Abstract
Membrane‐type 1 matrix metalloproteinase (MT1‐MMP), a powerful modulator of the pericellular environment, promotes migration, invasion, and proliferation of cells. To perform its potent proteolytic activity in a controlled manner, MT1‐MMP has to be regulated precisely. However, our knowledge about substrates and regulatory proteins is still very limited. In this study we identify a catalog of proteins that directly or indirectly interact with MT1‐MMP. We expressed a FLAG‐tagged MT1‐MMP stably in human malignant melanoma A375 cells. We prepared cell lysate using Brij98 and MT1‐MMP was affinity purified together with associating proteins using an anti‐FLAG antibody. A distinct set of membrane proteins was found to copurify with MT1‐MMP when biotin‐labeled proteins were monitored. The proteins were analyzed with an integrated system composed of nano‐flow liquid chromatography and tandem mass spectrometry. We identified 158 proteins including several previously reported to bind MT1‐MMP, although most had not previously been identified. Six of these membrane proteins, including one previously shown to interact with MT1‐MMP, were co‐expressed with MT1‐MMP in HT1080 cells. Five of the latter were found to associate with MT1‐MMP in an immunoprecipitation assay. Immunostaining of cells expressing each of these test proteins revealed that one colocalized with MT1‐MMP at the ruffling membrane and the other at the perinuclear vesicles. In contrast, another protein which did not coprecipitate with MT1‐MMP showed no colocalization. Recombinant MT1‐MMP cleaved two of the tested proteins at least in vitro. Thus, we provide a valuable resource to identify substrates and regulators of MT1‐MMP in tumor cells. (Cancer Sci 2009; 100: 1284–1290)
Abbreviations:
- Nano LC/MS/MS
nano‐flow liquid chromatography tandem mass spectrometry system
- MT1‐MMP
membrane‐type 1 matrix metalloproteinase
- CAT
catalytic domain
- HPX
hemopexin‐like domain
- ECM
extracellular matrix
- TIMPs
tissue inhibitor of metalloproteinases
- PMA
phorbol 12‐myristate 13‐acetate
Membrane‐type 1 matrix metalloproteinase (MT1‐MMP/MMP‐14) is a potent modulator of cell physiology shown to be involved in tumor growth, invasion, and metastasis.( 1 , 2 , 3 , 4 , 5 ) MT1‐MMP deficiency in mice causes early postnatal death associated with severe phenotypes such as dwarfism, osteopenia, arthritis, connective tissue disorder, and disturbed angiogenesis.( 6 , 7 ) The physiological roles of MT1‐MMP are mediated by its proteolytic activity in the pericellular space. Possible substrates include extracellular matrix (ECM) proteins (type I collagen, fibronectin, vitronectin, laminin‐1, ‐5, etc.), cell adhesion molecules (CD44, syndecan‐1, αv integrin), cytokines (SDF‐1, TGF‐β, etc.), and latent forms of proMMPs (proMMP‐2, proMMP‐13).( 1 , 2 , 8 , 9 ) However, our knowledge of the physiological substrates of MT1‐MMP is still limited and identification of these substrates should enable a better understanding of the biological functions of MT1‐MMP.
Proper physiological function of MT1‐MMP requires that its cell surface protease activity be active at the appropriate site and time. Uncontrolled protease activity is rather hazardous to cell physiology even in the presence of natural inhibitors such as TIMPs and reversion‐inducing cysteine‐rich protein with kazal motif (RECK).( 5 , 10 ) During the migration and invasion of cells, MT1‐MMP localizes to the leading edges and invadopodia of migrating and invading cells.( 11 , 12 ) Although the exact mechanisms that determine the localization of MT1‐MMP are not well understood, the binding of MT1‐MMP to cellular proteins linked to the actin cytoskeleton, such as CD44 or integrin, is thought to be a factor determining its localization.( 11 ) Thus, identification of the membrane proteins that interact with MT1‐MMP on the cell surface is important to understanding the mechanism of tumor cell invasion promoted by MT1‐MMP.
CD44 is also a substrate of MT1‐MMP and cleaves at different sites from those for ectodomain shedding by the A disintegrin and metalloproteinase (ADAM) family of proteases.( 13 ) Since the binding of proteins to MT1‐MMP brings them into close proximity to the catalytic site of the enzyme, such proteins are obvious candidates to be MT1‐MMP substrates. For example, type I collagen binds MT1‐MMP and is cleaved.( 14 ) MMP‐2, which is cleaved and activated by MT1‐MMP, also binds MT1‐MMP indirectly through TIMP‐2.( 15 ) Therefore, a survey of proteins associating with MT1‐MMP might identify new substrates.
In this study, we aimed to identify a catalog of proteins that associate either directly or indirectly with MT1‐MMP. To do this, we purified MT1‐MMP from cell lysate together with its associating proteins. We confirmed that a specific set of membrane proteins copurified with MT1‐MMP. The purified proteins were analyzed by nano‐flow liquid chromatography‐tandem mass spectrometry (nano‐LC‐MS/MS).( 16 ) We identified 158 proteins in the MT1‐MMP complex obtained from human melanoma A375 cells. Five of the six membrane proteins in the list were tested and demonstrated to co‐immunoprecipitate MT1‐MMP. Two proteins showed colocalization with MT1‐MMP in cells and they were cleaved by MT1‐MMP in vitro. Thus, the list of proteins associating with MT1‐MMP is a useful source to identify new substrates and regulators of MT1‐MMP in tumor cells.
