Abstract
In our previous study, mesenchymal–epithelial transition factor (c‐Met)‐binding peptides (cMBP) had been readily radiolabeled with radioactive iodide for glioma imaging because of five histidine amino acids. However, iodinated cMBP showed relatively unfavorable in vivo kinetics. For this reason, we tried to design dual peptide ligands that would be advantageous in recognizing both c‐Met receptor and integrin αvβ3. A cMBP‐click‐cRGDyk (cyclic Arg‐Gly‐Asp‐Tyr‐Lys) heterodimer was synthesized from mini polyethylene glycol‐conjugated cMBP‐3 glycine (GGG)‐a single name of amino acids (SC) (Ser‐Cys) and cRGDyk through a click (1 + 3 cycloaddition), and then labeled with iodine 125 (I‐125) via histidine in the cMBP and tyrosine in the cRGDyk. The receptor‐binding characteristics and tumor‐targeting efficacy of cMBP‐click‐cRGDyk were tested in vitro and in vivo. A cMBP‐click‐cRGDyk had comparable integrin αvβ3‐binding affinity with cRGDyk. The results of the biodistribution of 125I‐cMBP‐click‐cRGDyk at 4 h showed higher tumor‐to‐blood, tumor‐to‐liver, and tumor‐to‐muscle ratios: 10.07, 6.76, and 11.12, compared to 2.34, 1.99, and 5.18 of 125I‐cMBP‐GGG‐SC, respectively. U87MG tumor xenografts could be visualized by single photon emission computed tomography (SPECT)/CT using 125I‐cMBP‐click‐cRGDyk and also image contrast and overall quality were improved compared to 125I‐cMBP‐GGG‐SC. As the results of in vivo inhibition using free cRGDyk or cMBP‐GGG‐SC indicated, the tumoral uptake of 125I‐cMBP‐click‐cRGDyk decreased. This finding means that 125I‐cMBP‐click‐cRGDyk was specifically uptaken by integrin αvβ3 and the c‐Met receptor. Although imaging quality was improved, additional experiments are needed to acquire significant image‐quality improvement. (Cancer Sci 2011; 102: 1516–1521)
Many types of receptors are uniquely expressed or markedly overexpressed in tumors, and have been used as potential targets for cancer diagnosis and therapy. The ability to measure and identify specific receptor expression is crucial for the accurate diagnosis, staging, restaging, and classification of tumors, and for monitoring patient response to therapy.( 1 , 2 ) Many research groups are therefore interested in peptide radiopharmaceuticals. Compared to high molecular weight polymers, small peptides are structurally well defined and are generally cleared from circulation much faster, which might lead to a higher target‐to‐background ratio.
Multivalent ligands can be homomultivalent, with multiple copies of the same ligand, or heteromultivalent, with different types of ligands targeting different types of receptors. Multivalent ligands consist of multiple binding moieties (pharmacophores) that are bound together via chemical linkers. Multivalent binding can lead to increased functional affinity and binding specificity.( 3 , 4 , 5 ) A wide spectrum of binding moieties has been studied, including small peptide fragments, truncated versions of antibodies, and carbohydrate analogs.( 5 , 6 , 7 )
Integrin αvβ3 is highly expressed in invasive tumors, such as late‐stage glioblastomas, breast and prostate tumors, malignant melanomas, and ovarian carcinomas, as well as in newborn blood vessels.( 2 , 8 ) The expression level of integrin αvβ3 is an important factor in determining malignant invasiveness and metastatic potential in both preclinical animal models and cancer patients.( 9 ) Over the past several years, many researchers have successfully developed series RGD peptide radiotracers with favorable in vivo kinetics for tumor integrin αvβ3 imaging.( 10 , 11 ) Mesenchymal–epithelial transition factor (c‐Met) is a receptor tyrosine kinase known to stimulate invasive cancer cell growth and increase metastatic potential; it is also expressed and mutates in a variety of solid tumors.( 12 , 13 , 14 ) Mesenchymal–epithelial transition factor is overly expressed in human glioblastomas, with expression levels correlating to glioma malignancy grade and vascularity. The activated endothelial cells around c‐Met‐positive tumor tissues express high levels of integrin during tumor angiogenesis, invasion, and metastasis. We previously reported that iodine 125 (I‐125)‐radiolabeled, c‐Met‐binding peptides (cMBP) bound specifically to U87MG cells and in vivo tumors.( 14 ) A glioma tumor (U87MG) expresses both c‐Met and integrin αvβ3, and we hypothesize that a peptide ligand recognizing both receptors would be advantageous over a single receptor‐binding probe. Chen et al. previously studied this concept.( 7 )
Currently, the copper(I)‐catalyzed Huisgen 1,3‐dipolar cycloaddition of azides and alkynes, commonly called “click chemistry”, plays a crucial role in a wide range of biomedical applications.
