Abstract
Translocation t(11;18)(q21;q21) is the most frequent chromosomal aberration reported in gastric mucosa‐associated lymphoid tissue (MALT) lymphomas. Intriguingly, this translocation has been reported only rarely in diffuse large B‐cell lymphomas; it has been proposed that t(11;18)‐positive tumors rarely progress to diffuse large B‐cell lymphomas. We examined the frequency of chromosomal translocation t(11;18)(q21;q21) in mucosa‐associated lymphoid tissue lymphoma and diffuse large B‐cell lymphoma of the stomach. Paraffin‐embedded tissues from patients with gastric B‐cell lymphomas were selected retrospectively. The presence of the t(11;18)(q21;q21) was determined using reverse transcriptase–polymerase chain reaction and/or fluorescence in situ hybridization. β‐Actin transcript was also determined to evaluate the integrity and efficiency of RNA (cDNA) recovery from paraffin‐embedded tissues. We analyzed 53 gastric B‐cell lymphomas (33 diffuse large B‐cell and 20 mucosa‐associated lymphoid tissue) obtained from Italy, the USA, or Japan. β‐Actin transcript was amplified in 50 cases (94%), including 19 mucosa‐associated lymphoid tissue and 31 diffuse large B‐cell lymphomas (five with mucosa‐associated lymphoid tissue components). The t(11;18) translocation was detected in 19% (6 of 31) cases with diffuse large B‐cell lymphoma versus 26% (five of 19) with mucosa‐associated lymphoid tissue lymphoma (P = 0.72). One of five diffuse large B‐cell lymphomas with a mucosa‐associated lymphoid tissue component showed the t(11;18)(q21;q21). In conclusion, translocation t(11;18)(q21;q21) was found in both mucosa‐associated lymphoid tissue lymphomas and diffuse large B‐cell lymphomas of the stomach at approximately equivalent frequencies; its presence does not exclude progression to diffuse large B‐cell lymphoma. (Cancer Sci 2009; 100: 881–887)
More than 90% of gastric mucosa‐associated lymphoid tissue (MALT) lymphomas are associated with long‐standing antigenic stimulation by Helicobacter pylori (H. pylori) infection.( 1 ) This bacterial infection not only triggers the acquisition of MALT in the gastric mucosa, a prerequisite for lymphoma development,( 2 ) but also plays a critical role in malignant transformation and subsequent clonal expansion of the transformed cells. MALT lymphomas typically remain localized for long periods but can transform into high‐grade MALT lymphomas,( 3 , 4 , 5 ) which, according to the World Health Organization classification system are termed diffuse large B‐cell lymphomas (DLBCL) with features of MALT lymphomas.( 6 ) However, DLBCLs can also arise de novo in the stomach, and the proportion that transform from MALT lymphomas remains unclear.
The t(11;18)(q21;q21) translocation has been identified as the most common cytogenetic abnormality of MALT lymphomas.( 7 ) Although several studies have reported this translocation as the sole aberration present in lymphomas of MALT type, it has rarely been reported in gastric DLBCLs,( 7 , 8 , 9 , 10 ) and little is known of the association of this transformation with disease progression. It is currently recommended that testing for translocation t(11;18)(q21;q21) apoptosis inhibitor 2 (API2)/mucosa associated lymphoid tissue lymphoma translocation 1 (MALT1) be done to assist in prognosis as its presence is thought to predict a lower response rate to anti‐H. pylori treatment.( 11 , 12 )
This study examined for t(11;18)(q21;q21) translocation in MALT lymphomas and DLBCLs of the stomach with the goal of better defining the relationship between the translocation and clinical‐pathological characteristics of gastric B‐cell lymphomas.
Materials and Methods
Case selection. Paraffin‐embedded tissue of patients diagnosed with primary gastric B‐cell lymphoma from three institutions (Michael E. DeBakey VA Medical Center, Houston, TX; University of Padova, Padova, Italy; and Shinshu University, Nagano, Japan) obtained between 1987 and 2005 were included. These biopsies and resected specimens were diagnosed histopathologically according to the World Health Organization classification of lymphoid neoplasms.( 6 ) The cases were classified as lymphoma of MALT when there were dense diffuse infiltrates of centrocyte‐like cells with monocytoid differentiation and typical lymphoepithelial lesions. DLBCL was diagnosed if compact clusters, confluent aggregates, or sheets of transformed large centroblast‐ or immunoblast‐like cells with B‐cell immunophenotype were found.( 6 )
Sections were stained with hematoxylin–eosin (HE) and a triple stain for H. pylori,( 13 ) and scored for H. pylori infection, the degree of inflammatory response, lymphoepithelial lesions, and MALT with or without lymphoma. Immunocytochemistry for CD3, CD10, CD20, and Ki‐67 was performed on all cases with a modified streptavidin‐biotin complex method.
Tumor size was defined as the maximum diameter of the tumor by either radiographic imaging or pathologic analysis. The size of tumors ranged from 0.5 to 5 cm. The depth of tumor invasion was determined by histologic examination of the resected specimens or by endosonography. Typical staging of the study population included physical examination, chest X‐ray and/or computed tomography (CT)‐scan, abdominal CT‐scan, and bone marrow aspirate and biopsy. A combination of clinical, radiological, and surgical techniques was used to stage patients according to Musshoff's modification of the Ann Arbor staging system.( 14 )
This study was carried out in accordance with the ethical guidelines of the participating centres and countries. The pathologic review and the genetic studies of archived tumor tissues were approved by the Institutional Review Board at Baylor College of Medicine, Houston, TX, USA.
RNA extraction from paraffin sections. Two HE‐stained guide slides and 10 unstained slides were prepared from each paraffin block. Tumor areas were identified within deparaffinized sections lightly counterstained with hematoxylin and microdissected into 1.5 mL polypropylene vials, using an HE‐stained adjacent section from the same block as a guide.( 15 ) RNA extraction was extracted with a High Pure RNA Paraffin kit (Roche Diagnostics, Mannheim, Germany).
