Abstract
Hepatitis C virus (HCV) populations persist in vivo as a mixture of heterogeneous viruses called quasispecies. The relationship between the genetic heterogeneity of these variants and their responses to antiviral treatment remains to be elucidated. We have studied 26 virus strains to determine the influence of hypervariable region 1 (HVR-1) of the HCV genome on the effectiveness of alpha interferon (IFN-α) therapy. Following PCR amplification, we cloned and sequenced HVR-1. Pretreatment serum samples from 13 individuals with chronic hepatitis C whose virus was subsequently eradicated by treatment were compared with samples from 13 nonresponders matched according to the major factors known to influence the response, i.e., sex, genotype, and pretreatment serum HCV RNA concentration. The degree of virus variation was assessed by analyzing 20 clones per sample and by calculating nucleotide sequence entropy (complexity) and genetic distances (diversity). Types of mutational changes were also determined by calculating nonsynonymous substitutions per nonsynonymous site (Ka) and synonymous substitutions per synonymous site (Ks). The paired-comparison analysis of the nucleotide sequence entropy and genetic distance showed no statistical differences between responders and nonresponders. By contrast, nonsynonymous substitutions were more frequent than synonymous substitutions (P ≤ 0.05) in responders, but there was no significant difference in nonresponders. Nonsynonymous substitutions tended to be more frequent than synonymous substitutions in women (P = 0.06) but not in men. Nucleotide entropy and genetic distances were significantly related to serum RNA concentration (P ≤ 0.01). Our findings suggest that after controlling for the major determinants of interferon response, neither complexity nor diversity of the HVR-1 region is associated per se with virus eradication. Because a higher proportion of nonsynonymous substitutions than synonymous substitutions was found only in responders, host anti-HCV-specific immune response rather than viral factors may be playing an important role in the interferon response.
Hepatitis C virus (HCV), the causative agent of non-A, non-B hepatitis (1, 5), is a positive-strand RNA virus that exists within its host as pools of related genetic variants, referred to as quasispecies (19, 32). Its heterogeneous character is most evident in hypervariable region 1 (HVR-1) of the envelope gene, which mutates over time in response to host pressures (11, 18, 57). Recent data have suggested that the heterogeneity of quasispecies is involved in viral persistence (50), cellular tropism (48), the pathogenesis of hepatic disease (16, 37), and response to antiviral therapy (15, 31).
Alpha interferon (IFN-α) is the first approved drug therapy for hepatitis C virus infection (6, 20, 25). The standard treatment leads to a sustained clearance of HCV RNA in 15 to 20% of patients (21). There is evidence that the amount of HCV RNA in the patient's serum and the genotype of the HCV are both indicators of a sustained clearance of HCV (17, 22, 33). However, patients with the same genotype and similar RNA levels may respond differently, indicating that particular viral strains have characteristics conferring resistance or sensitivity to antiviral therapy. Several Japanese studies have found a relationship between mutations within the NS5A region of the HCV-1b genome and sensitivity to IFN-α (4, 9, 10, 13), but similar studies performed in other parts of the world have not (26, 60). In vitro experiments have shown that NS5A can interfere with IFN-α signaling pathways and cause resistance to therapy (14, 28, 54). Pawlotsky et al. recently showed that no NS5A sequence was intrinsically resistant or sensitive to IFN-α (43), nor does there appear to be any correlation between resistance to interferon treatment in patients infected with HCV-3 and the rate of mutation within the NS5A region (49, 53).
Several studies have suggested that the great heterogeneity of HVR-1 could be involved in the resistance to IFN-α (3, 25, 41), but this issue is controversial (38). Most of this work has suffered from an incomplete definition of the parameters studied (i.e., biochemical or virological responses), and the limited number of molecular clones sequenced (n ≤ 10) has raised concerns about sampling bias. In addition, viral factors such as genotype and serum RNA concentration, which are known to influence the effectiveness of IFN-α, have not been controlled. We have therefore performed a clonal analysis by sequencing more than 20 clones per sample from two groups of individuals with chronic HCV infection to determine more precisely the influence of pretreatment HVR-1 genetic heterogeneity on the response to IFN-α. The groups were matched according to the major determinants of virological response, including the HCV genotype and the serum HCV RNA concentration before treatment. We compared the genetic complexity and diversity and measured the proportions of synonymous and nonsynonymous mutations in the two groups.
MATERIALS AND METHODS
Patients and samples.
We retrospectively selected a group of 26 patients from the 136 patients given the standard IFN-α-2b treatment for chronic hepatitis C (3 MU three times per week for 6 to 12 months) and already enrolled in an observational study (22). They comprised 13 patients with a sustained virological response (R), defined by normal ALT activity and negative HCV RNA by PCR analysis 6 months after IFN-α withdrawal, and 13 non-sustained responders (NR), defined by elevated alanine aminotransferase (ALT) activity and detectable HCV viremia 6 months after IFN-α was stopped. The matching criteria were three factors independently associated with sustained virological response in a multivariate analysis, i.e., sex, HCV genotype, and pretreatment serum HCV RNA concentration (in each pair, ΔRNA was less than 0.5 log copies/ml). Interferon was given to 10 patients in each group for 12 months and to 3 patients for 6 months. There was no significant difference in the estimated duration of infection for the responders (129 months) and the nonresponders (131 months). The genetic heterogeneity of the virus was established by cloning and sequencing the HVR-1 region in samples taken just before the first dose of IFN-α. The demographic, histological (27), and virological features of these 26 patients are indicated in Table 1.
TABLE 1.
