Abstract
We previously reported the identification of natural human IgM antibodies, which recognize a M r 260 000 surface protein (NB‐p260) and induce both complement‐mediated cytotoxicity and apoptosis of human neuroblastoma cells. NB‐p260 was shown to belong to the family of filamin proteins. Filamin A is a high molecular weight actin‐binding protein, previously thought to be only located intracellularly. Here we show that NB cells as well as three NB‐unrelated human cell lines express filamin A also on the cell surface. Our findings suggest new biological functions for filamins, including a role as mediators in anti‐NB IgM‐induced apoptosis, and they add to the growing body of evidence of the interaction of cytoskeletal proteins with the extracellular matrix. (Cancer Sci 2006; 97: 1359–1365)
Abbreviations:
- BSA
bovine serum albumin
- DMEM
Dulbecco's modified Eagle's medium
- DTAF
dichlorotriazinylaminofluorescein
- FBS
fetal bovine serum
- FCS
fetal calf serum
- FLN
filamin
- FITC
fluorescein isothiocyanate
- HRP
horseradish peroxidase
- IgM
immunoglobulin M
- NB
neuroblastoma
- NCS
newborn calf serum
- PBS
phosphate‐buffered saline
- SDS‐PAGE
sodium dodecyl sulfate‐polyacrylamide gel electrophoresis
- TM
transmembrane domain.
Neuroblastoma is a tumor derived from primitive cells of the sympathetic nervous system and is the most common solid tumor in childhood.( 1 ) NB is usually referred to as the tumor with the highest rate of spontaneous regression. There are several theories explaining this phenomenon, including differentiation of tumor cells( 2 , 3 ) into benign ganglioneuroma or Schwann cell tumors,( 4 ) apoptosis,( 5 , 6 ) or complete lysis of the neoplastic cells either by cell‐mediated,( 7 , 8 ) or antibody‐mediated mechanisms.( 9 , 10 , 11 ) Antibody‐mediated mechanisms may include antibody‐dependent cellular cytotoxicity as well as complement‐mediated cytotoxicity. In our previous studies, we discovered the presence of anti‐NB antibodies in sera of healthy individuals including children and adults.( 12 , 13 ) These naturally occurring antibodies are of the IgM class and elicit effective killing of human NB cells in vitro by both complement activation and induction of apoptosis.( 13 , 14 ) Subsequent in vivo studies in nude rats with human NB xenografts demonstrated the inhibition of tumor growth by treatment with the IgM fraction from healthy adults with a high titer of natural anti‐NB IgM.( 15 , 16 ) In a Phase I/II study, NB patients were treated with plasmapheresis using donor plasma with a high anti‐NB IgM titer. Patients experienced pain in their tumors, and some patients showed radiographic evidence of tumor necrosis.( 17 ) No serious side‐effects were observed. The antigen recognized by the natural anti‐NB IgM was identified as a Mr 260 000 protein (NB‐p260).( 13 , 14 ) It was subsequently isolated and found to belong to the family of filamin proteins.( 18 , 19 )
Filamins are actin‐binding proteins, originally isolated from chicken gizzard( 20 ) that organize actin filaments into orthogonal networks and enhance the rigidity of the actin cortex.( 21 ) Three discrete FLNs have been described (FLNa, FLNb, and FLNc) and alternative splicing may allow for several additional isoforms.( 22 ) FLNa and FLNb contain an N‐terminal actin binding domain, followed by a semiflexible rod‐like domain consisting of 24 tandem repeats and two hinge (H) regions (H1 and H2), which confer flexibility to the otherwise rigid rod‐like structure (Fig. 1).( 23 ) Originally described in the context of its ability to organize the actin cytoskeleton, it is now known that FLNa interacts with a plethora of cytoplasmic, membrane, and nuclear proteins,( 23 , 24 , 25 ) although its biological functions remain largely unknown. Mutations in FLNa have been shown to cause diverse malformations in humans and periventricular heterotopia,( 26 , 27 ) whereas mutations in FLNb are associated with atelosteogenesis and boomerang dysplasia.( 28 )
Figure 1.
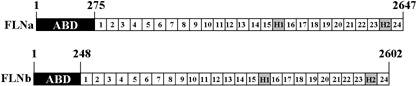
Structure of human filamin A (FLNa) and filamin B (FLNb). Filamins are cytoskeleton‐associated proteins and consist of an N‐terminal actin‐binding domain (ABD), 24 tandem repeats, two flexible hinge regions (H1 and H2) and a C–terminal self association domain (repeat 24).
