicity.( 26 ) However, in another study, this polymorphism confers a decreased risk of developing severe cumulative neuropathy.( 16 ) In another study, this polymorphism was not identified as having any correlation with oxaliplatin neurotoxicity.( 27 ) In some cases, these results should be interpreted with caution because some studies were analyzed on a retrospective basis with very limited sample size. Furthermore, concurrent medications and environmental factors may also play important roles in inter‐individual variability. Prospective studies with larger sample sizes and a homogenous population of patients treated with the same regimens are warranted in order to clarify this issue.
In the current study, patients with GSTP1 Val105 allele variants had a significantly higher response to FOLFOX‐4 (56.1%vs 37.6%, P = 0.04) (Table 2) and longer progression‐free and overall survivals (P < 0.01, Fig. 2), consistent with previous findings.( 12 , 13 ) GSTP1 is directly involved in the detoxification of cisplatin by the formation of cisplatin‐glutathione adducts,( 28 ) indicating that GSTP1 plays a role in the acquisition of resistance to this platinum compound.( 29 ) Patients with head and neck carcinoma with a lower level of intra‐tumor GST expression also have a better response and survival to platinum‐based treatment.( 30 ) However, discrepancies existed in the association between GSTP1‐105 variants and survival of CRC patients treated with oxaliplatin.( 26 , 27 ) The authors explained that these controversies might result from the exertion of effects through a multistep cascade by most chemotherapeutic drugs. In addition, reduced enzymatic activity in Val105 allele variants leading to increased toxicities might have influenced patient survival.( 26 )
Interestingly, a better effect in predicting treatment outcome was found in combined analysis with genes involved in detoxification and NER pathways (2, 3). In combined analysis with three risk genotypes, including GSTP1‐105 Ile/Ile, ERCC1‐118 C/T or T/T, and XPD‐751 Lys/Gln, patients without risk genotypes had a remarkably higher response rate than those who had any one of the genotypes (P < 0.01, Table 2). Accordingly, remarkably longer progression‐free and overall survivals (P < 0.01) are observed in patients without risk genotypes (HR = 5.73, Fig. 3). Because polymorphisms in genes involved in DNA repair also attributed to resistance to oxaliplatin,( 8 , 9 ) combined analysis of several polymorphisms in genes involved in NER and detoxification pathways may be more predictive of treatment outcome of oxaliplatin‐based chemotherapy.
Except for FOLFOX‐4, the GSTP1 I105V polymorphism also has effects on other types of chemotherapy. In non‐small‐cell lung cancer patients, GSTP1 I105V polymorphism increases the response to platinum‐based chemotherapy.( 31 ) In acute myeloid leukemia patients, a significantly longer relapse‐free, as well as overall survival, was found in patients with Val105 allele variants.( 32 ) For gastric cancer patients treated with cisplatin plus 5‐FU, carriage of the Val/Val genotype showed a significantly higher response rate and longer survival compared to those with at least one Ile allele.( 33 ) However, for metastatic breast cancer patients treated with high‐dose chemotherapy and autologous stem cell transplantation, the risks for disease progression and cancer specific death were higher in patients with AG or GG (Ile/Val or Val/Val) genotypes.( 34 ) Whether this is due to pre‐conditioning regimens that did not contain platinum drugs or simply due to different tumor type deserves further studies. With regard to irinotecan, this polymorphism was predictive of the response to irinotecan‐based chemotherapy in metastatic CRC patients, with the Val105 allele variants being associated with a better outcome.( 35 )
In summary, we have demonstrated that Asian populations have a lower prevalence of I105V polymorphism in the GSTP1 gene. I105V polymorphism of the GSTP1, by reducing its enzymatic activity and consequential detoxification to oxaliplatin, could be a key determinant for increased response but more neurotoxicity to FOLFOX‐4 treatment in Asian patients with metastatic CRC.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
National Science Council of the Republic of China grant no. NSC 96‐2320‐B‐010‐038 supported this study. We thank Mr. Chih‐Tung Ho for excellent technical assistance.