Clinical, histological, and virological features of the 26 patients studied and characteristics of the HVR-1 quasispecies before treatment
| Patienta | Sex | Age (yr) | Knodell score | Genotypeb | HCV RNA concn (log copies/ml)c | Normalized entropy
|
Mean genetic distance (SEM)d | Mutational changese
|
|||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide | Amino acid | Mean Ks (SEM) | Mean Ka (SEM) | Ka/Ks | |||||||
| R1 | Female | 34 | 7 | 1b | 5.79 | 0.525 | 0.525 | 0.2250 (0.0100) | 0.1200 (0.0076) | 0.2890 (0.0196) | 2.41 |
| R2 | Male | 25 | 10 | 3a | 3.95 | 0.132 | 0.066 | 0.0035 (0.0003) | 0.0050 (0.0011) | 0.0030 (0.0005) | 0.60 |
| R3 | Female | 29 | 6 | 3a | 4.91 | 0.328 | 0.291 | 0.0247 (0.0020) | 0.0050 (0.0010) | 0.0330 (0.0029) | 6.60 |
| R4 | Female | 33 | 10 | 3a | 5.86 | 0.494 | 0.385 | 0.1280 (0.0070) | 0.2260 (0.0244) | 0.1240 (0.0127) | 0.55 |
| R5 | Male | 32 | 9 | 3a | 5.58 | 0.322 | 0.322 | 0.0630 (0.0050) | 0.0320 (0.0043) | 0.0780 (0.0089) | 2.44 |
| R6 | Male | 37 | 10 | 3a | 4.63 | 0.507 | 0.275 | 0.0135 (0.0010) | 0.0225 (0.0020) | 0.0110 (0.0012) | 0.50 |
| R7 | Male | 32 | 4 | 3a | 5.97 | 0.647 | 0.586 | 0.0544 (0.0050) | 0.0180 (0.0031) | 0.0670 (0.0091) | 3.72 |
| R8 | Female | 34 | 4 | 3a | 4.94 | 0.196 | 0.063 | 0.0023 (0.0002) | 0.0000 (0.0000) | 0.0004 (0.0005) | 1.04 |
| R9 | Female | 33 | 3 | 3a | 4.98 | 0.786 | 0.549 | 0.1360 (0.0050) | 0.0581 (0.0034) | 0.1820 (0.0096) | 3.13 |
| R10 | Female | 30 | 8 | 2 | 5.67 | 0.618 | 0.483 | 0.1390 (0.0070) | 0.1250 (0.0090) | 0.1440 (0.0114) | 1.15 |
| R11 | Female | 39 | 10 | 2 | 4.19 | 0.461 | 0.461 | 0.0135 (0.0010) | 0.0040 (0.0008) | 0.0180 (0.0009) | 4.50 |
| R12 | Male | 24 | 4 | 2 | 4.19 | 0.496 | 0.496 | 0.1020 (0.0070) | 0.0450 (0.0047) | 0.1370 (0.0135) | 3.04 |
| R13 | Male | 57 | 12 | 2 | 4.11 | 0.365 | 0.328 | 0.0110 (0.0010) | 0.0090 (0.0014) | 0.0120 (0.0007) | 1.33 |
| Mean (group) | 34 | 7 | 4.98 | 0.452 | 0.372 | 0.0705 (0.0040) | 0.0515 (0.0048) | 0.0845 (0.0070) | 2.39 | ||
| NR1 | Female | 55 | 10 | 1b | 6.08 | 0.837 | 0.712 | 0.1850 (0.0080) | 0.1080 (0.0063) | 0.2220 (0.0136) | 2.06 |
| NR2 | Male | 28 | 4 | 3a | 4.80 | 0.899 | 0.876 | 0.1400 (0.0040) | 0.2490 (0.0110) | 0.1200 (0.0042) | 0.48 |
| NR3 | Female | 41 | 13 | 3a | 4.65 | 0.440 | 0.440 | 0.0300 (0.0010) | 0.0300 (0.0021) | 0.0330 (0.0015) | 1.10 |
| NR4 | Female | 28 | 6 | 3a | 5.91 | 0.812 | 0.789 | 0.0675 (0.0020) | 0.0510 (0.0049) | 0.0635 (0.0027) | 1.24 |
| NR5 | Male | 31 | 8 | 3a | 5.13 | 0.236 | 0.132 | 0.0063 (0.0004) | 0.0190 (0.0019) | 0.0030 (0.0048) | 0.16 |
| NR6 | Male | 57 | 8 | 3a | 4.99 | 0.514 | 0.296 | 0.0525 (0.0020) | 0.0540 (0.0040) | 0.0560 (0.0040) | 1.04 |
| NR7 | Male | 41 | 7 | 3a | 5.48 | 0.593 | 0.204 | 0.0180 (0.0010) | 0.0000 (0.0000) | 0.0250 (0.0015) | 3.50 |
| NR8 | Female | 32 | 6 | 3a | 6.14 | 0.769 | 0.769 | 0.1260 (0.0050) | 0.1960 (0.0099) | 0.1040 (0.0062) | 0.53 |
| NR9 | Female | 35 | 8 | 3a | 6.01 | 0.708 | 0.631 | 0.1460 (0.0070) | 0.2019 (0.0156) | 0.2018 (0.0138) | 1.00 |
| NR10 | Female | 53 | 6 | 2 | 6.08 | 0.858 | 0.786 | 0.1150 (0.0040) | 0.1180 (0.0070) | 0.1230 (0.0061) | 1.04 |
| NR11 | Female | 59 | 11 | 2 | 3.95 | 0.167 | 0.167 | 0.0250 (0.0040) | 0.0000 (0.0000) | 0.0400 (0.0090) | 5.00 |
| NR12 | Male | 43 | 6 | 2 | 4.07 | 0.569 | 0.479 | 0.0550 (0.0040) | 0.0650 (0.0082) | 0.0690 (0.0069) | 1.06 |
| NR13 | Male | 54 | 8 | 2 | 3.60 | 0.288 | 0.193 | 0.0240 (0.0020) | 0.0140 (0.0021) | 0.0280 (0.0041) | 1.65 |
| Mean (group) | 43 | 8 | 5.15 | 0.591 | 0.498 | 0.0762 (0.0034) | 0.0844 (0.0060) | 0.0848 (0.0066) | 1.53 | ||
R, responders, characterized by viral eradication 6 months after IFN-α therapy was stopped; NR, nonresponders, characterized by insufficient effect of IFN-α to obtain viral eradication.
Genotype was determined by the Inno-Lipa II HCV method.
HCV RNA was measured by the Amplicor Monitor assay.
The genetic distances within samples were calculated by using the DNADIST program in the PHYLIP package version 3.752.
The proportions of synonymous substitutions per synonymous site (Ks) and of nonsynonymous substitutions per nonsynonymous site (Ka) were calculated by means of the MEGA program.
HCV RNA quantification.
HCV RNA levels were measured in pretreatment sera by the standardized quantitative reverse transcription (RT)-PCR assay Amplicor HCV Monitor (Roche Molecular Systems, Branchburg, N.J.) according to the manufacturer's instructions.
HCV genotyping.
HCV genotype was determined by the Inno-LiPA II HCV method (Innogenetics S.A., Gent, Belgium). After RT-PCR amplification, the amplified products were hybridized to immobilized probes specific for the different genotypes and subtypes.
Cloning and sequencing of HVR-1. (i) Primers.