In this study, we show that FLNa is expressed in NB cells and several other human cell lines. Significantly, this is the first demonstration that FLNa is not only expressed intracellularly but also on the cell surface. Using computer‐assisted transmembrane prediction analyses, we identified a transmembrane region in the N‐terminal part of FLNa. Based on our data, we propose a model in which the C‐terminal part of FLNa is exposed to the extracellular environment and thereby allows the interaction with natural human anti‐NB IgM or anti‐FLNa antibodies.
Materials and Methods
Cell lines and chemicals. The human NB cell lines were from Dr R. Seeger (LAN‐1; University of California Los Angeles, USA), Dr R. Wada (LAN‐5; Cancer Research Center of Hawaii, USA), and Dr N.‐K. Cheung (NMB‐7; Memorial Sloan‐Kettering Cancer Center, USA). The human melanoma cell line M2 (deficient in FLNa) was provided by Dr T. Stossel (Brigham and Women's Hospital, Harvard Medical School, USA), and the melanoma cell line A7 (M2 cells with reconstituted FLNa) was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). The cervical cancer cell line Henrietta Lacks (HeLa) and the human embryonic kidney cell line 293 (HEK293) were provided by Dr A. Lau and the ovarian cancer cell line SKOV‐3 by Dr B. Warn‐Cramer (Cancer Research Center of Hawaii, USA).
Mouse monoclonal FLNa antibodies MAB1678 and MAB1680 were obtained from Chemicon (Temecula, CA, USA), AB‐1 and AB‐2 from Laboratory Vision (Fremont, CA, USA), and ABP‐280 from BD Biosciences (San Jose, CA, USA). A delipidized goat antiserum against FLNa (F2762) was purchased from Sigma Chemicals (St Louis, MO, USA), and a goat polyclonal antibody against β‐catenin (SC‐1496) was from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The secondary antimouse HRP antibody was purchased from Amersham Biosciences (Piscatawa, NJ, USA), antigoat HRP antibody from Pierce (Rockford, IL, USA), antimouse Alexa Fluor 488 antibody from Molecular Probes (Eugene, OR, USA), antimouse and antigoat DTAF antibodies from Dianova (Hamburg, Germany), and antimouse FITC antibody from Sigma. HRP‐conjugated streptavidin was from Amersham Biosciences. Cell surface impermeable EZ‐Link Sulfo‐NHS‐Biotin was purchased from Pierce.
Cell culture conditions. The human NB cell lines LAN‐1, LAN‐5, and NMB‐7 were grown in RPMI 1640 medium (Biosource, Rockville, MD, USA) containing 10% heat‐inactivated FBS (Invitrogen, Carlsbad, CA, USA) as previously described.( 29 ) The human melanoma cell lines M2 and A7 were grown in Eagle's Minimum Essential Medium (Mediatech, Herndon, VA, USA) containing 2% FBS and 8% NCS. G418 (Gibco, Carlsbad, CA, USA) was added to A7 cells at 0.3 mg/mL. The human cell lines HeLa and HEK293 were cultured in high glucose DMEM (Gibco, Carlsbad, CA, USA) supplemented with 10% FCS (HyClone, Walkersville, MD, USA) and 2 mM L‐glutamine (Gibco). All above cell culture media were supplemented with penicillin (100 IU/mL) and streptomycin (100 µg/mL). The human cell line SKOV‐3 was maintained in McCoys 5 A medium (Sigma) supplemented with 5% (v/v) FBS and 100 µg/mL gentamicin.
Preparation of membrane fractions. To isolate membrane proteins, NMB‐7 cells were harvested and processed using the Mem‐PER Eukaryotic Membrane Protein Extraction Kit (Pierce) according to the manufacturer's instructions. Briefly, cells (5 × 106) were suspended in 0.15 mL of reagent A, incubated for 10 min at room temperature, and then placed on ice. A mixture of reagent B (1 volume) and reagent C (2 volumes) was added to the cells in reagent A and incubated on ice for 30 min and occasionally mixed by vortex. All reagents contained protease inhibitors leupeptin (10 µg/mL), EGTA (1 mM), and PMSF (1 mM). After centrifugation at 10 000 g for 3 min, the supernatant was incubated for 10 min at 37°C allowing the formation of two phases. After centrifugation at 10 000 g for 2 min, the upper phase (hydrophilic protein fraction) was transferred to a new tube, the intermediate layer was carefully removed, and the lower phase (solubilized membrane protein fraction) was recovered. Samples were analyzed by SDS‐PAGE (7.5%) and western blot.