To minimize contamination and false‐positive results, all RNA isolation procedures were performed under a UV‐sterilized laminar flow hood, in a room physically separated from that used to set up nucleic acid amplification reaction mixes and from the “post–polymerase chain reaction (PCR)” room. Filter tips, powder‐free gloves, new disposable blades, and separate set of pipettes were used.
Reverse transcriptase–PCR (RT‐PCR). cDNA was generated using the Superscript III first‐strand synthesis system (Invitrogen, Carlsbad, CA, USA) and random hexamers as primers using the protocol provided by the manufacturer. β‐Actin transcript was used as an internal control to confirm cDNA integrity and PCR suitability. Translocation t(11;18) presence was examined by three different seminested PCRs. The analyses were performed to evaluate the following breakpoints: 1446 of API2 gene (GenBank accession no. L49432) and the four breakpoints (541, 814, 1123, and 1150) within the MALT1 gene (accession no. AF130356). We used oligonucleotides previously reported by Inagaki et al.( 16 ) with some modifications. Inagaki et al.( 16 ) designed a multiplex one‐tube RT‐PCR (the first round) followed by three parallel multiplex nested PCRs (the second round). We converted total RNA to cDNA, and then t(11;18) (q21;q21) was detected by seminested PCRs as separate reactions for each breakpoint using cDNA as template for the first reaction and the DNA amplified from the first reaction for the second reaction. We also designed new primers to amplify small fragments, allowing analysis of highly degraded RNA available in paraffin‐embedded tissues (Table 1). PCRs were carried out in 20 µL volumes with 10× PCR buffer, 2.5 mM MgCl2, 200 µM dNTPs, 10 pM of each primer, 1.5 units of AmpliTaq Gold (Applied Biosystems, Foster City, CA, USA), and 2 µL of cDNA. External PCR conditions included initial denaturation at 95°C for 10 min followed by 42 cycles of 95°C for 30 s, 56°C (60°C for MALT1 541 and 58°C for MALT1 814) for 30 s, and 72°C for 30 s. Final extension was at 72°C for 7 min. For internal PCRs (26 cycles) annealing was at 60°C (56°C for β‐actin and 58°C for MALT1 1123/1150). PCR products were analyzed by electrophoresis on a 4% agarose gel. All samples were tested in at least two separate experiments.
Table 1.
Oligonucleotide sequences of primers
| Primer | Sequence (5′→3′) | Product size (bp) | Reference |
|---|---|---|---|
| β‐Actin | |||
| AC1 | GAGCAAGAGAGGCATCCT | Inagaki et al. (2001)( 16 ) | |
| A5 | AGCCACACGCAGCTCATT | 115 | Present study |
| A6 | GCTCATTGTAGAAGGTGT | 104 | Present study |
| API2 1446, MALT1 541 | |||
| PA4 | GAAATAAGGGAAGAGGAGAG | Inagaki et al. (2001)( 16 ) | |
| PM1 | CAGCCAAGACTGCCTTTGAC | 94 | Inagaki et al. (2001)( 16 ) |
| R1 | CCAAGACTGCCTTTGACTCT | 91 | Present study |
| API2 1446, MALT1 814 | |||
| PA4 | GAAATAAGGGAAGAGGAGAG | Inagaki et al. (2001)( 16 ) | |
| R2 | GCTTTTGGGAAGTTGGTTCA | 112 | Present study |
| R3 | GGATTCAGAGACGCCATCAA | 75 | Present study |
| API2 1446, MALT1 1123/1150 | |||
| PA3 | TTACTCAATGCAGAAGATGA | Inagaki et al. (2001)( 16 ) | |
| PM5 | AAAGGCTGGTCAGTTGTTTG | 125/98 | Inagaki et al. (2001)( 16 ) |
| PA4 | GAAATAAGGGAAGAGGAGAG | 107/80 | Inagaki et al. (2001)( 16 ) |
Various negative controls were included to confirm the specificity of the RT‐PCR assay. In the synthesis of cDNA we included a negative control without reverse transcriptase. A negative extraction control was also processed to detect contamination. Finally, in the PCR assay we included previous negative controls plus a PCR blank (PCR reagents without template cDNA).
Before screening unknown cases for t(11;18)(q21;q21), we validated the efficiency of our RT‐PCR assay on five paraffin‐embedded samples of gastric MALT lymphoma known to possess the API2–MALT1 fusion transcript (blocks were received from Dr Wotherspoon and Dr de Jong). Five t(11;18)(q21;q21)‐negative paraffin‐embedded specimens were also analyzed.
Fluorescence in situ hybridization (FISH). FISH was performed on paraffin sections of four DLBCL cases to validate the presence of t(11;18) translocation as detected by RT‐PCR. The locus specific dual‐color, dual‐fusion probe for t(11;18) was purchased from Abbott Molecular, Abbott Park, IL, USA. The probe set is comprised of SpectrumGreen and SpectrumOrange labeled API2 (11q21) and MALT1 (18q21), respectively. The slides were pretreated according to the previously published method.( 17 ) Briefly, the slides were incubated overnight at 70°C and then deparaffinized by serial soaks in Hemo‐D (Fisher) and absolute ethanol. After drying, the slides were soaked for 45 min in 1 M sodium thiocyanate (Sigma, St Louis, MO, USA) at 45°C, rinsed once with running distilled water, and transferred to a digestion buffer containing pronase 10 mg/mL phosphate buffered saline (Protease type XXIV; Sigma) at 45°C for 30 min. Then the slides were rinsed with distilled water and checked under the phase microscope for clearing of the cytoplasm. After the digestion, slides were rinsed twice in distilled water and then dried down through serial ethanol rinses, 70%, 80%, and 100% for 2 min each. Probe hybridization was performed as recommended by the manufacturer's protocol with some modifications. The images were captured on a Nikon E800 microscope using Quips Pathvysion (Applied Imaging, Santa Clara, CA, USA) imaging software. A total of 200 interphase cells were scored.
Statistical methods. Statistical evaluation of data was performed using Fisher's exact test, Student's t‐test, Mann–Whitney Rank Sum test or, when appropriate, the χ2‐test (both two‐tailed). A P‐value of less than 0.05 for each test was regarded as statistically significant.
