Abstract
During the last few decades, many studies have been performed on the early detection of cancer using noninvasive or minimally invasive techniques in lieu of traditional excisional biopsy. Early detection can make an immense difference because cancer treatment is often simpler and more effective when diagnosed at an early stage. Cancer detecting methods may help physicians to diagnose cancer, to dissect the malignant region with a safe margin, and to evaluate the tumor bed after resection. In this paper, the advanced hyperspectral imaging system has been assessed using infrared wavelengths region for tumor detection. We were able to identify an appropriate wavelength region for cancer detection, spatially resolved images, and highlight the differences in reflectance properties of cancerous versus non‐cancerous tissues. The capability of this instrument was demonstrated by observing gastric tumors in 10 human subjects. The spectral signatures were extracted and evaluated in cancerous and non‐cancerous tissues. Processing means with the standard deviation of the spectral diagram, support vector machine, and first derivatives and integral of in spectral diagram were proposed to enhance and detect the cancerous regions. The first derivatives in spectral region between 1226–1251 nm and 1288–1370 nm were proposed as criteria that successfully distinguish between non‐cancerous and cancerous tissue. The results of this study will lead to advances in the optical diagnosis of cancer. (Cancer Sci 2011; 102: 852–857)
To reduce the morbidity and mortality of the cancer, many efforts have been made to improve early diagnostic and therapeutic procedures. One of the most challenging concerns in medical procedures in diagnosis or management of tumors is the segmentation of tumors because of the ambiguity between the cancerous and the non‐cancerous adjacent tissues. In almost all endoscopic and laparoscopic procedures, surgeons face the problem of detecting and localizing tumors before a biopsy, to determine the border for resection, and to evaluate the resection bed after tumor extraction. Imaging techniques such as radiography and endoscopy are routinely used as diagnostic tools for patients suspected of having gastric cancer. However, diagnosis is subjective and thus requires expert judgment. Gene detection in the target organ is a promising method for cancer diagnosis.( 1 , 2 ) However, the problem of low specificity of this method due to the large variety of gene expression patterns must be resolved before the method can be used clinically.( 1 , 3 ) Histopathology is the current gold standard for diagnosis of cancer. On the other hand, even this method has caveats such as its invasiveness and great dependence on the judgment of the pathologists. Therefore, new techniques that are simple and enable rapid detection of cancer would be very helpful.( 1 ) Recently, Fourier transform infrared spectroscopy has widely been used as a cancer diagnostic technique that can only be applied to point measurements.( 1 , 4 , 5 , 6 , 7 , 8 , 9 )
The hyperspectral imaging that operates as a tunable optical band pass filter has been utilized to discriminate between non‐cancerous and cancerous tissue.( 10 ) Nonetheless, this method is limited to the visible wavelength range and requires injecting a fluorescent material, porphyrin, which can be potentially hazardous. The hyperspectral imaging was evaluated for cytologic diagnosis of cancer cells( 11 ) and the high‐resolution hyperspectral imaging microscopy was evaluated to detect abnormalities in skin tissue using H&E stained preparations of normal and abnormal skin, benign nevi and melanomas.( 12 ) Conversely, these methods are limited to the use at the cytological level, visible wavelength range, and require sample preparation.
The goal of this research is to develop an advanced hyperspectral image processing technique to obtain spatially resolved images and highlight the differences in luminescent properties of malignant tumors versus non‐cancerous tissues. This paper lays the groundwork for these goals by demonstrating the instrument’s capabilities for gastric cancer detection. Three image processing methods were proposed and evaluated. We tried to find an appropriate wavelength region for discrimination of cancerous and non‐cancerous tissue in ten patients.