Cell surface biotinylation. Subconfluent (60%) LAN‐1 cells were incubated with cell membrane impermeable EZ‐Link Sulfo‐NHS‐Biotin (0.5 mg/mL) (Pierce) or with PBS (control) for 60 min at 4°C. Cell viability was determined before and after biotinylation using trypan blue and was higher than 95%. Cells were washed with PBS and 100 mM glycine/PBS, and incubated in 100 mM glycine/PBS for 90 min at 4°C. After several washes in PBS, cells were lyzed in RIPA buffer (20 mM Tris‐HCl, pH 7.5, 0.1% sodium lauryl sulfate, 0.5% sodium deoxycholate, 135 mM NaCl, 1% Triton X‐100, 10% glycerol, 2 mM EDTA), supplemented with leupeptin (10 µg/mL), EGTA (1 mM), PMSF (1 mM), and the Complete protease inhibitor cocktail (Roche, Indianapolis, IN, USA). Precleared cell lysates were then incubated with the FLNa‐recognizing antibody MAB1678 (60 min at 4°C), immunoprecipitated using protein G beads (60 min at 4°C) (Amersham Biosciences), and analyzed by SDS‐PAGE (7.5%) and western blot.
Western blot analysis and immunodetection. Precipitated proteins or total cell lysates were loaded in reducing sample buffer (Bio‐Rad, Hercules, CA, USA), separated by 7.5% SDS‐PAGE, and electroblotted onto a PVDF membrane (200 mA, 3 h at 4°C). Membranes were probed with individual primary antibodies as indicated, washed several times, and then incubated with secondary HRP‐conjugated antibodies or HRP‐conjugated streptavidin. Bands were detected using the ECL reagent (Amersham Biosciences) and Kodak BioMax XAR film (Fisher Scientific, Pittsburgh, PA, USA).
Immunocytochemistry. Cells were grown on cover slips to subconfluency in 12‐well plates and FLNa visualized as follows: Cells were washed, fixed in 100% methanol for 20 min, and exposed to 0.1% Triton X‐100 for 5 min. Fixed cells were washed again, blocked in 1% BSA/PBS for 30 min and incubated with the FLNa‐recognizing antibody MAB1680, followed by repeated washes and incubation with antimouse Alexa Fluor 488 antibody. Washed cover slips were fixed onto slides with ProLong Antifade (Molecular Probes) and samples analyzed with an LSM 510 confocal laser microscope (Carl Zeiss, Goettingen, Germany).
Flow cytometry. The cytofluorometric binding assay was performed as described previously.( 13 ) Cell lines (0.25–5 × 105) were incubated with primary antibody in PBS for 45–60 min at 4°C and washed, followed by incubation with secondary DTAF or FITC‐conjugated antimouse antibodies or DTAF‐conjugated antigoat antibody in PBS for 30–60 min at 4°C. Controls included the incubation of cells with PBS or with secondary antibody only. After several washes in PBS, cells were suspended in 0.3 mL PBS containing propidium iodide (Sigma) for exclusion of dead cells. The binding studies were performed using the FACSscan instrument (Becton Dickinson, San Jose, CA, USA) and data were evaluated with the program CellQuest.
Computational analyses. The primary amino acid sequence of FLNa was analyzed with the Dense Alignment Surface transmembrane prediction server (http://www.sbc.su.se/~miklos/DAS/).( 30 ) Profile scores of 1.7 and 2.2 indicate a loose cutoff and a strict cutoff, respectively. The presence of the transmembrane domain was confirmed with the HMMTOP server, Version 2.0 (http://www.enzim.hu/hmmtop/index.html).( 31 , 32 ) The prediction program ScanProsite, available at the ExPASy server (http://c.expasy.org/tools/scanprosite/),( 33 ) was used to identify the RGD integrin binding sites, and these sites were confirmed using the Eukaryotic Linear Motif server (http://elm.eu.org/).( 34 )
Results
To test whether human NB cells express endogenous FLNa, and to study its localization within the cells, we visualized FLNa in NB cell lines LAN‐1 and LAN‐5 using a confocal laser microscope (Fig. 2). In both cell lines, FLNa was mainly expressed at the cell membrane and in neurite extensions, and was also present in the cytoplasm. As expected, FLNa was detected in the FLNa‐overexpressing cell line A7, and no FLNa staining was observed in the FLNa‐deficient M2 cells (Fig. 2). These results demonstrate that FLNa is expressed in NB cells, and predominantly localizes to the cell membrane.
Figure 2.