Materials and Methods
Detailed information about materials, cell culture conditions, expression constructs, and experimental protocols is described in the Supplementary Information.
Isolation of MT1‐MMP associated proteins by FLAG affinity column. Stable transfectants of A375 subclones were cultured for four days in the presence or absence of doxycycline. The cells were lyzed with buffer containing 1% Brij98, 25 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MgCl2, and proteinase inhibitor cocktail III (Calbiochem, La Jolla, CA, USA). After removal of insoluble materials, each sample was subjected to a column filled with agarose beads conjugated to anti‐FLAG M2 antibody. After washing with the lysis buffer, the column was further washed with buffer containing 0.15, 0.5 and 1.0 M NaCl. The proteins retained in the column were eluted with lysis buffer containing FLAG peptide (200 µg/mL). Each fraction was analyzed by SDS–PAGE with a linear 10–20% gel gradient under reducing conditions. To label the cell surface proteins with biotin, cells were incubated with sulfo‐LC‐NHS‐biotin (Pierce, Rockford, IL, USA) at 0.1 mg/mL for 30 min at 4°C.
Immunoprecipitation. HT1080 cells were transfected using FuGENETM (Roche Applied Science, Indianapolis, IN, USA) with expression plasmids for proteins tagged with FLAG at the C‐terminus (5T4 antigen, CD9, CLA‐1, CD63, CD98, CPD) and c‐Myc‐tagged at the N‐terminus MT1‐MMP. Cells were lyzed using 1% CHAPS containing 25 mM HEPES (pH 7.5), 150 mM NaCl, 5 mM MgCl2, and a proteinase inhibitor cocktail III. The supernatant of cell lysate was incubated with mouse IgG agarose for 1 h at 4°C, and then the supernatant was incubated with anti‐FLAG M2 antibody‐conjugated agarose (Sigma, St Louis, MO, USA) overnight at 4°C. The proteins trapped on the beads were eluted with FLAG peptide (200 µg/mL).
Immunostaining. HT1080 cells were transfected with expression plasmids for proteins tagged with FLAG at the C‐terminus in test proteins (5T4 antigen, CPD, CLA‐1) or c‐Myc at the N‐terminus in MT1‐MMP transiently. Transfected cells were cultured on a glass coverslip for 24 h in the presence of 1 µM BB94. After cells were fixed and permeabilized, immunostaining was carried out as described previously using anti‐FLAG polyclonal antibody or antic‐Myc monoclonal antibody.( 11 )
Results
Inducible expression of MT1‐MMP in A375 cells. To isolate a protein complex containing MT1‐MMP, we FLAG‐tagged the protein at either the N‐ (MT1‐NF) or C‐terminus (MT1‐CF) as indicated in Fig. 1(a). To prevent possible degradation of the protein components in the complex by the proteolytic activity of MT1‐MMP, a catalytically inactive mutant with a substitution of glutamic acid in the catalytic domain to alanine (E/A) was used. These tagged MT1‐MMPs were stably expressed in A375 cells under the control of the tetracycline‐inducible promoter. Although A375 cells express endogenous MT1‐MMP (not shown), expression of the FLAG‐tagged MT1‐MMP was almost negligible when the cells were cultured in the presence of doxycycline (+Dox). Depletion of the drug (– Dox) induced several‐fold expression of MT1‐MMP as demonstrated by Western blot analysis using an anti‐FLAG antibody (Fig. 1b). The expression levels were slightly higher for E/A mutants compared to the wild‐type enzyme (WT). Some degradation forms were detected with MT1‐CF even for the E/A mutant. This is presumably because of the activity of endogenous MT1‐MMP.
Figure 1.
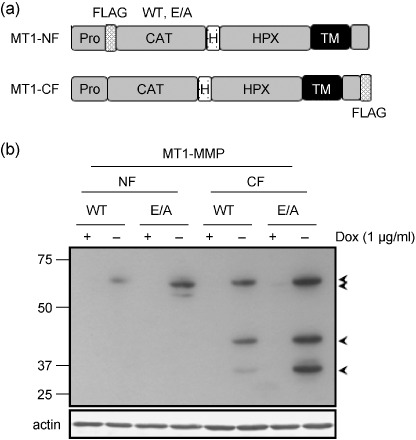
Inducible expression of MT1‐MMP in A375 cells. (a) Schematic illustration of MT1‐MMP proteins tagged with FLAG. WT, wild type; E/A, catalytically inactive mutant; Pro, propeptide; FLAG, FLAG epitope; CAT, catalytic domain; H, hinge domain; HPX, hemopexin‐like domain; TM, transmembrane domain. (b) Stable transfectants of A375 cells cultured in the presence (+) or absence (–) of 1.0 µg/mL doxycycline (Dox) at 37°C for 4 days. Cell lysates were subjected to Western blot analysis using anti‐FLAG antibody (upper panel). Actin was detected with antiactin monoclonal antibody (bottom panel). Arrows indicate specific bands to MT1‐MMP. Molecular weight is indicated on the left.
