Abstract
The occurrence of aberrations in cell adhesion is a critical phase in the invasion and metastasis of human cancer. A tumor suppressor gene, TSLC1/IGSF4, from chromosomal region 11q23 was identified in non‐small cell lung cancer (NSCLC) by its tumor suppressor activity in nude mice. TSLC1/IGSF4 is expressed in most tissues except for peripheral blood lymphocytes, but it is inactivated in 44% of NSCLC and 30–60% of various cancers, including liver, pancreatic, and prostate cancers, especially in those with invasion or metastasis. Inactivation occurs by two hits: through promoter methylation, and through loss of heterozygosity at the gene locus. TSLC1/IGSF4 encodes an immunoglobulin superfamily cell adhesion molecule and associates with an actin‐binding protein, DAL‐1/4.1B, and members of the membrane‐associated guanylate kinase homologue (MAGuK) group, providing a novel tumor suppressor cascade that is inactivated in more than 80% of NSCLC. TSLC1/IGSF4 appears to be involved in the formation of an epithelial cell structure with DAL‐1/4.1B and MAGuK. Furthermore, TSLC1/IGSF4 may act as a tumor antigen recognized by activated NK or CD8+ T cells. These two distinct mechanisms based on homophilic and heterophilic interactions would be responsible for tumor suppression by TSLC1/IGSF4. TSLC1/IGSF4 is ectopically expressed in adult T‐cell leukemia (ATL) cells, providing not only a diagnostic marker for ATL, but also a possible therapeutic target against its invasion. The distinct roles of TSLC1/IGSF4 in the oncogenesis of carcinomas and ATL could be due to tissue‐specific differences in the downstream cascades, and is a novel concept with respect to cell adhesion in human oncogenesis. (Cancer Sci 2005; 96: 543 –552)
Over the past two decades, a number of tumor suppressor genes have been identified, providing new insights into human oncogenesis as well as into the growth, differentiation, and maintenance of the normal cell population.( 1 ) So far, most human tumor suppressor genes have been identified by linkage analysis of the pedigrees associated with hereditary tumors. However, no linkage studies are available for the cloning of tumor suppressors in non‐small cell lung cancer (NSCLC) because, like most other human cancers, NSCLC develops as a sporadic disease. One approach for identifying a tumor suppressor gene in a sporadic tumor is using positional cloning of the common region showing a loss of heterozygosity (LOH) in the tumor DNA. Most cancer cells, however, harbor genetic and epigenetic instability, and have extremely complicated alterations in their chromosomes, genes, nucleotides, and methylation status. Therefore, it is usually difficult to identify a single responsible gene from many candidate genes by a structural analysis alone. An alternative approach for the identification of a tumor suppressor gene is using functional cloning based on the genetic complementation of malignant phenotype with normal DNA fragments. In addition to the transfer of single chromosomal fragments, more restricted DNA fragments cloned into yeast artificial chromosomes (YAC) have been made available for expression cloning of the gene.( 2 ) By combinatorial analyses of YAC transfer into human NSCLC cells with a tumorigenicity assay in nude mice, the TSLC1/IGSF4 gene at chromosomal region 11q23 was identified as a tumor suppressor in NSCLC.( 3 , 4 )
It should be noted, however, that functional complementation is not an exact alternative to a linkage study, because the features of a ‘tumor suppressor’ are dependent on the assays used for gene cloning.( 5 ) For example, tumorigenicity is assayed by injecting a number of cells grown in culture into nude mice. Thus, a gene that does not affect cell growth in vitro, but strongly suppresses growth in vivo would be selected as a ‘tumor suppressor’. In fact, the TSLC1/IGSF4 gene encodes an immunoglobulin superfamily cell adhesion molecule (IgCAM), which is a membrane protein involved in cell–cell interactions.( 4 , 6 ) The functions of a number of cell adhesion molecules are altered or affected during the process of invasion and metastasis of human cancers.( 7 ) As a cell adhesion molecule between epithelial cells, TSLC1/IGSF4 acts as a tumor suppressor in a variety of cancers of epithelial origin.( 4 ) In contrast, TSLC1/IGSF4 has recently been found to be ectopically expressed in adult T‐cell leukemia (ATL), and could be implicated in tumor invasion through interactions of the leukemia cells with the endothelial cells.( 8 ) Furthermore, molecules that are identical to human TSLC1/IGSF4 or its murine orthologues have been found to play key roles in the adhesion of spermatogenic cells to Sertoli cells, mast cells to fibroblasts, presynaptic cells to postsynaptic cells, tumor cells to natural killer (NK) cells, and antigen‐presenting cells to CD8+ T cells through homophilic or heterophilic interactions.( 9 , 10 , 11 , 12 , 13 ) TSLC1/IGSF4 is also termed Necl‐2/SgIGSF/RA175/SynCAM1 depending on the biological situation.( 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 )
Here, I aim to introduce a functional cloning approach to identify TSLC1/IGSF4 as a tumor suppressor in NSCLC and review the unique features of TSLC1/IGSF4 in human oncogenesis as a tumor suppressor in epithelial cancer and a possible oncoprotein in ATL, paying attention to cell adhesion. Furthermore, additional functions of the same molecules in specific types of cell–cell interactions are briefly summarized. We primarily use TSLC1/IGSF4 as the name of the gene because the HUGO Gene Nomenclature Committee has approved both IGSF4 and TSLC1 (http://www.gene.ucl.ac.uk/nomenclature). The dual roles of TSLC1/IGSF4 in epithelial cancers and ATL is a novel concept with respect to cell adhesion in human oncogenesis.