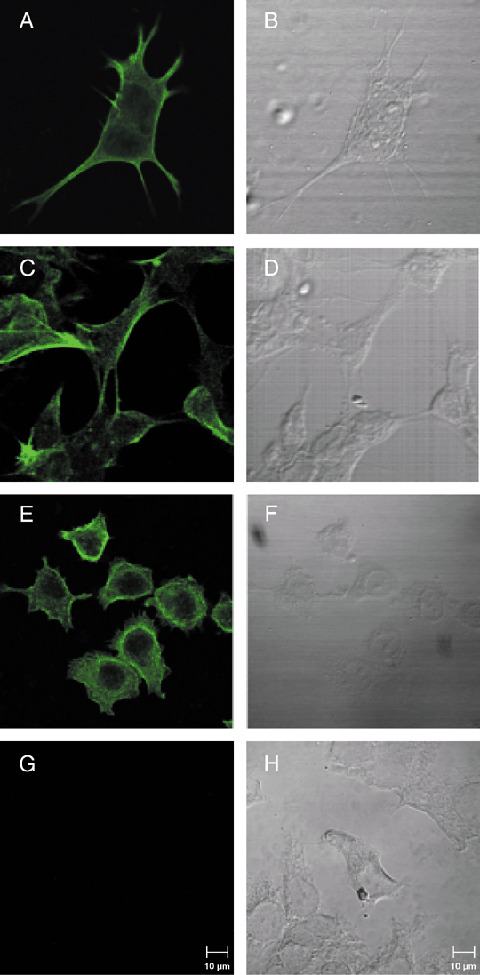
Detection of endogenous filamin A (FLNa) by confocal laser microscopy (A, C, E, G) and corresponding differential interference contrast images (B, D, F, H) of human NB cell lines LAN‐1 (A, B) and LAN‐5 (C, D), and human melanoma cell lines A7 (E, F) and FLNa‐deficient M2 (G, H).
To address whether FLNa is exposed on the cell surface, LAN‐1 cells were surface biotinylated, lyzed, and cell lysates were immunoprecipitated with the monoclonal anti‐FLNa antibody MAB1678. Both cell lysates and immunoprecipitates were separated on SDS‐PAGE, electroblotted onto PVDF membranes, and probed with anti‐FLNa antibody MAB1678 or streptavidin (Fig. 3). The result indicates that FLNa is exposed on the cell surface. Two smaller bands of approximately 73 kDa and 65 kDa were also observed in the immunoprecipitates. These smaller bands stained particularly strong with streptavidin, suggesting the additional presence of FLNa fragments on the cell surface.
Figure 3.
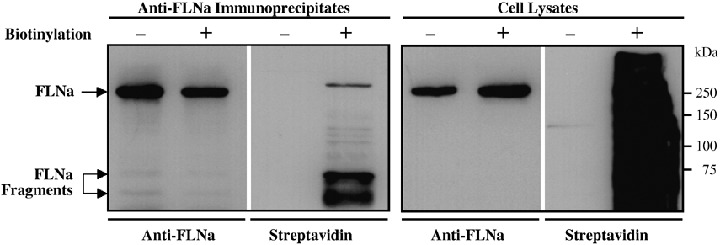
Detection of surface‐expressed filamin A (FLNa) in LAN‐1 cells. Subconfluent cells were surface‐biotinylated, lyzed, and immunoprecipitated with the anti‐FLNa antibody MAB1678. Cell lysates (right panel) or precipitated proteins (left panel) were resolved by 7.5% SDS‐PAGE under reducing conditions, electroblotted onto a PVDF membrane, and probed with the MAB1678 antibody or streptavidin.
Next, we tested whether FLNa can be detected in membrane fractions of NB cells. As shown in Fig. 4, the solubilized integral membrane protein fraction of NMB‐7 cells contained full length FLNa (280 kDa), indicating that FLNa is tightly associated with the cell membrane of NB cells.
Figure 4.
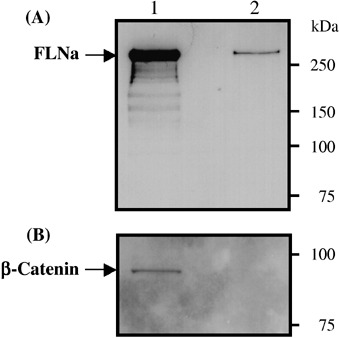
Detection of endogenous filamin A (FLNa) in the integral membrane protein fraction of NMB‐7 NB cells. Integral membrane proteins of NMB‐7 cells were extracted into a solubilized membrane protein fraction. Both the integral membrane protein fraction (lane 2) and the hydrophilic protein fraction containing cytoplasmic and peripheral membrane proteins (lane 1) were subjected to 7.5% SDS‐PAGE and western blotting using (A) the anti‐FLNa antibody MAB1678 or (B) an anti‐β‐catenin antibody (as a cytoplasmic control protein).