Affinity purification of complexes containing FLAG‐tagged MT1‐MMP. We first identified an appropriate detergent to isolate MT1‐MMP in a form containing its associated proteins. We used CD44 as a control protein for this experiment, since it is known to bind MT1‐MMP.( 11 , 17 ) Cell lysis with Brij98 did not disrupt the association of CD44 with MT1‐NF in an immunoprecipitation assay (data not shown). After cell lysis, insoluble materials were removed by centrifugation and the supernatant was applied to an affinity column filled with agarose beads conjugated to an anti‐FLAG antibody. The proteins retained in the column were then eluted with increasing concentrations of NaCl and the eluate at each step was analyzed as shown in Fig. 2(a). Proteins retained in the column in a non‐specific manner were removed mostly with wash buffer containing 0.5 M NaCl and this was confirmed when negligible protein was eluted with 1.0 M NaCl from a column to which was applied lysate prepared from cells cultured in the presence of Dox (+Dox). The proteins still bound to the column even in 1.0 M NaCl buffer were subsequently eluted with a buffer containing the FLAG peptide. Two major bands were detected by Coomassie Brilliant Blue staining and a representative result using MT1‐NF(E/A) is presented in Fig. 2(a). The two bands reacted with anti‐FLAG antibody by Western blot analysis (Fig. 2b).
Figure 2.
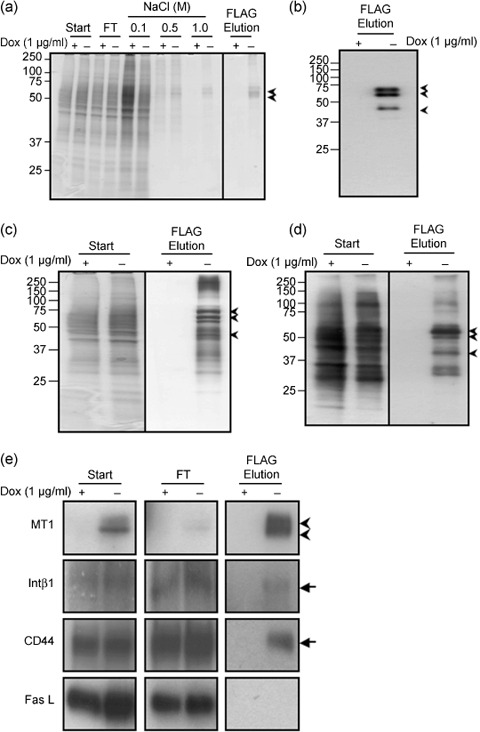
Affinity purification of a complex containing MT1‐MMP in A375 cells. (a) Cell lysates (Start) were applied to an anti‐FLAG antibody affinity column and eluted. The fractions of each step and flowthrough (FT) were analyzed by SDS–PAGE with 10–20% gel gradient. Gels were stained by Coomassie Brilliant Blue R‐250. (b) MT1‐NF in the final fraction was confirmed by Western blot analysis using anti‐MT1‐MMP antibody. Arrows indicate specific bands for MT1‐NF(E/A) corresponding to proMT1‐NF (top), MT1‐NF (middle), and a degraded fragment (bottom) from the size, respectively. (c) Silver staining of the gels. (d) Surface proteins of A375 cells were biotinylated purified similarly to (a). Labeled proteins were detected by Avidin‐HRP. (e) CD44, β1 integrin (Intβ1), and Fas ligand (Fas L) in the indicated fractions were detected by Western blot analysis using a specific antibody to each protein. Arrowheads indicate pro‐ and processed MT1‐NF, respectively, from the top. Arrows indicate specific bands for CD44, Intβ1, and Fas L, respectively.
Although proteins copurified with MT1‐NF(E/A) were not visible following such staining, multiple proteins were visible following silver staining (Fig. 2c, –Dox). The pattern of staining of the proteins that copurified with MT1‐MMP clearly differed from that produced by the starting material (Fig. 2c, Start). Almost no proteins were detected in the final fraction of the lysate prepared from non‐induced cells (+Dox). We further confirmed that membrane proteins were not trapped in the column in a non‐specific manner and eluted in the final preparation. Membrane proteins on the cell surface were labeled with biotin before cell lysis and subjected to affinity purification. The labeled proteins were detected by Western blot analysis using Avidin‐HRP (Fig. 2d). Only a distinct subset of cell surface proteins was copurified with MT1‐NF(E/A).
We next examined whether membrane proteins reported to associate with MT1‐MMP were enriched in the final eluted fraction. These include integrin β1 (Intβ1) and CD44.( 11 , 18 , 19 ) The starting material (Start), flowthrough fraction (FT), and the final eluate (FLAG Elution) were subjected to Western blot analysis using specific antibodies (Fig. 2e). MT1‐NF(E/A) bound to the column and eluted efficiently at the final step (Fig. 2e, MT1). Both Intβ1 and CD44 were detected in the FT fraction, but a substantial amount of these proteins bound to the column and was recovered in the final FLAG elution fraction (Fig. 2e, Intβ1, CD44). We also tested the binding of the Fas ligand (Fas L), which was detected mostly in the FT and not in the final fraction (Fig. 2e, Fas L). Thus, the proteins eluted in the final fraction likely represent those that interact with MT1‐NF(E/A). Proteins associating with MT1‐CF were similarly isolated (data not shown).