This reaction can be carried out in high yields under mild conditions, and the 1,2,3‐triazole formed has a similar polarity and size with an amide bond. Due to these favorable aspects, the use of click chemistry to conjugate two (bio)molecular components has recently been reported.( 12 , 15 , 16 )
In this study, we hypothesized that the introduction of cRGDyk using click reaction methodology would help reduce radioactive accumulation in major organs by enhancing elimination, while increasing tumoral uptake. We developed a 1,2,3‐triazole‐associated, cMBP‐click‐cRGDyk peptide heterodimer that recognizes c‐Met receptors through the cMBP motif and integrin αvβ3 through the RGD motif. After radiolabeling this synthesized heterodimer, we conducted studies targeting integrin and c‐Met receptors both in vitro and in vivo.
Materials and Methods
Regents and chemicals. Na iodine 125 (Na125I) and 125I‐echistatin were obtained from Perkin Elmer Life Science (Boston, MA, USA). The KSLSRHDHIHHH‐3 glycine (GGG)‐βA‐K mini polyethylene glycol azide (cMBP‐GGG‐N3) was synthesized by Peptron (Seoul, Korea) using standard Fmoc chemistry. Propiolic–cyclic RGDyk was purchased from Future Chem (Seoul, Korea). The purity of all samples was >90%.
Preparation of cMBP‐click‐cRGDyk. The synthesis of azidoacetic acid and peptides was described previously.( 12 ) Compound 7 was synthesized on an automated, solid‐phase peptide synthesizer (ASP48S; Peptron, Korea) using traditional Fmoc chemistry and Rink amide 4‐methylbenzhydrylamine hydrochloride salt (MBHA) resin (25 μmol). Each coupling step was performed using N,N,N′,N′‐tetramethyl‐O‐(1H‐benzotriazol‐1‐yl)uronium hexafluorophosphate (HBTU)/1‐hydroxybenzotriazole (HOBt) and appropriate Fmoc‐protected amino acids (100 μmol), including the following side‐chain‐protecting groups: Gln(Trt), Trp(Boc), His(Trt), and Ser(tBu). After washing the resin (with DMF, iPrOH), 2 eq propiolic–cyclic RGDyk, 0.2 eq Cu(I)Br, and 2 eq N,N‐diisopropylethylamine (DIPEA) were added to the resin. The click reaction was shaken for 4 h at room temperature. The final product was cleaved by a standard procedure using a cocktail containing triisopropylsilane, thioanisol, water, ethanedithiol, and trifluoroacetic acid (mixing ratio of 2.5/2.5/2.5/2.5/90%, respectively). Typical yields of the crude peptide were 75–85%. The crude material was purified by reverse phase (RP)‐HPLC, and electrospray ionization mass spectrometry (ESI‐MS) was used to determine the molecular mass of the prepared peptide (HPLC: Retention time = 6.78 m for 7; ESI‐MS (m/z) found (calculation): 2772 (2771.57) for 7).
Radioiodinated peptide. For in vivo imaging, cMBP‐click‐cRGDyk was radiolabeled with Na125I using chloramine‐T. Briefly, 10 μg (≒3.6 nmol) cMBP‐click‐cRGDyk was labeled with approximately 0.4 mCi Na125I (23 ng 125I) using 30 μg chloramine‐T in 10 μL phosphate‐buffered saline (PBS; 0.5 M, pH 7.5). The reaction was terminated with sodium bissulfite (30 μg in 10 μL PBS). Radiochemical purity was assessed by instant thin layer chromatography‐silica gel (0.9% normal saline). Radioactivity was determined with a radio‐TLC scanner (Bioscan, Washington, DC, USA). The partition coefficient (logP) value of the 125I‐cMBP‐click‐cRGDyk and 125I‐cMBP‐GGG‐a single name of amino acids (SC) was previously described.( 14 )
In vitro binding assays. In vitro c‐Met‐binding affinity and the specificity of cMBP‐click‐cRGDyk were assessed via a displacement cell‐binding assay using 125I‐cMBP (100 ng/well) as the radioligand. Experiments were performed on U87MG cells by a previously‐published method.( 14 ) For the in vitro integrin‐binding affinity analysis, U87MG cells were seeded onto 96‐well plates at 1 × 104 cells per well, and incubated overnight at 37 °C. Serial dilutions of cMBP‐click‐cRGDyk and 125I‐echistatin (corresponding to concentrations of 0.04 nM–150 μM) were added to the 96‐well plates. The plates were then incubated for 1 h at 37 °C, washed, and dried; 0.1 mL of 2 N NaOH was then added to each well to facilitate cell lysis. The lysates were collected and counted in a gamma counter (Packard, Meriden, CT, USA). Binding affinities (IC50) for both receptors were calculated by non‐linear regression analysis (sigmoidal dose‐response equation) using the GraphPad Prism 4.0 computer‐fitting program (Graph‐Pad Software, San Diego, CA, USA).
Cell inhibition study. For the blocking study, U87MG cells were seeded onto 24‐well plates at 1 × 105 cells per well. The next day, the cells were co‐treated with 125I‐cMBP‐click‐cRGDyk (0.18 MBq/2 pmol/well) and cMBP‐GGG‐SC (60 nmol) or cRGDyk (60 nmol) as blockers. The cells were incubated for 40 min at 37 °C. After incubation, the radioactive medium was aspirated, and the cells were washed with acid buffer (PBS, 0.1 M, pH 7.5). After washing, the plate was dried, and 0.1 mL of 2 N NaOH was added to the wells to facilitate cell lysis. The lysates were collected and counted in a gamma counter. The experiment was repeated five times.