To further investigate the localization of FLNa, a cytofluorometric binding study was performed with five monoclonal antibodies and one polyclonal antiserum against FLNa (Fig. 5). The polyclonal antiserum and two of the five monoclonal antibodies detected FLNa on the surface of LAN‐1 cells. This result provides further evidence that FLNa is exposed on the cell surface of NB cells. The observation that not all monoclonal antibodies recognized FLNa might be a consequence of the fact that not all epitopes are exposed or freely accessible to an antibody on the cell surface.
Figure 5.
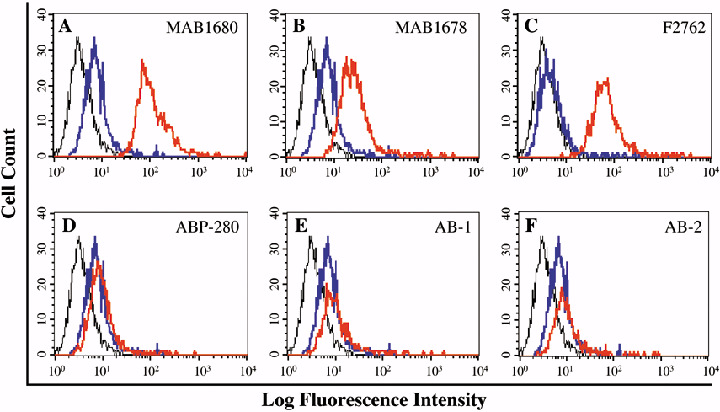
Detection of filamin A (FLNa) on the cell surface of NB cells by flow cytometry. The binding analysis was performed with LAN‐1 cells using five different mouse monoclonal antibodies (A, B, D, E, F) and one goat polyclonal antiserum (C) against FLNa. The red line represents the FLNa‐specific antibody. Controls include secondary antibody alone (blue line), and cells incubated with PBS in the absence of antibodies (black line). The two monoclonal antibodies (MAB1680 and MAB1678) and the goat polyclonal antiserum (F2762) showed strong binding to the NB cells. No binding was detected with the monoclonal antibodies ABP‐280, AB‐1, and AB‐2.
To test whether FLNa is also expressed on the cell surface of NB‐unrelated cells, we selected the cervical cancer cell line HeLa, the ovarian cancer cell line SKOV‐3, and the human embryonic kidney cell line HEK293 for comparative studies. For these experiments, MAB1680 was chosen as this FLNa‐recognizing monoclonal antibody successfully recognized FLNa in LAN‐1 cells (see Fig. 5). As shown in Fig. 6, FLNa was also expressed on the surface of all three cell lines, thus indicating that FLNa surface exposure is not unique to NB cells.
Figure 6.
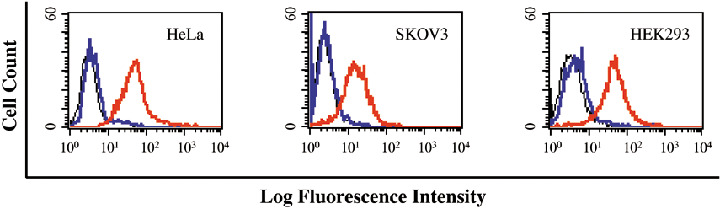
Detection of filamin A (FLNa) on the cell surface of human cell lines by flow cytometry. The binding analysis was performed with HeLa, SKOV‐3, and HEK293 cells using the FLNa‐recognizing mouse monoclonal antibody MAB1680 (red line). Controls include secondary antibody alone (blue line), and cells incubated with PBS in the absence of primary and secondary antibodies (black line).
To further examine these observations, a computer‐assisted analysis of the FLNa primary amino acid sequence using two different transmembrane prediction programs was performed. Both programs predicted a transmembrane domain near the N‐terminus of FLNa (residues 137 and 155) (Fig. 7). The programs further predicted that the N‐terminus of FLNa is in the intracellular space. The profile for FLNb was nearly identical (not shown).
Figure 7.