Functional cloning of a novel tumor suppressor gene, TSLC1/IGSF4, in human NSCLC
Non‐small cell lung cancer is a malignant human tumor that is refractory to most therapeutic approaches. Numerous genetic alterations, including mutations and amplifications of oncogenes, such as the K‐ras and EGFR genes, and inactivation of tumor suppressor genes, including the TP53, CDKN4A, and RB1 genes, have been found to play roles in the biological and clinical aspects of NSCLC.( 17 , 18 ) In addition, LOH in numerous chromosomal regions, including 3p, 9p, 11q, 13q, and 17p, is observed at high frequencies in primary NSCLC, suggesting that additional tumor suppressor genes are involved in the development or progression of NSCLC. It has been demonstrated that LOH at 9p, 13q, and 17p is responsible for the inactivation of the CDKN4A, RB1, and TP53 genes, respectively. The RASSF1A gene was later identified as a tumor suppressor at 3p by extensive structural analysis at 3p in a large number of primary NSCLC.( 19 ) However, a tumor suppressor gene, TSLC1/IGSF4, at 11q23 was identified by functional cloning.( 3 , 4 )
Functional complementation has greatly contributed to research on cancer genetics, as discussed in a previous review.( 5 ) In 1969, suppression of the malignant phenotype of cancer cells by fusing them with normal fibroblasts was the first demonstration of the recessive phenotype of cancer cells.( 20 ) Evidence of tumor suppressor genes on specific chromosomes was later provided by functional complementation of the malignant phenotype through microcell‐mediated chromosome transfer (MMCT).( 21 ) Modified MMCT using fragmented chromosomes by irradiation limited the region harboring possible tumor suppressor genes to 2–20 cM.( 22 , 23 ) The next breakthrough was made by transferring YAC clones carrying more restricted DNA fragments of approximately 1 Mb into mammalian cells by spheroplast fusions, microinjections, or lipofections.( 2 ) One of the advantages of MMCT or YAC transfer is that a gene on the fragment can be expressed in a manner similar to endogenous gene expression because only a single copy of a DNA fragment is usually integrated into the recipient cells, and most genes on the fragments are expressed under the control of their endogenous promoters. This is particularly important when negative cell growth caused by tumor suppressors is assayed.( 5 )
The presence of a tumor suppressor gene(s) in NSCLC on chromosome 11 was initially demonstrated by Satoh et al. through MMCT using a human lung adenocarcinoma cell line, A549, as a recipient.( 24 ) A549 is strongly tumorigenic in nude mice, and has lost one allele of chromosome 11. An LOH study in 79 primary NSCLC tumors has revealed that a fragment from 11q23 of about 5 cM is commonly deleted.( 25 ) Thus, five continuous YAC clones located within this region were transferred into A549 cells one by one through spheroplast fusion. Analysis of the tumorigenicity of these hybrid cells revealed that the suppressor activity was present within a YAC clone of a 1.6‐Mb insert from 11q23.2. Subsequent transfer of the fragmented YAC into A549, coupled with an assessment of tumorigenicity, localized the tumor suppressor activity to a 100‐kb fragment.( 3 ) A predicted tumor suppressor gene, TSLC1 (Tumor Suppressor in Lung Cancer 1), was cloned from this region (Fig. 1). TSLC1/IGSF4 encodes a membrane protein belonging to the immunoglobulin (Ig) superfamily of molecules. Expression of the full‐length TSLC1/IGSF4, but not of the truncated TSLC1/IGSF4, strongly suppressed tumor formation by A549 in nude mice. Furthermore, two‐hit inactivation of the TSLC1/IGSF4 gene by frameshift or nonsense mutations associated with LOH at 11q23 was observed in a subset of primary NSCLC. In addition, loss of TSLC1/IGSF4 expression by promoter methylation was detected in more than 80% of primary NSCLC tumors showing LOH at 11q23. Thus, it was concluded that TSLC1/IGSF4 was a tumor suppressor gene in human NSCLC.( 4 )
Figure 1.
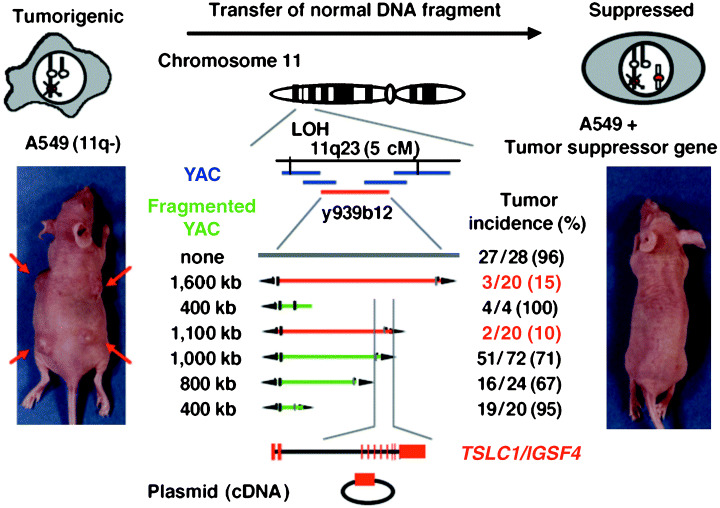
Functional complementation for identifying a lung tumor suppressor gene at 11q23. A549 cells lost one copy of chromosome 11q. A549 cells (left) and A549 cells with the tumor suppressor gene (TSG) (right) were injected into the back of BALB/c nu/nµ mice. Tumor incidence was determined 10 weeks after injection, as described previously.( 3 , 4 ) Red bars indicate YAC with tumor suppressor activity, and blue bars indicate YAC without tumor suppressor activity.
Inactivation of TSLC1/IGSF4 in NSCLC and various cancers
The TSLC1/IGSF4 gene consists of a total of 12 exons at 11q23.2, of which exons 8a, 8b, and 8c have a variety of splicing isoforms mainly observed in the brain. Normal epithelial cells, however, have a single isoform containing exon 8a. In the 5′ region of the TSLC1 gene, there are a couple of typical sequences matching the CpG island. A 5′ CpG island of approximately 900 bp covers the upstream region, exon 1, and intron 1 of TSLC1/IGSF4. Bisulfite sequencing analyses revealed that methylation of the 6–12 CpG sites around the transcription initiation site was strongly correlated with the loss of gene expression in NSCLC and several other cancer cells, as well as primary NSCLC tumors.( 4 , 26 ) The quantitative state, as well as the allelic pattern of methylation, was more exactly analyzed by bisulfite single‐strand conformation polymorphism (SSCP), in which the polymerase chain reaction (PCR) product from the bisulfite‐treated DNA is directly subjected to SSCP analysis as shown in Figure 2.( 26 ) Using these methods, promoter methylation of the TSLC1/IGSF4 gene was demonstrated in 44% of NSCLC, 27% of pancreatic cancers, 29% of hepatocellular carcinomas, and 32% of prostate cancers.( 4 , 5 , 26 , 27 ) In addition, promoter methylation and/or loss of expression of the TSLC1/IGSF4 gene was reported in 20–60% of cancers from the esophagus, stomach, pancreas, nasopharynx, breast, and uterine cervix, as well as meningiomas, as summarized in Table 1.( 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 ) As most of these tumors are known to have LOH at 11q23, the TSLC1/IGSF4 gene appears to be the major target of inactivation in tumors with loss of 11q23.
Figure 2.
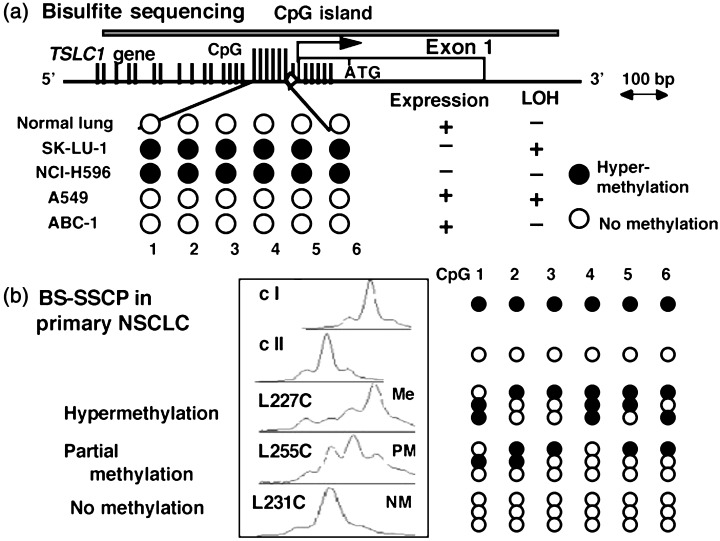
Analysis of promoter methylation of the TSLC1/IGSF4 gene by bisulfite single‐strand conformation polymorphism (SSCP). (a) Structure of the TSLC1/IGSF4 gene promoter. Vertical bars indicate CpG sites within the CpG island of the TSLC1 gene, in which long vertical bars indicate the six CpG sites examined. (□), exon 1; (◊), the predicted TATA box sequence. ATG indicates the initial codon for methionine. The results of bisulfite sequencing of DNA from four non‐small cell lung cancer (NSCLC) cell lines and the normal lung are shown.( 4 ) (b) Bisulfite SSCP analysis of the DNA fragments from primary NSCLC tumors.( 26 ) The results of a completely methylated fragment (clone I) and an unmethylated fragment (clone II) are shown at the top. C indicates DNA from a primary lung cancer. Sequences of three independent clones were determined in each sample and are schematically represented in the right panel. (•), Methylated CpG; (○), unmethylated CpGs; M, hypermethylation; PM, partial methylation; NM, no methylation.