Results
Patients. In total, 53 patients with gastric B‐cell lymphomas from three different institutions were analyzed. Three cases where we failed to amplify β‐actin transcript were excluded. The final study group comprised 50 subjects, 31 men (62%) and 19 women (38%) with a mean age of 62.7 years (range, 16–93). Table 2 presents the clinical profiles of the patients.
Table 2.
Clinical and histopathologic features of patients
| Country | No. of cases | Mean age, years (range) | Sex | Diseases | |||
|---|---|---|---|---|---|---|---|
| M | F | MALT | DLBCL + MALT | DLBCL | |||
| Italy | 30 | 62.4 (38–84) | 16 | 14 | 8 | 4 | 18 |
| USA | 12 | 65.8 (42–93) | 11 | 1 | 6 | 1 | 5 |
| Japan | 8 | 59.1 (16–75) | 4 | 4 | 5 | 0 | 3 |
| Total | 50 | 62.7 (16–93) | 31 | 19 | 19 | 5 | 26 |
DLBCL, diffuse large B‐cell lymphomas, MALT, mucosa‐associated lymphoid tissue.
Nineteen cases were classified as marginal zone B‐cell lymphoma of the MALT type and 31 cases as gastric DLBCL according to the World Health Organization classification.( 6 ) In five patients with gastric DLBCL, a simultaneous occurrence of marginal zone lymphoma was observed. All cases classified as MALT lymphomas showed no sheets of blasts, and immunophenotyping revealed the characteristic phenotype (positive for CD20, and negative for CD3 and CD10, all antibodies from Dako, Carpinteria, CA, USA). The number of Ki‐67‐positive cells ranged from 5 to 20%. DLBCLs on the other hand, showed sheets of blast cells with a cytologic spectrum ranging from centroblasts to immunoblasts or plasmablasts with high Ki‐67 indices (60–90%). When a sheet‐like pattern of large cells was associated with lymphoepithelial lesions characteristic of MALT lymphoma, DLBCL with MALT component was diagnosed.
API2–MALT1 detection. β‐Actin transcript (used as an internal control) was amplified by RT‐PCR in 50 of 53 cases (94.3%); thus, 19 MALT lymphomas, five of DLBCL with a MALT component, and 26 DLBCLs were considered suitable for API2–MALT1 fusion transcripts detection. All five cases known harboring the t(11;18) (internal positive controls) showed significant bands at expected locations (75 bp for three cases with breakpoint at 814; 80 bp for one case with breakpoint at 1150) while no band was observed in the negative controls.
The t(11;18)(q21;q21) translocation was identified in five of 19 (26.3%) with MALT lymphoma and in six of 31 (19.4%) with DLBCL of the stomach (P = 0.72), including one case with a MALT component (Fig. 1). The translocation breakpoint for MALT1 gene was at 814 bp in eight patients, at 1123 bp breakpoint in two patients, and at 1150 bp breakpoint in one patient. To validate t(11;18) translocation identified by RT‐PCR, we performed FISH using the API2–MALT1 dual color, dual fusion probe. All four DLBCL cases were positive for t(11;18). Two yellow signals were noted in three cases, indicating the fusion of API2–MALT1 and loss of normal chromosome 11 and 18 (Fig. 2a,b). In case USA‐5443, only one yellow signal was displayed in more than 4% of cells examined indicating the loss of one of the derivative chromosomes, der(11) or der(18) (Fig. 2c). Histologic features from representative sections are also shown in Figure 3.
Figure 1.
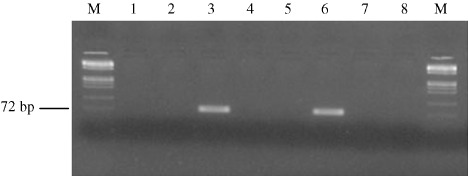
Detection of API2–mucosa‐associated lymphoid tissue‐1 (MALT1) fusion transcript. The figure shows representative photograph of seminested polymerase chain reaction (PCR) with primers PA4‐R3 for fusion transcript API2 1446–MALT1 814. M, DNA Marker IX (72–1353 bp; Roche Applied‐Science, Indianapolis, IN, USA); lanes 1, 2, 4 and 5, cases that did not show any amplified fragments; lane 3, case of diffuse large B‐cell lymphoma showing a 75‐bp band in the PCR, indicating that API2 1446–MALT1 814 type fusion is present (case USA‐2170 from USA); lane 6, positive control for fusion transcript API2 1446–MALT1 814; lane 7, negative control for RNA extraction; lane 8, negative control for PCR reaction.
Figure 2.
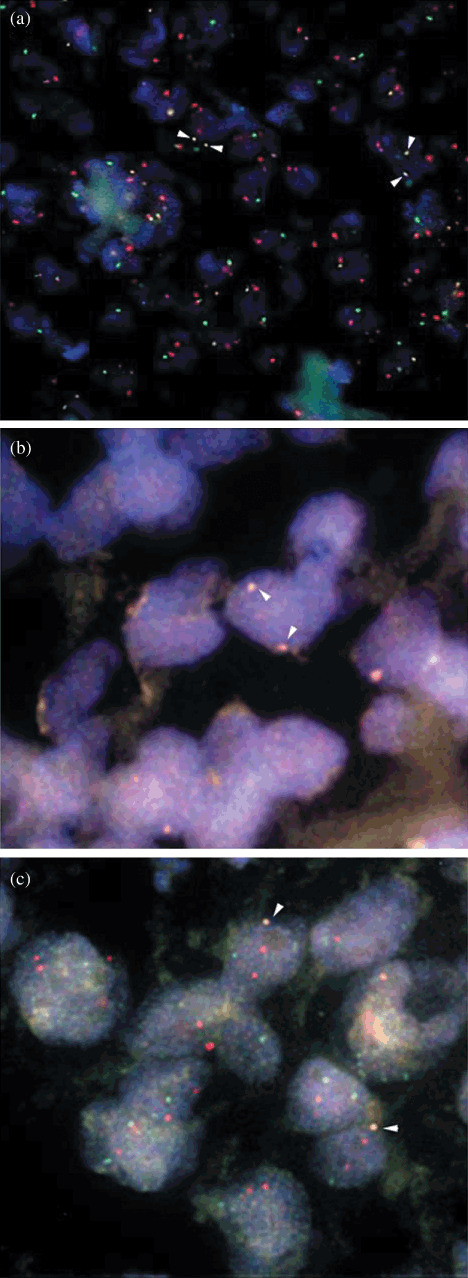
Fluorescence in situ hybridization (FISH) patterns in representative cases of t(11;18)(q21;q21) polymerase chain reaction (PCR)‐positive diffuse large B‐cell lymphomas (DLBCL). Arrows indicate lymphoma cells showing fusion signals with the apoptosis inhibitor 2 (API2)/mucosa associated lymphoid tissue lymphoma translocation 1 (MALT1) probes. (a,b) One normal API2 green signal, one normal MALT1 orange signal, and two yellow fusion signals (one from each of the derivative chromosomes) are shown. (c) Only one yellow signal was displayed indicating the loss of one of the derivative chromosomes, der(11) or der(18).