Materials and Methods
To capture the hyperspectral image data, the ImSpector Spectral Camera, SW ver 2.739 (Spectral Imaging Ltd, Oulu, Finland) is employed. This model has the spectral range 1000–2500 nm and spectral resolution 6.29 nm. Figure 1 shows the acquisition setup. The light source consists of two 150 W halogen lamps with diffusing reflectors that were fixed on a frame with the camera and illuminate the object to be captured. The two lamps provide a fairly uniform illumination on the subject. Also, the camera was calibrated and the frame was fixed. The frame was installed on a computer‐controlled linear actuator. The linear actuator was fixed on a bridge that was itself installed over the surgical bed. Therefore, the distance between the lens and the sample was kept constant.
Figure 1.
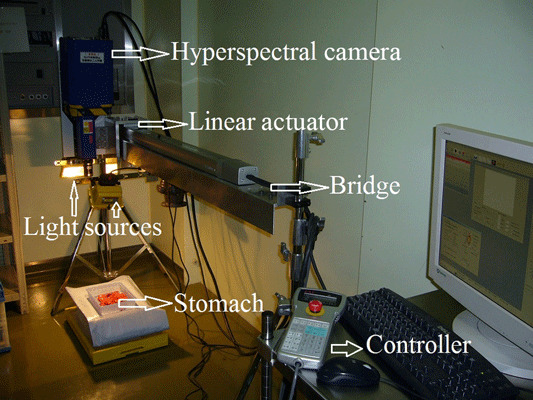
The acquisition setup.
Prior to this study, approval was obtained from the Institution of Ethics Committee, Tokyo Medical and Dental University. Ten patients whom were suffering from a gastric cancer underwent a total gastrectomy. After the surgical operation, for pathological inspection, the gastric cancer samples were taken from randomly selected patients. Immediately after gastrectomy, the stomachs were prepared for hyperspectral imaging from the resected stomachs. The hyperspectral images of stomach from the mucosal surface of the tumor were collected. These resected stomachs consist of normal tissue and the tumor. It was believed that the region with 5–10‐cm distance from the tumor was to be healthy and that was confirmed by pathologic results. A total of 101 hyperspectral images were captured from these ten resected stomach. After image the stomachs were sent to the pathological laboratory. Pathologic diagnosis was carried out by a professional pathologist and the results were compared with the image processing results.
In a hyperspectral image, each pixel has a sequence of reflectance in different spectral wavelengths that can display the spectral diagram of that pixel. Figure 2 shows a schematic view of the hyperspectral image. The differences in spectral diagrams of the normal and cancerous tissues were evaluated and four different image processing methods were compared to classify the tumor and normal tissue. The methods that were utilized for cancer detection were support vector machine (SVM), a filter based on the standard deviation, a filter designed based on first derivative, and a wavelet decomposition method that are explained in the following sections. We tried to find a wavelength region that has more functional information for cancer detection.
Figure 2.
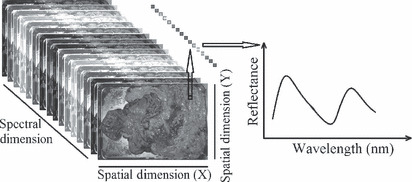
Left side, a schematic view of a hyperspectral image of stomach is shown. In right side, a spectral graph of a sample pixel is shown. The graph depicts the normalized reflectance for each wavelength in that pixel.
Hyperspectral image capturing. The halogen lamps illuminate the stomach and the camera objective lens collects the radiation from the tissue and displays an image on the entrance slit plane. The slit determines the field of imaging in spatial directions. The radiation from the slit is projected to the prism‐grating‐prism components (ImSpector optics); therefore the direction of propagation of the radiation changes on its wavelength. Every point of the tissue is represented on the matrix detector by a series of monochromatic points that makes a continuous spectrum in the direction of the spectral axis. The imaging technique used with this camera is a pushbroom scanner; the slit entrance limits the imaging field. By moving the camera between subsequent images, eventually all regions and wavelengths of the object are captured. By reconstructing the subsequent images, a monochromatic spectral image for each wavelength can be constructed.