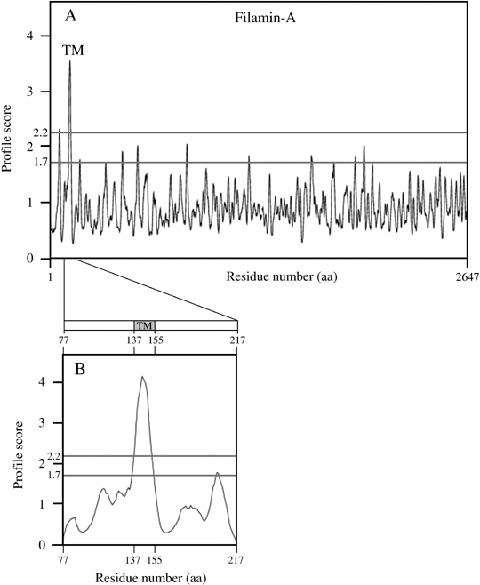
Computational analysis of the filamin A (FLNa) structure. (A) Using the Dense Alignment Surface transmembrane prediction program, a transmembrane domain was predicted in the N‐terminal part of FLNa. (B) A detailed analysis of residues 77–217 of FLNa localized the predicted transmembrane domain between residues 137 and 155. TM, transmembrane domain; aa, amino acids.
Discussion
This is the first report that shows the expression of FLNa on the cell surface. Based on our biochemical data and combined with our computational analyses, we propose a model in which the C‐terminus of FLNa is exposed to the extracellular matrix and the N‐terminus of FLNa is located in the cytoplasm (Fig. 8). In this model, the actin‐binding domain of the N‐terminus continues to associate with the actin cytoskeleton, while the surface‐displayed portion allows for the interaction with various extracellular ligands, including natural anti‐NB IgM or anti‐FLNa antibodies. Consistent with this model is our previous observation that the treatment of human NB cells with proteinase K abolished the binding of natural anti‐NB IgM.( 13 ) Our model is also consistent with the observed binding of the two FLNa‐recognizing monoclonal antibodies MAB1678 and MAB1680. Although the exact epitopes recognized by these antibodies are not known, they are directed against the calpain‐cleaved 190‐kDa and 90‐kDa fragments of FLNa, respectively, of which the N‐terminal 190‐kDa would be partially extracellular and the 90‐kDa C‐terminal fragment completely extracellular. It will also be of interest to test whether FLNb is displayed on the cell surface as FLNb‐specific antibodies become available.
Figure 8.

Hypothetical structure model for filamin A (FLNa). The actin binding domain‐containing N‐terminus of FLNa is in the intracellular space and continues to interact with the actin cytoskeleton, while the C‐terminus is exposed to the extracellular environment. Possible integrin binding sites (RGD) are indicated (residues 437–439 and 1312–1314).
Interestingly, computational motif searches and 3‐D structure predictions of FLNa revealed two loop‐forming arginine‐glycine‐aspartate (RGD) motifs. These motifs are known to be present, for example, in vitronectin and fibronectin, both extracellular proteins, which interact with integrin receptors.( 35 ) Similarly, FLNb contained one RGD motif. The functional relevance of this observation is currently unclear and warrants further investigation.
In summary, our data suggest that some cell types expressing FLNa possess a mechanism that enables the translocation of FLNa (and possibly FLNb) to the cell surface while remaining attached to the plasma membrane via an N‐terminal transmembrane anchor. The spectrum of human cells expressing FLNa on the surface is currently not known. Although surface expression has now been demonstrated for various human NB cells, cervical cancer cells (HeLa), ovarian cancer cells (SKOV‐3), human embryonic kidney cells (HEK293), and human keratinocytes (HaCaT),( 36 ) our previous work indicates that other human tumor and normal cell lines do not express FLNa (NB‐p260) on the surface.( 13 ) The prediction that a transmembrane domain is located close to the N‐terminus is further supported by previous findings that FLNa contains two different lipid‐binding sites, namely residues 49–71 and 131–155, which insert into membranes under in vitro conditions.( 37 ) These sites are also present in FLNb with approximately 90% homology. It is also possible that FLN isoforms and splice variants,( 22 , 38 ) or post‐translational modifications of FLNa, localize to different sites within the cell and thereby assume new functions. For example, it was recently shown that FLNa or fragments of FLNa localize to the nucleus where they regulate the function of FOXC1 transcription factors and androgen receptors.( 24 , 39 ) Furthermore, decorin, a small extracellular matrix proteoglycan, was found to bind to the C‐terminus of FLNa, thus supporting our model in which the C‐terminus is accessible for interactions with extracellular proteins and antibodies.( 40 ) Our results expand the spectrum of cellular localization of FLNa to include the cell surface. The differential localization of FLNs and their isoforms or fragments is expanding our understanding of the variety of functions that these multifaceted proteins can perform within the intracellular and extracellular environment.