Primers were designed to amplify fragments containing the amino-terminal region of the E2 gene (HVR-1). All the primer sequences are listed in Table 2.
TABLE 2.
PCR primers
| Genotype | Primer set | Polarity | Sequence | Positionsa |
|---|---|---|---|---|
| 1 | External | Sense | 5′-TTGCAGTTTAAGGCAGTCC-3′ | 1245–1266 |
| Antisense | 5′-CAGGACTGCAATTGCTCAATCAT-3′ | 1612–1630 | ||
| Nested | Sense | 5′-ATGTGCCAGCTGCCATTGGT-3′ | 1395–1414 | |
| Antisense | 5′-CACTGGGGAGTCCTGGCGGG-3′ | 1587–1606 | ||
| 2 | External | Sense | 5′-TGGGTAGCGCTCACTCCC-3′ | 1055–1072 |
| Antisense | 5′-TAGAACAGCGCGGCGAGGAA-3′ | 1649–1670 | ||
| Nested | Sense | 5′-GCTTGGGATATGATGATGAACTGGTC-3′ | 1295–1320 | |
| Antisense | 5′-TTGATGTGCCAGCTGCCATTGGTGT-3′ | 1584–1608 | ||
| 3 | External | Sense | 5′-GGGATATGATGATGAATTGGT-3′ | 1298–1318 |
| Antisense | 5′-AGGACATCCAGTAGAGTTGAACT-3′ | 1676–1698 | ||
| Nested | Sense | 5′-TGCAAGGCAACTGGGCCAAGGT-3′ | 1429–1451 | |
| Antisense | 5′-GCTATGAACCCGGTGTTTAT-3′ | 1637–1655 |
Positions according to HCV-J (genotype 1b), HCV-D504109 (genotype 2), and HCV-NZL1 (genotype 3a).
(ii) RNA extraction, RT, and nested-PCR amplification.
Virus RNA was first extracted from 100 μl of serum by the guanidinium thiocyanate-phenol-chloroform method. The RNA was eluted in 30 μl of sterile water and stored at −80°C. The RNA (10 μl) was then reverse transcribed at 37°C for 60 min with 1 pmol of the outer antisense primer in the presence of 20 U of Moloney murine leukemia virus reverse transcriptase (Boehringer GmbH, Mannheim, Germany), and nested PCR was performed. The first round was done with 5 pmol of outer primers, and the second round was done with 5 pmol of inner primers. The two rounds were performed with 2.5 U of Taq DNA polymerase (AmpliTaq; Perkin-Elmer Cetus, Norwalk, Conn.) under the same conditions, i.e., 5 min of denaturation at 95°C followed by 35 cycles of 95°C for 30 s, 55 to 60°C for 30 s, and 72°C for 90 s, and then by a final extension at 72°C for 10 min. The amplified products were analyzed by electrophoresis through a 2% agarose gel (Gibco BRL, Paisley, Scotland) and staining with ethidium bromide.
(iii) Plasmid cloning.
PCR products were purified with QIAamp columns (Qiagen, Courtaboeuf, France) as specified by the manufacturer. The purified products were quantified by spectrophotometry; 10 ng was directly ligated into 50 ng of PCR II vector (Original TA Cloning Kit, Invitrogen BV, Leek, The Netherlands) at 14°C overnight. Recombinant plasmids were used to transform Escherichia coli competent cells according to the manufacturer's protocol, and transformants were grown on ampicillin plates.
(iv) Nucleotide sequencing.
Twenty independently isolated cDNA clones from PCR products were selected. Plasmid DNAs containing HVR-1 inserts were prepared and sequenced on both strands by the dideoxy chain termination method (PRISM Ready Reaction AmpliTaq Fs and Dye Deoxy primers; Applied Biosystems, Paris, France) on a model 377 automated DNA sequencer (Applied Biosystems, Foster City, Calif.). Electropherogram data were analyzed by the Sequence Navigator program.
(v) Calculation of genetic complexity and diversity.
Nucleotide sequences were aligned with the CLUSTAL W program version 1.5. We quantified the complexity of the HCV strain in the region of interest by calculating the Shannon entropy as follows: S = − Σi (pi ln pi), where pi is the frequency of each sequence in the viral quasispecies (58). The normalized entropy, Sn, was calculated at the nucleotide level as follows: Sn = S/ln N, where N is the total number of sequences analyzed. It was calculated at both the nucleotide and amino acid levels. We quantified diversity as the mean genetic distance calculated for all pairs of nucleotide sequences by using the DNADIST module in the PHYLIP package version 3.572. The calculation was based on a Kimura two-parameter distance matrix with a transition-to-transversion ratio of 2.0. The mean and standard error of the mean (SEM) within-sample genetic distances were calculated for the quasispecies in each of the 26 patients before treatment. The numbers of synonymous (Ks) and nonsynonymous (Ka) substitutions per synonymous and nonsynonymous site, respectively, were calculated with the Jukes-Cantor correction for multiple substitutions (24) by using the MEGA program (30).
Statistical analysis.
Comparisons between the responders and nonresponders were performed by using Student's paired t test. Correlations among quantitative variables were computed with Pearson's rank correlation test, and linear regression was used to illustrate these correlations. P values of less than 0.05 were considered to be significant.
Nucleotide sequence accession numbers.
The sequences have been submitted to EMBL with accession no. AF 166548 to AF 166589.
RESULTS
Patient characteristics.
The patients' demographic and biological characteristics are indicated in Table 1. No significant differences in matching criteria (i.e., sex, genotype, and pretreatment HCV RNA levels) were found between responders (4.98 ± 0.20 log copies/ml) and nonresponders (5.15 ± 0.25 log copies/ml). There was also no significant difference between the Knodell histological scores of the two groups. By contrast, responders were significantly younger (34 ± 2 years) than nonresponders (43 ± 3 years; P ≤ 0.05).
Analysis of HCV quasispecies distribution.