Tumor model and in vivo gamma imaging. All animal experiments were performed in compliance with the policies and procedures of the Institutional Animal Care and Use Committee for animal treatment of Chonbuk National University (Jeonbuk, South Korea). Female athymic, nude mice (4 weeks, nu/nu) were obtained from Orient‐Bio (Seoul, Korea). Glioblastoma tumors were established by subcutaneous injection of 5 × 106 cells mixed with Matrigel into the right flank. The mice underwent single photon emission computed tomography (SPECT)/computed tomography (CT) studies when the tumor volume reached 250–350 mm3 (tumor volume eq. = length × wide2 × 0.5, 4–5 weeks after inoculation). Iodine‐125 gamma camera imaging and imaging processing were performed using a small animal imaging system with pin‐hole collimation (aperture diameter = 1 mm, focal length = 9 cm) and a 15–45 keV photopeak energy window (X‐SPECT/CT; GE Healthcare, Uppsala, Sweden). For the U87MG tumor model, mice were injected via the tail vein with approximately 14.8 MBq (400 μCi) of 125I‐cMBP‐click‐RGDyk under isoflurane anesthesia. For the in vivo inhibition studies, 125I‐cMBP‐click‐cRGDyk, free cRGDyk (0.16 μmol), and cMBP‐GGG‐SC (0.16 μmol) were co‐injected into a U87MG tumor model via a lateral tail vein (n = 2). Potassium iodide was not administered. Five‐minute pin‐hole planar images were acquired at 30 min, 1, 2, and 4 h after injection. At 1 h, SPECT scans were performed at 32 projections over 360 degrees (radius of rotation = 7.6 cm, 60 s/projection). Reconstructed data from SPECT and CT were visualized and co‐registered using AMIRA 3.1 (GE Healthcare).
Biodistribution study. Biodistribution studies were carried out on U87MG xenografted female athymic, nude mice. Tumors were allowed to grow for 4–5 weeks after inoculation, at which time the animals received 0.01 mCi of 125I‐cMBP‐click‐cRGDyk (≒0.5 pmol) in 100 μL PBS via lateral tail vein injection. The animals were then euthanized at specified time points (30 min, 1 h, 4 h, n = 3). After the mice were killed, selected tissues and organs of interest were removed and weighed, and their activity measured in a gamma counter. The percentage of the injected dose per organ weight (%ID/g) was then calculated.
Statistical analysis. Data are expressed as mean ± SD. Paired t‐test (SPSS12.0; SPSS, Chicago, IL, USA) was used to determine statistical significance at the 95% confidence level, with P < 0.05 considered statistically significant.
Results
Radiolabeling. The structure of the cMBP‐click‐cRGDyk target peptide is shown in Figure 1. The radiochemical purity of the 125I‐labeled peptide conjugates were 90–95% at 24 h post‐labeling. Radiochemical purity following incubation in human serum was over 90% at 4 h. The logP value of the 125I‐cMBP‐click‐cRGDyk was −2.40 ± 0.05, and 125I‐cMBP‐GGG‐SC was −2.75 ± 0.05.
Figure 1.
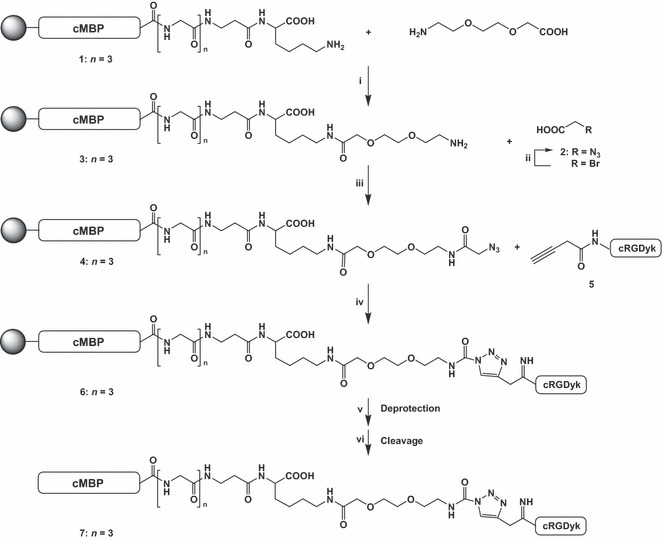
Synthesis of mesenchymal–epithelial transition factor‐binding peptide‐click‐cRGDyk (cMBP‐click‐cRGDyk) heterodimer. Reagents and conditions: (i) 2‐(2‐[2‐Fmoc aminoethoxy]ethoxy)acetic acid, DIC, HOBT, DMF, rt, 2 h; (ii) NaN3, water, 4°C, 16 h; (iii) DIC, HOBT, DMF, rt, 2 h; (iv) Cu(I)Br, DIPEA, DMF, propiolic cRGDyk, rt, 4 h; (v) 20% piperidine in DMF, rt, 10 min (Fmoc deprotection); (vi) triisopropylsilane, thioanisol, water, ethanedithiol, trifluoroacetic acid (2.5/2.5/2.5/2.5/90), rt, 3 h (cleavage). DIC, N,N′‐diisopropylcarbodiimide; DIPEA, N,N‐diisopropylethylamine; DMF, N,N‐dimethylformamide; HOBT, hydroxybenzotriazole.