Identification of proteins with systemic mass‐spectrometry. To identify proteins associating with MT1‐MMP, the final preparation was digested with a protease (Lys‐C) and applied to nano‐flow liquid chromatography (nano‐LC). Then the separated peptide fragments were automatically analyzed by tandem mass spectrometry (MS/MS) connected online a search engine, Mascot (Matrixscience, Boston, MA, USA), against the non‐redundant protein sequence database at the National Center for Biotechnology Information. Independent preparations of MT1‐NF(E/A) and MT1‐CF(E/A) were analyzed at least three times. These analyses collectively identified 68 proteins for MT1‐NF(E/A) and 137 proteins for MT1‐CF(E/A) (Fig. 3a). Forty‐seven proteins were commonly identified as putative binding partners for MT1‐NF(E/A) and MT1‐CF(E/A). Combining the results, a total of 158 proteins were identified as MT1‐MMP‐associated proteins and these were classified into membrane, cytoplasmic, and secretory proteins, and unknown proteins as summarized in Fig. 3(b) (Supplementary Information Table S1). Cytoplasmic proteins were the most abundant (57% of the total), including those related to vesicle transport and signaling molecules, and others. Membrane proteins (28%) include adhesion molecules, receptors, transporters, and others as listed in Fig. 3(c). Proteins known to associate or bind MT1‐MMP were also included: αv( 20 ) and Intβ1,( 18 ) CD63,( 21 ) and TIMP‐3;( 22 ) some of these are listed in Fig. 3(c).
Figure 3.
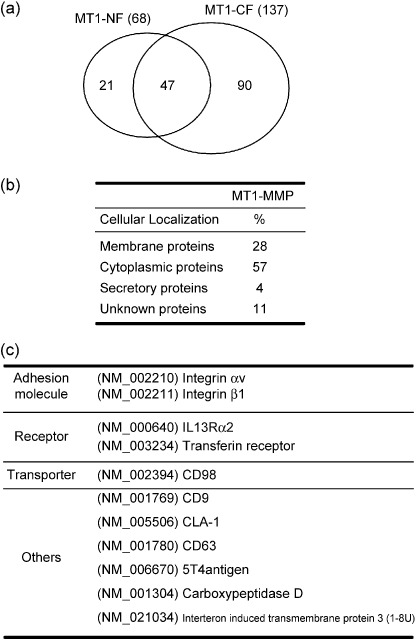
Identification and classification of the associated‐proteins with MT1‐MMP. (a) Proteins in the MT1‐MMP complex (158 proteins) were identified by nano‐LC‐MS/MS analyses for independent preparations. These were 68 and 137 proteins for MT1‐NF and MT1‐CF complexes, respectively, as presented by the circles. (b) The identified proteins in the MT1‐MMP complex were classified into four groups of cellular localization, membrane, cytoplasmic, secretory, and unknown proteins. (c) Membrane proteins were further classified into adhesion molecules, receptors, transporters, and other functional proteins. Some are listed here and the rest in the supplemental data. Integrins αv,( 20 )β1,( 18 ) and CD63,( 21 ) are reported to bind MT1‐MMP.
Evaluation of identified proteins. To confirm the association of the membrane proteins we identified with MT1‐MMP, we selected six proteins that exhibit ostensible relevance to tumor progression based on the results of previous studies.( 23 , 24 , 25 ) We expressed each of these proteins with a FLAG‐tag fused to the C‐terminus (Fig. 4a). The tagged proteins were individually coexpressed with Myc‐tagged MT1‐MMP (MT1‐WT) in HT1080 cells and immunoprecipitation of cell lysates prepared from the transfected cells was carried out using anti‐FLAG antibody. The cells were cultured in the presence of a synthetic MMP inhibitor BB94 to prevent possible degradation of the test proteins by MT1‐MMP. For this assay we used 1% CHAPS to prepare the cell lysate, which represents more stringent conditions than the use of Brij98. Coprecipitation of MT1‐MMP with the FLAG‐tagged proteins was analyzed by Western blot analysis (Fig. 4b). Among the six test proteins, CD63 was previously reported to bind MT1‐MMP,( 21 ) and thus served as a positive control. The other five interacting proteins are 5T4 antigen,( 26 ) CD9,( 24 ) LIMPII Analog‐1 (CLA‐1),( 27 ) CD98,( 28 ) and carboxypeptidase D (CPD).( 29 ) With the exception of CLA‐1, all of the test proteins precipitated MT1‐MMP (Fig. 4b). Thus, the results represent the specificity of the method to purify these proteins.
Figure 4.