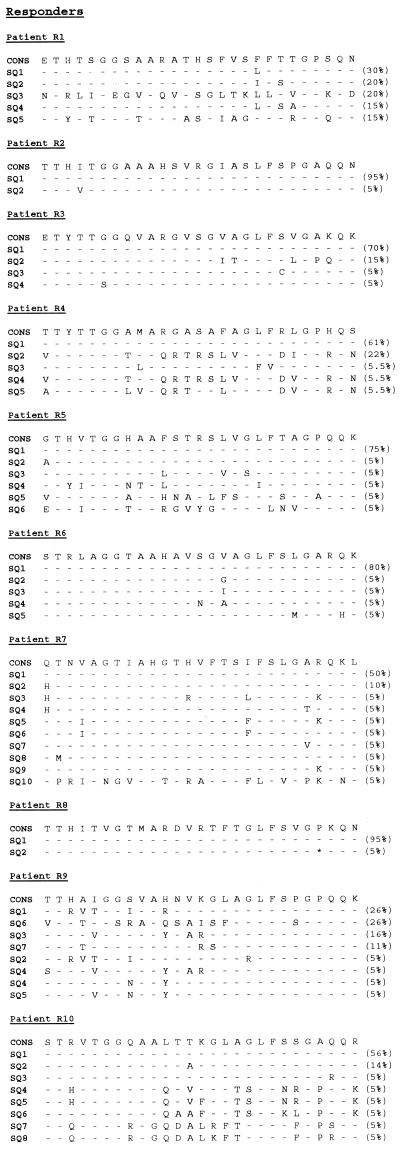
The quasispecies distribution of HVR-1 was analyzed by cloning and sequencing the PCR products of pretreatment samples from the 26 selected patients (Table 1). A total of 515 pretreatment HVR-1 clones were generated and sequenced. Amino acid sequences and their relative frequencies in the virus population are shown in Fig. 1. The complexity of the HVR-1 region was estimated by normalized nucleotide sequence entropy in the 26 pretreatment samples on the basis of the first 20 clones isolated. The normalized nucleotide entropy was 0.522 ± 0.044. The normalized amino acid entropy was 0.435 ± 0.046. The diversity of HVR-1 was evaluated as the average genetic distance within the quasispecies. The mean within-sample genetic distance in the 26 patients was 0.0733 ± 0.0124. Complexity and diversity were significantly related to each other (r = 0.74; P ≤ 0.001). The proportion of nonsynonymous substitutions (0.0837 ± 0.0146) was significantly higher than the proportion of synonymous substitutions (0.0677 ± 0.0149; P ≤ 0.05), suggesting that HVR-1 mutations were more products of an immune selection pressure than of random genetic drift.
FIG. 1.
Amino acid sequences of HVR-1 in samples obtained before treatment from 26 patients receiving IFN-α therapy, 13 who presented viral eradication (R) and 13 who did not (NR). Relative frequencies are shown in parentheses on the right.
Correlation between HCV quasispecies heterogeneity and response to IFN-α.
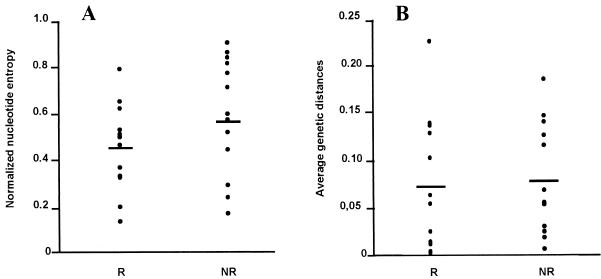
We have compared HVR-1 genetic heterogeneity in responders and nonresponders. We first analyzed the repertoire sizes of the two groups. The mean nucleotide sequence entropy of responders (0.452 ± 0.050) did not differ significantly from that of nonresponders (0.592 ± 0.069) (Fig. 2). The mean amino acid sequence entropies of responders (0.372 ± 0.047) and nonresponders (0.498 ± 0.077) also did not differ. The average within-sample genetic distances were similar in responders (0.0704 ± 0.0194) and in nonresponders (0.0762 ± 0.0163) (Fig. 2). We then analyzed the types of mutational changes. The proportion of synonymous and nonsynonymous substitutions in responders (0.0509 ± 0.0187 and 0.0848 ± 0.0241, respectively) did not differ significantly from those observed in nonresponders (0.0838 ± 0.0229 and 0.0826 ± 0.0177). The Ka/Ks ratio was higher in responders (Ka/Ks = 2.4) than in nonresponders (Ka/Ks = 1.5), but the difference did not reach statistical significance. By contrast, nonsynonymous substitutions were significantly more frequent than synonymous substitutions in the responders (P ≤ 0.05) while there was no significant difference in the nonresponders. As highlighted in Fig. 1, the nonsynonymous changes occurred throughout the HVR-1 domain.
FIG. 2.
Normalized nucleotide sequence entropy (A) and average genetic distances (B) within the HVR-1 quasispecies in the 13 patients who had viral eradication (responders [R]) and in the 13 who did not (nonresponders [NR]); P > 0.05.
Relationship between HVR-1 genetic heterogeneity and clinical, histological, and virological features.
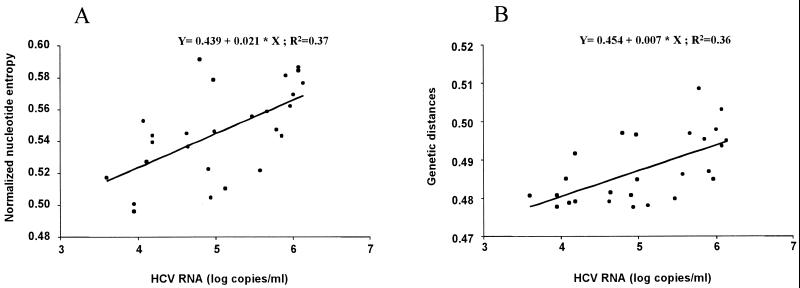
Nucleotide sequence entropy did not differ significantly between women (0.571 ± 0.063) and men (0.464 ± 0.060), but the genetic distances were higher in women (0.0974 ± 0.0187) than in men (0.0453 ± 0.0121) (P ≤ 0.05). The proportion of nonsynonymous substitutions in women (0.1117 ± 0.0227) was significantly higher than in men (0.0511 ± 0.0129; P ≤ 0.05). The Ka/Ks ratio was higher in women (Ka/Ks = 2.2) than in men (Ka/Ks = 1.6), although the difference was not significant. Lastly, nonsynonymous substitutions tended to be more frequent than synonymous substitutions in women (P = 0.06) but not in men. Nucleotide and amino acid entropy, genetic distances, the proportion of synonymous and nonsynonymous substitutions, and Ka/Ks ratios did not differ among the different genotypes. None of the parameters used to assess HVR-1 heterogeneity correlated with age. However, we did find positive correlations between pretreatment HCV RNA concentrations and nucleotide sequence entropy (r = 0.64; P ≤ 0.01) or genetic distance (r = 0.60; P ≤ 0.01) (Fig. 3). The proportions of synonymous and nonsynonymous substitutions were also related to HCV RNA concentrations (rKs = 0.57; rKa = 0.56; P ≤ 0.01).