Figure 3.
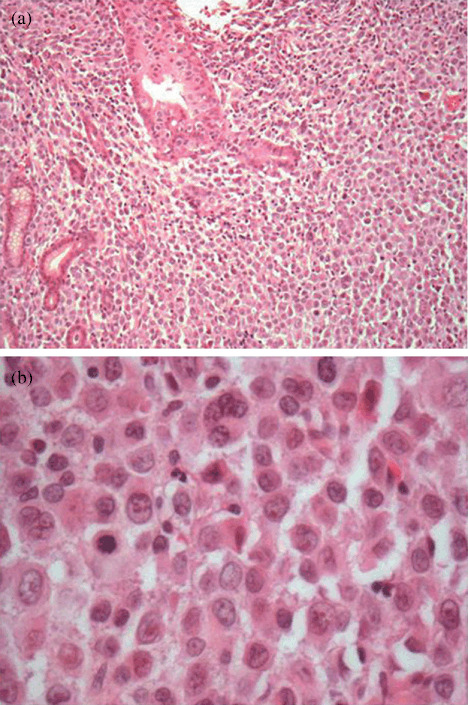
The designation gastric diffuse large B‐cell lymphoma (DLBCL) is given to lymphomas with > 80% blasts (b), hematoxylin–eosin, original magnification ×60. Figure (a) shows focal lymphoepithelial lesions (LELs) (a), hematoxylin–eosin, original magnification ×10.
Interestingly, the t(11;18) presence was only identified in patients older than 60 years (35.5%) compared to none (0%) of patients younger than 60 years (P = 0.003). As shown in Table 3, patient age also differed between t(11;18)‐positive and ‐negative DLBCL patients (P = 0.053). Other factors, such as sex, geographic factors, H. pylori status, and clinical stage showed no differences with regard to t(11;18)(q21;q21) status (Table 3). Complete remissions after H. pylori eradication were frequent among cases without MALT lymphoma and t(11;18)‐negative (Table 3). Complete remission was defined as complete disappearance of clinical and histologic evidence of lymphoma. We found no relationship between tumor size and presence of chromosomal alteration (data not shown).
Table 3.
The relevance of API2–MALT1 positivity to the clinicopathologic and geographical factors in 50 gastric B‐cell lymphomas
| Factor | MALT n = 19 | P‐values | DLBCL + MALT n = 5 | P‐values | DLBCL n = 26 | P‐values | Total cases n = 50 | P‐values | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t(11;18)+ | t(11;18)− | t(11;18)+ | t(11;18)− | t(11;18)+ | t(11;18)− | t(11;18)+ | t(11;18)− | |||||
| Age (years) | ||||||||||||
| ≤60 | 0 (0%) | 5 (100%) | 0.256 | 0 (0%) | 3 (100%) | 0.400 | 0 (0%) | 11 (100%) | 0.053 | 0 (0%) | 19 (100%) | 0.003 |
| >60 | 5 (35.7%) | 9 (64.3%) | 1 (50%) | 1 (50%) | 5 (33.3%) | 10 (66.7%) | 11 (35.5%) | 20 (64.5%) | ||||
| Sex | ||||||||||||
| Female | 3 (33.3%) | 6 (66.7%) | 0.628 | 0 (0%) | 3 (100%) | 0.400 | 2 (28.6%) | 5 (71.4%) | 0.588 | 5 (26.3%) | 14 (73.7%) | 0.727 |
| Male | 2 (20%) | 8 (80%) | 1 (50%) | 1 (50%) | 3 (15.8%) | 16 (84.2%) | 6 (19.4%) | 25 (80.6%) | ||||
| Geographical | ||||||||||||
| Italy | 2 (25%) | 6 (75%) | 0.62 | 0 (0%) | 4 (100%) | NA | 4 (22.2%) | 14 (77.8%) | 0.66 | 6 (20%) | 24 (80%) | 0.912 |
| USA | 1 (16.7%) | 5 (83.3%) | 1 (100%) | 0 (0%) | 1 (20%) | 4 (80%) | 3 (25%) | 9 (75%) | ||||
| Japan | 2 (40%) | 3 (60%) | 0 (0%) | 0 (0%) | 0 (0%) | 3 (100%) | 2 (25%) | 6 (75%) | ||||
| Depth of invasion † | ||||||||||||
| Mucosa/submucosa | 4 (26.6%) | 11 (73.3%) | 1.000 | 1 (50%) | 1 (50%) | NA | 0 (0%) | 5 (100%) | 0.544 | 5 (22.7%) | 17 (77.2%) | 1.000 |
| Muscularis propria | 0 (0%) | 1 (100%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 1 (100%) | ||||
| Serosa | 0 (0%) | 2 (100%) | 0 (0%) | 2 (100%) | 1 (16.7%) | 5 (83.3%) | 1 (10%) | 9 (90%) | ||||
| Adjacent organes | 1 (100%) | 0 (0%) | 0 (0%) | 1 (100%) | 3 (23.1%) | 10 (76.9%) | 4 (26.7%) | 11 (73.3%) | ||||
| H. pylori ‡ | ||||||||||||
| Positive | 4 (23.5%) | 13 (76.5%) | 1.000 | 1 (20%) | 4 (80%) | NA | 4 (21%) | 15 (79%) | 1.000 | 9 (22%) | 32 (78%) | 1.000 |
| Negative | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 2 (100%) | 0 (0%) | 1 (100%) | ||||
| Response to H. pylori eradication § | ||||||||||||
| Complete remission | 0 (0%) | 10 (100%) | NA | 0 (0%) | 0 (0%) | NA | 0 (0%) | 0 (0%) | NA | 0 (0%) | 10 (100%) | NA |
| Partial remission | 2 (66.7%) | 1 (33.3%) | 0 (0%) | 1 (100%) | 0 (0%) | 1 (100%) | 2 (40%) | 3 (60%) | ||||
| No change | 2 (50%) | 2 (50%) | 1 (25%) | 3 (75%) | 4 (23.5%) | 13 (76.5%) | 7 (28%) | 18 (72%) | ||||
| Stage ¶ | ||||||||||||
| IE1 | 4 (33.3%) | 8 (66.7%) | NA | 1 (100%) | 0 (0%) | NA | 0 (0%) | 4 (100%) | NA | 5 (29.4%) | 12 (70.6%) | NA |
| IE2 | 0 (0%) | 1 (100%) | 0 (0%) | 1 (100%) | 2 (33.3%) | 4 (66.7%) | 2 (25%) | 6 (75%) | ||||
| IIE1 | 0 (0%) | 3 (100%) | 0 (0%) | 1 (100%) | 0 (0%) | 1 (100%) | 0 (0%) | 5 (100%) | ||||
| IIE2 | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | 0 (0%) | ||||
| IIIE | 0 (0%) | 2 (100%) | 0 (0%) | 1 (100%) | 2 (18.2%) | 9 (81.8%) | 2 (14.3%) | 12 (85.7%) | ||||
| IVE | 1 (100%) | 0 (0%) | 0 (0%) | 1 (100%) | 1 (33.3%) | 2 (66.7%) | 2 (40%) | 3 (60%) | ||||
Depth of invasion was unknown in 2 cases.
H. pylori infection was determined in 42 cases only.
The tumor response to H. pylori eradication was evaluated in H. pylori‐positive cases treated with antibiotics.