Table 1.
Inactivation of the TSLC1/IGSF4 gene in human cancers: incidence of methylation or loss of expression
| Tumors | Promoter methylation (%) (primary tumors) | Loss of expression (%) (cell lines) | Suppression of tumorigenicity (Cell lines) | References |
|---|---|---|---|---|
| NSCLC | 21/48 (44) | 6/12 (50) | A549 | ( 4 , 26 ) |
| SCLC | ND | 2/10 (20) | ( 26 ) | |
| Nasopharyngeal cancer | 13/38 (34) | 2/5 (40) | ( 31 , 32 ) | |
| Esophageal cancer* | 28/56 (50)* | 3/3 (100) | KYSE520 | ( 28 ) |
| Gastric cancer | 15/97 (15) | 8/9 (89) | ( 29 ) | |
| Hepatocellular cancer | 4/14 (29) | 3/8 (38) | ( 4 , 5 ) | |
| Pancreatic cancer | 25/91 (27) | 8/11 (73) | ( 30 ) | |
| Breast cancer | 10/30 (33) | 1/3 (33) | ( 26 , 33 ) | |
| Cervical cancer | 30/52 (58) | 9/10 (90) | SiHa | ( 34 ) |
| Prostate cancer | 7/22 (32) | 2/5 (40) | PPC‐1 | ( 27 ) |
| Meningioma* | 26/41 (63)* | ND | ( 35 ) | |
| Medulloblastoma | 0/30 (0) | ND | ( 55 ) | |
| Total | 44/76 (58) |
Loss of TSLC1/IGSF4 expression. ND, not done; NSCLC, non‐small cell lung cancer; SCLC, small cell lung cancer.
Clinicopathological analyses have revealed that the inactivation of TSLC1/IGSF4 occurs more frequently in tumors in advanced stages (Table 2). For example, TSLC1/IGSF4 methylation in primary NSCLC was observed preferentially in tumors at pathological stages Ib to IV rather than in tumors at stage Ia.( 26 ) Loss of TSLC1/IGSF4 expression in primary esophageal cancers was also detected more frequently in tumors at pathological stages II to IV in comparison with those at stage I.( 28 ) In an immunohistochemical study using a specific antibody against TSLC1/IGSF4, Uchino et al. demonstrated that TSLC1/IGSF4 expression was inversely correlated with advanced disease stage, lymph node involvement, lymphatic permeation, and vascular invasion.( 36 ) Furthermore, 4‐year survival and disease‐free survival are significantly shorter in patients with lung adenocarcinomas lacking TSLC1/IGSF4 expression.( 36 ) Ito et al. and Goto et al. also demonstrated that TSLC1/IGSF4 expression was downregulated in the invasive components of pulmonary adenocarcinoma, but not in bronchioloalveolar carcinoma.( 37 , 38 ) Therefore, TSLC1/IGSF4 is likely to be involved in the biological aggressiveness of tumors, including invasion or metastasis.
Table 2.
Inactivation of the TSLC1/IGSF4 gene in human cancers: clinicopathological parameters and TSLC1/IGSF4 methylation
| Tumors | Parameters | Loss of expression (%) | References | |
|---|---|---|---|---|
| NSCLC | pStage | I | 1/16 (6) | ( 36 ) |
| II | 5/6 (83) | |||
| III | 8/16 (50) P < 0.0004 | |||
| Histology | BAC | 0/16 (0) | ( 37 ) | |
| Papillary | 10/15 (67) | |||
| Solid | 3/4 (75) | |||
| Mixed | 6/12 (50) P < 0.0001 | |||
| Esophageal cancer | pStage | I | 1/11 (9) | ( 28 ) |
| II | 12/15 (80) | |||
| III | 7/20 (35) | |||
| IV | 8/10 (80) P < 0.002 | |||
Identification of the TSLC1/IGSF4 cascade in epithelial tissues
The TSLC1/IGSF4 gene encodes a class I membrane protein of 442 amino acids, containing three immunoglobulin (Ig) loops in the extracellular domain, a transmembrane domain, and a short cytoplasmic domain (Fig. 3a).( 4 , 6 ) The overall structure of TSLC1/IGSF4 is similar to that of another important family of IgCAM, namely the nectins, and TSLC1/IGSF4 is also called nectin‐like molecule 2 (necl‐2). Unlike nectins, however, necl‐2 does not associate with afadin.( 14 ) TSLC1/IGSF4 is an N‐linked and O‐linked glycoprotein with a molecular weight of 75 kD in normal epithelium including the lung, 70 kD in the brain, and approximately 100 kD in the testis.( 9 , 11 , 39 ) Post‐translational modification, in addition to a variation in splicing, appears to be involved in the differences in molecular weight in the tissues. As a typical member of IgCAM, TSLC1/IGSF4 forms homodimers through cis‐interactions, expresses in the lateral membrane in polarized cells, and has cell aggregation activity through homophilic trans interactions in a Ca++‐ and Mg++‐independent manner (Fig. 3b).( 39 ) Furthermore, necl‐2/IGSF4 undergoes heterophilic interactions with necl‐1/TSLL1/IGSF4B, nectin‐3, and CRTAM.( 12 , 13 , 40 , 41 ) Using immunoelectron microscopic analyses, Shingai et al. demonstrated that necl‐2 was present in the basolateral membrane of mouse gall bladder cells, but not in the tight junctions, adherence junctions, or desmosomes.( 42 )
Figure 3.
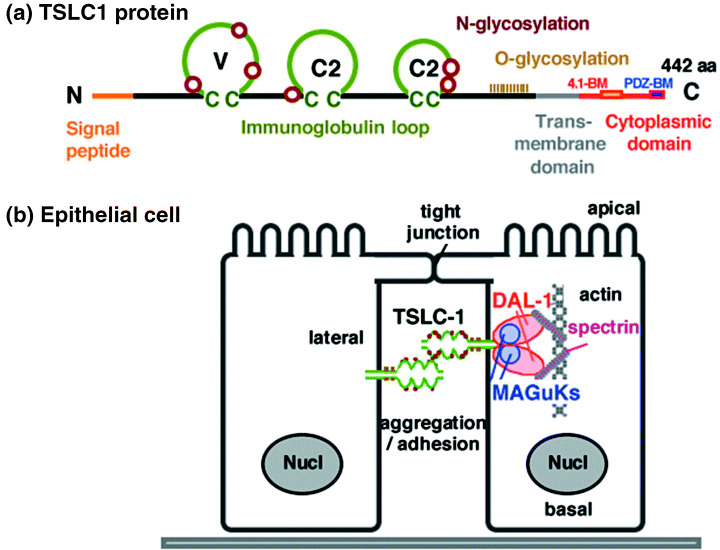
Schematic representation of the TSLC1/IGSF4 protein and its cascade. (a) A predicted structure of TSLC1/IGSF4. V, variable type immunoglobulin loop; C2, constant 2‐type immunoglobulin loop; 4.1‐BM, protein 4.1‐binding motif; PDZ‐BM, PDZ‐binding motif. Brown circles and khaki perpendicular lines indicate possible N‐ and O‐glycosylation sites, respectively. (b) A cascade of TSLC1/IGSF4, DAL‐1/4.1B, and membrane‐associated guanylate kinase homologue (MAGuK) in polarized epithelial cells. TSLC1/IGSF4 forms cis‐homodimers, expressed in the lateral membrane, and has cell adhesion activity through trans‐homomultimers. DAL‐1/4.1B and MAGuK associate with TSLC1/IGSF4 through 4.1‐BM and PDZ‐BM, respectively. DAL‐1/4.1B interacts with actin filaments and spectrin.