FIG. 3.
Correlation between pretreatment serum HCV RNA and normalized sequence nucleotide entropy (A) (R2 = 0.37; P ≤ 0.01) and genetic distances (B) (R2 = 0.36; P ≤ 0.01) within the HVR-1 quasispecies in 26 patients who received IFN-α therapy.
DISCUSSION
Many attempts have been made to establish a correlation between the genetic heterogeneity of HCV RNA and the sensitivity of HCV to IFN-α, but published studies have given conflicting results. This may be because the authors have used different techniques with different performances to assess virus heterogeneity, such as single-strand conformational polymorphism analysis (29, 36, 44), heteroduplex analysis (HDA) (45), and combined HDA and single-strand conformational polymorphism analysis (56). For this study, we have cloned and sequenced a sufficiently large number of molecular clones per sample (more than 20) to prevent sample bias (8, 15). We have also checked for virus factors known to influence the effectiveness of IFN-α by analyzing the HVR-1 sequences in two groups of subjects, matched for sex, HCV genotype, and pretreatment serum HCV RNA concentration. To the best of our knowledge, this is the first case-control study designed to assess the influence of the genetic heterogeneity of HVR-1 on virus sensitivity to IFN-α.
Genetic variation in responders and nonresponders was characterized by complexity and diversity. The complexity, the distribution of variants in the population, was estimated by calculating the Shannon entropy. Diversity was measured as the mean genetic distance calculated for all pairs of sequences. Our data show no significant difference between responders and nonresponders for either measure of HVR-1 genetic heterogeneity. A similar observation was also reported by Polyak et al. with HDA (45). By contrast, other studies based on cloning and sequencing techniques have reported that patients with heterogeneous virus populations are less responsive than patients with homogeneous virus populations (3, 25, 41). However, the number of molecular clones analyzed per sample was less than 10 and the influence of HCV RNA concentration on the response to IFN-α was not controlled. We found that sustained response was significantly associated with youth, whereas previous multivariate analyses have found the predictive value of age to be less than those of virus characteristics (22). Thus, by checking for the influence of major factors, genotype and HCV RNA concentration, we could show the impact of other factors such as age and therefore could measure the effect of virus genetic heterogeneity on the sustained response to IFN-α. We found a correlation among complexity, diversity, and serum HCV RNA concentrations, in contrast to the absence of a relationship between quasispecies distribution and virus eradication. This supports the possibility that viral quasispecies arise as a consequence of the limited fidelity of HCV replication and the highly dynamic process of virus production (40).
An important finding of this study is that pretreatment nonsynonymous substitutions in HVR-1 were significantly more frequent than synonymous substitutions in patients who had cleared their HCV after IFN-α therapy, suggesting a stronger selective pressure for changes in amino acids in these patients. It has been demonstrated that the 27-amino-acid segment located in the N-terminal portion of the HCV envelope protein, HVR-1, contains linear neutralizing B-cell epitopes (11, 50, 51, 61). There is recent evidence that HVR-1 is also a helper T-cell recognition site (52) and may be responsible for antagonism to T cells by influencing the priming of a CD4+-T-cell response toward HVR-1 immunogenic variants (12). Thus, the selection pressure driving the genetic variations of the virus may come from the T cells themselves or from neutralizing antibodies whose production depends on T cells specific for this region. A collaboration between helper T cells and cytotoxic T lymphocytes (CTLs) is a common feature of many virus infections, and it has been reported that intrahepatic HCV-specific CTL activity may affect the subsequent response to IFN-α therapy (39). In vitro experiments have shown that the T-cell response to HCV peptides and recombinant core protein during IFN-α treatment was significantly more vigorous in patients infected with genotype 2c than in those infected with genotype 1b (35). The authors suggest that this could be one of the factors contributing to the different susceptibilities of genotype 1 and genotype 2 to IFN-α treatment. Different humoral responses to the HVR-1 region in HCV-1b- and HCV-2c-infected patients have also been reported (59). The antiviral effect of IFN-α results from both inhibition of viral replication and modulation of the immune response to viral epitopes. As recently suggested by mathematical models, the major initial effect of IFN-α is to block virion production or release (40), but the subsequent second-phase decline is thought to reflect the death rate of productively infected cells, where HCV-specific CTL activity could be a major contributor. Other recent studies demonstrated that IFN-α therapy was associated with an increased rate of fixation of mutations in the HVR-1 region compared to the same region in untreated patients, supporting the idea that IFN-α acts partially via immunomodulation (42, 46). Thus, in addition to genotype and HCV RNA concentration, the pretreatment host immune status could influence IFN-α-stimulated immune responses and ultimately virus eradication. Interestingly, women had a higher proportion of nonsynonymous substitutions than did men. It has previously been suggested that the greater response of women to IFN-α could be a consequence of their being given relatively higher doses because of their lower body weight (2, 23). Differences in the immunological status of men and women may represent another factor.
In summary, in our case control study, which included a majority of HCV genotype 2- and 3-infected patients, the complexity and diversity of HVR-1 were correlated with the serum HCV RNA concentration but were not associated per se with virus eradication after IFN-α therapy. By contrast, the higher proportion of nonsynonymous substitutions found in responders suggests that the HCV-specific immune response is involved in the clearing of HCV by IFN-α. Pretreatment immune status, and especially the intensity and the quality of the anti-HCV immune responses of individuals, could be a major determinant of HCV RNA clearance. The beneficial effect of combining ribavirin with IFN-α to treat HCV infection (7, 34, 47) could be due to the modulation of several immune cell functions, but the influence of the envelope sequence, apart from HVR-1, on the early antiviral effect of IFN-α cannot be excluded. It has been suggested recently that a short amino acid sequence in the E2 C terminus region containing phosphorylation sites interacts with PKR (55). Studies combining the characterization of HCV quasispecies heterogeneity and anti-HCV immune responses should be useful for determining how therapeutic agents act and for further optimizing the treatment of chronic hepatitis C.
ACKNOWLEDGMENTS
We thank Jean-Paul Charlet (Service d'Epidémiologie Hôtel-Dieu, Toulouse, France) for help with statistical analysis. We thank Corine Tourne-Peteilh for technical assistance. We also thank Eliane Coutanceau for secretarial assistance.