The linear actuator, a ROBO Cylinder Slider, model RCS‐SM‐A‐100‐H‐1000‐T1‐S (IAI Corporation, Shizuoka, Japan), is used to move the camera. This actuator is controlled by an XSEL‐J‐1‐100A‐N3‐EEE‐2‐1 type controller (IAI Corporation). The movement and velocity are adjusted by a setting tool that is connected to the controller. The actuator moves the camera with a constant velocity (10 mm/s).
Data normalization. The problem with the spectral non‐uniformity of the illumination device and the influence of the dark current are eliminated through normalization of the image data to find the normalized reflectance of the specimen. A standard reference white board was placed in the scene of imaging and its data were utilized as the white reference. This white reference is a standard reflectance that should be used for data normalization which shows maximum standard reflectance in each wavelength and in capturing time temperature. The reflectance from the board provides an estimate of the incident light on the organs at each wavelength and normalizes the temperature changes, which is used in normalization of the spectrum. The dark current was captured by keeping the camera shutter closed. Then the data were normalized to find a relative reflectance using the following equation:
| (1) |
where 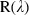 is the calculated relative reflectance value for each wavelength,
is the calculated relative reflectance value for each wavelength, 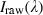 is the raw data radiance value of a given pixel, and
is the raw data radiance value of a given pixel, and 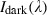 and
and 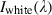 are, respectively, the dark current and the white board radiance acquired for each line and spectral band of the sensor.
are, respectively, the dark current and the white board radiance acquired for each line and spectral band of the sensor.
Spectral diagrams. Each point in the stomach tissue has a sequence of relative reflectance in different wavelength that makes the spectral diagram or spectral signature of a point. The spectral diagrams in all tumor and normal tissues in all 10 cases were evaluated to find a difference between tumor and normal tissue. Figure 3 shows nine samples of spectral signatures of normal tissue and nine samples of spectral signatures of the tumors were captured of several regions.
Figure 3.
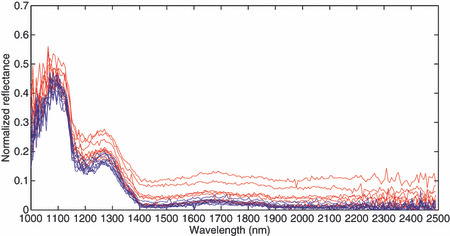
Normalized reflectance spectra. The horizontal axis shows different wavelengths in nanometers and the vertical axis shows the normalized reflectance. Tumor pixels are shown in red and normal pixels are shown in blue.
The differences of spectral signatures in tumors and normal tissues that are consistent in all cases and regions are evaluated to find the best classification method. It is obvious in Fig. 3 that one of the main differences between normal tissues and tumors is in standard deviations of the spectral signatures. The standard deviations of spectral signature of the tumor spatially and spectrally are much higher compared to normal tissues. The area under the spectral diagram looks higher in tumor compared to normal tissue and the slops about 1200–1400 nm seem lower in tumor regions. Therefore, these differences were employed in finding the best method for tumor classification in hyperspectral images.
Support vector machines. SVM have been employed in hyperspectral image segmentation and have shown superior performance to the other available segmentation methods.( 13 , 14 , 15 , 16 , 17 ) Therefore, an SVM method was compared with the other proposed method. The least squares SVM with kernel method, a modified configuration of the SVM, was employed in this paper.( 13 , 18 ) The optimization function of the SVM was expressed as follows:
| (2) |
subject to the equality constraints
| (3) |
where w is the weighting vector, b is the bias term, e is for misclassifications, and γ is the tuning parameter. This constrained optimization problem can be solved by determining the saddle points in the Lagrange functional as:
| (4) |
where 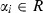 are Lagrange multipliers that can be positive or negative in the least squares SVM formulation. The RBF kernel was as follows:
are Lagrange multipliers that can be positive or negative in the least squares SVM formulation. The RBF kernel was as follows:
| (5) |
where σ is constant.
The grid search method was used to select the kernel parameter.( 19 ) Leave‐one‐out cross‐validation was employed in this method.