The cytoplasmic domain of TSLC1/IGSF4 of 46 amino acids includes a couple of important motifs that interact with other proteins. One of these is a protein 4.1‐binding motif in the juxtamembrane portion of TSLC1/IGSF4. Through this motif, TSLC1/IGSF4 directly interacts with DAL‐1/4.1B, a member of the protein 4.1 family of molecules, which are known to be spectrin‐actin‐binding proteins.( 43 ) TSLC1/IGSF4 further interacts with the actin filament and appears to transmit the signal of cell adhesion towards the cytoskeleton organization.( 44 ) The other motif in TSLC1/IGSF4 is a PDZ‐binding motif at its carboxyl end, through which a group of proteins belonging to the membrane‐associated guanylate kinase homologs (MAGuK) interacts with TSLC1/IGSF4. CASK, MPP3, Pals2, and syntenin are the MAGuK so far identified as binding partners of TSLC1/IGSF4.( 11 , 42 , 45 ) These proteins appear to function as molecular scaffolds that can regulate cell polarity, at least in part.( 46 ) In addition, TSLC1/IGSF4, DAL‐1/4.1B, and an MAGuK appear to form a ternary complex and participate in epithelial‐like cell structures associated with cell adhesion (Fig. 3b).
Notably, DAL‐1/4.1B at 18p11.3 is considered to be another tumor suppressor in NSCLC because DAL‐1/4.1B was originally isolated as a molecule whose expression is downregulated in adenocarcinoma of the lung.( 47 ) Restoration of DAL‐1/4.1B expression into cancer cells in which it is lacking significantly suppresses cell growth.( 47 ) Moreover, hypermethylation of the DAL‐1/4.1B gene is observed in 57% of primary NSCLC and provides a marker of poor prognosis in patients with lung adenocarcinoma.( 48 ) Taken together, these findings indicate that the TSLC1/IGSF4–DAL‐1/4.1B cascade is deeply involved in NSCLC because more than 80% of NSCLC have inactivation of one of the genes (Table 3).( 48 ) The TSLC1/IGSF4–DAL‐1/4.1B cascade is also affected in primary meningiomas.( 35 , 49 ) The role of MAGuK in human tumors remains unclear, although loss of MPP3 expression has been observed in a subset of NSCLC cell lines.( 45 )
Table 3.
Promoter methylation of the TSLC1/IGSF4 and DAL‐1/4.1B genes in human non‐small cell lung cancer( 48 )
| DAL‐1/4.1B | Total | |||
|---|---|---|---|---|
| Methylated | Unmethylated | |||
| TSLC1/IGSF4 | Methylated | 11 (23%) | 10 (21%) | 21 (44%) |
| Unmethylated | 18 (37%) | 9 (19%) | 27 (56%) | |
| Total | 29 (60%) | 19 (40%) | 48 (100%) | |
Possible mechanisms of tumor suppression by TSLC1/IGSF4
Disruption of cell adhesion in the primary tumor is an initial step of cancer invasion and metastasis.( 7 ) It is likely that as a cell adhesion molecule, TSLC1/IGSF4 is primarily involved in epithelial cell adhesion, and loss of its function could lead cancer cells to invasion or metastasis. In fact, loss of TSLC1/IGSF4 is associated with the invasion or metastasis of lung adenocarcinomas, as shown by immunohistochemical studies.( 36 ) In culture, NSCLC cells lacking TSLC1/IGSF4 have a typical transformed morphology with immature cell adhesion, whereas NSCLC cells expressing TSLC1/IGSF4 show relatively mature cell adhesion. Moreover, our preliminary findings using RNAi suggest that the suppression of TSLC1/IGSF4 expression could lead epithelial cells to a more transformed cell morphology, with immature cell adhesion. Therefore, TSLC1/IGSF4 appears to be involved in the formation of an epithelial cell structure with mature cell adhesion.
In addition to simple adhesion between adjacent cells, however, several observations suggest that TSLC1/IGSF4 plays a more active role in suppressing tumor formation with its binding partners, DAL‐1/4.1B and the MAGuK. It has been demonstrated that full‐length TSLC1/IGSF4 suppresses the tumorigenic phenotype of an NSCLC cell line, A549.( 4 ) However, the tumor suppressor activity of TSLC1/IGSF4 was abrogated by truncating the cytoplasmic domain, the 4.1‐binding motif, or the PDZ‐binding motif, although the cell–cell adhesion activity was partially retained in vitro. ( 50 ) These results also support the hypothesis that not only TSLC1/IGSF4 but also DAL‐1/4.1B or MAGuK participate in tumor suppressor functions. Mao et al. reported that high expression levels of TSLC1/IGSF4 from a recombinant adenovirus vector (Ad‐TSLC1) inhibited cell proliferation and induced apoptosis in A549 cells.( 51 ) Furthermore, the intratumoral injection of Ad‐TSLC1 strongly suppressed the subcutaneous tumor growth of A549 cells in nude mice. In this process, the restoration of TSLC1/IGSF4 expression activates the apoptotic protease caspase‐3, which is accompanied by cleavage of its substrate poly‐(ADP‐ribose) polymerase (PARP). This antiproliferative and pro‐apoptotic activity of TSLC1/IGSF4 again requires the presence of the 4.1‐binding motif and the PDZ‐binding motif in the cytoplasmic domain. These findings suggest that apoptosis is a possible mechanism of tumor suppression by TSLC1/IGSF4, as shown in Figure 4a, although it is not clear whether the physiological level of TSLC1/IGSF4 expression also induces apoptosis in cancer cells.
Figure 4.