REFERENCES
- 1.Alter H, Purcell R, Shih J, Melpolder J, Houghton M, Choo Q, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 2.Causse X, Godinot H, Chevallier M, Chossegros P, Zoulim F, Ouzan D. Comparison of 1 or 3 MU of interferon alpha-2b and placebo in patients with chronic non-A, non-B hepatitis. Gastroenterology. 1991;101:497–502. doi: 10.1016/0016-5085(91)90030-o. [DOI] [PubMed] [Google Scholar]
- 3.Chayama K, Tsubota A, Arase Y, Saitoh S, Ikeda K, Matsumoto T, Hashimoto M, Kobayashi M, Kanda M, Morinaga T, Kumada H. Genotype, slow decrease in virus titer during interferon treatment and high degree of sequence variability of hypervariable region are indicative of poor response to interferon treatment in patients with chronic hepatitis type C. J Hepatol. 1995;23:684–653. doi: 10.1016/0168-8278(95)80029-8. [DOI] [PubMed] [Google Scholar]
- 4.Chayama K, Tsubota A, Kobayashi M, Okamoto K, Hashimoto M, Miyano Y, Koibe H, Kobayashi M, Koida I, Arase Y, Saitoh S, Suzuki Y, Murashima N, Ikeda K, Kumada H. Pretreatment virus load and multiple amino acid substitutions in the interferon sensitivity-determining region predict the outcome of interferon treatment in patients with chronic genotype 1b hepatitis C virus infection. Hepatology. 1997;25:745–749. doi: 10.1002/hep.510250342. [DOI] [PubMed] [Google Scholar]
- 5.Choo Q, Kuo G, Weiner A, Overby L, Bradley D, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 6.Davis G, Balart L, Schiff E, Lindsay K, Bodenheimer H, Perrillo R, Carey W, Jacobson I, Payne J, Dienstag J, Van Thiel D, Tamburro C, Lefkowitch J, Albrecht J, Meschievitz C, Ortego T, Gibas A. Treatment of chronic hepatitis C with recombinant interferon alpha. A multicentre randomized, controlled trial. The Hepatitis Interventional Therapy Group. N Engl J Med. 1989;22:1501–1505. doi: 10.1056/NEJM198911303212203. [DOI] [PubMed] [Google Scholar]
- 7.Davis G, Esteban-Mur R, Rustgi V, Hoefs J, Gordon S C, Trepo C, Shiffman M L, Zeuzem S, Craxi A, Ling M H, Albrecht J. Interferon alfa-2b alone or in combination with ribavirin for the treatment of relapse of chronic hepatitis C. International Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1493–1499. doi: 10.1056/NEJM199811193392102. [DOI] [PubMed] [Google Scholar]
- 8.Domingo E. Biological significance of viral quasispecies. Viral Hepat Rev. 1996;2:247–261. [Google Scholar]
- 9.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 11.Farci P, Alter H, Wong D, Miller R, Govindarajan S, Engle R, Shapiro M, Purcell R. Prevention of hepatitis C virus infection in chimpanzees after antibody-mediated in vitro neutralization. Proc Natl Acad Sci USA. 1994;91:7792–7796. doi: 10.1073/pnas.91.16.7792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frasca M, Del Porto P, Tuosto L, Marinari B, Scotta C, Carbonari M, Nicosia A, Piccolella E. Hypervariable region 1 variants act as TCR antagonists for hepatitis C virus-specific CD4+ T cells. J Immunol. 1999;163:650–658. [PubMed] [Google Scholar]
- 13.Gale M, Korth M, Tang N, Tan S, Hopkins D, Dever T, Polyak S, Gretch D, Katze M. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 14.Gale M, Jr, Kwieciszewski B, Dossett M, Nakao H, Katze M G. Antiapoptotic and oncogenic potentials of hepatitis C virus are linked to interferon resistance by viral repression of the PKR protein kinase. J Virol. 1999;73:6506–6516. doi: 10.1128/jvi.73.8.6506-6516.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gretch D, Polyak S. The quasispecies nature of hepatitis C virus: research methods and biological implications. In: Eurotext J L, editor. Hepatitis C virus: genetic heterogeneity and viral load. Paris, France: Groupe Français d'Etudes Moléculaires des Hépatites (GEMHEP); 1997. pp. 57–69. [Google Scholar]
- 16.Gretch D, Polyak S, Wilson J, Carithers R, Perkins J, Corey L. Tracking hepatitis C virus quasispecies major and minor variants in symptomatic and asymptomatic liver transplant recipients. J Virol. 1996;70:7622–7631. doi: 10.1128/jvi.70.11.7622-7631.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hagiwara H, Hayashi N, Mita E, Takehara T, Kasahara A, Fusamoto H, Kamada T. Quantitative analysis of hepatitis C virus RNA in serum during interferon alpha therapy. Gastroenterology. 1993;104:877–883. doi: 10.1016/0016-5085(93)91025-d. [DOI] [PubMed] [Google Scholar]
- 18.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holland J, De La Torre J, Steinhauer D. RNA virus populations as quasispecies. Curr Top Microbiol Immunol. 1992;176:1–20. doi: 10.1007/978-3-642-77011-1_1. [DOI] [PubMed] [Google Scholar]
- 20.Hoofnagle J, Di Bisceglie A. The treatment of chronic viral hepatitis. N Engl J Med. 1997;336:347–356. doi: 10.1056/NEJM199701303360507. [DOI] [PubMed] [Google Scholar]
- 21.Hoofnagle J, Lau D. Chronic viral hepatitis—benefits of current therapies. N Engl J Med. 1996;334:1470–1471. doi: 10.1056/NEJM199605303342210. [DOI] [PubMed] [Google Scholar]
- 22.Izopet J, Payen J, Alric L, Sandres K, Charlet J, Vinel J, Duffaut M, Pascal J, Puel J. Baseline level and early suppression of serum HCV RNA for predicting sustained response to alpha-interferon therapy. J Med Virol. 1998;54:86–91. doi: 10.1002/(sici)1096-9071(199802)54:2<86::aid-jmv3>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 23.Jouet P, Roudot-Thoraval F, Dhumeaux D, Metreau J. Comparative efficacy of interferon alpha in cirrhotic and noncirrhotic patients with non-A, non-B,C hepatitis. Gastroenterology. 1994;106:686–690. doi: 10.1016/0016-5085(94)90703-x. [DOI] [PubMed] [Google Scholar]
- 24.Jukes T H, Cantor T R. Evolution of protein molecules. In: Munro I H N, editor. Mammalian protein metabolism. New York, N.Y: Academic Press, Inc.; 1969. pp. 21–132. [Google Scholar]