References
- 1. Rixe O, Ortuzar W, Alvarez M et al. Oxaliplatin, tetraplatin, cisplatin, and carboplatin: spectrum of activity in drug‐resistant cell lines and in the cell lines of the National Cancer Institute’s Anticancer Drug Screen panel. Biochem Pharmacol 1996; 52: 1855–65. [DOI] [PubMed] [Google Scholar]
- 2. De Gramont A, Figer A, Seymour M et al. Leucovorin and fluorouracil with or without oxaliplatin as first‐line treatment in advanced colorectal cancer. J Clin Oncol 2000; 18: 2938–47. [DOI] [PubMed] [Google Scholar]
- 3. Andre T, Boni C, Mounedji‐Boudiaf L et al. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N Engl J Med 2004; 350: 2343–51. [DOI] [PubMed] [Google Scholar]
- 4. Cassidy J, Tabernero J, Twelves C et al. XELOX (capecitabine plus oxaliplatin): active first‐line therapy for patients with metastatic colorectal cancer. J Clin Oncol 2004; 22: 2084–91. [DOI] [PubMed] [Google Scholar]
- 5. Tournigand C, Cervantes A, Figer A et al. OPTIMOX1: a randomized study of FOLFOX4 or FOLFOX7 with oxaliplatin in a stop‐and‐go fashion in advanced colorectal cancer – a GERCOR study. J Clin Oncol 2006; 24: 394–400. [DOI] [PubMed] [Google Scholar]
- 6. Wilson RH, Lehky T, Thomas RR et al. Acute oxaliplatin‐induced peripheral nerve hyperexcitability. J Clin Oncol 2002; 20: 1767–74. [DOI] [PubMed] [Google Scholar]
- 7. Extra JM, Marty M, Brienza S et al. Pharmacokinetics and safety profile of oxaliplatin. Semin Oncol 1998; 25: 13–22. [PubMed] [Google Scholar]
- 8. Chang PM, Tzeng CH, Chen PM et al. ERCC1 codon 118 C→T polymorphism associated with ERCC1 expression and outcome of FOLFOX‐4 treatment in Asian patients with metastatic colorectal carcinoma. Cancer Sci 2009; 100: 278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lai JI, Tzeng CH, Chen PM et al. Very low prevalence of XPD K751Q polymorphism and its association with XPD expression and outcomes of FOLFOX‐4 treatment in Asian patients with colorectal carcinoma. Cancer Sci 2009; 100: 1261–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Board PG, Baker RT, Chelvanayagam G et al. A novel class of glutathione transferases in a range of species from plants to humans. Biochem J 1997; 328: 929–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watson MA, Stewart RK, Smith GB, Massey TE, Bell DA. Human glutathione S‐transferase P1 polymorphisms: relationship to lung tissue enzyme activity and population frequency distribution. Carcinogenesis 1998; 19: 275–80. [DOI] [PubMed] [Google Scholar]
- 12. Stoehlmacher J, Park DJ, Zhang W et al. A multivariate analysis of genomic polymorphisms: prediction of clinical outcome to 5‐FU/oxaliplatin combination chemotherapy in refractory colorectal cancer. Br J Cancer 2004; 91: 344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stoehlmacher J, Park DJ, Zhang W et al. Association between glutathione S‐transferase P1, T1, and M1 genetic polymorphism and survival of patients with metastatic colorectal cancer. J Natl Cancer Inst 2002; 94: 936–42. [DOI] [PubMed] [Google Scholar]
- 14. Sato K. Glutathione S‐transferases as markers of preneoplasia and neoplasia. Adv Cancer Res 1989; 52: 205–55. [DOI] [PubMed] [Google Scholar]
- 15. Mir O, Alexandre J, Tran A et al. Relationship between GSTP1 Ile(105)Val polymorphism and docetaxel‐induced peripheral neuropathy: clinical evidence of a role of oxidative stress in taxane toxicity. Ann Oncol 2009; 20: 736–40. [DOI] [PubMed] [Google Scholar]
- 16. Lecomte T, Landi B, Beaune P, Laurent‐Puig P, Loriot MA. Glutathione S‐transferase P1 polymorphism (Ile105Val) predicts cumulative neuropathy in patients receiving oxaliplatin‐based chemotherapy. Clin Cancer Res 2006; 12: 3050–6. [DOI] [PubMed] [Google Scholar]
- 17. Harries LW, Stubbins MJ, Forman D, Howard GC, Wolf CR. Identification of genetic polymorphisms at the glutathione S‐transferase Pi locus and association with susceptibility to bladder, testicular and prostate cancer. Carcinogenesis 1997; 18: 641–4. [DOI] [PubMed] [Google Scholar]
- 18. Chang A, Parikh P, Thongprasert S et al. Gefitinib (IRESSA) in patients of Asian origin with refractory advanced non‐small cell lung cancer: subset analysis from the ISEL study. J Thorac Oncol 2006; 1: 847–55. [PubMed] [Google Scholar]