The stage of disease was available in 49 patients. API2, apoptosis inhibitor 2; DLBCL, diffuse large B‐cell lymphomas, H. pylori, Helicobacter pylori; MALT, mucosa‐associated lymphoid tissue; NA, not applicable.
For eight patients we also have available clinical follow‐up data including initial treatment regimens (H. pylori eradication, chemotherapy, and/or radiation), length of remission, time to recurrence, therapeutic regimens used for recurrence, and date of death, if any. Eradication of H. pylori was performed in three patients with MALT lymphoma who also received resection therapy. A combined therapy of resection and chemotherapy was performed on five patients (four with DLBCL and one with MALT). During the follow‐up period of 24 months, relapse of lymphoma occurred in none of four (0%) patients with MALT lymphoma and in one of four (25%) patients with DLBCL lymphoma. No significant difference was found between patients with and without translocation.
Discussion
This study showed t(11;18)(q21;q21) translocation was detectable in both MALT lymphomas and DLBCLs of the stomach. The prevalence was highest in patients over the age of 60. Translocation t(11;18)(q21;q21) was found in 26% of gastric MALT lymphoma which is in agreement with other large series studies examining the translocation in gastric MALT lymphomas using various methods including FISH, RT‐PCR, or cytogenetics.( 7 , 12 , 18 , 19 , 20 , 21 , 22 , 23 ) Our results differ from prior studies where the translocation was absent or rare in DLBCLs with/without a MALT component as we detected the translocation in approximately the same proportion of gastric MALT lymphomas and DLBCLs (26% and 19%, respectively). Other studies also reported finding API2–MALT1 fusion transcripts in one of 46 DLBCL of the stomach,( 24 ) one of 13 colorectum,( 25 ) and two of 14 in the ocular adnexa.( 26 ) The t(11;18)(q21;q21) translocation has also been described in transformed diffuse gastric large B‐cell lymphoma of MALT (two of three,( 10 ) one of 22,( 27 ) and two of two( 28 )). In the present study, we examined 31 cases of DLBCL and six cases (19%) were t(11;18)‐positive. This suggests that the frequency and importance of the translocation has been underestimated in the past. Interestingly, the API2–MALT1 fusion transcript was also found in one of the five DLBCLs with MALT component. Based on our findings, an alternative explanation for the presence of the t(11;18) in DLBCLs could be that the origin of these DLBCLs are MALT lymphomas harboring the translocation. This hypothesis is also supported by the recent observation that translocation or rearrangements of the MALT1 gene can be found not only in gastric MALT lymphomas (26%) but also in transformed MALT lymphomas (13%) and in de novo DLBCLs (20%).( 29 ) Our data suggest that some DLBCLs may develop by clonal evolution from a pre‐exiting MALT lymphoma. A clonal link between small‐ and large‐cell components in gastric DLBCL was already shown by immunohistochemical studies of Ig light chain restriction and sequence analysis of the rearranged IgH.( 4 , 30 ) Recently, de Boer et al. reported that the transformation of non‐gastric MALT lymphoma to DLBCL was present in 6% patients at presentation and in 7% patients during follow‐up with a median time to transformation of 66.5 (range 14–146) months.( 31 ) Similar findings were also observed for the gastric series (7/115, 6%).( 32 ) The exact mechanisms underlying the transition of MALT lymphoma to an aggressive lymphoma including DLBCL remain speculative. Our results, together with previous findings, support the notion that the oncogenic role of t(11;18)(q21;q21) is related to inhibition of apoptosis and is mediated by deregulated nuclear factor‐kappa B (NF‐κB) signaling pathways.( 33 ) We also hypothesize that the presence of the t(11;18)(q21;q21) in cells of MALT may facilitate the emergence of other molecular events such as p53 alterations and dysregulation of other oncogenes,( 34 ) c‐myc activation,( 35 ) or epigenetic changes,( 36 ) that result in the progression to DLBCL. Prospective follow‐up of individuals with MALT lymphomas will be required to resolve whether the time to progression from MALT to DLBCLs differs depending on this primary chromosomal event. Experiments along the current lines may ultimately lead to the identification of reliable predictive factors that allow patients at risk for development of DLBCLs to be identified early.