- 25.Kanazawa Y, Hayashi N, Mita E, Li T, Hagiwara H, Kasahara A, Fusamoto H, Kamada T. Influence of viral quasispecies on effectiveness of interferon therapy in chronic hepatitis C patients. Hepatology. 1994;20:1121–1130. [PubMed] [Google Scholar]
- 26.Khorsi H, Castelain S, Wyseur A, Izopet J, Canva V, Rombout A, Capron D, Capron J, Lunel F, Stuyver L, Duverlie G. Mutations of hepatitis C virus 1b NS5A 2209-2248 amino acid sequence do not predict the response to recombinant interferon-alpha therapy in French patients. J Hepatol. 1997;27:72–77. doi: 10.1016/s0168-8278(97)80282-6. [DOI] [PubMed] [Google Scholar]
- 27.Knodell K, Ishak K, Black W, Chen T, Craig R, Kaplowitz N. Formulation and application of a numerical scoring system for assessing histological activity in asymptomatic chronic active hepatitis. Hepatology. 1981;5:431–435. doi: 10.1002/hep.1840010511. [DOI] [PubMed] [Google Scholar]
- 28.Koch J O, Bartenschlager R. Modulation of hepatitis C virus NS5A hyperphosphorylation by nonstructural proteins NS3, NS4A, and NS4B. J Virol. 1999;73:7138–7146. doi: 10.1128/jvi.73.9.7138-7146.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koizumi K, Enomoto N, Kurosaki M, Murakami T, Izumi N, Marumo F, Sato C. Diversity of quasispecies in various disease stages of chronic hepatitis C virus infection and its significance in interferon treatment. Hepatology. 1995;22:30–35. [PubMed] [Google Scholar]
- 30.Kumar S, Tamura K, Nei M. MEGA: molecular evolutionary genetic analysis for microcomputers. Comput Appl Biosci. 1993;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- 31.Le Guen B, Squadrito G, Nalpas B, Berthelot P, Pol S, Brechot C. Hepatitis C virus genome complexity correlates with response to interferon therapy: a study in French patients with chronic hepatitis C. Hepatology. 1997;25:1250–1254. doi: 10.1002/hep.510250531. [DOI] [PubMed] [Google Scholar]
- 32.Martell M, Esteban J, Quer J, Genesca J, Weiner A, Esteban R, Guardia J, Gomez J. Hepatitis C virus (HCV) circulates as a population of different but closely related genomes: quasispecies nature of HCV genome distribution. J Virol. 1992;66:3225–3229. doi: 10.1128/jvi.66.5.3225-3229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinot-Peignoux M, Marcellin P, Pouteau M, Castelnau C, Boyer N, Poliquin M, Degott C, Descombes I, Le Breton V, Milotova V, Benhamou J, Erlinger S. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22:1050–1056. [PubMed] [Google Scholar]
- 34.McHutchison J G, Gordon S C, Schiff E R, Shiffman M L, Lee W M, Rustgi V K, Goodman Z D, Ling M H, Cort S, Albrecht J K. Interferon alfa-2b alone or in combination with ribavirin as initial treatment for chronic hepatitis C. Hepatitis Interventional Therapy Group. N Engl J Med. 1998;339:1485–1492. doi: 10.1056/NEJM199811193392101. [DOI] [PubMed] [Google Scholar]
- 35.Missale G, Cariani E, Lamonaca V, Ravaggi A, Rossini A, Bertoni R, Houghton M, Matsuura Y, Fiaccadori F, Ferrari C. Effects of interferon treatment on the antiviral T-cell response in hepatitis C virus 1b- and genotype 2c-infected patients. Hepatology. 1997;26:792–797. doi: 10.1053/jhep.1997.v26.pm0009303515. [DOI] [PubMed] [Google Scholar]
- 36.Moribe T, Hayashi N, Kanazawa Y, Mita E, Fusamoto H, Negi M, Kaneshige T, Igimi H, Kamada T, Uchida K. Hepatitis C viral complexity detected by single-strand conformation polymorphism and response to interferon therapy. Gastroenterology. 1995;108:789–795. doi: 10.1016/0016-5085(95)90452-2. [DOI] [PubMed] [Google Scholar]
- 37.Naito M, Hayashi N, Moribe T, Hagiwara H, Mita E, Kanazawa Y, Kasahara A, Fusamoto H, Kamada T. Hepatitis C viral quasispecies in hepatitis C virus carriers with normal liver enzymes and patients with type C chronic liver disease. Hepatology. 1995;22:407–412. [PubMed] [Google Scholar]
- 38.Nakazawa T, Kato N, Ohkoshi S, Shibuya A, Shimotohno K. Characterization of the 5′ noncoding and structural region of the hepatitis C virus genome from patients with non-A, non-B hepatitis responding differently to interferon treatment. J Hepatol. 1994;20:623–629. doi: 10.1016/s0168-8278(05)80350-2. [DOI] [PubMed] [Google Scholar]
- 39.Nelson D, Marousis C, Ohno T, Davis G, Lau J. Intrahepatic hepatitis C virus-specific cytotoxic T lymphocyte activity and response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1998;28:225–230. doi: 10.1002/hep.510280129. [DOI] [PubMed] [Google Scholar]
- 40.Neumann A, Lam N, Dahari H, Grecht D, Wiley T, Layden T, Perelson A. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon alfa therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 41.Okada S, Akahane Y, Suzuki H, Okamoto H, Mishiro S. The degree of variability in the amino terminal region of the E2/NS1 protein of hepatitis C virus correlated with responsiveness to interferon therapy in viremic patients. Hepatology. 1992;16:619–624. doi: 10.1002/hep.1840160302. [DOI] [PubMed] [Google Scholar]
- 42.Pawlotsky J, Germanidis G, Frainais P, Bouvier M, Soulier A, Pellerin M, Dhumeaux D. Evolution of the hepatitis C virus second envelope protein hypervariable region in chronically infected patients receiving alpha interferon therapy. J Virol. 1999;73:6490–6499. doi: 10.1128/jvi.73.8.6490-6499.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pawlotsky J, Germanidis G, Neumann A, Pellerin M, Frainais P, Dhumeaux D. Interferon resistance of hepatitis C virus genotype 1b: relationship to nonstructural 5a gene quasispecies mutations. J Virol. 1998;72:2795–2805. doi: 10.1128/jvi.72.4.2795-2805.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pawlotsky J, Pellerin M, Bouvier M, Roudot-Thoraval F, Germanidis G, Bastie A, Darthuy F, Remire J, Soussy C, Dhumeaux D. Genetic complexity of the hypervariable region 1 (HVR1) of hepatitis C virus (HCV): influence on the characteristics of the infection and responses to interferon alfa therapy in patients with chronic hepatitis C. J Med Virol. 1998;54:256–264. [PubMed] [Google Scholar]