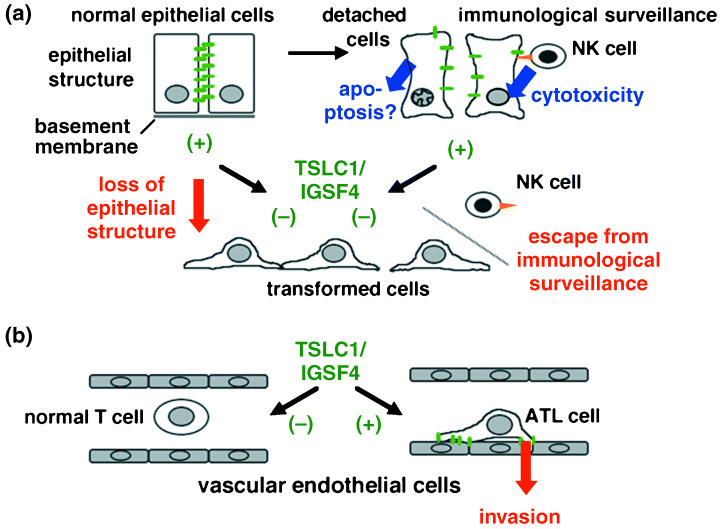
Dual roles of TSLC1/IGSF4 in human oncogenesis. (a) Possible mechanisms of tumor suppression in carcinomas of epithelial origin by TSLC1/IGSF4. The green bars and orange arrowhead indicate TSLC1/IGSF4 and CRTAM, respectively. Normal epithelial cells expressing TSLC1/IGSF4 have polarized morphology with mature cell adhesion through the homophilic interaction of TSLC1/IGSF4. Detached cells expressing TSLC1/IGSF4 are recognized through the heterophilic interaction of TSLC1/IGSF4 and CRTAM and attacked by natural killer cells. Apoptosis is another possible mechanism of tumor suppression. Cells lacking TSLC1/IGSF4 expression lose their epithelial cell structure and escape the immunological surveillance conducted by the host system, although tumor‐specific modification of TSLC1/IGSF4 is not demonstrated. (b) Possible enhancement of the invasion of adult T‐cell leukemia (ATL) cells by TSLC1/IGSF4. Ectopic expression of TSLC1/IGSF4 may promote invasion into the vascular endothelial cells, possibly through heterophilic interactions. The downstream cascade specific to ATL is under investigation.
Accumulating evidence suggests that the malignant transformation of cancer cells leading to invasion or metastasis is based, at least in part, on the phenotypic changes caused by the endogenous program of cells, called an epithelial‐mesenchymal transition (EMT). Our preliminary data suggest that the full‐length TSLC1/IGSF4, but not the truncated TSLC1/IGSF4, inhibits the EMT in experimental systems to maintain an epithelial phenotype. Intervention in the EMT might be another possible molecular mechanism of tumor suppression by TSLC1/IGSF4.
Escape from immunological surveillance by masking TSLC1/IGSF4
In addition to the phenotypic regulation of epithelial cells, it has been recently reported that TSLC1/IGSF4 is involved in the immunological responses between tumor cells and NK cells, as well as antigen‐presenting cells and CD8+ T cells. Boles et al. and Galibert et al. have demonstrated that the TSLC1/IGSF4 in tumor cells forms highly specific heterodimers with another IgCAM, class I‐restricted T‐cell‐associated molecule (CRTAM), whose expression is only induced on the membrane of activated NK cells or CD8+ T cells.( 12 , 13 ) The most distal Ig‐V‐type loop of TSLC1/IGSF4 is responsible for this highly specific interaction with CRTAM. This heterophilic trans interaction between TSLC1/IGSF4 and CRTAM enhances the cytotoxicity of NK cells and the secretion of γ‐interferon from CD8+ T cells to attack the TSLC1/IGSF4‐expressing cells. Furthermore, tumor cells expressing TSLC1/IGSF4 are efficiently rejected by NK cells in the early stages of inoculation into the peritoneal cavity of nude mice.( 13 ) Thus, it is possible to speculate that TSLC1/IGSF4 could act as a tumor antigen, whereas loss of TSLC1/IGSF4 expression might be advantageous for cancer cells to escape the immunological surveillance conducted by the host system. Preferential loss of TSLC1/IGSF4 expression in invasive cancers also supports this speculation.( 26 , 28 , 30 , 34 , 35 , 36 , 37 ) It is not likely, however, that NK or CD8+ T cells recognize the physiologically expressed TSLC1/IGSF4 as an antigen, because TSLC1/IGSF4 is involved in various normal epithelial cell adhesions through homophilic trans multimers. Post‐translational modifications or aberrantly exposed forms of TSLC1/IGSF4 specific to cancer cells might make the function of tumor antigens on this molecule recognizable by NK or CD8+ T cells. Taken together, these intriguing findings provide an additional mechanism of tumor suppression mediated by TSLC1/IGSF4 that involves cytotoxic lymphocytes.
It should be noted, however, that these immunological reactions are triggered through the distal Ig‐V‐type loop of TSLC1/IGSF4, whereas its cytoplasmic domain mediates other types of reactions related to cytoskeleton organization, cell morphology, and apoptosis, as described in the previous section. The possible mechanisms of TSLC1/IGSF4 in tumor suppression, integrating the aspects so far demonstrated, are schematically represented in Figure 4a. TSLC1/IGSF4 is a unique tumor suppressor that is involved in both adhesion‐mediated epithelial cell structure and immunological responses.
Ectopic expression of TSLC1/IGSF4 in adult T‐cell leukemia
The function of TSLC/IGSF4 as a tumor suppressor in carcinomas of epithelial origin has been described. However, an unexpected result was recently obtained during expression profiling of more than 12 000 genes in primary adult T‐cell leukemia (ATL) cells. Sasaki et al. demonstrated that TSLC1/IGSF4 is one of four genes that are upregulated more than 30‐fold in comparison with normal CD4+ lymphocytes from the same person.( 8 ) Subsequent studies detected the ectopic expression of TSLC1/IGSF4 in eight of eight primary ATL cells and five of seven cultured cell lines from ATL. In contrast, TSLC1/IGSF4 expression was not detected in two T‐ALL cell lines or 34 other leukemia cell lines unrelated to HTLV‐1 infection, or 10 normal CD4+ T cells from healthy individuals. It is noteworthy that ectopic expression of TSLC1/IGSF4 was also detected in two of three HTLV‐1‐infected T‐cell lines. It is well known that ATL is triggered by the retroviral infection of HTLV‐1, although ATL develops only 2–5% of the carriers of HTLV‐1 after more than 35 years of latency.( 52 ) The finding of TSLC1/IGSF4 expression in both HTLV‐1‐infected cells and tax‐negative ATL cells strongly suggests that TSLC1/IGSF4 is deeply involved in the development and progression of ATL in multistage leukemogenesis. Furthermore, when TSLC1/IGSF4 was introduced into a human ATL cell line, ED, both self‐aggregation and adhesion ability to vascular endothelial cells were enhanced (Fig. 4b).( 8 ) These findings suggest that the ectopic expression of TSLC1/IGSF4 could provide a novel molecular marker for ATL, and may participate in tissue invasion, a characteristic feature of malignant ATL cells.
When its tumor suppressor activity in various cancers is taken into consideration, TSLC1/IGSF4 appears to play dual roles in human oncogenesis: it is a tumor suppressor in epithelial cancers, and a possible oncoprotein in ATL. These opposite functions might be caused by the different downstream cascades of TSLC1/IGSF4 in epithelial cells and T‐lymphocytes. The absence of 4.1B/DAL‐1 in ATL cells supports this hypothesis. Furthermore, the state of cell adhesion as well as the pattern of molecular interaction of TSLC1/IGSF4 is different in epithelial cells and T‐lymphocytes: TSLC1/IGSF4 creates homophilic interactions between adjacent cells in the epithelia, whereas it might generate heterophilic interactions with vascular endothelial cells because they do not express TSLC1/IGSF4, as summarized in Table 4. These molecular mechanisms are being investigated, in order to better understand the dual roles of this unique cell adhesion molecule.
