Abstract
Diffuse large B‐cell lymphoma (DLBCL) accounts for 30% of non‐Hodgkin's lymphomas and is known to comprise heterogeneous groups. We previously reported that CD5+ DLBCL is a clinically distinct subgroup of these tumors that is associated with poor prognosis. In our current study, we have used gene expression profiling technology in an attempt to identify new markers and to further characterize the biological features of CD5+ DLBCL. Candidate genes, which showed the greatest difference in expression between 22 CD5+ and 26 CD5− DLBCL cases, were selected from our screening and subjected to clustering analysis. This resulted in identification of a specific mRNA profile (a CD5 signature) for CD5+ DLBCL. The CD5 signature included downregulated extracellular matrix genes such as POSTN, SPARC, COL1A1, COL3A1, CTSK, MMP9 and LAMB3, and comprised upregulated genes including TRPM4. We tested this CD5 signature for its potential use as a relevant marker for CD5+ DLBCL and found that it did indeed recognize this subgroup. The tumors identified by the CD5 signature contained most of the CD5+ DLBCL cases and some CD5− DLBCL cases. Moreover, the subgroup of cases with this CD5 signature showed a poorer prognosis. The subsequent application of the CD5 signature to the analysis of an independent series of DLBCL microarray data resulted in identification of a subgroup of DLBCL cases with a similar clinical outcome, further suggesting that the CD5 signature can be used as a clinically relevant marker of this disease. (Cancer Sci 2006; 97: 868–874)
Diffuse large B‐cell lymphoma (DLBCL) is the most common form of non‐Hodgkin's lymphoma.( 1 ) Because it is known to include pathophysiologically and clinically heterogeneous groups, the proper identification of well‐defined DLBCL subgroups has become an urgent requirement for clinicians. We previously reported that CD5+ DLBCL is a clinically distinct subgroup of DLBCL, accounting for 5–10% of all DLBCL.( 2 , 3 ) Moreover, this subgroup of cases is characterized by more aggressive clinical features and poorer prognosis, compared with CD5− DLBCL.( 3 ) Genomic aberrations that are characteristic of CD5+ DLBCL have also been identified.( 4 , 5 , 6 ) CD5 positivity is essential for the detection of CD5+ DLBCL cases but is also a marker of mantle cell lymphoma (MCL), and indeed some instances of CD5+ DLBCL possess similar histological features to MCL. For such cases, the use of other markers such as cyclin D1 has become important,( 1 ) and this indicates that the detection of CD5 alone is not sufficient to define the entire spectrum of CD5+ DLBCL tumors. Hence, CD5 can serve as an effective marker when it is used in combination with other biological indicators but it is clear that more effective markers for CD5+ DLBCL will need to be identified.
Expression profiling of mRNA has been used previously as a marker for subgroups of DLBCL.( 7 , 8 , 9 , 10 ) Activated B‐cell‐like (ABC) and germinal‐center B‐cell‐like (GCB) DLBCL are examples of subgroups of DLBCL that have been identified in this way.( 7 , 8 ) It is thus possible that the use of expression profiling can yield novel and effective markers for defining subgroups of DLBCL, in conjunction with established markers such as CD5. With these findings in mind, we compared the expression profiles of 22 cases of CD5+ and 26 cases of CD5− DLBCL in the present study to identify better markers that may provide new insights into our understanding of the pathobiology of CD5+ DLBCL.
Materials and Methods
Patients and samples
The lymph node samples and clinical data used in our present analyses were obtained through an Institutional Review Board approved protocol from 22 patients with CD5+ DLBCL, and a further 26 individuals with CD5− DLBCL. Within these subject groups, 74% of the CD5+ DLBCL cases (14/17) and 29% of the CD5− DLBCL cases (7/24) were associated with extranodal sites (at least one site). None of these patients had a previous history of lymphoma and all 48 individuals received adequate treatment with cyclophosphamide, adriamycin, vincristine and predonine (CHOP)‐like regimens. The median follow‐up time was 2.4 years and the 5‐year overall survival rate of the DLBCL patients was 40%. Each of the 22 CD5+ DLBCL cases was diagnosed for CD5 positivity by immunohistology and four of these cases were confirmed by fluorescence‐activated cell sorter (FACS) analysis. All of the 26 CD5− DLBCL cases were diagnosed for CD5 negativity also by immunohistology and 23 of these were confirmed by FACS analysis. Either Leu1 (Becton Dickinson, Mountain View, CA, USA) or 4C7 (Novocastra, Newcastle, UK) was used as the monoclonal antibody for detection of CD5 antigen. The lymphomas were judged to be CD5 positive when more than 20% of the tumor cells showed positive staining.( 3 ) Classical cytogenetics was used to confirm a negative t(11;14) in each case to exclude the diagnosis of a large cell variant of mantle cell lymphoma.