Competitive binding assay and cell inhibition study. The binding affinities of cMBP‐click‐cRGDyk and cMBP‐GGG‐SC for c‐Met and integrin were evaluated for U87MG cells (Fig. 2). The results of the cell‐binding assays were plotted in sigmoid curves for the displacement of 125I‐cMBP and 125I‐echistatin from U87MG cells as a function of increasing the concentration of the cMBP‐click‐cRGDyk analogs. The IC50 values for integrin affinity were determined to be 2.38 μM for cRGDyk and 3.42 μM for cMBP‐click‐cRGDyk on U87MG cells. A cRGDyk and cMBP‐click‐cRGDyk shows similar binding affinities for integrin. The IC50 values for the c‐Met affinity of cMBP GGG‐SC and cMBP‐click‐cRGDyk were 1.53 and 3.84 μM, respectively (n = 2). The comparable IC50 values from these two sets of experiments suggest that the cMBP‐click‐cRGDyk peptide possesses comparable c‐Met and integrin αvβ3 receptor‐binding affinities as the corresponding monomer. A blocking study with 125I‐cMBP‐click‐cRGDyk in U87MG cells showed that 125I‐cMBP‐click‐cRGDyk binding was significantly inhibited by cMBP‐GGG and cRGDyk (Fig. 3).
Figure 2.
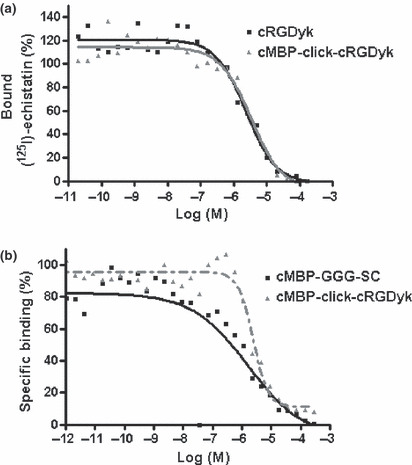
Inhibition of iodine 125 (125I)‐echistatin binding to integrin αvβ3 on U87MG cells by cRGDyk and mesenchymal–epithelial transition factor (c‐Met)‐binding peptide (cMBP)‐click cRGDyk (a). Mesenchymal–epithelial transition factor receptor tyrosine kinase binding of cMBP 3 glycine (GGG)‐a single name of amino acids (SC) and cMBP‐click cRGDyk on U87MG cells (b).
Figure 3.
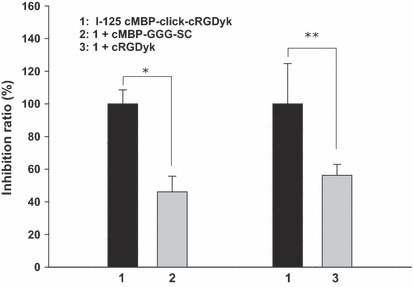
Inhibition study of iodine 125 (125I)‐mesenchymal–epithelial transition factor‐binding peptide (cMBP)‐click cRGDyk (0.18 MBq) with cMBP‐3 glycine (GGG)‐a single name of amino acids (SC) and cRGDyk in U87MG cells. All of the points were performed five times. Data are presented as the mean ± SD. *P = 0.000, **P = 0.012.
Biodistribution studies. The biodistribution of 125I‐cMBP‐click‐cRGDyk was determined in a U87MG xenograft model (Table 1). The tumor uptake was 5.34 ± 0.49%ID/g at 30 min, and decreased over time to 4.60 ± 2.08%ID/g at 1 h and 3.13 ± 0.27%ID/g at 4 h. Kidney accumulation peaked at 30 min (40.67 ± 4.67%ID/g), quickly cleared by 4 h (15.83 ± 1.56%ID/g), and exhibited a higher signal intensity than 125I‐cMBP‐GGG‐SC.( 14 ) The biodistribution of both 125I‐cMBP‐click‐cRGDyk and 125I‐cMBP‐GGG‐SC showed similar patterns at 30 min, but 125I‐cMBP‐click‐cRGDyk was eliminated sooner from the whole body by 4 h compared to 125I‐cMBP‐GGG‐SC (Fig. 4a,b). Tumor‐to‐blood (T/B), tumor‐to‐kidney (T/K), tumor‐to‐liver (T/L) and tumor‐to‐muscle (T/M) ratios of 125I‐cMBP‐click‐cRGDyk at 30 min post‐injection were 2.34, 0.13, 1.99, and 5.18, and at 4 h post‐injection, they were 10.07, 0.33, 6.75, and 11.12, respectively. However, the T/B, T/K, T/L, and T/M ratios of 125I‐cMBP‐GGG‐SC at 30 min post‐injection were 1.26, 0.19, 5.23, and 2.08, and at 4 h post‐injection, they were 2.23, 0.86, 2.36, and 1.71, respectively (Fig. 4c,d).( 14 )
Table 1.