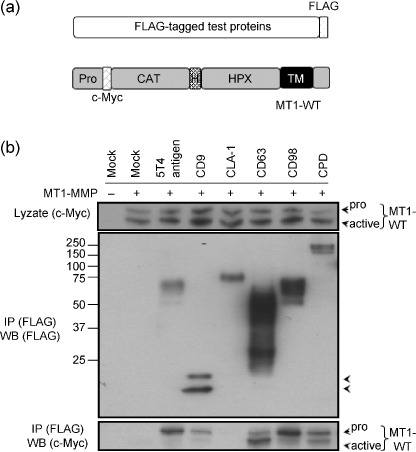
Evaluation of the test proteins for association with MT1‐MMP. (a) Schematic illustration of the test proteins tagged with FLAG at the C‐terminus and c‐Myc‐tagged MT1‐MMP at the N‐terminus (MT1‐WT). FLAG, FLAG Epitope; c‐Myc, c‐Myc epitope; CAT, catalytic domain; H, hinge domain; HPX, hemopexin‐like domain; TM, transmembrane domain. (b) HT1080 cells were transiently cotransfected with the expression plasmids for the indicated test proteins and MT1‐WT. After cell lysis, an immunoprecipitation assay was performed by using anti‐FLAG antibody‐conjugated agarose. The precipitates were analyzed by Western blot analysis using indicated antibodies. Arrowheads indicate pro‐ and processed MT1‐WT.
It is of particular interest that the test proteins precipitated two forms of MT1‐MMP corresponding to the intact (pro) and the processed (active) forms. MT1‐MMP is translated as a latent form (proMT1‐MMP) and its propeptide portion is cleaved off intracellularly and transported to the surface.( 30 ) Thus, the different efficiencies of the test proteins to precipitate the two forms of MT1‐MMP presumably reflect different modes and sites of interaction of those proteins with MT1‐MMP. Indeed, CD63, which is reported to bind internalized MT1‐MMP,( 21 ) preferentially precipitated the active form of MT1‐MMP compared to the pro form.
Localization of test proteins and MT1‐MMP. We next performed immunohistochemistry in cells coexpressing the FLAG‐tagged text proteins and MT1‐MMP to provide further evidence in favor of interaction of the test proteins with MT1‐MMP in vivo. As previously reported, MT1‐MMP localized to the ruffling membrane and perinuclear vesicles presumably corresponding to the Golgi apparatus (Fig. 5).( 11 ) The 5T4 antigen also localized to the ruffling membrane in these cells, although some localization was observed within intracellular vesicles (Fig. 5a, arrows). On the other hand, CPD, which is a Golgi‐resident protein,( 29 ) was detected mostly within intracellular vesicles and did not localize to the ruffling membrane (Fig. 5b). CLA‐1, which failed to precipitate MT1‐MMP, was detected in cytoplasmic vesicles as reported previously,( 31 ) but it did not colocalize with MT1‐MMP in the perinuclear region (Fig. 5c).
Figure 5.
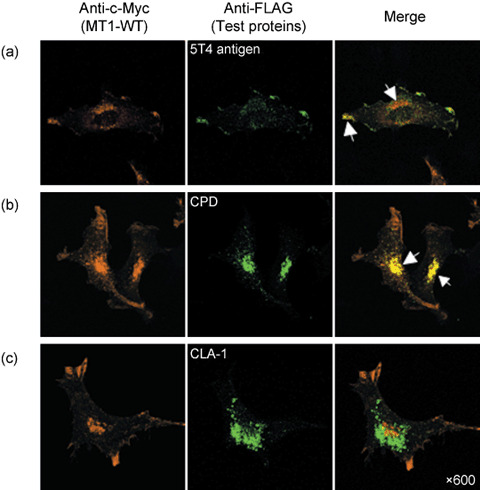
Immuno‐localization assay of the test proteins. HT1080 cells were transfected with plasmids for c‐Myc tagged MT1‐MMP (MT1‐WT) and test FLAG‐tagged proteins. Cells were then stained with antic‐Myc antibody (orange) and anti‐FLAG polyclonal antibody (green) after permeabilization, and analyzed by confocal laser microscopy. (a) 5T4 antigen colocalized with MT1‐MMP at the ruffling membrane area, (b) CPD was detected mainly in the intracellular vesicles but not at the ruffling membrane, and (c) CLA‐1 failed to precipitate MT1‐MMP, and was detected in the cytoplasmic vesicles. Arrows show colocalized MT1‐MMP and FLAG‐tagged test proteins.
Cleavage of test proteins by MT1‐MMP in vitro. Since MT1‐MMP is a protease, wild‐type MT1‐MMP might cleave the test proteins after association. Therefore, we examined whether some of the test proteins could be cleaved by MT1‐MMP in vitro. The FLAG‐tagged test proteins were expressed in COS‐1 cells, which lack expression of endogenous MT1‐MMP, and were purified using affinity beads conjugated to anti‐FLAG antibody. The isolated proteins were then incubated with increasing doses of a catalytic fragment of recombinant MT1‐MMP (rMT1‐MMP) and analyzed by Western blot analysis (Fig. 6). The amount of intact 5T4 antigen decreased by incubation with rMT1‐MMP and the appearance of new fragments was observed (Fig. 6a). BB94 nearly completely inhibited the formation of these fragments. The amount of CPD was also decreased following incubation with rMT1‐MMP, although we could not detect the presence of the cleaved fragments (Fig. 6b). BB94 also inhibited the loss of intact CPD completely. On the other hand, CLA‐1 and CD9 were not affected by incubation with rMT1‐MMP (Fig. 6c).