Our data are also consistent with geographic variations in the occurrence of a given chromosome abnormality that have been described in other hematological malignancies.( 37 ) For example, in Japanese cases we found the t(11;18) translocation only among those with MALT lymphomas (i.e. the prevalence was 40%) where in the USA and Italian cases it was present in both cases of MALT lymphoma and DLBCL. Larger studies will be required to confirm or refute our findings in relation to geographic site. It is also tempting to speculate whether the difference is related to chance or to differences in the typical H. pylori strain in circulation which are know to vary markedly between East‐Asia and the USA.( 38 ) To address this question, future studies will need to control for country, age, and infecting strain.
A possible explanation for different results and conclusions between studies include studies of different populations, classification bias,( 39 ) and differences in methodology. Several authors have tested for the t(11;18)(q21;q21) in gastric DLBCL. Indeed, most of the studies performed on this topic analyzed the role of the chromosomal aberration only in few cases (n < 10),( 23 , 40 , 41 , 42 ) or using less sensitive techniques.( 7 , 43 ) A recent large study by Nakamura et al.( 44 ) detected by FISH analysis the t(11;18) or MALT1‐involved translocation in none of 105 cases of gastric DLBCL, except for one case with a MALT component. The failure to detect t(11;18)(q21;q21) in these cases can be related to the different patient populations studied or to the use of FISH with low sensitivity compared to molecular assays, or both. For example, PCR is known to be a useful method for detecting small copy numbers of DNA. Our study design was geared to achieve maximal sensitivity from archival samples; some specimens had been stored for more than 15 years. The RT‐PCR assay was designed with the knowledge that the total RNA extracted from formalin‐fixed, paraffin‐embedded tissue is generally significantly degraded. We performed cDNA synthesis with random hexamers and then nested PCRs to amplify very small fragments (<130 bp). This accuracy of the method was confirmed by FISH and confirmed the presence of the translocation in lymphoma tissue. To date, few methods have been reported for the identification of API–MALT1 fusion transcript by RT‐PCR in archival specimens. Inagaki et al.( 16 ) designed performed multiplex one‐tube RT‐PCR (the first round) followed by multiplex nested PCRs (the second round), reporting a β‐actin transcript detection rate of 88%. In this study, the API2–MALT1 fusion was detected only in one of eight MALT lymphomas and in none of the 14 high‐grade lymphomas. This low incidence of the t(11;18) is probably due to the sensitivity of the method and/or to the small number of cases. Streubel et al.( 45 ) obtained a 100% detection rate using same method with one difference: first‐round RT‐PCR products were amplified in a second round separately and not as multiplex‐nested PCRs. The t(11;18)(q21;q21) was found in six of nine gastric MALT lymphoma in patients with Sjögren's syndrome; unfortunately no case of DLBCL was analyzed. To detect mRNA transcripts Liu et al.( 12 ) performed a ‘hot‐start touch‐down’ PCR method using cDNA synthesized with oligo(dT) primer. The glucose‐6‐phosphate dehydrogenase gene was used as internal control and was amplified in all cases. The series included only MALT lymphomas and storage time was not given. That RT‐PCR methodology missed 7% of the total API2–MALT1 fusions in paraffin‐embedded tissues which may therefore underestimate the true frequency of t(11;18)(q21;q21). We used samples collected from 1987 to 2005 and used β‐actin transcript detection to identify suitable samples. The detection rate was 94.3% which is high for decades‐old archival specimens and proved suitable for identification of API–MALT1 fusion transcript in both analyzed gastric MALT lymphomas and DLBCL. These data were then validated by the FISH analysis done on isolated nuclei from a subset of DLBCL cases.
The presence of the translocation in a higher proportion of patients over age 60 awaits confirmation. However, the finding is consistent with the notion that the disease progresses and transforms over time with continued antigenic stimulation. Given that the translocation predicts a worse outcome with poor response to H. pylori treatment,( 12 ) it seems unlikely that the lymphoma would lose the translocation once transformed. Our findings suggest that translocation t(11;18) occurs frequently and may be involved in the tumorigenesis of MALT lymphomas as well as the progression of at least a subset of tumors. Overall, our results are consistent with the opinions recommending screening for t(11;18)(q21;q21) in gastric B‐cell lymphomas.
Acknowledgments
This material is based upon work supported in part by the Office of Research and Development Medical Research Service Department of Veterans Affairs and by Public Health Service grant DK56338 which funds the Texas Medical Center Digestive Disease Center. Dr. Sonia Toracchio was supported by a fellowship from Fondazione Italiana per la Ricerca sul Cancro (FIRC), Milan, Italy. We thank Michael Wu for the preparation of histological sections and the Texas Children's Cancer Center Cytogenetic Core Laboratory, Houston, TX, USA, for FISH analysis.