Biodistribution of iodine 125‐mesenchymal–epithelial transition factor‐binding peptide‐click cRGDyk (125I‐cMBP‐click cRGDyk)
| Tissue | 125I‐cMBP‐click‐cRGDyk (%ID/g) | ||
|---|---|---|---|
| 30 min | 1 h | 4 h | |
| Blood | 2.28 ± 0.32 | 2.35 ± 0.63 | 0.53 ± 0.02 |
| Heart | 1.64 ± 0.16 | 1.75 ± 0.59 | 0.44 ± 0.02 |
| Lung | 2.80 ± 0.28 | 1.81 ± 0.28 | 0.64 ± 0.04 |
| Pancreas | 7.86 ± 0.70 | 16.07 ± 5.07 | 2.27 ± 0.40 |
| Intestine | 2.76 ± 0.50 | 2.97 ± 0.84 | 1.04 ± 0.12 |
| Stomach | 2.63 ± 0.89 | 9.77 ± 3.73 | 4.65 ± 0.56 |
| Liver | 2.68 ± 0.22 | 2.54 ± 0.71 | 0.79 ± 0.02 |
| Spleen | 2.65 ± 0.05 | 2.64 ± 0.76 | 1.09 ± 0.17 |
| Kidney | 40.67 ± 4.67 | 20.72 ± 7.71 | 15.83 ± 1.56 |
| Muscle | 1.03 ± 0.20 | 2.20 ± 0.55 | 0.48 ± 0.01 |
| Bone | 1.21 ± 0.37 | 1.63 ± 0.50 | 0.46 ± 0.03 |
| Tumor | 5.34 ± 0.49 | 4.60 ± 2.08 | 3.13 ± 0.27 |
Activity concentrations (mean ± SD, n = 3) at various times, post‐injection. %ID/g, percentage of the injected dose per organ weight.
Figure 4.
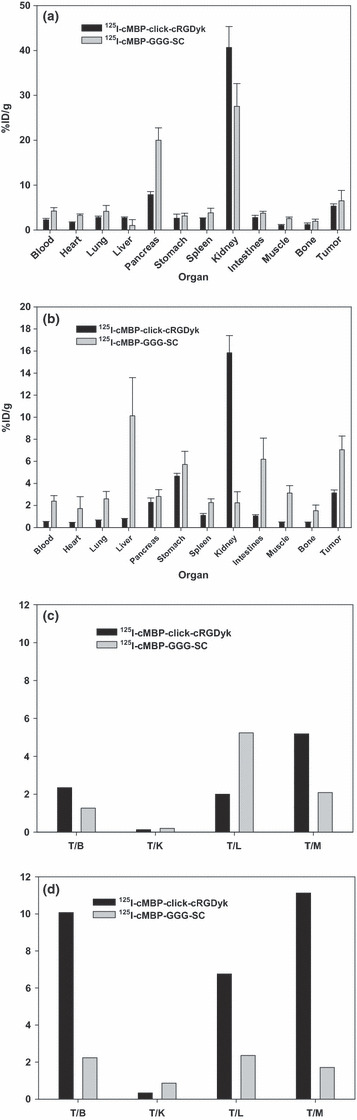
Comparison of tissue distribution at 30 min (a) and 4 h (b) after post‐injection between iodine 125 (125I)‐mesenchymal–epithelial transition factor‐binding peptide (cMBP)‐click cRGDyk and 125I‐cMBP 3 glycine (GGG)‐a single name of amino acids (SC) in athymic nude mice bearing U87MG tumors (mean ± SD). Comparison of tumor (T) with muscle, kidney, and liver ratios of 125I‐cMBP‐click‐cRGDyk and 125I‐cMBP‐GGG‐SC at 30 min (c) and 4 h (d) after injection (i.v.) for athymic nude mice bearing U87MG tumors. T/B, tumor to blood; T/K, tumor to kidney; T/L, tumor to liver; T/M, tumor to muscle. %ID/g, percentage of the injected dose per organ weight.