Microarray procedures
Total RNA extracts were isolated from each specimen by cesium chloride centrifugation, as described previously.( 11 ) An oligonucleotide array, custom‐made for the Cancer Institute of the Japanese Foundation for Cancer Research and on which 21 619 genes had been spotted, was used for analysis according to the manufacturer's protocol (Agilent Technologies, Palo Alto, CA, USA). The probe consisted of a mixture of an experimental Cy5‐labeled cRNA and control Cy3‐labeled cRNA, with the latter prepared from a pool of total RNA from 10 hyperplastic lymph node samples. Non‐flagged array elements with a fluorescent intensity greater than 300 (one standard deviation below the mean of all fluorescent data) were considered well measured. Ratios of the fluorescence of the experimental Cy5‐labeled samples to that of the Cy3‐labeled controls were then log transformed (base 2).
Clustering analyses of microarray data
Genes that showed the greatest average difference in expression between 22 cases of CD5+ and 26 cases of CD5− DLBCL after log transformation were selected from the screening. A hierarchical clustering algorithm was then applied to the DLBCL cases, according to the expression level of these genes, with the aid of Cluster and TreeView programs (http://rana.lbl.gov/EisenSoftware.htm).( 12 ) The classification of either ABC or GCB DLBCL was based on the analysis of 100 genes identified in a previous study.( 8 ) Our array slides contained 67 of these genes, which we used in a simple clustering method. Log‐transformed ratios were centered by subtracting the median observed value of each of the genes for clustering analysis.( 8 )
Analysis of the published microarray data
The DLBCL gene expression profile data generated by the Cancer Genomics group were obtained from the supplemental data listed in at http://www.broad.mit.edu/cancer/pub/dlbcl.( 10 ) None of the 176 DLBCL cases listed here had a previous history of lymphoma. The array fluorescence of these genes was log‐transformed and centered by subtracting the median observed value of each of the genes for clustering analysis.
Results
Identification of CD5 signature genes for the CD5+ DLBCL subgroup
Genes showing differential expression between 22 CD5+ and 26 CD5− DLBCL were identified and 24 of these candidates showed an average expression difference of more than 2.5‐fold between these tumor subgroups (Fig. 1a). These genes were therefore selected for clustering analysis and are referred to as CD5 signature genes. A further 70 genes showing a difference of more than 2‐fold were also subjected to the same analysis.
Figure 1.
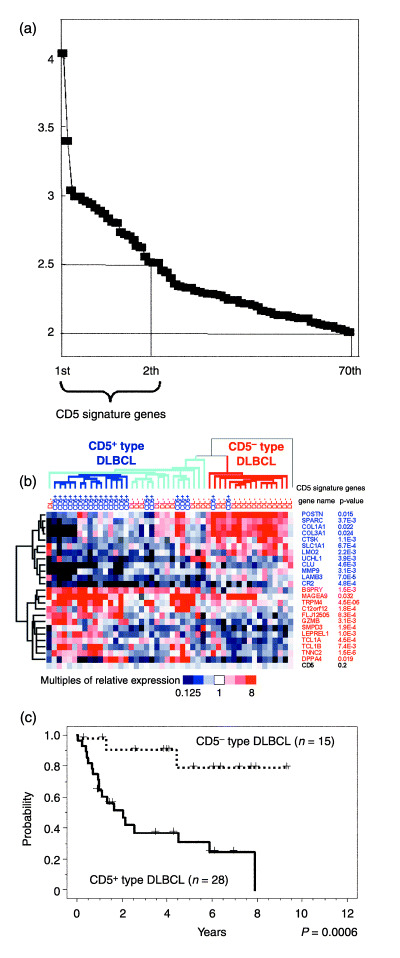
CD5 signature genes define CD5+‐type and CD5−‐type diffuse large B‐cell lymphoma (DLBCL). (a) Differences in the expression of CD5 signature genes. Genes showing the highest level of differential expression between 22 CD5+ and 26 CD5− DLBCL cases are aligned, and multiples of the average differences in their expression levels are shown on the vertical axis. Twenty‐four genes showing an average difference of more than 2.5‐fold between the subgroups were designated as the CD5 signature genes. Seventy genes in total showed a difference of more than 2.0‐fold. (b) Hierarchical clustering of 48 DLBCL cases via the expression levels of the 24 CD5 signature genes. Each row represents one gene. The dendrogram on the left shows the degree to which each gene is related to the others. Half of the CD5 signature genes in CD5+ DLBCL showed low expression