References
- 1. Parsonnet J, Hansen S, Rodriguez L et al . Helicobacter pylori infection and gastric lymphoma. N Engl J Med 1994; 330: 1267–71. [DOI] [PubMed] [Google Scholar]
- 2. Wotherspoon AC, Ortiz Hidalgo C, Falzon MR, Isaacson PG. Helicobacter pylori‐associated gastritis and primary B‐cell gastric lymphoma. Lancet 1991; 338: 1175–6. [DOI] [PubMed] [Google Scholar]
- 3. Carlson SJ, Yokoo H, Vanagunas A. Progression of gastritis to monoclonal B‐cell lymphoma with resolution and recurrence following eradication of Helicobacter pylori . JAMA 1996; 275: 937–9. [PubMed] [Google Scholar]
- 4. Chan JK, Ng CS, Isaacson PG. Relationship between high‐grade lymphoma and low‐grade B‐cell mucosa‐associated lymphoid tissue lymphoma (MALToma) of the stomach. Am J Pathol 1990; 136: 1153–64. [PMC free article] [PubMed] [Google Scholar]
- 5. Murayama H, Kikuchi M, Eimoto T, Doki T, Doki K. Early lymphoma coexisting with reactive lymphoid hyperplasia of the stomach. Acta Pathol Jpn 1984; 34: 679–86. [DOI] [PubMed] [Google Scholar]
- 6. Isaacson PG, Müller‐Hermelink HK, Piris MA et al . Extranodal marginal zone B‐cell lymphoma of mucosa‐associated lymphoid tissue (MALT lymphoma). In: Jaffe ES, Harris NL, Stein H, Vardiman JW, eds. World Health Organization Classification of tumours. Pathology, Genetics of Tumours of Haematopoietic, Lymphoid Tissues. Lyon: IARC Press, 2001: 157–60. [Google Scholar]
- 7. Ott G, Katzenberger T, Greiner A et al . The t(11;18) (q21;q21) chromosome translocation is a frequent and specific aberration in low‐grade but not high‐grade malignant non‐Hodgkin's lymphomas of the mucosa‐associated lymphoid tissue (MALT–) type. Cancer Res 1997; 57: 3944–8. [PubMed] [Google Scholar]
- 8. Akagi T, Motegi M, Tamura A et al . A novel gene, MALT1 at 18q21, is involved in t(11;18) (q21;q21) found in low‐grade B‐cell lymphoma of mucosa‐associated lymphoid tissue. Oncogene 1999; 18: 5785–94. [DOI] [PubMed] [Google Scholar]
- 9. Starostik P, Patzner J, Greiner A et al . Gastric marginal zone B‐cell lymphomas of MALT type develop along 2 distinct pathogenetic pathways. Blood 2002; 99: 3–9. [DOI] [PubMed] [Google Scholar]
- 10. Chuang SS, Lee C, Hamoudi RA et al . High frequency of t(11;18) in gastric mucosa‐associated lymphoid tissue lymphomas in Taiwan, including one patient with high‐grade transformation. Br J Haematol 2003; 120: 97–100. [DOI] [PubMed] [Google Scholar]
- 11. Bertoni F, Zucca E. State‐of‐the‐art therapeutics: marginal‐zone lymphoma. J Clin Oncol 2005; 23: 6415–20. [DOI] [PubMed] [Google Scholar]
- 12. Liu H, Ye H, Ruskone‐Fourmestraux A et al . T(11;18) is a marker for all stage gastric MALT lymphomas that will not respond to H. pylori eradication. Gastroenterology 2002; 122: 1286–94. [DOI] [PubMed] [Google Scholar]
- 13. El‐Zimaity HM, Ota H, Scott S, Killen DE, Graham DY. A new triple stain for Helicobacter pylori suitable for the autostainer: carbol fuchsin/Alcian blue/hematoxylin‐eosin. Arch Pathol Lab Med 1998; 122: 732–6. [PubMed] [Google Scholar]
- 14. Musshoff K. Klinische Stadieneinteilung der Nicht‐Hodgkin‐Lymphome. [Classification of the clinical stages of non‐Hodgkin's lymphoma]. Strahlentherapie 1977; 153: 218–21. [PubMed] [Google Scholar]
- 15. Toracchio S, El‐Zimaity HMT, Urmacher C, Katz S, Graham DY. Mycobacterium avium subspecies paratuberculosis and Crohn's disease granulomas. Scand J Gastroenterol 2008; 43: 1108–11. [DOI] [PubMed] [Google Scholar]
- 16. Inagaki H, Okabe M, Seto M, Nakamura S, Ueda R, Eimoto T. API2‐MALT1 fusion transcripts involved in mucosa‐associated lymphoid tissue lymphoma: multiplex RT‐PCR detection using formalin‐fixed paraffin‐embedded specimens. Am J Pathol 2001; 158: 699–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jazaeri AA, Lu K, Schmandt R et al . Molecular determinants of tumor differentiation in papillary serous ovarian carcinoma. Mol Carcinog 2003; 36: 53–9. [DOI] [PubMed] [Google Scholar]
- 18. Streubel B, Simonitsch‐Klupp I, Müllauer L et al . Variable frequencies of MALT lymphoma‐associated genetic aberrations in MALT lymphomas of different sites. Leukemia 2004; 18: 1722–6. [DOI] [PubMed] [Google Scholar]
- 19. Dierlamm J, Baens M, Stefanova‐Ouzounova M et al . Detection of t(11;18) (q21;q21) by interphase fluorescence in situ hybridization using API2 and MLT specific probes. Blood 2000; 96: 2215–8. [PubMed] [Google Scholar]
- 20. Remstein ED, James CD, Kurtin PJ. Incidence and subtype specificity of API2‐MALT1 fusion translocations in extranodal, nodal, and splenic marginal zone lymphomas. Am J Pathol 2000; 156: 1183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Yonezumi M, Suzuki R, Suzuki H et al . Detection of AP12‐MALT1 chimaeric gene in extranodal and nodal marginal zone B‐cell lymphoma by reverse transcription polymerase chain reaction (PCR) and genomic long and accurate PCR analyses. Br J Haematol 2001; 115: 588–94. [DOI] [PubMed] [Google Scholar]