Acknowledgments
This work was supported in part by funds from the American Cancer Society (Institutional Support Grant IRG‐92‐025‐08) to Dr A.S. Bachmann. We are grateful to Drs T. Stossel (Brigham and Women's Hospital, Harvard Medical School), S. Shapiro (Thomas Jefferson University), A. Lau, B. Warn‐Cramer, and R. Wada (Cancer Research Center of Hawaii) for providing cell lines and helpful advice. We also thank Dr D. Park for helpful discussions and Mr D. Albert for technical contributions (Cancer Research Center of Hawaii). Dr T. Koropatnick (University of Hawaii) is thanked for technical assistance with the confocal laser microscope.
References
- 1. Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer 2003; 3: 203–16. [DOI] [PubMed] [Google Scholar]
- 2. Pahlman S, Mamaeva S, Meyerson G et al. Human neuroblastoma cells in culture: a model for neuronal cell differentiation and function. Acta Physiol Scand Suppl 1990; 592: 25–37. [PubMed] [Google Scholar]
- 3. Hellstrom KE, Hellstrom I. Spontaneous tumor regression: possible relationship to in vitro parameters of tumor immunity. Natl Cancer Inst Monogr 1976; 44: 131–4. [PubMed] [Google Scholar]
- 4. Adam A, Hochholzer L. Ganglioneuroblastoma of the posterior mediastinum: a clinicopathologic review of 80 cases. Cancer 1981; 47: 373–81. [DOI] [PubMed] [Google Scholar]
- 5. Pritchard J, Hickman JA. Why does stage 4s neuroblastoma regress spontaneously? Lancet 1994; 344: 869–70. [DOI] [PubMed] [Google Scholar]
- 6. Papac RJ. Spontaneous regression of cancer: possible mechanisms. In Vivo 1998; 12: 571–8. [PubMed] [Google Scholar]
- 7. Reynolds JV, Shou J, Choi H, Sigal R, Ziegler MM, Daly JM. The influence of natural killer cells in neuroblastoma. Arch Surg 1989; 124: 235–9. [DOI] [PubMed] [Google Scholar]
- 8. D’Uscio CH, Jungi TW, Blaser K. Cellular cytotoxicity mediated by isotype‐switch variants of a monoclonal antibody to human neuroblastoma. Br J Cancer 1991; 64: 445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hellstrom IE, Hellstrom KE, Pierce GE, Bill AH. Demonstration of cell‐bound and humoral immunity against neuroblastoma cells. Proc Natl Acad Sci USA 1968; 60: 1231–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bolande RP. The spontaneous regression of neuroblastoma. Experimental evidence for a natural host immunity. Pathol Annu 1991; 26: 187–99. [PubMed] [Google Scholar]
- 11. Bolande RP, Mayer DC. The cytolysis of human neuroblastoma cells by a natural IgM ‘antibody’‐complement system in pregnancy serum. Cancer Invest 1990; 8: 603–11. [DOI] [PubMed] [Google Scholar]
- 12. Erttmann R, Schmitt C, Ollert MW, David K, Bredehorst R, Vogel CW. Naturally occurring humoral cytotoxicity against neuroblastoma (NB) cells in healthy persons and NB patients. Pediatr Hematol Oncol 1996; 13: 545–8. [DOI] [PubMed] [Google Scholar]
- 13. Ollert MW, David K, Schmitt C et al. Normal human serum contains a natural IgM antibody cytotoxic for human neuroblastoma cells. Proc Natl Acad Sci USA 1996; 93: 4498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. David K, Ollert MW, Vollmert C et al. Human natural immunoglobulin M antibodies induce apoptosis of human neuroblastoma cells by binding to a Mr 260 000 antigen. Cancer Res 1999; 59: 3768–75. [PubMed] [Google Scholar]
- 15. David K, Ollert MW, Juhl H et al. Growth arrest of solid human neuroblastoma xenografts in nude rats by natural IgM from healthy humans. Nat Med 1996; 2: 686–9. [DOI] [PubMed] [Google Scholar]
- 16. Ollert MW, David K, Vollmert C et al. Mechanisms of in vivo anti‐neuroblastoma activity of human natural IgM. Eur J Cancer 1997; 33: 1942–8. [DOI] [PubMed] [Google Scholar]
- 17. Schmitt C, David K, Hiller J et al. Natural human IgM‐antibodies in neuroblastoma therapy: preliminary findings of a phase I/II clinical trial. Klin Padiatr 1999; 211: 314–8. [DOI] [PubMed] [Google Scholar]
- 18. Bachmann AS, Heiligtag S, Howard JP, David K, Vogel CW. A possible new role for filamins in natural human IgM‐mediated apoptosis in neuroblastoma. In: Proceedings of the Conference on Advances in Neuroblastoma Research; 17–19 June 2002, Paris, France. 2002; PB–28.