- 45.Polyak S, Faulkner G, Carithers R, Corey L, Gretch D. Assessment of hepatitis C virus quasispecies heterogeneity by gel shift analysis: correlation with response to interferon therapy. J Infect Dis. 1997;175:1101–1107. doi: 10.1086/516448. [DOI] [PubMed] [Google Scholar]
- 46.Polyak S, McArdle S, Liu S, Sullivan D, Chung M, Hofgartner W, Carithers R J, McMahon B, Mullins J, Corey L, Grecht D. Evolution of hepatitis C virus quasispecies in hypervariable region 1 and the putative interferon sensitivity-determining region during interferon therapy and natural infection. J Virol. 1998;72:4288–4296. doi: 10.1128/jvi.72.5.4288-4296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poynard T, Marcellin P, Lee S S, Niederau C, Minuk G S, Ideo G, Bain V, Heathcote J, Zeuzem S, Trepo C, Albrecht J. Randomised trial of interferon alpha 2b plus ribavirin for 48 weeks or for 24 weeks versus interferon alpha 2b plus placebo for 48 weeks for treatment of chronic infection with hepatitis C virus. International Hepatitis Interventional Therapy Group. Lancet. 1998;352:1426–1432. doi: 10.1016/s0140-6736(98)07124-4. [DOI] [PubMed] [Google Scholar]
- 48.Qian C, Camps J, Maluenda M, Civeira M, Prieto J. Replication of hepatitis C virus in peripheral blood mononuclear cells. Effect of alpha-interferon therapy. J Hepatol. 1992;16:380–383. doi: 10.1016/s0168-8278(05)80674-9. [DOI] [PubMed] [Google Scholar]
- 49.Saiz J, Lopez-Labrador F, Ampurdanes S, Dopazo J, Forns X, Sanchez-Tapias J, Rodes J. The prognostic relevance of the nonstructural 5A gene interferon sensitivity determining region is different in infections with genotype 1b and 3a isolates of hepatitis C virus. J Infect Dis. 1998;177:839–847. doi: 10.1086/515243. [DOI] [PubMed] [Google Scholar]
- 50.Shimizu Y, Hijikata M, Iwamoto A, Alter H, Purcell R, Yoshikura H. Neutralizing antibodies against hepatitis C virus and the emergence of neutralization escape mutant viruses. J Virol. 1994;68:1494–1500. doi: 10.1128/jvi.68.3.1494-1500.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shimizu Y K, Igarashi H, Kiyohara T, Cabezon T, Farci P, Purcell R H. A hyperimmune serum against a synthetic peptide corresponding to the hypervariable region 1 of hepatitis C virus can prevent viral infection in cell cultures. Virology. 1996;223:409–412. doi: 10.1006/viro.1996.0497. [DOI] [PubMed] [Google Scholar]
- 52.Shirai M, Arichi T, Chen M, Nishioka M, Ikeda K, Takahashi H, Enomoto N, Saito T, Major M E, Nakazawa T, Akatsuka T, Feinstone S M, Berzofsky J A. T cell recognition of hypervariable region-1 from hepatitis C virus envelope protein with multiple class II MHC molecules in mice and humans: preferential help for induction of antibodies to the hypervariable region. J Immunol. 1999;162:568–576. [PubMed] [Google Scholar]
- 53.Squadrito G, Leone F, Sartori M, Nalpas B, Berthelot P, Raimondo G, Pol S, Brechot C. Mutations in the nonstructural 5A region of hepatitis C virus and response of chronic hepatitis C to interferon alfa. Gastroenterology. 1997;113:567–572. doi: 10.1053/gast.1997.v113.pm9247477. [DOI] [PubMed] [Google Scholar]
- 54.Tan S L, Nakao H, He Y, Vijaysri S, Neddermann P, Jacobs B L, Mayer B J, Katze M G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533–5558. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor D R, Shi S T, Romano P R, Barber G N, Lai M M. Inhibition of the interferon-inducible protein kinase PKR by HCV E2 protein. Science. 1999;285:107–109. doi: 10.1126/science.285.5424.107. [DOI] [PubMed] [Google Scholar]
- 56.Wang Y, Ray S, Laeyendecker O, Ticehurst J, Thomas D. Assessment of hepatitis C virus sequence complexity by electrophoretic mobilities of both single- and double-stranded DNAs. J Clin Microbiol. 1998;36:2982–2989. doi: 10.1128/jcm.36.10.2982-2989.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weiner A J, Geysen H M, Christopherson C, Hall J E, Mason T J, Saracco G, Bonino F, Crawford K, Marion C D, Crawford K A, Brunetto M, Barr P, McHutchinson M T J, Houghton M. Evidence for immune selection of hepatitis C virus (HCV) putative envelope glycoprotein variants: potential role in chronic HCV infections. Proc Natl Acad Sci USA. 1992;89:3468–3472. doi: 10.1073/pnas.89.8.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolinsky S M, Korber B T M, Neumann A U, Daniels M, Kunstman K J, Whestell A J, Furtado M R, Cao Y, Ho D D, Safrit J T, Koup R A. Adaptive evolution of human immunodeficiency virus-type 1 during the natural course of infection. Science. 1996;272:537–542. doi: 10.1126/science.272.5261.537. [DOI] [PubMed] [Google Scholar]
- 59.Yoshioka K, Aiyama T, Okumura A, Takayanagi M, Iwata K, Ishikawa T, Nagai Y, Kakumu S. Humoral immune response to the hypervariable region of hepatitis C virus differs between genotypes 1b and 2a. J Infect Dis. 1997;175:505–510. doi: 10.1093/infdis/175.3.505. [DOI] [PubMed] [Google Scholar]
- 60.Zeuzem S, Lee J, Roth W. Mutations in the nonstructural 5A gene of European hepatitis C virus isolates and response to interferon alfa. Hepatology. 1997;25:740–744. doi: 10.1002/hep.510250341. [DOI] [PubMed] [Google Scholar]
- 61.Zibert A, Schreier E, Roggendorf M. Antibodies in human sera specific to hypervariable region 1 of hepatitis C virus can block viral attachment. Virology. 1995;208:653–661. doi: 10.1006/viro.1995.1196. [DOI] [PubMed] [Google Scholar]