levels (upper portion of the figure, shown in blue), some of which were related to the extracellular matrix (POSTN, SPARC, COL1A1, COL3A1, CTSK, MMP9 and LAMB3). The remaining CD5 signature genes in CD5+ DLBCL showed high expression (lower portion of the figure, shown in red). The relative expression of CD5 for each sample is indicated at the bottom of the figure, which also shows the P‐values of these genes, determined using the Student's t‐test. Each column represents one DLBCL case and the dendrogram on the top shows the degree to which each DLBCL is related to the other tumor samples in terms of gene expression. The DLBCL cases were divided into two subgroups: CD5+‐type (left) and CD5−‐type DLBCL (right). There was a cluster of 15 CD5+ DLBCL cases at the core of the CD5+‐type DLBCL group (marked with dark blue lines). One CD5− DLBCL case, located in a row on the extreme right, belonged to neither of the subgroups. (c) Kaplan–Meier analysis of the CD5+‐type and the CD5−‐type DLBCL cases in this study. The CD5+‐type DLBCL patients showed a significantly poorer prognosis than their CD5−‐type DLBCL counterparts. The P‐values for these subgroups were analyzed using the log‐rank test.
Clustering analysis of CD5 signature genes in CD5+‐type and CD5−‐type DLBCL
Hierarchical clustering analysis with each of the identified 24 CD5 signature genes, applied to all 48 DLBCL cases, is shown in Fig. 1b. A further series of analyses using the 70 genes produced almost identical results (data not shown). It was noted that many of the genes that were found to be downregulated in CD5+ DLBCL, such as POSTN, SPARC, COL1A1, COL3A1, CTSK, MMP9 and LAMB3, are associated with the extracellular matrix (Fig. 1b). UCHL1 and CR2 were also found to be downregulated in CD5+ DLBCL tumors and are known to be expressed in T cells, macrophages and follicular dendritic cells, which are subsets of cells that function during the immune response (Fig. 1b).
Our clustering analysis further enabled us to classify the DLBCL cases under study into two groups, one comprising 20 CD5+ and 11 CD5− DLBCL cases, and the other consisting of 14 CD5− and two CD5+ DLBCL samples (Fig. 1b). This indicated that the CD5 signature genes could potentially serve as markers for subgroups that are related to CD5+ DLBCL. All except two cases of CD5+ DLBCL could be included in the first of these two groups, which we designated as CD5+‐type DLBCL, in order to distinguish it from CD5+ DLBCL (Table 1). The second group is referred to as CD5−‐type DLBCL (Table 1). One CD5− DLBCL case could not be assigned to either of these subgroups. The clinical features of CD5+‐type and CD5−‐type DLBCL are shown in Table 2. Patients with CD5+‐type DLBCL showed a more advanced tumor stage at diagnosis, compared with the CD5−‐type DLBCL cases (stage III/IV: 90% and 50%, respectively; P = 0.0062), and also displayed a higher international prognostic index( 13 ) (IPI score 3–5: 66% and 27%, respectively; P = 0.0398). In addition, the overall survival of patients with CD5+‐type DLBCL after treatment with CHOP‐like regimens was significantly poorer than patients with CD5−‐type DLBCL (Fig. 1c; P = 0.0006).
Table 1.
Diffuse large B‐cell lymphoma (DLBCL) subgroups defined by different markers
| Surface marker | mRNA profiling marker | |
|---|---|---|
| CD5+‐type DLBCL (n = 31) | CD5−‐type DLBCL (n = 16) | |
| CD5+ DLBCL (n = 22) | 20 | 2 |
| CD5− DLBCL (n = 25) | 11 | 14 |
| P = 0.0008 † | ||
P‐values were calculated with Fisher's exact test.
Table 2.