Spectral standard deviation. To enhance the differences of the standard deviations in normal tissue and tumors, two methods were used and compared. In the first method, the standard deviation is calculated in a square around each pixel in each wavelength. In the other words, the standard deviation in two spatial dimensions is calculated as follows.
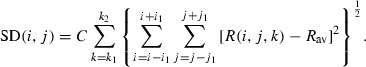 |
(6) |
In second method, the three‐dimensional (two spatial and one spectral dimension) standard deviation is calculated as follows.
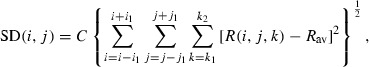 |
(7) |
where SD is the standard deviation, i and j are spatial coordinates, i 1 and j 1 are predefined neighboring area size, k is the wavelength band number, k 1 and k 2 are the rang of the wavelength bands, C is a coefficient, R is the reflectance, and R av is the mean of the reflectance. By changing i 1, j 1, k 1, and k 2, the contrast between tumor regions and normal tissues was tried to maximize. The equation( 7 ) could enhance the tumor visualization better than equation.( 6 ) By changing k 1 and k 2, a key wavelengths rang was chosen from all wavelengths that provides the best differentiation between normal and malignant tissues while potentially reducing the amount of data collected in subsequent work. The experiments showed that k 1 = 1251 nm and k 2 = 1302 nm results were the best enhanced. In each spatial dimension, different filter size can be assigned. By changing the filter size, through i 1 and j 1, the best enhancement was defined as i 1, j 1 = 3 which was used for detection. The pixels that were detected as tumor tissue due to the glare were eliminated. The surfaces under the spectral diagram in glare pixels have a high magnitude that can easily be detected. These pixels were detected by defining a threshold equal to the average intensity of each image multiplied by seven that was enough to eliminate the glare regions.( 20 )
Cancer index. The normalized difference index is a fast and simple method to evaluate hyperspectral data. The large‐sized hyperspectral image can be processed and visualized by this method in a short time. Many normalized difference indices have been proposed in different studies.( 21 , 22 , 23 , 24 ) To find the optimum difference index, the spectral diagrams of the tumor regions, and normal tissues were compared through different methods. To increase the signal to noise ratio, a median noise removal filter was employed. The subtraction of the spectral diagrams, the absolute value of the subtraction of the spectral diagrams, the squared value of the subtraction of the spectral diagrams, first and second derivatives, and integrals differences are compared in all wavelengths. These methods were evaluated for all regions in hyperspectral images. The optimum contrasts between the tumors and normal tissue were result from the first derivative method in between 1226–1251 and 1288–1370 nm wavelength regions. The following equation was designed to calculate the normalized difference cancer index:
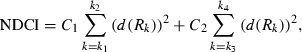 |
(8) |
where NDCI is the normalized cancer index, C 1 and C 2 are coefficients, R k is the normalized reflectance in wavelength k after smoothing, d(R k ) is the first derivative of R k with respect to wavelength, k 1, k 2, k 3 and k 4 are the range of the wavelength bands which were defined as k 1 = 1226 nm, k 2 = 1251 nm, k 3 = 1288, and k 4 = 1370 nm. The integral of the spectral signatures in different wavelength regions were evaluated to find the maximum contrast between tumor and normal tissues. The maximum contrast was found in the integral of the spectral diagram between 1057 and 2440 nm.
Results
The experiment was performed using the stomachs after total gastrectomy. The captured hyperspectral images were analyzed through the methods that were explained above. The standard deviation methods, SVM, integral filters, and NDCI equation were employed as filters to enhance the tumor region in images captured with the hyperspectral camera. To obtain a normal/tumor classification, the grayscale images were binarized by defining a threshold value. The capturing time for each image is about 15 s and calculation time is <20 s using Matlab software (MathWorks, Inc., Natick, MA, USA) on a 3.6 GHz computer with a 2 GB RAM. A generic methodology for target‐detection and performance evaluation was applied. The thresholds were adjusted in order to maximize the tumor detection to ensure detection of all the tumor regions. The performance was evaluated using the quality of detection with respect to the pathology.