Figure 6.
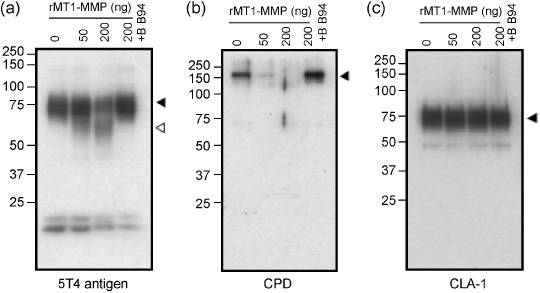
Cleavage of the associating proteins by MT1‐MMP. FLAG‐tagged test proteins were expressed in COS‐1 cells and purified using anti‐FLAG antibody‐conjugated beads. The proteins were incubated with rMT1‐MMP in the presence or absence of BB94. The samples were then resolved on SDS–PAGE, followed by Western blot analysis. Black arrowheads indicate mature proteins and white arrowheads, processed fragments. Molecular weight markers are shown on the left.
Discussion
In this study, we aimed to generate a catalog of proteins that associate with MT1‐MMP either directly or indirectly. Since MT1‐MMP is an integral membrane protein, we took care to avoid inclusion of non‐specific membrane proteins in the preparation. Thus, we do not think membrane proteins that have no relevance to MT1‐MMP were present at a significant level in the final fraction eluted with the FLAG peptide. Biotin‐labeled membrane proteins within the final column eluate differed considerably from the original membrane proteins as analyzed by SDS–PAGE (Fig. 2d), and Fas L detected as a control of membrane proteins did not copurify with MT1‐MMP (Fig. 2e).
Preparations containing MT1‐MMP‐associated proteins were subjected to an integrated system for proteomics (nano‐LC/MS/MS) that is able to analyze multiple protein components simultaneously. Repeated analysis of independent preparations identified a total of 158 proteins that apparently interact either directly or indirectly with MT1‐MMP. The number of newly identified proteins decreased with each repetition of the assay and the total number appears to converge to two hundred according to the repetition of analysis. The identified proteins comprised both membrane (44 proteins) and cytoplasmic protein (90 proteins). Six of the membrane proteins were selected and their association with MT1‐MMP was further analyzed. An immunoprecipitation assay revealed that five of the six test proteins, including CD63 as a positive control, associated with MT1‐MMP. Among these, both 5T4 antigen and CPD colocalized with MT1‐MMP in cells, but CLA‐1, which did not coprecipitate MT1‐MMP, did not colocalize.
Although we believe the proteins identified in our analysis represent those localizing closely to MT1‐MMP in cells, some proteins that have been reported to bind MT1‐MMP were not detected here. For example, TIMP‐2 and MMP‐2, which form a tri‐molecular complex with MT1‐MMP,( 32 , 33 , 34 ) were not identified in this assay. This result may be due to the low level of expression of TIMP‐2 in A375 cells (not shown). CD44 was also not identified in this assay even though it was clearly present in the final elution fraction (Fig. 2e). This may be due to the ionization efficiency of the fragments and complexity of the samples. Some proteins expected to interact with MT1‐MMP were also missing from the result. These are components of the cytoskeleton that are connected to adhesion molecules such as CD44 or integrins. ECM molecules such as type I collagen were also not detected. MT1‐MMP proteins firmly connected to macromolecules, and which form an insoluble matrix, may not be purified by this method.
At least two of the MT1‐MMP associating proteins, 5T4 antigen and CPD, were cleaved by MT1‐MMP in vitro. Since these proteins colocalized with MT1‐MMP in cells, these are candidate physiological substrates of MT1‐MMP. Thus, we think the list of interacting proteins presented here is a valuable resource to identify putative in vivo substrates of MT1‐MMP. Recently, another systematic approach to identify protease substrates has been reported.( 35 ) In this method, isotope‐coded affinity tag (ICAT) MS was applied to analyze cleaved fragments released from cells expressing MT1‐MMP. Use of this method has led to the efficient identification of soluble substrates in the extracellular space. We believe that our approach complements the ICAT method,( 35 ) particularly in identifying membrane proteins whose fragments are relatively difficult to release from the cell surface.
The identified membrane proteins include adhesion molecules, receptor proteins, and a transporter. These proteins may regulate tumor cell functions together with MT1‐MMP. Among these, 5T4 antigen is an oncofetal glycoprotein first identified in trophoblasts using 5T4 mAb,( 36 ) and CD9 is a member of the transmembrane‐4 super family (TM4SF, tetraspanins), which has been reported to associate with integrins.( 37 ) We believe the catalog of proteins associating with MT1‐MMP directly or indirectly is a valuable resource to understand the roles of MT1‐MMP in tumor cells.
Supporting information
Materials and Methods
Detailed information about materials, cell culture conditions, expression constructs, and experimental protocols is described.
Table S1. MT1‐MMP associating proteins identified by LC/MS/MS system in A375 cells
List of 158 proteins is presented.