Characteristics of patients with diffuse large B‐cell lymphoma (DLBCL)
| Characteristic | CD5+‐type DLBCL (n = 31) | CD5−‐type DLBCL (n = 31) | P‐value † | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| IPI factor | |||||
| Age > 60 years | 20 | 65 | 11 | 69 | >0.99 |
| Stage > 2 | 27 | 90 | 7 | 50 | 0.0062 |
| LDH > normal | 23 | 79 | 10 | 91 | 0.65 |
| Performance status > 1 | 8 | 28 | 0 | 0 | 0.08 |
| Extranodal site > 1 | 8 | 30 | 1 | 7 | 0.13 |
| IPI score 3–5 (high) | 19 | 66 | 3 | 27 | 0.0398 |
P‐values were calculated with Fisher's exact test. IPI, international prognostic index; LDH, lactate dehydrogenase.
In the CD5+‐type DLBCL group, we assigned 11 CD5− DLBCL cases (Table 1), and these were further examined to explore whether in fact any similarities to CD5+ DLBCL existed. Both groups showed similar clinical features except for performance status (Table 3) and no significant differences in survival were observed between the two (P = 0.31, log‐rank test; data not shown). However, the CD5− DLBCL cases in the CD5+‐type DLBCL group did show a significantly poorer prognosis than their CD5− DLBCL counterparts in the CD5−‐type DLBCL group (P = 0.0333; data not shown), indicating that these 11 CD5− DLBCL cases had some clinical features that could be regarded as similar to CD5+ DLBCL.
Table 3.
Characteristics of patients with CD5+ diffuse large B‐cell lymphoma (DLBCL) and CD5− cases in the CD5+‐type DLBCL subgroup
| Characteristic | CD5+ DLBCL (n = 22) | CD5−DLBCL cases in the DLBCL subgroup (n = 22) | P‐value † | ||
|---|---|---|---|---|---|
| n | % | n | % | ||
| IPI factor | |||||
| Age > 60 years | 13 | 59 | 8 | 73 | 0.70 |
| Stage > 2 | 19 | 95 | 9 | 82 | 0.28 |
| LDH > normal | 17 | 85 | 7 | 70 | 0.37 |
| Performance status > 1 | 8 | 40 | 0 | 0 | 0.0288 |
| Extranodal site > 1 | 6 | 32 | 3 | 30 | >0.99 |
| IPI score 3–5 (high) | 14 | 70 | 6 | 60 | 0.69 |
P‐values were calculated with Fisher's exact test. IPI, international prognostic index; LDH, lactate dehydrogenase.
Incidence of ABC and GCB DLBCL among the 48 DLBCL subject cases
Clustering analysis using 67 of the 100 established ABC and GCB markers( 8 ) was applied to our current 48 DLBCL cases, and it was found that these cases could be classified as either ABC or GCB DLBCL (Fig. 2a). Kaplan–Meier analysis further revealed that the ABC DLBCL cases under analysis showed a significantly poorer prognosis, compared with the GCB DLBCL samples (Fig. 2b; P = 0.0028). This is in agreement with results reported previously.( 7 , 8 )
Figure 2.
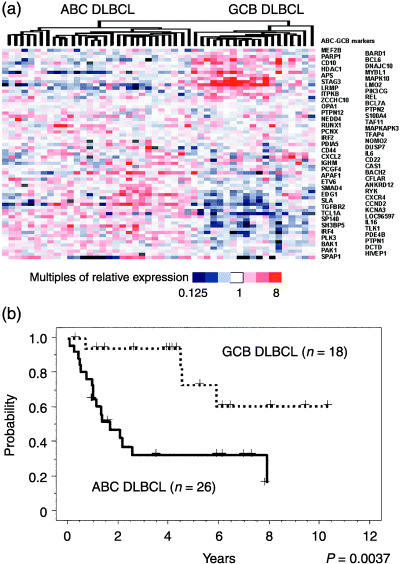
Designation of activated B‐cell‐like (ABC) and germinal‐center B‐cell‐like (GCB) diffuse large B‐cell lymphoma (DLBCL) subtypes among the 48 DLBCL cases in this study. (a) Hierarchical clustering of the 48 DLBCL subject cases based on the expression levels of 67 known ABC and GCB marker genes that are also common to our dataset. (8 ) Our DLBCL samples were classified as ABC (left) and GCB (right) DLBCL in terms of the specific mRNA profiles of these marker genes. (b) Kaplan–Meier analysis of the ABC and GCB DLBCL samples among the current 48 DLBCL cases. This survival analysis showed significant differences between these subgroups.