- 19. Spillner E, Deckers S, Grunwald T, Bredehorst R. Paratope‐based protein identification by antibody and peptide phage display. Anal Biochem 2003; 321: 96–104. [DOI] [PubMed] [Google Scholar]
- 20. Wang K. Filamin, a new high‐molecular‐weight protein found in smooth muscle and nonmuscle cells. Purification and properties of chicken gizzard filamin. Biochemistry 1977; 16: 1857–65. [DOI] [PubMed] [Google Scholar]
- 21. Cunningham CC, Gorlin JB, Kwiatkowski DJ et al. Actin‐binding protein requirement for cortical stability and efficient locomotion. Science 1992; 255: 325–7. [DOI] [PubMed] [Google Scholar]
- 22. Van Der Flier A, Kuikman I, Kramer D et al. Different splice variants of filamin‐B affect myogenesis, subcellular distribution, and determine binding to integrin β‐subunits. J Cell Biol 2002; 156: 361–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stossel TP, Condeelis J, Cooley L et al. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol 2001; 2: 138–45. [DOI] [PubMed] [Google Scholar]
- 24. Berry FB, O’Neill MA, Coca‐Prados M, Walter MA. FOXC1 transcriptional regulatory activity is impaired by PBX1 in a filamin A‐mediated manner. Mol Cell Biol 2005; 25: 1415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol 2004; 6: 1034–8. [DOI] [PubMed] [Google Scholar]
- 26. Fox JW, Lamperti ED, Eksioglu YZ et al. Mutations in filamin 1 prevent migration of cerebral cortical neurons in human periventricular heterotopia. Neuron 1998; 21: 1315–25. [DOI] [PubMed] [Google Scholar]
- 27. Robertson SP, Twigg SR, Sutherland‐Smith AJ et al. Localized mutations in the gene encoding the cytoskeletal protein filamin A cause diverse malformations in humans. Nat Genet 2003; 33: 487–91. [DOI] [PubMed] [Google Scholar]
- 28. Bicknell LS, Morgan T, Bonafe L et al. Mutations in FLNB cause boomerang dysplasia. J Medical Genet 2005: 42: e43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wallick CJ, Gamper I, Thorne M et al. Key role for p27Kip1, retinoblastoma protein Rb, and MYCN in polyamine inhibitor‐induced G1 cell cycle arrest in MYCN‐amplified human neuroblastoma cells. Oncogene 2005; 24: 5606–18. [DOI] [PubMed] [Google Scholar]
- 30. Cserzo M, Wallin E, Simon I, Von Heijne G, Elofsson A. Prediction of transmembrane alpha‐helices in prokaryotic membrane proteins: the dense alignment surface method. Protein Eng 1997; 10: 673–6. [DOI] [PubMed] [Google Scholar]
- 31. Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol 1998; 283: 489–506. [DOI] [PubMed] [Google Scholar]
- 32. Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics 2001; 17: 849–50. [DOI] [PubMed] [Google Scholar]
- 33. Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in‐depth protein knowledge and analysis. Nucl Acids Res 2003; 31: 3784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Puntervoll P, Linding R, Gemund C et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucl Acids Res 2003; 31: 3625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Takagi J, Strokovich K, Springer TA, Walz T. Structure of integrin alpha5beta1 in complex with fibronectin. EMBO J 2003; 22: 4607–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. David K, Bredehorst R, Vogel CW, Ollert MW. Natural IgM to a 260 kD cell surface antigen is cytotoxic for the human keratinocyte cell line HaCaT. Arch Dermatol Res 1995; 287: 402. [Google Scholar]
- 37. Goldmann WH, Teodoridis JM, Sharma CP, Hu B, Isenberg G. Fragments from actin binding protein (ABP‐280; filamin) insert into reconstituted lipid layers. Biochem Biophys Res Commun 1999; 259: 108–12. [DOI] [PubMed] [Google Scholar]
- 38. Chakarova C, Wehnert MS, Uhl K et al. Genomic structure and fine mapping of the two human filamin gene paralogues FLNB and FLNC and comparative analysis of the filamin gene family. Hum Genet 2000; 107: 597–611. [DOI] [PubMed] [Google Scholar]
- 39. Loy CJ, Sim KS, Yong EL. Filamin‐A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc Natl Acad Sci USA 2003; 100: 4562–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshida K, Suzuki Y, Honda E et al. Leucine‐rich repeat region of decorin binds to filamin‐A. Biochimie 2002; 84: 303–8. [DOI] [PubMed] [Google Scholar]