Please note: Wiley‐Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item
Acknowledgments
We thank Dr I. Yana and Dr A. Matsuda for valuable discussions. This work was supported by a grant for the Integrated Proteomics System Project and Pioneer Research on Genome the Frontier, and by a grant‐in‐aid for Scientific Research on Priority Area Cancer from the Ministry of Education, Culture, Sports, Science and Technology Japan.
References
- 1. Seiki M, Yana I. Roles of pericellular proteolysis by membrane type‐1 matrix metalloproteinase in cancer invasion and angiogenesis. Cancer Sci 2003; 94: 569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sato H, Takino T, Miyamori H. Roles of membrane‐type matrix metalloproteinase‐1 in tumor invasion and metastasis. Cancer Sci 2005; 96: 212–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Uekita T, Gotoh I, Kinoshita T et al . Membrane‐type 1 matrix metalloproteinase cytoplasmic tail‐binding protein‐1 is a new member of the Cupin superfamily. A possible multifunctional protein acting as an invasion suppressor down‐regulated in tumors. J Biol Chem 2004; 279: 12 734–43. [DOI] [PubMed] [Google Scholar]
- 4. Hotary K, Allen E, Punturieri A, Yana I, Weiss SJ. Regulation of cell invasion and morphogenesis in a three‐dimensional type I collagen matrix by membrane‐type matrix metalloproteinases 1, 2, and 3. J Cell Biol 2000; 149: 1309–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Itoh Y, Seiki M. MT1‐MMP: a potent modifier of pericellular microenvironment. J Cell Physiol 2006; 206: 1–8. [DOI] [PubMed] [Google Scholar]
- 6. Holmbeck K, Bianco P, Caterina J et al . MT1‐MMP‐deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 1999; 99: 81–92. [DOI] [PubMed] [Google Scholar]
- 7. Zhou Z, Apte SS, Soininen R et al . Impaired endochondral ossification and angiogenesis in mice deficient in membrane‐type matrix metalloproteinase I. Proc Natl Acad Sci USA 2000; 97: 4052–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer 2002; 2: 161–74. [DOI] [PubMed] [Google Scholar]
- 9. Overall CM, McQuibban GA, Clark‐Lewis I. Discovery of chemokine substrates for matrix metalloproteinases by exosite scanning: a new tool for degradomics. Biol Chem 2002; 383: 1059–66. [DOI] [PubMed] [Google Scholar]
- 10. Noda M, Takahashi C. Recklessness as a hallmark of aggressive cancer. Cancer Sci 2007; 98: 1659–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mori H, Tomari T, Koshikawa N et al . CD44 directs membrane‐type 1 matrix metalloproteinase to lamellipodia by associating with its hemopexin‐like domain. Embo J 2002; 21: 3949–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakahara H, Howard L, Thompson EW et al . Transmembrane/cytoplasmic domain‐mediated membrane type 1‐matrix metalloprotease docking to invadopodia is required for cell invasion. Proc Natl Acad Sci USA 1997; 94: 7959–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nakamura H, Suenaga N, Taniwaki K et al . Constitutive and induced CD44 shedding by ADAM‐like proteases and membrane‐type 1 matrix metalloproteinase. Cancer Res 2004; 64: 876–82. [DOI] [PubMed] [Google Scholar]
- 14. Tam EM, Wu YI, Butler GS, Stack MS, Overall CM. Collagen binding properties of the membrane type‐1 matrix metalloproteinase (MT1‐MMP) hemopexin C domain. The ectodomain of the 44‐kDa autocatalytic product of MT1‐MMP inhibits cell invasion by disrupting native type I collagen cleavage. J Biol Chem 2002; 277: 39 005–14. [DOI] [PubMed] [Google Scholar]
- 15. Itoh Y, Seiki M. MT1‐MMP: an enzyme with multidimensional regulation. Trends Biochem Sci 2004; 29: 285–9. [DOI] [PubMed] [Google Scholar]
- 16. Natsume T, Yamauchi Y, Nakayama H et al . A direct nanoflow liquid chromatography‐tandem mass spectrometry system for interaction proteomics. Anal Chem 2002; 74: 4725–33. [DOI] [PubMed] [Google Scholar]
- 17. Suenaga N, Mori H, Itoh Y, Seiki M. CD44 binding through the hemopexin‐like domain is critical for its shedding by membrane‐type 1 matrix metalloproteinase. Oncogene 2005; 24: 859–68. [DOI] [PubMed] [Google Scholar]
- 18. Galvez BG, Matias‐Roman S, Yanez‐Mo M, Sanchez‐Madrid F, Arroyo AG. ECM regulates MT1‐MMP localization with beta1 or alphavbeta3 integrins at distinct cell compartments modulating its internalization and activity on human endothelial cells. J Cell Biol 2002; 159: 509–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ellerbroek SM, Fishman DA, Kearns AS, Bafetti LM, Stack MS. Ovarian carcinoma regulation of matrix metalloproteinase‐2 and membrane type 1 matrix metalloproteinase through beta1 integrin. Cancer Res 1999; 59: 1635–41. [PubMed] [Google Scholar]