Table 4.
Multiple functions of the TSLC1/IGSF4 protein in various tissues
| Tissues | TSLC1/IGSF4‐expressing cells (pattern) | Interacting cells (pattern of cell contact) | Interacting molecules (pattern of expression) | Type of interaction | Downstream binding proteins | Coupled function | Pathological/medical significance | Aliases used by investigators | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Epithelium | Epithelial cells (constitutive) | Epithelial cells (stable) | TSLC1 /IGSF4 (constitutive) | Homophilic | 4.1B MPP3 Pals‐2 | Epithelial‐structure formation | Tumor suppression | TSLC1 Necl‐2 RA175 | ( 4, 14, 15 ) |
| ATL | ATL cells (ectopic) | Vascular endothelial cell, etc. (transient) | Not identified | Heterophilic (?) | Not identified | Invasion | Invasion of ATL | TSLC1 | ( 8 ) |
| Immune system | Tumor cells (constitutive) | NK cells (transient) | CRTAM (induced) | Heterophilic | 4.1B MPP3 Pals‐2 | Cytotoxicity | Immuno‐surveillance and Tumor rejection | TSLC1 Necl‐2 | ( 12 , 13 ) |
| Antigen‐presenting cells (transient) | CD8+ cells (transient) | CRTAM (induced) | Heterophilic | Not identified | γ‐Interferon secretion | Immuno‐surveillance and Tumor rejection | |||
| Synapse | Pre‐/post synapse (constitutive) | Pre‐/post synapse (stable) | SynCAM1 /IGSF4 (constitutive) | Homophilic | CASK syntenin | Synapse formation | Not identified | SynCAM1 | ( 11 ) |
| Testis | Spermatogenic cells (constitutive) | Sertoli cells (stable) | Not identified | Heterophilic (?) | Not identified | Spermatogenesis | Not identified | SgIGSF | ( 9 ) |
| Peritoneum | Mast cells (constitutive) | Fibroblasts (transient) | Not identified | Heterophilic (?) | Not identified | Cell survival | Not identified | SgIGSF | ( 10 ) |
ATL, adult T‐cell leukemia; NK, natural killer.
Involvement of TSLC1/IGSF4 in the cell adhesion of other tissues
It has been reported that TSLC1/IGSF4 also plays critical roles in various types of cell adhesion in different tissues. From mouse testes, Wakayama et al. cloned a novel Ig superfamily molecule, the spermatogenic Ig superfamily (SgIGSF), which turned out to be a murine orthologue of TSLC1/IGSF4.( 9 ) In mouse testes, SgIGSF is expressed in the plasma membrane of spermatogenic cells from the spermatogonia to the zygotene spermatocytes along the attachment site to the Sertoli cells. Because maturation of the spermatogenic cells is regulated by cell–cell interaction with Sertoli cells, SgIGSF/IGSF4 might be involved in cell adhesion coupled with differentiation of spermatogenic cells.( 53 )
Biederer et al. cloned SynCAM1, an identical molecule to TSLC1/IGSF4, as a brain‐specific Ig‐domain‐containing protein that binds to the intracellular PDZ‐domain protein.( 11 ) SynCAM1 is expressed in both the pre‐ and postsynaptic compartments in the brain, and plays an essential role in synapse formation with its cytoplasmic binding partners, CASK and syntenin. The introduction of the cytoplasmic fragment of SynCAM1 into neuron cells inhibits synapse assembly. Moreover, the expression of a full‐length SynCAM1 and a glutamatergic receptor in a non‐neuronal cell, HEK293, was sufficient by itself to reconstitute an artificial synapse between a hippocampal neuron and a non‐neuronal cell. These results strongly suggest that SynCAM1/IGSF4 is a cell adhesion molecule that is necessary and sufficient for synapse formation.( 11 )
Furthermore, Ito et al. reported that SgIGSF is involved in the cell adhesion of mast cells to fibroblasts in the mouse peritoneal cavity.( 10 ) They found that the number of peritoneal mast cells was greatly reduced in mutant mice lacking the transcriptional factor MITF. Subsequent analysis by mRNA subtraction identified SgIGSF as a downregulated molecule in MITF‐deficient mast cells. The introduction of SgIGSF restored cell adhesion to the fibroblasts, and resulted in the survival of the peritoneal mast cells. These findings suggest that SgIGSF/IGSF4 is involved in cell adhesion coupled with the survival of mast cells.( 54 )
The various functions of TSLC1/IGSF4 so far reported are summarized in Table 4. It is extremely interesting that TSLC1/IGSF4 plays a key role in each physiological or pathological situation in which adhesion between the same or different types of cells is coupled with various specific functions.
Conclusions and perspectives
Assays for tumor suppressors by growth suppression in vivo rather than in vitro resulted in the discovery of a unique cell adhesion molecule, TSLC1/IGSF4. As a multifunctional membrane protein that regulates the entrance of the signal transduction of cells, TSLC1/IGSF4 plays dual roles in oncogenesis as a tumor suppressor in carcinomas of epithelial origin and a possible oncoprotein in ATL. Apoptosis induction, as well as the formation or maintenance of an epithelial cell structure through homophilic interaction, is one of the possible molecular mechanisms of TSLC1/IGSF4 in the tumor suppression of carcinomas. The critical involvement of TSLC1/IGSF4 in immunosurveillance by cytotoxic lymphocytes as a tumor antigen through heterophilic interactions is another possible mechanism of tumor suppression in vivo. TSLC1/IGSF4 is the first example of a tumor suppressor that involves both epithelial cell adhesion and immunological surveillance. In ATL, however, cell adhesion coupled with invasion through heterophilic interactions would confer on this molecule a possible function as an oncoprotein in vivo. TSLC1/IGSF4 would provide a new concept of cell adhesion in the formation of human tumors. In addition, it is possible to speculate that the dual roles of TSLC1/IGSF4 could also be implicated in the multiple steps of cancer metastasis. This could then explain why the TSLC1/IGSF4 gene is mostly inactivated by promoter methylation, a relatively unstable and potentially reversible mechanism compared with mutational inactivation, although further studies are required to verify this hypothesis. TSLC1/IGSF4 also acts as a key component of cell adhesion in spermatogenesis, synapse formation, and mast cell survival.
Detailed analyses of the function of TSLC1/IGSF4 and its cascade in each biological situation, including the transcriptional regulation of the gene, would be required for further understanding the roles of this multifunctional cell adhesion molecule. In cancer research, the ectopic expression of TSLC1/IGSF4 and its downstream proteins would provide promising molecular targets for not only the diagnosis but also the treatment of ATL, a fatal disorder endemic to the western part of Japan as well as the Caribbean basin. Finally, an investigation into TSLC1/IGSF4 and its cascade would provide new insights for our understanding of cancer as a common disease that develops in the human body.
Acknowledgments
The author thanks all the present and previous members of the Tumor Suppression and Functional Genomics Project, National Cancer Center Research Institute, and all the collaborators for their helpful suggestions and participation in fruitful discussions. The work carried out by my laboratory was supported, in part, by a Grant‐in‐Aid for the Third‐Term Comprehensive Control Research for Cancer from the Ministry of Health, Labor, and Welfare of Japan; a Grant‐in‐Aid for Scientific Research on Priority Areas from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, and a Grant from the Program for Promotion of Fundamental Studies in Health Sciences of the Pharmaceuticals and Medical Devices Agency (PMDA) of Japan.