Application of the CD5 signature to published microarray data
In order to further test the validity of our CD5 signature, we applied it to published microarray data. Among the data from the Cancer Genomics group, comprising the expression level of 44 792 genes from 176 DLBCL cases, we were able to locate 22 (91%) of the 24 CD5 signature genes. Clustering analysis of these DLBCL cases, carried out with 22 of the CD5 signature genes, identified two subgroups of DLBCL, showing downregulation and upregulation of extracellular matrix genes, respectively (Fig. 3a). In addition, the mRNA profiles of these subgroups were similar to the profiles obtained from our current DLBCL cases, such that the first subgroup could be identified as CD5+‐type and the second subgroup as CD5−‐type. The CD5+‐type DLBCL cases again showed a trend toward a poorer prognosis than their CD5−‐type counterparts, although this difference was not statistically significant (P = 0.0762; Fig. 3b).
Figure 3.
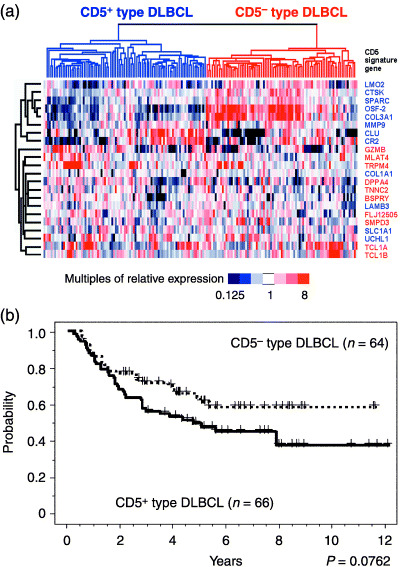
Characterization of CD5+‐type and CD5−‐type diffuse large B‐cell lymphoma (DLBCL) with existing published microarray data. (a) Clustering analysis of DLBCL samples from the Cancer Genomics group in terms of the expression levels of 22 of the CD5 signature genes. The DLBCL samples from published microarray data were classified as CD5+‐type (left) and CD5−‐type (right) DLBCL. The CD5+‐type DLBCL was again characterized by downregulation of the genes that are related to the extracellular matrix, but there were some exceptions. The genes that showed a low expression level in our CD5+‐type DLBCL cases (Fig. 1b) are indicated in blue and those with high expression (Fig. 1b) are shown in red. (b) Kaplan–Meier analysis of the Cancer Genomics group DLBCL patients that were subgrouped by means of the CD5 signature. The CD5+‐type DLBCL patients again showed poorer prognosis than the CD5−‐type DLBCL patients, although this difference was found not to be statistically significant. The P‐values for these subgroups were analyzed with the log‐rank test.
We investigated the relationship between CD5+‐type and CD5−‐type DLBCL, and also between ABC and GCB DLBCL. The 176 DLBCL cases were divided into 34 cases of ABC DLBCL, 85 cases of GCB DLBCL and 57 cases of classless leftovers on the basis of published data.( 10 , 14 ) These ABC and GCB DLBCL cases could be divided into CD5+‐type and CD5−‐type DLBCL groups, so that the DLBCL cases could be classified into four subgroups by combining these two different modes of expression profiling. In the ABC DLBCL group, the CD5+‐type cases showed a poorer prognosis than the CD5−‐type cases (Fig. 4a; P = 0.0397). However, in the GCB DLBCL group, there was no significant difference in survival outcome between the CD5+‐type and the CD5−‐type cases (Fig. 4b; P = 0.5073). Thus, the clinical outcome for the CD5+‐type cases in the ABC DLBCL group was poorer than the outcomes in the other three subgroups.
Figure 4.