- 20. Ratnikov BI, Rozanov DV, Postnova TI et al . An alternative processing of integrin alpha(v) subunit in tumor cells by membrane type‐1 matrix metalloproteinase. J Biol Chem 2002; 277: 7377–85. [DOI] [PubMed] [Google Scholar]
- 21. Takino T, Miyamori H, Kawaguchi N, Uekita T, Seiki M, Sato H. Tetraspanin CD63 promotes targeting and lysosomal proteolysis of membrane‐type 1 matrix metalloproteinase. Biochem Biophys Res Commun 2003; 304: 160–6. [DOI] [PubMed] [Google Scholar]
- 22. Will H, Atkinson SJ, Butler GS, Smith B, Murphy G. The soluble catalytic domain of membrane type 1 matrix metalloproteinase cleaves the propeptide of progelatinase A and initiates autoproteolytic activation. Regulation by TIMP‐2 and TIMP‐3. J Biol Chem 1996; 271: 17 119–23. [DOI] [PubMed] [Google Scholar]
- 23. Carsberg CJ, Myers KA, Evans GS, Allen TD, Stern PL. Metastasis‐associated 5T4 oncofoetal antigen is concentrated at microvillus projections of the plasma membrane. J Cell Sci 1995; 108 (Pt 8): 2905–16. [DOI] [PubMed] [Google Scholar]
- 24. Miyake M, Nakano K, Itoi SI, Koh T, Taki T. Motility‐related protein‐1 (MRP‐1/CD9) reduction as a factor of poor prognosis in breast cancer. Cancer Res 1996; 56: 1244–9. [PubMed] [Google Scholar]
- 25. Hara K, Kudoh H, Enomoto T, Hashimoto Y, Masuko T. Malignant transformation of NIH3T3 cells by overexpression of early lymphocyte activation antigen CD98. Biochem Biophys Res Commun 1999; 262: 720–5. [DOI] [PubMed] [Google Scholar]
- 26. Carsberg CJ, Myers KA, Stern PL. Metastasis‐associated 5T4 antigen disrupts cell‐cell contacts and induces cellular motility in epithelial cells. Int J Cancer 1996; 68: 84–92. [DOI] [PubMed] [Google Scholar]
- 27. Armesilla AL, Vega MA. Structural organization of the gene for human CD36 glycoprotein. J Biol Chem 1994; 269: 18 985–91. [PubMed] [Google Scholar]
- 28. Rintoul RC, Buttery RC, Mackinnon AC et al . Cross‐linking CD98 promotes integrin‐like signaling and anchorage‐independent growth. Mol Biol Cell 2002; 13: 2841–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalinina EV, Fricker LD. Palmitoylation of carboxypeptidase D. Implications for intracellular trafficking. J Biol Chem 2003; 278: 9244–9. [DOI] [PubMed] [Google Scholar]
- 30. Seiki M. Membrane‐type 1 matrix metalloproteinase: a key enzyme for tumor invasion. Cancer Lett 2003; 194: 1–11. [DOI] [PubMed] [Google Scholar]
- 31. Baranova IN, Vishnyakova TG, Bocharov AV et al . Serum amyloid A binding to CLA‐1 (CD36 and LIMPII analogous‐1) mediates serum amyloid A protein‐induced activation of ERK1/2 and p38 mitogen‐activated protein kinases. J Biol Chem 2005; 280: 8031–40. [DOI] [PubMed] [Google Scholar]
- 32. Kinoshita T, Sato H, Okada A et al . TIMP‐2 promotes activation of progelatinase A by membrane‐type 1 matrix metalloproteinase immobilized on agarose beads. J Biol Chem 1998; 273: 16 098–103. [DOI] [PubMed] [Google Scholar]
- 33. Strongin AY, Collier I, Bannikov G, Marmer BL, Grant GA, Goldberg GI. Mechanism of cell surface activation of 72‐kDa type IV collagenase. Isolation of the activated form of the membrane metalloprotease. J Biol Chem 1995; 270: 5331–8. [DOI] [PubMed] [Google Scholar]
- 34. Butler GS, Butler MJ, Atkinson SJ et al . The TIMP2 membrane type 1 metalloproteinase ‘receptor’ regulates the concentration and efficient activation of progelatinase A. A kinetic study. J Biol Chem 1998; 273: 871–80. [DOI] [PubMed] [Google Scholar]
- 35. Tam EM, Morrison CJ, Wu YI, Stack MS, Overall CM. Membrane protease proteomics: Isotope‐coded affinity tag MS identification of undescribed MT1‐matrix metalloproteinase substrates. Proc Natl Acad Sci USA 2004; 101: 6917–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hole N, Stern PL. A 72 kD trophoblast glycoprotein defined by a monoclonal antibody. Br J Cancer 1988; 57: 239–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Berditchevski F, Zutter MM, Hemler ME. Characterization of novel complexes on the cell surface between integrins and proteins with 4 transmembrane domains (TM4 proteins). Mol Biol Cell 1996; 7: 193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Materials and Methods
Detailed information about materials, cell culture conditions, expression constructs, and experimental protocols is described.
Table S1. MT1‐MMP associating proteins identified by LC/MS/MS system in A375 cells
List of 158 proteins is presented.
Please note: Wiley‐Blackwell is not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
Supporting info item
Supporting info item