A performance criterion for cancer detection was a false negative rate (FNR) and the false positive rate (FPR), which was calculated for each hyperspectral image. When a pixel was not detected as a tumor pixel, the detection was considered as a false negative if the pixel was an actual tumor pixel in pathological results. The FNR was defined as the number of false negative pixels divided by the total number of the tumor pixel. When a pixel was detected as a tumor tissue, the detection was a false positive if the pixel was not a tumor. The FPR was defined as the number of false positive pixels divided by the total number of normal tissue. The numerical results of the FPR and FNR and a comparison among standard deviation method, SVM, NDCI, and integral method are given in Table 1. Figure 4 shows the tumor enhancement and detection through integral and NDCI methods. As it is obvious in this image and Table 1, NDCI is the best method for cancer detection in hyperspectral images. These results were compared with pathological data. The method worked well even in tumors with a depth <2–3 mm and was covered with normal mucosa.
Table 1.
Evaluation results
| Method | SD (%) | SVM (%) | Integral (%) | NDCI (%) |
|---|---|---|---|---|
| FPR | 18 | 20 | 12 | 7 |
| FNR | 46 | 27 | 16 | 9 |
FNR, false negative rate; FPR, false positive rate; NDCI, normalized cancer index; SD, standard deviation; SVM, support vector machine.
Figure 4.
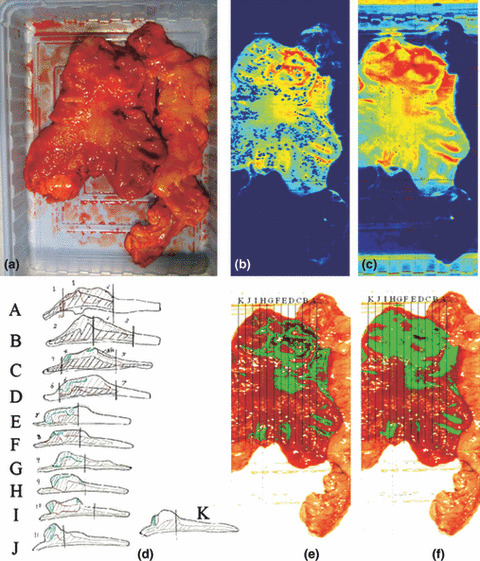
Cancer detection using the normalized cancer index (NDCI) and integral filter and comparison with pathological results. (a) the RGB image; (b) the cancer enhanced regions using integral filter in the hyperspectral image (1057–2440 nm), the tissues are shown in a blue to red spectrum and the red regions represent the tumor; (c) the cancer enhanced regions using NDCI; (d) pathological results; (e) detected tumor using integral filter (f) detected tumor using NDCI.
Discussion
Cancer detection is one of most important challenges in medicine today. The incidence of gastric cancer remains the second most common cancer worldwide. The highest incidence rates are found in Asia (especially Japan and Korea).( 25 , 26 ) In many cancer cases, if the cancer is diagnosed earlier and treated, the patient will have a better prognosis and a much greater chance of a full recovery. The initial diagnosis of gastric cancer often is delayed as up to 80% of patients are asymptomatic during the early stages of gastric cancer. Esophagogastroduodenoscopy is the diagnostic imaging procedure of choice in the work up of gastric cancer.( 27 ) In this paper, the hyperspectral imaging technique is employed to detect cancer. While previous studies that employed hyperspectral in visible wavelength and microscopically,( 10 , 12 ) in current paper invisible light at the macroscopic level was utilized for cancer detection. Several methods were evaluated to find the best method for cancer detection. Among the proposed methods, NDCI resulted in the lowest FPR and FNR. The FPR in NDCI method is 7% which shows 93% specificity. The FNR is 9% that shows 91% sensitivity. It is a great improvement compared to the reported specificity of 74.2% for identifying normal fibroblast cell type based on spatial and spectral algorithms.( 11 ) The sensitivity of Fourier transform infrared spectroscopy for diagnosis of cervical cancer and gastric cancer has reached 79% and 88.6%, respectively.( 1 , 8 ) Various research groups have developed laser‐induced fluorescence techniques for diagnosis of cancer.( 10 , 28 , 29 ) It is well known that biological tissues exhibit fluorescent properties when excited with ultra‐violet light.( 10 ) However, in current method the cancer can be detected without injecting a fluorescent material. Esophagogastroduodenoscopy is a sensitive and specific diagnostic test, especially when combined with endoscopic biopsy. Multiple biopsy specimens should be obtained from any visually suspicious areas; this step involves repeated sampling at the same tissue site, so that each subsequent biopsy reaches deeper into the gastric wall.( 30 ) Histopathology is the current gold standard for diagnosis of cancer. However, even this method has problems such as invasiveness, great dependence on the judgment of pathologists, and needs time for results preparing. Moreover, the biopsy specimens can only be captured form a few points. A simple, noninvasive, and reliable technique that enables rapid detection of cancer would aid many physicians.