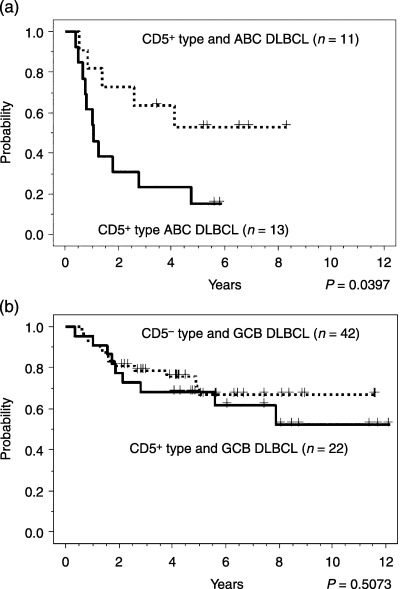
Comparison of the CD5+‐type and the CD5−‐type diffuse large B‐cell lymphoma (DLBCL) in combination with activated B‐cell‐like (ABC) and germinal‐center B‐cell‐like (GCB) DLBCL. Kaplan–Meier analysis of (a) ABC DLBCL and (b) GCB DLBCL patients from the Cancer Genomics group subgrouped by means of the CD5 signature. The clinical outcomes were poorest for ABC patients with CD5+‐type DLBCL. In contrast, there was no significant difference in survival outcome between the CD5+‐type and the CD5−‐type GCB subgroups. The P‐values for these subgroups were analyzed with the log‐rank test.
We also attempted to apply the CD5 signature to other published microarray data. However, as only eight (33%) of the 24 CD5 signature genes could be found among the Lymphochip microarray data,( 8 ) we considered that the results of any analysis using so few genes would not be meaningful.
Discussion
In a Japanese study, CD5+ DLBCL was identified as a known subgroup of DLBCL that is associated with poor prognosis.( 3 ) Furthermore, CD5+ malignancies in Asian countries are different from those in Western countries, as evidenced by the incidence of chronic lymphocytic leukemia (CLL), which is the most frequent leukemia in Europe and the USA, but occurs at one‐fifth of this rate in Japan.( 1 , 15 , 16 ) In this regard, clinicians in Asian countries are in a better situation to investigate CD5+ DLBCL due to a lower level of noise that would be caused by high incidence of CLL and CLL‐related malignancies. In our current study, we attempted to elucidate novel markers and further characterize the biological features of CD5+ DLBCL by means of expression profiling. Differentially expressed genes between CD5+ and CD5− DLBCL samples were selected and subjected to clustering analyses, resulting in the identification of a specific mRNA profile of CD5+ DLBCL, which we refer to as a CD5 signature.
The CD5 signature shows a characteristic profile featuring downregulated genes that are associated with the extracellular matrix. This profile was also found in the DLBCL samples from independent array data. The lack of an extracellular matrix may well be partially responsible for the aggressive clinical features that are characteristic of CD5+ DLBCL. Our CD5 signature was compared with some reported gene sets that are concerned with the DLBCL subgroups. (1) Gascoyne et al. have reported 10 genes to be differentially expressed in CD5+ and CD5− DLBCL.( 17 ) MMP9 is one of these genes and was in fact included in our CD5 signature gene set. (2) Kobayashi et al. have previously described an mRNA profile for CD5+ DLBCL,( 18 ) but our current findings are somewhat discordant with these previous data. The study of Kobayashi et al. reported ITGB1 as one of the strong classifier genes showing high expression in CD5+ DLBCL, whereas our present analysis shows its expression in CD5+ DLBCL to be 1.28 times lower on average, compared with CD5− DLBCL. The classifier gene CD36 was also reported by Kobayashi et al. to show high expression levels in CD5+ DLBCL, but was found to be only 1.29 times higher in our present analysis. Significantly, none of the CD5 signature genes that we identified overlap with these previously characterized classifiers. These differences may be partly due to the fact that the study of Kobayashi et al. incorporated a 2400 spotted array, whereas we have used a 21 619 gene array. (3) The lymph‐node signature is a profile of DLBCL reportedly related to clinical outcome and includes many extracellular matrix‐associated genes.( 8 ) Of these 375 lymph‐node signature genes, 124 were included in our present data set and a further eight of our 24 CD5 signature genes can be found in their data set.( 8 ) However, only three of these CD5 signature genes overlap, whereas the remaining five differ from the lymph‐node signature genes, suggesting that our CD5 signature is likely to be different from the lymph‐node signature. (4) We previously reported that ABC DLBCL was closely related to CD5+ DLBCL,( 6 ) but none of the ABC and GCB markers( 7 ) overlap with the CD5 signature genes. However, IRF4, which is one of the markers that shows high expression in ABC DLBCL, was found in our current analysis to be expressed at a level that was on average 1.56 times higher in CD5+ DLBCL than in CD5− DLBCL. Other markers that have low expression in ABC DLBCL include CD10 and BCL6, expressed 1.39 times and 1.88 times lower, respectively, in CD5+ DLBCL, and this is in agreement with results reported previously.( 6 ) (5) Array comparative genomic hybridization (CGH) analyses have previously uncovered some genomic imbalances that characterize CD5+ DLBCL,( 5 , 6 ) one of which is a loss of 9p21. This is consistent with our present mRNA profile analysis, which shows that the expression of CDKN2A, located on 9p21, is on average 1.5 times lower in CD5+ DLBCL, compared with CD5− DLBCL. However, we did not include this gene in the CD5 signature because its differential expression was lower than the 2.5‐fold cut‐off threshold. This threshold for inclusion in the CD5 signature is probably one of the reasons why none of the CD5 signature genes is located on chromosomal loci that have been shown to be lost or gained in lymphoma.