Single photon emission computed tomography/CT. Representative SPECT/CT images of 125I‐cMBP‐click‐cRGDyk are illustrated in Figure 4. The static pin‐hole image shows tumoral uptake and kidney and bladder activity at 1 h (Fig. 5a). Tumor uptake was clearly visualized by CT and CT fusion imaging (Fig. 5b,c). The planar image of 125I‐cMBP‐click‐cRGDyk at 4 h shows higher tumoral uptake than the 1 h image, and also shows thyroid uptake (Fig. 5d). For the in vivo inhibition studies, 125I‐cMBP‐click‐cRGDyk and free cRGDyk were co‐injected into the U87MG tumor mouse. As shown in Figure 5e, tumoral uptake was significantly decreased by free cRGDyk at 4 h post‐injection. The image‐based region of interest (ROI) ratios for both the tumor and non‐tumor regions in 125I‐cMBP‐click‐cRGDyk‐injected mice at 30 min and 4 h post‐injection were 24 ± 8 and 4 ± 3, and 30 ± 7 and 4 ± 2.9 (mean of count/pixel), respectively. The ROI ratios in inhibited mice at 30 min and 4 h post‐injection were 14 ± 5 and 12 ± 3, and 12 ± 4 and 6 ± 2 (mean of count/pixel), respectively. Moreover, whole body activity was quickly eliminated through the kidney, but accumulated in the bladder, from 10 min post‐injection due to the residual receptor presence of free cRGDyk in the whole body. Iodine‐125‐cMBP‐click‐cRGDyk and free cMBP‐GGG‐SC were co‐injected into the U87MG tumor mouse. The ROI ratios at 30 min and 4 h post‐injection were 46 ± 6.7 and 19 ± 5.9, and 23 ± 6.5 and 16 ± 3.8 (mean of count/pixel), respectively (Fig. 5f).
Figure 5.
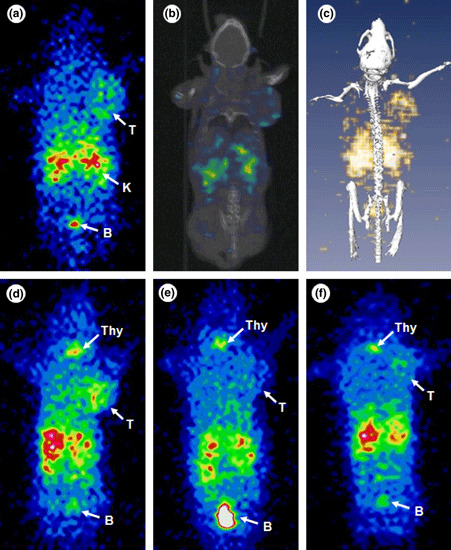
Pin‐hole planar and reconstructed co‐registered transverse single photon emission computed tomography (SPECT)/computed tomography (CT) images of iodine 125 (125I)‐mesenchymal–epithelial transition factor‐binding peptide (cMBP)‐click cRGDyk in an U87MG xenograft mouse at 1 h. Pinhole gamma camera image (a), reconstructed, co‐registered coronal slice image (b), and maximum‐intensity‐projection SPECT/CT fusion image (c) of 125I‐cMBP‐click‐cRGDyk. Planar image of 125I‐cMBP‐click‐cRGDyk in the U87MG xenograft mouse at 4 h (d). Inhibition image at 4 h post‐co‐injection of 125I‐cMBP‐click‐cRGDyk and free cRGDyk (48 nmol) in the U87MG mouse (e). Free cMBP 3 glycine‐a single name of amino acids (SC) (0.16 μmol) inhibition image at 4 h (f). B, bladder; K, kidney; Thy, thyroid, T, tumor.
Discussion
Since U87MG tumor cells express both c‐Met and integrin αvβ3, we explored whether a dual c‐Met‐ and integrin αvβ3‐targeting approach would allow us to develop improved imaging probes over 125I‐cMBP‐GGG‐SC that only recognizes one receptor type. In a previous study, cMBP was conjugated with two types of linkers, such as GGG and 8‐aminooctanoic acid (AOC). The tissue distribution of three different peptides, 125I‐cMBP, 125I‐cMBP‐GGG‐SC, and 125I‐cMBP‐AOC‐C, were determined in U87MG‐xenografted mice.( 14 )
As determined by a biodistribution study, 125I‐cMBP‐GGG‐SC exhibited the highest T/B at 4 h. However, static pin‐hole images of 125I‐cMBP‐GGG‐SC showed a relatively low tumor uptake and high body background activity at 1 and 4 h, and even higher pancreatic and renal activities at all time points. Therefore, a modification of cMBP‐GGG‐SC through heterodimerization of two ligands, one targeting c‐Met and the other targeting integrin, was needed to improve the targetability for an in vivo cancer model.
Peptide modifications to be used as imaging agents can be achieved by various methods. In order to improve binding affinity and receptor selectivity and to avoid the fast degradation of peptides in vivo, several researchers introduced cyclization methods.( 17 ) Heteromultimerization and homomultimerization of ligands with multitargeting properties have also been developed by many researchers.( 2 , 3 , 4 , 5 , 6 ) These approaches have successfully improved tumor‐targeting efficacy and pharmacokinetics compared to single analog methodologies. We therefore synthesized radiolabeled 125I‐cMBP‐click‐cRGDyk (Fig. 1). The linking groups in small peptide receptor‐targeted radiopharmaceutical designs have largely been viewed as merely convenient ways to adjust blood retention and overall clearance of radiopharmaceuticals, without making substantial alterations to the targeting vector. In this study, two different peptides were connected, forming a 1,2,3‐trizole ring between an alkyne and an azide terminal.