Each patient’s sample was imaged about 10 times that may be an evidence for reproducibility of the imaging and the method. The proposed method detects a cancerous tissue pixel by pixel. The detection of one pixel as a cancerous pixel does not depend to adjacent pixels. Therefore, the cancer size or shape would not cause a problem. It means this method can detect the tumors with a size of <0.5 mm.
Using the white reference and dark currents could normalize the image intensities. The white reference shows the maximum reflectance in each wavelength and current temperature. Although the cancerous and non‐cancerous spectra presented in Fig. 3 seem to have similar spectral shape, after noise removal using a median filter, the difference could be extracted using the proposed method. The changes of the spectral signature are strongly related to the proteins changes as shown in Tsenkova’s paper.( 31 ) Excessive expression of p53, expression of Trefoil factor family 1, overexpression of c‐erb B‐2 (HER‐2/neu), high intranuclear levels of β‐catenin protein( 32 ) are some of changes in molecular level that can perform a key role in spectral changes. The normalized spectra for each pixel were employed for cancer detection. Therefore, the sample size did not take effect on the results. The difference between cancerous and non‐cancerous region due to higher reflectance at all region was extracted using integral method. Although this difference is meaningful after data normalization, the first derivative in the mentioned wavelength regions resulted in better detection.
The hyperspectral camera in the current shape cannot be used in endoscopic diagnosis. However, using the more useful bands that are extracted would be used in laparoscopic and endoscopic procedures. The mentioned wavelength region may be utilized to insert narrow band filters to the laparoscope or optic fibers may be utilized which are two possibilities for using this method to access organs within the body with minimal invasiveness. Moreover, this modification would decrease the detection time to be converted to endoscopic procedure. This method may be replaced by biopsy and pathological diagnosis.
The spatial resolution can increase by decreasing the distance between the camera and the sample and by decreasing the velocity of the camera. Although the idea of the spectral signature may be utilize in microscopic level, a macroscopic method was proposed in the current paper. Although more evaluation with more patient data may result in a higher statistical power, the 10 patients imaged represent an evidence to start up a bigger project with more patients. This patient number was defined based on similar research project for evaluation of the cancer detection methods.( 10 , 11 , 12 )
Infrared spectroscopy is based on molecular overtone and combination vibrations. One advantage is that NIR can typically penetrate much farther into a sample than visible wavelength. Therefore, infrared spectroscopy is a sensitive technique that can be very useful in detecting bulk material with no sample preparation. The molecular overtone and combination bands seen in the infrared (IR) are typically very broad, leading to complex spectra; it can be difficult to assign specific features to specific chemical components. However, it is possible to employ multiple wavelengths and signal processing techniques to analyze the IR data for detection of specific properties or component.( 33 )
Hyperspectral image aids cancer detection and can be used not only for diagnosis but also for determining the tumor margin. The extent of gastric resection is an important concern for surgeons. Safe margin resection accompanied with low operative mortality and morbidity will remove the field change of gastric mucosa, reduces the risk of resection margin involvement, and therefore yields better long‐term survival. Moreover, it may be employed to evaluate the tumor base after tumor resection to ensure that the tumor resection was complete. Hyperspectral imaging allows physicians to survey and examine a vast area less invasively and has the potential to be employed in other types of cancer.