Although the CD5 signature genes are statistically representative of CD5+ DLBCL, there were some CD5− DLBCL cases found to express the CD5 signature. Eleven of 26 CD5− DLBCL cases expressed the CD5 signature, according to our microarray data, and although only 5–10% of the published cases were expected to be CD5+ DLBCL,( 3 ) 50% of all DLBCL cases from the Cancer Genomics group were found to express the CD5 signature. The incidence of CD5− DLBCL cases expressing the CD5 signature can be partly explained by the results of the clustering analysis of our current cases (Fig. 1b). The CD5+‐type DLBCL group that we identified is composed of two parts: a central cluster consisting of only CD5+ DLBCL cases and the rest comprising mainly CD5− DLBCL patients. This indicates that the CD5 signature provides a rough identification of CD5− DLBCL with expression profiles that are similar to those of CD5+ DLBCL, and that these cases may in fact resemble CD5+ DLBCL. Indeed, these same CD5− DLBCL cases did show clinical features that resemble CD5+ DLBCL. We speculate that CD5− DLBCL cases that show clinical features similar to CD5+ DLBCL will become evident also in independent array data sets.
We utilized CD5 mRNA expression levels to determine which samples could be assigned to the CD5+ DLBCL subgroup among the available DLBCL cases of the Cancer Genomics group. However, this turned out to be ineffective because the CD5 expression levels evaluated by microarray did not correlate with the results obtained by immunostaining or FACS analysis. This discrepancy was most likely caused by background cells such as T cells that express CD5 more strongly than CD5+ DLBCL. However, when the CD5 signature was applied to independent data sets, it was found to be a useful tool that could be used to assign CD5+ DLBCL cases.
The CD5+‐type DLBCL tumors showed a significantly poorer clinical outcome than the CD5−‐type DLBCL in our present analysis. The significance of the differences in survival outcome between CD5+‐type and CD5−‐type DLBCL was further examined by analysis of the independent data set from the Cancer Genomics group and the P‐value for this set was calculated as 0.0762, whereas that for our current study was 0.0037. Although the independent data set did not show significance, there was a measurable trend toward a poor prognosis, which should be noted. Interestingly, however, the CD5+‐type DLBCL of the ABC type showed the worst prognosis, whereas the CD5−‐type DLBCL of the ABC type showed a better prognosis that was almost equivalent to the GCB DLBCL cases. The two types of DLBCL of the GCB type, however, showed no significant difference in clinical outcome. We contend therefore that the CD5 signature could serve as an effective future marker of DLBCL when used in combination with ABC/GCB profiling subtypes. Hence, both ABC/GCB and the CD5 signatures are likely to be effective in identifying the DLBCL subtype with the poorest prognosis and to thus help determine the most appropriate treatment for DLBCL patients.
Acknowledgments
We wish to express our appreciation to Miss H. Suzuki and Miss Y. Kasugai for their outstanding technical assistance. We also thank Dr Ryuzo Ohno, the president of the Aichi Cancer Center, for his support. This work was supported in part by Grants‐in‐Aid from the Japan New Energy and Industrial Technology Development Organization (NEDO) and the Ministry of Economy, Trade and Industry (METI), the Ministry of Health, Labor and Welfare, the Ministry of Education, Culture, Sports Science and Technology, the Japan Society for the Promotion of Science, and the Foundation of Promotion of Cancer Research. Additional support was also obtained via a Research Grant from the Princess Takamatsu Cancer Research Fund (03‐23503).