Our data from the receptor‐binding assays demonstrated that cMBP‐click‐cRGDyk is similar to cMBP‐GGG‐SC for c‐Met binding and to cRGDyk for integrin αvβ3 binding (Fig. 2). However, the binding affinity of cMBP‐click‐cRGDyk for both receptors was not improved. As shown in Table 1, 125I‐cMBP‐click‐cRGDyk tends to have a faster washout than 125I‐cMBP‐GGG‐SC, but logP values of the two radiolabeled compounds were similar (−2.40 ± 0.05 and −2.75 ± 0.05, respectively). In another previous heterodimer study, 18F‐FB‐BBN‐RGD tended to have a slower washout than 18F‐FB‐BBN, which might be the result of enhanced, effective binding due to dual targeting.( 7 ) In comparing our results, although 125I‐cMBP‐click‐cRGDyk and 125I‐cMBP‐GGG‐SC showed similar binding affinity and hydrophilicity, 125I‐cMBP‐click‐cRGDyk tended to have a faster clearance rate in vivo than 125I‐cMBP‐GGG‐SC.
The tumoral uptake decreased with time compared to 125I‐cMBP‐GGG‐SC (6.53 ± 2.29, 6.85 ± 1.89, and 7.05 ± 1.24 at 30 min, 2 and 4 h). Moreover, high kidney uptake appeared in three time points compared to 125I‐cMBP‐GGG‐SC (27.54 ± 5.11, 9.54 ± 1.91, and 6.18 ± 1.92 at 30 min, 2 and 4 h), slowly decreasing from 40.67 ± 4.67 to 15.83 ± 1.56. As shown Figure 4(c,d), the T/B, T/L, and T/M ratios of 125I‐cMBP‐click‐cRGDyk were higher than those of 125I‐cMBP‐GGG‐SC. While the initial tumor accumulation of 125I‐cMBP‐click‐cRGDyk was less than 125I‐cMBP‐GGG‐SC, the T/B, T/L, and T/M ratios increased with time due to the relatively low activities in major organs, such as the heart, lung, blood, liver, intestines, muscles, and spleen. Iodine‐125‐cMBP‐click‐cRGDyk demonstrated a quicker clearance rate than 125I‐cMBP‐GGG‐SC (blood activity: 4.22 ± 0.75, 2.77 ± 0.38 and 2.39 ± 0.50 at 30 min, 2 and 4 h) and lower background activity.
The imaging quality of 125I‐cMBP‐click‐cRGDyk was evaluated in a U87MG tumor xenografted model. The quality was also improved by 125I‐cMBP‐click‐cRGDyk compared to 125I‐cMBP‐GGG‐SC in the liver, heart, pancreas, and kidneys at 1 and 4 h (Fig. 5). However, thyroid uptake was observed in the 4 h post‐injection image (Fig. 5d), but this might be due to the deiodination of 125I‐cMBP‐click‐cRGDyk. In a previous study, the labeling stability of 125I‐cMBP‐GGG‐SC was stable, and cMBP‐GGG‐SC did not internalize into the U87MG cells; thyroid uptake did not appear in the in vivo study. However, as labeled radioiodine on the tyrosine ring of RGD peptides was found to be unstable in vivo,( 18 , 19 ) 125I‐cMBP‐click‐cRGDyk was thought to be internalized like 125I‐cMBP‐AOC‐C and then deiodinated. Taking this into consideration, a direct comparison between two compounds with radioiodine activity might be difficult to achieve, since there is the possibility of underestimating cMBP‐click‐cRGDyk binding with integrin due to deiodination from the heterodimer compound, compared to cMBP‐GGG‐SC for c‐Met only. When labeling efficiency and the stability of 125I‐cRGDyk was estimated, the in vitro labeling stability of 125I‐cRGDyk quickly decreases with time. Consequently, 125I‐cMBP‐click‐cRGDyk might demonstrate faster clearance than 125I‐cMBP‐GGG‐SC.
Moreover, although the tumor volumes used in this study were smaller than those of 125I‐cMBP‐GGG‐SC, they were well visualized. For the identification of integrin receptor targetability, free cRGDyk (48 nmol) and cMBP‐GGG‐SC were co‐injected, and images were acquired (Fig. 5e). These inhibition results suggest that 125I‐cMBP‐click‐cRGDyk was specifically taken up by integrin αvβ3 and the c‐Met receptor.
In conclusion, we successfully developed a heterodimeric peptide that binds to both the c‐Met receptor and integrin αvβ3 using click reaction methodology. Dual integrin and/or c‐Met receptor recognition showed slightly improved tumor‐targeting efficacy and imaging quality compared to 125I‐cMBP‐GGG‐SC single analogs, despite high renal clearance activity and decreasing tumoral uptake over time. However, additional experiments are necessary to improve clinical pharmacokinetics, such as decreased kidney uptake and prolonged tumor uptake.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgments
This study was supported grants from Chonbuk National University Hospital Research Institute of Clinical Medicine and the National R&D Program for Cancer Control, Ministry for Health, Welfare and Family Affairs, Republic of Korea (No. 0620220).