- 22. Baens M, Maes B, Steyls A, Geboes K, Marynen P, De Wolf‐Peeters C. The product of the t(11;18), an API2‐MLT fusion, marks nearly half of gastric MALT type lymphomas without large cell proliferation. Am J Pathol 2000; 156: 1433–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nakamura S, Matsumoto T, Nakamura S et al . Chromosomal translocation t(11;18) (q21;q21) in gastrointestinal mucosa associated lymphoid tissue lymphoma. J Clin Pathol 2003; 56: 36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huang X, Zhang Z, Liu H et al . T(11;18) (q21;q21) in gastric MALT lymphoma and diffuse large B‐cell lymphoma of Chinese patients. Hematol J 2003; 4: 342–5. [DOI] [PubMed] [Google Scholar]
- 25. Sakugawa ST, Yoshino T, Nakamura S et al . API2‐MALT1 fusion gene in colorectal lymphoma. Mod Pathol 2003; 16: 1232–41. [DOI] [PubMed] [Google Scholar]
- 26. Takada S, Yoshino T, Taniwaki M et al . Involvement of the chromosomal translocation t(11;18) in some mucosa‐associated lymphoid tissue lymphomas and diffuse large B/cell lymphomas of the ocular adnexa evidence from multiplex reverse transcriptase‐polymerase chain reaction and fluorescence in situ hybridization on using formalin‐fixed, paraffin‐embedded specimens. Mod Pathol 2003; 16: 445–52. [DOI] [PubMed] [Google Scholar]
- 27. Kuo SH, Chen LT, Yeh KH et al . Nuclear expression of BCL10 or Nuclear Factor Kappa B predicts Helicobacter pylori‐independent status of early‐stage, high‐grade gastric mucosa‐associated lymphoid tissue lymphomas. J Clin Oncol 2004; 22: 3491–7. [DOI] [PubMed] [Google Scholar]
- 28. Tan SY, Ye H, Liu H et al . t(11;18) (q21;q21)‐positive transformed MALT lymphoma. Histopathology 2008; 52:777–80. [DOI] [PubMed] [Google Scholar]
- 29. Bernasconi B, Karamitopolou‐Diamantiis E, Tornillo L et al . Chromosomal instability in gastric mucosa‐associated lymphoid tissue lymphomas: a fluorescent in situ hybridization study using a tissue microarray approach. Hum Pathol 2008; 39: 536–42. [DOI] [PubMed] [Google Scholar]
- 30. Peng HM, Diss TC, Isaacson PG, Pan L. Genetic evidence for a clonal link between low and high‐grade components in gastric MALT B‐cell lymphoma. Histopathology 1997; 30: 425–9. [DOI] [PubMed] [Google Scholar]
- 31. de Boer JP, Hiddink RF, Raderer M et al . Dissemination patterns in non‐gastric MALT lymphoma. Haematologica 2008; 93: 201–6. [DOI] [PubMed] [Google Scholar]
- 32. Vrieling C, de Jong D, Boot H, de Boer JP, Wegman F, Aleman BM. Long‐term results of stomach‐conserving therapy in gastric MALT lymphoma. Radiother Oncol 2008; 87: 405–11. [DOI] [PubMed] [Google Scholar]
- 33. Jost PJ, Ruland J. Aberrant NF‐κB signaling in lymphoma: mechanisms, consequences, and therapeutic implications. Blood 2007; 109: 2700–7. [DOI] [PubMed] [Google Scholar]
- 34. Du M, Peng H, Singh N, Isaacson PG, Pan L. The accumulation of p53 abnormalities is associated with progression of mucosa‐associated lymphoid tissue lymphoma. Blood 1995; 86: 4587–93. [PubMed] [Google Scholar]
- 35. Peng H, Diss TC, Isaacson PG, Pan L. c‐myc gene abnormalities in mucosa‐associated lymphoid tissue (MALT) lymphomas. J Pathol 1997; 181: 381–6. [DOI] [PubMed] [Google Scholar]
- 36. Min KO, Seo EJ, Kwon HJ et al . Methylation of p16INK4A and p57KIP2 are involved in the development and progression of gastric MALT lymphomas. Mod Pathol 2006; 19: 141–8. [DOI] [PubMed] [Google Scholar]
- 37. Johansson B, Mertens F, Mitelman F. Geographic heterogeneity of neoplasia‐associated chromosome aberrations. Genes Chromosomes Cancer 1991; 3: 1–7. [DOI] [PubMed] [Google Scholar]
- 38. Falush D, Wirth T, Linz B et al . Traces of human migrations in Helicobacter pylori populations. Science 2003; 299: 1582–5. [DOI] [PubMed] [Google Scholar]
- 39. El‐Zimaity HM, Wotherspoon A, de Jong D. Interobserver variation in the histopathological assessment of malt/malt lymphoma: towards a consensus. Blood Cells Mol Dis 2005; 34: 6–16. [DOI] [PubMed] [Google Scholar]
- 40. Iwano M, Watanabe N, Matsushima Y et al . Rapid development of diffuse large B‐cell lymphoma after successful eradication of Helicobacter pylori for gastric MALT lymphoma. Am J Gastroenterol 2006; 101: 2878–83. [DOI] [PubMed] [Google Scholar]
- 41. Kalla J, Stilgenbauer S, Schaffner C et al . Heterogeneity of the API2‐MALT1 gene rearrangement in MALT‐type lymphoma. Leukemia 2000; 14: 1967–74. [DOI] [PubMed] [Google Scholar]
- 42. Motegi M, Yonezumi M, Suzuki H et al . API2‐MALT1 chimeric transcripts involved in mucosa‐associated lymphoid tissue type lymphoma predict heterogeneous products. Am J Pathol 2000; 156: 807–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rosenwald A, Ott G, Stilgenbauer S et al . Exclusive detection of the t (11;18) (q21;q21) in extranodal marginal zone B cell lymphomas (MZBL) of MALT type in contrast to other MZBL and extranodal large B cell lymphomas. Am J Pathol 1999; 155: 1817–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nakamura S, Ye H, Bacon CM et al . Translocations involving the immunoglobulin heavy chain gene locus predict better survival in gastric diffuse large B‐cell lymphoma. Clin Cancer Res 2008; 14: 3002–10. [DOI] [PubMed] [Google Scholar]
- 45. Streubel B, Huber D, Wohrer S, Chott A, Raderer M. Frequency of chromosomal aberrations involving MALT1 in mucosa‐associated lymphoid tissue lymphoma in patients with Sjogren's syndrome. Clin Cancer Res 2004; 10: 476–80. [DOI] [PubMed] [Google Scholar]