References
- 1. Gatter KC, Warnke RA. Diffuse large B‐cell lymphoma. In: Jaffe ES, Harris NL, Stein H, Vardiman JW, eds. World Health Classification of Tumors. Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Washington: IARC Press, 2001; 171–4. [Google Scholar]
- 2. Harada S, Suzuki R, Uehira K et al. Molecular and immunological dissection of diffuse large B cell lymphoma: CD5+, and CD5 with CD10+ groups may constitute clinically relevant subtypes. Leukemia 1999; 13: 1441–7. [DOI] [PubMed] [Google Scholar]
- 3. Yamaguchi M, Seto M, Okamoto M et al. De novo CD5+ diffuse large B‐cell lymphoma: a clinicopathologic study of 109 patients. Blood 2002; 99: 815–21. [DOI] [PubMed] [Google Scholar]
- 4. Karnan S, Tagawa H, Suzuki R et al. Analysis of chromosomal imbalances in de novo CD5‐positive diffuse large‐B‐cell lymphoma detected by comparative genomic hybridization. Genes Chromosomes Cancer 2004; 39: 77–81. [DOI] [PubMed] [Google Scholar]
- 5. Tagawa H, Tsuzuki S, Suzuki R et al. Genome‐wide array‐based comparative genomic hybridization of diffuse large B‐cell lymphoma: Comparison between CD5‐positive and CD5‐negative cases. Cancer Res 2004; 64: 5948–55. [DOI] [PubMed] [Google Scholar]
- 6. Tagawa H, Suguro M, Tsuzuki S et al. Comparison of genome profiles for identification of distinct subgroups of diffuse large B‐cell lymphoma. Blood 2005; 106: 1770–7. [DOI] [PubMed] [Google Scholar]
- 7. Alizadeh AA, Eisen MB, Davis RE et al. Distinct types of diffuse large B‐cell lymphoma identified by gene expression profiling. Nature 2000; 403: 503–11. [DOI] [PubMed] [Google Scholar]
- 8. Rosenwald A, Wright G, Chan WC et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large‐B‐cell lymphoma. N Engl J Med 2002; 346: 1937–47. [DOI] [PubMed] [Google Scholar]
- 9. Shipp MA, Ross KN, Tamayo P et al. Diffuse large B‐cell lymphoma outcome prediction by gene‐expression profiling and supervised machine learning. Nat Med 2002; 8: 68–74. [DOI] [PubMed] [Google Scholar]
- 10. Monti S, Savage KJ, Kutok JL et al. Molecular profiling of diffuse large B‐cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 2005; 105: 1851–61. [DOI] [PubMed] [Google Scholar]
- 11. Seto M, Yamamoto K, Iida S et al. Gene rearrangement and overexpression of PRAD1 in lymphoid malignancy with +(11;14)(q13; q32) translocation. Oncogene 1992; 7: 1401–6. [PubMed] [Google Scholar]
- 12. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome‐wide expression patterns. Proc Natl Acad Sci USA 1998; 95: 14863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. The International Non‐Hodgkin's Lymphoma Prognostic Factors Project. A predictive model for aggressive non‐Hodgkin's lymphoma. N Engl J Med 1993; 329: 987–94. [DOI] [PubMed] [Google Scholar]
- 14. Wright G, Tan B, Rosenwald A et al. A gene expression‐based method to diagnose clinically distinct subgroups of diffuse large B cell lymphoma. Proc Natl Acad Sci USA 2003; 100: 9991–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. The World Health Organization classification of malignant lymphomas in Japan: Incidence of recently recognized entities. Lymphoma Study Group of Japanese Pathologists. Pathol Int 2000; 50: 696–702. [DOI] [PubMed] [Google Scholar]
- 16. Tamura K, Sawada H, Izumi Y et al. Chronic lymphocytic leukemia (CLL) is rare, but the proportion of T‐CLL is high in Japan. Eur J Haematol 2001; 67: 152–7. [DOI] [PubMed] [Google Scholar]
- 17. Gascoyne RD, Dave S, Zettl A et al. Gene expression microarray analysis of de novo CD5+ diffuse large B‐cell lymphoma (LLMPP Study): a distinct entity? Blood 2003; 102: 178a. [Google Scholar]
- 18. Kobayashi T, Yamaguchi M, Kim S et al. Microarray reveals differences in both tumors and vascular specific gene expression in de novo CD5+ and CD5− diffuse large B‐cell lymphomas. Cancer Res 2003; 63: 60–6. [PubMed] [Google Scholar]