- 19. Liu CY, Chen PM, Chiou TJ et al. UGT1A1*28 polymorphism predicts irinotecan‐induced severe toxicities without affecting treatment outcome and survival in patients with metastatic colorectal carcinoma. Cancer 2008; 112: 1932–40. [DOI] [PubMed] [Google Scholar]
- 20. Marsh S, Collie‐Duguid ES, Li T et al. Ethnic variation in the thymidylate synthase enhancer region polymorphism among Caucasian and Asian populations. Genomics 1999; 58: 310–2. [DOI] [PubMed] [Google Scholar]
- 21. Horie N, Aiba H, Oguro K, Hojo H, Takeishi K. Functional analysis and DNA polymorphism of the tandemly repeated sequences in the 5’‐terminal regulatory region of the human gene for thymidylate synthase. Cell Struct Funct 1995; 20: 191–7. [DOI] [PubMed] [Google Scholar]
- 22. Chenevix‐Trench G, Young J, Coggan M, Board P. (1995) Glutathione S‐transferase M1 and T1 polymorphisms: susceptibility to colon cancer and age at onset. Carcinogenesis 1995; 16: 1655–7. [DOI] [PubMed] [Google Scholar]
- 23. Lee WH, Morton RA, Epstein JL et al. Cytidine methylation of regulatory sequences near the Pi class glutathione S transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci USA 1994; 91: 11733–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ates NA, Tamer L, Ates C et al. Glutathione S‐transferase M1, T1, P1 genotypes and risk for development of colorectal cancer. Biochem Genet 2005; 43: 149–63. [DOI] [PubMed] [Google Scholar]
- 25. Grolleau F, Gamelin L, Boisdron‐Celle M et al. A possible explanation for a neurotoxic effect of the anticancer agent oxaliplatin on neuronal voltage‐gated sodium channels. J Neurophysiol 2001; 85: 2293–7. [DOI] [PubMed] [Google Scholar]
- 26. Ruzzo A, Graziano F, Loupakis F et al. Pharmacogenetic profiling in patients with advanced colorectal cancer treated with first‐line FOLFOX‐4 chemotherapy. J Clin Oncol 2007; 25: 1247–54. [DOI] [PubMed] [Google Scholar]
- 27. Kweekel DM, Gelderblom H, Antonini NF et al. Glutathione‐S‐transferase pi (GSTP1) codon 105 polymorphism is not associated with oxaliplatin efficacy or toxicity in advanced colorectal cancer patients. Eur J Cancer 2009; 45: 572–8. [DOI] [PubMed] [Google Scholar]
- 28. Goto S, Iida T, Cho S, Oka M, Kohno S, Kondo T. Overexpression of glutathione S‐transferase pi enhances the adduct formation of cisplatin with glutathione in human cancer cells. Free Radic Res 1999; 31: 549–58. [DOI] [PubMed] [Google Scholar]
- 29. Hayes JD, Wolf CR, Townsend DM et al. The role of glutathione‐S‐transferase in anti‐cancer drug resistance. Oncogene 2003; 22: 7369–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nishimura T, Newkirk K, Sessions RB et al. Immunohistochemical staining for glutathione S‐transferase predicts response to platinum‐based chemotherapy in head and neck cancer. Clin Cancer Res 1996; 2: 1859–65. [PubMed] [Google Scholar]
- 31. Sun N, Sun X, Chen B et al. MRP2 and GSTP1 polymorphisms and chemotherapy response in advanced non‐small cell lung cancer. Cancer Chemother Pharmacol 2009. Jul 1. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Voso MT, Hohaus S, Guidi F et al. Prognostic role of glutathione S‐transferase polymorphisms in acute myeloid leukemia. Leukemia 2008; 22: 1685–91. [DOI] [PubMed] [Google Scholar]
- 33. Goekkurt E, Hoehn S, Wolschke C et al. Polymorphisms of glutathione S‐transferases (GST) and thymidylate synthase (TS) – novel predictors for response and survival in gastric cancer patients. Br J Cancer 2006; 94: 281–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bewick MA, Conlon MS, Lafrenie RM. Polymorphisms in manganese superoxide dismutase, myeloperoxidase and glutathione‐S‐transferase and survival after treatment for metastatic breast cancer. Breast Cancer Res Treat 2008; 111: 93–101. [DOI] [PubMed] [Google Scholar]
- 35. Kweekel DM, Koopman M, Antonini NF et al. GSTP1 Ile105Val polymorphism correlates with progression‐free survival in MCRC patients treated with or without irinotecan: a study of the Dutch Colorectal Cancer Group. Br J Cancer 2008; 99: 1316–21. [DOI] [PMC free article] [PubMed] [Google Scholar]