References
- 1. Cavenee WK, White RL. The genetic basis of cancer. Sci Am 1995; 272: 72–9. [DOI] [PubMed] [Google Scholar]
- 2. Pavan WJ, Hieter P, Reeves RH. Modification and transfer into an embryonal carcinoma cell line of a 360‐kilobase human‐derived yeast artificial chromosome. Mol Cell Biol 1990; 10: 4163–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murakami Y, Nobukuni T, Tamura K et al. Localization of tumor suppressor activity important in non‐small cell lung carcinoma on chromosome 11q. Proc Natl Acad Sci USA 1998; 95: 8153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kuramochi M, Fukuhara H, Nobukuni T et al. TSLC1 is a tumor suppressor gene in human non‐small cell lung cancer. Nat Genet 2001; 27: 427–30. [DOI] [PubMed] [Google Scholar]
- 5. Murakami Y. Functional cloning of a tumor suppressor gene, TSLC1, in human non‐small cell lung cancer. Oncogene 2002; 21: 6936–48. [DOI] [PubMed] [Google Scholar]
- 6. Gomyo H, Arai Y, Tanigami A et al. A 2MB sequence‐ready contig map and a novel immunoglobulin superfamily gene IGSF4 in the LOH region of chromosome 11q23.2. Genomics 1999; 62: 139–46. [DOI] [PubMed] [Google Scholar]
- 7. Hirohashi S, Kanai Y. Cell adhesion system and human cancer morphogenesis. Cancer Sci 2003; 94: 575–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sasaki H, Nishikata I, Shiraga T et al. Overexpression of a cell adhesion molecule, TSLC1, as a possible molecular marker for acute type of adult T‐cell leukemia. Blood 2005; 105: 1204–13. [DOI] [PubMed] [Google Scholar]
- 9. Wakayama T, Ohashi K, Mizuno K, Iseki S. Cloning and characterization of a novel mouse immunoglobulin superfamily gene expressed in early spermatogenic cells. Mol Reprod Dev 2001; 60: 158–64. [DOI] [PubMed] [Google Scholar]
- 10. Ito A, Jippo T, Wakayama T et al. SgIGSF: a new mast‐cell adhesion molecule used for attachment to fibroblasts and transcriptionally regulated by MITF. Blood 2003; 101: 2601–8. [DOI] [PubMed] [Google Scholar]
- 11. Biederer T, Sara Y, Mozhayeva M et al. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 2002; 297: 1525–31. [DOI] [PubMed] [Google Scholar]
- 12. Galibert L, Diemer GS, Liu Z et al. Nectin‐like protein 2 defines a subset of T‐cell zone dendritic cells and is a ligand for class‐I restricted T‐cell associated molecule. J Biol Chem 2005; 280: 21 955–64. [DOI] [PubMed] [Google Scholar]
- 13. Boles KS, Barchet W, Diacovo T, Cella M, Colonna M. The tumor suppressor TSLC1/NECL‐2 triggers NK cell and CD8+ T cell responses through the cell surface receptor CRTAM. Blood 2005; 106: 779–86. [DOI] [PubMed] [Google Scholar]
- 14. Takai Y, Irie K, Shimizu K, Sakisaka T, Ikeda W. Nectins and nectin‐like molecules. Roles in cell adhesion, migration, and polarization. Cancer Sci 2003; 94: 655–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fujita E, Soyama A, Momoi T. RA175, which is the mouse ortholog of TSLC1, a tumor suppressor gene in human lung cancer, is a cell adhesion molecule. Exp Cell Res 2001; 287: 57–66. [DOI] [PubMed] [Google Scholar]
- 16. Watabe K, Ito A, Koma YI, Kitamura Y. IGSF4: a new intercellular adhesion molecule that is called by three names, TSLC1, SgIGSF and SynCAM, by virtue of its diverse function. Histol Histopathol 2003; 18: 1321–9. [DOI] [PubMed] [Google Scholar]
- 17. Yokota J, Kohno T. Molecular footprints of human lung cancer progression. Cancer Sci 2004; 95: 197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tomida S, Yatabe Y, Yanagisawa K, Mitsudomi T, Takahashi T. Throwing new light on lung cancer pathogenesis: updates on three recent topics. Cancer Sci 2005; 96: 63–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zabarovsky ER, Lerman MI, Minna JD. Tumor suppressor genes on chromosome 3p involved in the pathogenesis of lung and other cancers. Oncogene 2002; 21: 6915–35. [DOI] [PubMed] [Google Scholar]
- 20. Harris H, Miller OJ, Klein G, Worst P, Tachibana T. Suppression of malignancy by cell fusion. Nature 1969; 223: 363–8. [DOI] [PubMed] [Google Scholar]
- 21. Weissman BE, Saxon PJ, Pasquale SR, Jones GR, Geiser AG, Stanbridge EJ. Introduction of a normal human chromosome 11 into a Wilms’ tumor cell line controls its tumorigenic expression. Science 1987; 236: 175–80. [DOI] [PubMed] [Google Scholar]
- 22. Koi M, Johnson LA, Kalikin LM, Little PF, Nakamura Y, Feinberg AP. Tumor cell growth arrest caused by subchromosomal transferable DNA fragments from chromosome 11. Science 1993; 260: 361–4. [DOI] [PubMed] [Google Scholar]
- 23. Murakami Y, Brothman AR, Leach RJ, White RL. Suppression of malignant phenotype in a human prostate cancer cell line by fragments of normal chromosome 17q. Cancer Res 1995; 55: 3389–94. [PubMed] [Google Scholar]
- 24. Satoh H, Lamb PW, Dong JT et al. Suppression of tumorigenicity of A549 lung adenocarcinoma cells by human chromosomes 3 and 11 introduced via microcell‐mediated chromosome transfer. Mol Carcinog 1993; 7: 157–64. [DOI] [PubMed] [Google Scholar]
- 25. Iizuka M, Sugiyama Y, Shiraishi M, Jones C, Sekiya T. Allelic losses in human chromosome 11 in lung cancers. Genes Chromosomes Cancer 1995; 13: 40–6. [DOI] [PubMed] [Google Scholar]
- 26. Fukami F, Fukuhara H, Kuramochi M et al. Promoter methylation of the TSLC1 gene in advanced lung tumors and various cancer cell lines. Int J Cancer 2003; 107: 53–9. [DOI] [PubMed] [Google Scholar]
- 27. Fukuhara H, Kuramochi M, Fukami T et al. Promoter methylation of the TSLC1 and tumor suppression by its gene product in human prostate cancer. Jpn J Cancer Res 2002; 93: 605–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ito T, Shimada Y, Hashimoto Y et al. Involvement of TSLC1 in progression of esophageal squamous cell carcinoma. Cancer Res 2003; 63: 6320–6. [PubMed] [Google Scholar]