References
- 1. Mankoff DA, Link JM, Linden HM, Sundararajan L, Krohn KA. Tumor receptor imaging. J Nucl Med 2008; 49: 149S–63S. [DOI] [PubMed] [Google Scholar]
- 2. Liu Z, Yan Y, Chin FT, Wang F, Chen X. Dual integrin and gastrin‐releasing peptide receptor targeted tumor imaging using 18F‐labeled PEGylated RGD‐bombesin heterodimer 18F‐FB‐PEG3‐Glu‐RGD‐BBN. J Med Chem 2009; 52: 425–32. [DOI] [PubMed] [Google Scholar]
- 3. Mammen M, Choi S‐K, Whitesides GM. Polyvalent interactions in biological systems: implications for design and use of multivalent ligands and inhibitors. Angew Chem Int Ed Engl 1998; 37: 2754–94. [DOI] [PubMed] [Google Scholar]
- 4. Kiessling LL, Gestwicki JE, Strong LE. Synthetic multivalent ligands in the exploration of cell–surface interactions. Curr Opin Chem Biol 2000; 4: 696–703. [DOI] [PubMed] [Google Scholar]
- 5. Xu L, Vagner J, Josan J et al. Enhanced targeting with heterobivalent ligands. Mol Cancer Ther 2009; 8: 2356–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vagner J, Xu L, Handl HL et al. Heterobivalent ligands crosslink multiple cell‐surface receptors: the human melanocortin‐4 and deltaopioid receptors. Angew Chem Int Ed 2008; 47: 1685–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li ZB, Wu Z, Chen K, Ryu EK, Chen X. 18F‐Labeled BBN‐RGD heterodimer for prostate cancer imaging. J Nucl Med 2008; 49: 453–61. [DOI] [PubMed] [Google Scholar]
- 8. Hynes RO. Integrins: versatility, modulation, and signaling in cell adhesion. Cell 1992; 69: 11–25. [DOI] [PubMed] [Google Scholar]
- 9. Brooks PC, Clark RA, Cheresh DA. Requirement of vascular integrin alpha v beta 3 for angiogenesis. Science 1994; 64: 569–71. [DOI] [PubMed] [Google Scholar]
- 10. Chen X, Park R, Tohme M, Shahinian AH, Bading JR, Conti PS. MicroPET and autoradiographic imaging of breast cancer alpha v‐integrin expression using 18F‐ and 64Cu‐labeled RGD peptide. Bioconjug Chem 2004; 15: 41–9. [DOI] [PubMed] [Google Scholar]
- 11. Wu Z, Li ZB, Chen K et al. microPET of tumor integrin αvβ3 expression using 18F‐labeled PEGylated tetrameric RGD peptide (18F‐FPRGD4). J Nucl Med 2007; 48: 1536–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim EM, Joung MH, Lee CM et al. Synthesis of Tc‐99m labeled 1,2,3‐triazole‐4‐yl c‐met binding peptide as a potential c‐met receptor kinase positive tumor imaging agent. Bioorg Med Chem Lett 2010; 20: 4240–3. [DOI] [PubMed] [Google Scholar]
- 13. Kim EM, Park EH, Cheong SJ et al. In vivo imaging of mesenchymal‐epithelial transition factor (c‐Met) expression using an optical imaging system. Bioconjug Chem 2009; 20: 1299–306. [DOI] [PubMed] [Google Scholar]
- 14. Kim EM, Park EH, Cheong SJ et al. Characterization, biodistribution and small‐animal SPECT of I‐125‐labeled c‐Met binding peptide in mice bearing c‐Met receptor tyrosine kinase‐positive tumor xenografts. Nucl Med Biol 2009; 36: 371–8. [DOI] [PubMed] [Google Scholar]
- 15. Rostovtsev VV, Green LG, Fokin VV, Sharpless KBA. stepwise huisgen cycloaddition process: copper(I)‐catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl 2002; 41: 2596–9. [DOI] [PubMed] [Google Scholar]
- 16. Li ZB, Wu Z, Chen K, Chin FT, Chen X. Click chemistry for (18)F‐labeling of RGD peptides and microPET imaging of tumor integrin alphavbeta3 expression. Bioconjug Chem 2007; 18: 1987–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Miao Y, Gallazzi F, Guo H, Quinn TP. 111In‐labeled lactam bridge‐cyclized alpha‐melanocyte stimulating hormone peptide analogues for melanoma imaging. Bioconjug Chem 2008; 19: 539–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Haubner R, Wester HJ, Reuning U et al. Radiolabeled alpha(v)beta3 integrin antagonists: a new class of tracers for tumor targeting. J Nucl Med 1999; 40: 1061–71. [PubMed] [Google Scholar]
- 19. Chen X, Park R, Shahinian AH, Bading JR, Conti PS. Pharmacokinetics and tumor retention of 125I‐labeled RGD peptide are improved by PEGylation. Nucl Med Biol 2004; 31: 11–9. [DOI] [PubMed] [Google Scholar]