- 29. Honda T, Tamura G, Waki T et al. Hypermethylation of the TSLC1 gene promoter in primary gastric cancers and gastric cancer cell lines. Jpn J Cancer Res 2002; 93: 857–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jansen M, Fukushima N, Rosty C et al. Aberrant methylation of the 5′ CpG island of TSLC1 is common in pancreatic ductal adenocarcinoma and is first manifest in high‐grade PanlNs. Cancer Biol Ther 2002; 1: 293–6. [DOI] [PubMed] [Google Scholar]
- 31. Hui AB, Lo KW, Kwong J et al. Epigenetic inactivation of TSLC1 gene in nasopharyngeal carcinoma. Mol Carcinog 2003; 38: 170–8. [DOI] [PubMed] [Google Scholar]
- 32. Lung H‐L, Cheng Y, Kumaran MK et al. Fine mapping of 11q22–23 tumor suppressive region and involvement of TSLC1 in nasopharyngeal carcinoma. Int J Cancer 2004; 112: 628–35. [DOI] [PubMed] [Google Scholar]
- 33. Allinen M, Peri L, Kujala S et al. Analysis of 11q21–24 loss of heterozygosity candidate target genes in breast cancer: indications of TSLC1 promoter hypermethylation. Genes Chromosomes Cancer 2002; 34: 384–9. [DOI] [PubMed] [Google Scholar]
- 34. Steenbergen RD, Kramer D, Braakhuis BJ et al. TSLC1 gene silencing in cervical cancer cell lines and cervical neoplasia. J Natl Cancer Inst 2004; 96: 294–305. [DOI] [PubMed] [Google Scholar]
- 35. Surace EI, Murakami Y, Scheithauer BW, Perry A, Gutmann DH. Tumor suppressor in lung cancer‐1 (TSLC1) expression is lost in sporadic meningioma. J Neuropathol Exp Neurol 2004; 63: 1015–27. [DOI] [PubMed] [Google Scholar]
- 36. Uchino K, Ito A, Wakayama T et al. Clinical implication and prognostic significance of the tumor suppressor TSLC1 gene detected in adenocarcinoma of the lung. Cancer 2003; 98: 1002–7. [DOI] [PubMed] [Google Scholar]
- 37. Ito A, Okada M, Uchino K et al. Expression of the TSLC1 adhesion molecule in pulmonary epithelium and its down‐regulation in pulmonary adenocarcinoma other than bronchioloalveolar carcinoma. Lab Invest 2003; 83: 1175–83. [DOI] [PubMed] [Google Scholar]
- 38. Goto A, Niki T, Chi‐pin L et al. Loss of TSLC expression in lung adenocarcinoma: Relationships with histological subtypes, sex and prognostic significance. Cancer Sci 2005; 96: 480–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Masuda M, Yageta M, Fukuhara H et al. The tumor suppressor protein TSLC1 is involved in cell‐cell adhesion. J Biol Chem 2002; 277: 31 014–9. [DOI] [PubMed] [Google Scholar]
- 40. Kakunaga S, Ikeda W, Itoh S et al. Nectin‐like molecule‐1/TSLL1/SynCAM3: a neural tissue‐specific immunoglobulin‐like cell‐cell adhesion molecule localizing at non‐junctional contact sites of presynaptic nerve terminals, axons and glia cell processes. J Cell Sci 2005; 118: 1267–7. [DOI] [PubMed] [Google Scholar]
- 41. Fukuhara H, Kuramochi M, Nobukuni T et al. Isolation of the TSLL1 and TSLL2 genes, members of the tumor suppressor TSLC1 gene family encoding transmembrane proteins. Oncogene 2001; 20: 5401–7. [DOI] [PubMed] [Google Scholar]
- 42. Shingai T, Ikeda W, Kakunaga S et al. Implications of nectin‐like molecule‐2/IGSF4/RA175/SgIGSF/TSLC1/SynCAM1 in cell‐cell adhesion and transmembrane protein localization in epithelial cells. J Biol Chem 2003; 278: 35 421–7. [DOI] [PubMed] [Google Scholar]
- 43. Yageta M, Kuramochi M, Masuda M et al. Direct association of TSLC1 and DAL‐1, two distinct tumor suppressor proteins in lung cancer. Cancer Res 2002; 62: 5129–33. [PubMed] [Google Scholar]
- 44. Sun CX, Robb VA, Gutmann DH. Protein 4.1 tumor suppressors: getting a FERM grip on growth regulation. J Cell Sci 2002; 115: 3991–4000. [DOI] [PubMed] [Google Scholar]
- 45. Fukuhara H, Masuda M, Yageta M et al. Association of a lung tumor suppressor TSLC1 with MPP3, a human homologue of drosophila tumor suppressor Dlg. Oncogene 2003; 22: 6160–5. [DOI] [PubMed] [Google Scholar]
- 46. Bachmann A, Schneider M, Theilenberg E, Grawe F, Knust E. Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature 2001; 414: 638–43. [DOI] [PubMed] [Google Scholar]
- 47. Tran YK, Bogler O, Gorse KM, Wieland I, Green MR, Newsham IF. A novel member of the NF2/ERM/4.1 superfamily with growth suppressing properties in lung cancer. Cancer Res 1999; 59: 35–43. [PubMed] [Google Scholar]
- 48. Kikuchi S, Yamada D, Fukami T et al. Promoter methylation of the DAL‐1/4.1B predicts poor prognosis in non‐small cell lung cancer. Clin Cancer Res 2005; 11: 2954–61. [DOI] [PubMed] [Google Scholar]
- 49. Gutmann DH, Donahoe J, Perry A et al. Loss of DAL‐1, a protein 4.1‐related tumor suppressor, is an important early event in the pathogenesis of meningiomas. Hum Mol Genet 2000; 9: 1495–500. [DOI] [PubMed] [Google Scholar]
- 50. Mao X, Seidlitz E, Ghosh K, Murakami Y, Ghosh HP. The cytoplasmic domain is critical to the tumour suppressor activity of TSLC1 in non‐small cell lung cancer. Cancer Res 2003; 63: 7979–85. [PubMed] [Google Scholar]
- 51. Mao X, Seidlitz E, Truant R, Hitt M, Ghosh HP. Re‐expression of TSLC1 in a non‐small‐cell lung cancer cell line induces apoptosis and inhibits tumor growth. Oncogene 2004; 23: 5632–42. [DOI] [PubMed] [Google Scholar]
- 52. Takatsuki K. Discovery of adult T‐cell leukemia: reminiscences. In: Sugamura K, Uchiyama T, Matsuoka M, Kannagi M, eds. Two decades of adult T‐cell leukemia and HTLV‐1 research. Japanese Cancer Association Gann Monograph on Cancer Research No. 50. Tokyo: Japan Scientific Society Press; 2003; 7–9. [Google Scholar]
- 53. Wakayama T, Koami H, Ariga H et al. Expression and functional characterization of the adhesion molecule spermatogenic immunoglobulin superfamily in the mouse testis. Biol Reprod 2003; 68: 1755–63. [DOI] [PubMed] [Google Scholar]
- 54. Morii E, Ito A, Jippo T et al. Number of mast cells in the peritoneal cavity of mice: influence of microphthalmia transcription factor through transcription of newly found mast cell adhesion molecule, spermatogenic immunoglobulin superfamily. Am J Pathol 2004; 165: 491–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lindsey JC, Lusher ME, Anderton JA et al. Identification of tumour‐specific epigenetic events in medulloblastoma development by hypermethylation profiling. Carcinogenesis 2004; 25: 661–8. [DOI] [PubMed] [Google Scholar]