Disclosure Statement
None.
Acknowledgments
The authors would like to thank Mr. Tatsuhiko Saito and Masato Tanaka of Sumitomo Electric Industries, Ltd. for providing the hyperspectral sensor. This research was partly supported by the Grant in Aid for Scientific Research (A) #20246069.
References
- 1. Fujioka N, Morimoto Y, Arai T, Kikuchi M. Discrimination between normal and malignant human gastric tissues by Fourier transform infrared spectroscopy. Cancer Detect Prev 2004; 28(1): 32–6. [DOI] [PubMed] [Google Scholar]
- 2. Tannapfel A, Schmelzer S, Benicke M et al. Expression of the p53 homologues p63 and p73 in multiple simultaneous gastric cancer. J Pathol 2001; 195: 163–70. [DOI] [PubMed] [Google Scholar]
- 3. Tolbert D, Fenoglio‐Preiser C, Noffsinger A et al. The relation of p53 gene mutations to gastric cancer subsite and phenotype. Cancer Causes Control 1999; 10: 227–31. [DOI] [PubMed] [Google Scholar]
- 4. Rigas B, Morgello S, Goldman IS, Wong PT. Human colorectal cancers display abnormal Fourier‐transform infrared spectra. Proc Natl Acad Sci USA 1990; 87: 8140–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wong PT, Goldstein SM, Grekin RC, Godwin TA, Pivik C, Rigas B. Distinct infrared spectroscopic patterns of human basal cell carcinoma of the skin. Cancer Res 1993; 53: 762–5. [PubMed] [Google Scholar]
- 6. Cohenford MA, Rigas B. Cytologically normal cells from neoplastic cervical samples display extensive structural abnormalities on IR spectroscopy: implications for tumor biology. Proc Natl Acad Sci USA 1998; 95: 15327–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rigas B, Wong PT. Human colon adenocarcinoma cell lines display infrared spectroscopic features of malignant colon tissues. Cancer Res 1992; 52: 84–8. [PubMed] [Google Scholar]
- 8. Cohenford MA, Godwin TA, Cahn F, Bhandare P, Caputo TA, Rigas B. Infrared spectroscopy of normal and abnormal cervical smears: evaluation by principal component analysis. Gynecol Oncol 1997; 66: 59–65. [DOI] [PubMed] [Google Scholar]
- 9. McIntosh LM, Jackson M, Mantsch HH, Stranc MF, Pilavdzic D, Crowson AN. Infrared spectra of basal cell carcinomas are distinct from non‐tumor‐bearing skin components. J Invest Dermatol 1999; 112: 951–6. [DOI] [PubMed] [Google Scholar]
- 10. Martin ME, Wabuyele MB, Chen K et al. Development of an advanced hyperspectral imaging (HSI) system with applications for cancer detection. Ann Biomed Eng 2006; 34(6): 1061–8. [DOI] [PubMed] [Google Scholar]
- 11. Siddiqi AM, Li H, Faruque F et al. Use of hyperspectral imaging to distinguish normal, precancerous, and cancerous cells. Cancer Cytopathol 2008; 114(1): 13–21. [DOI] [PubMed] [Google Scholar]
- 12. Dicker DT, Lerner J, Belle PV et al. Differentiation of normal skin and melanoma using high resolution hyperspectral imaging. Cancer Biol Ther 2006; 5(8): 1033–8. [DOI] [PubMed] [Google Scholar]
- 13. Camps‐Valls G, Bruzzone L. Kernel‐based methods for hyperspectral image classification. IEEE Trans Geosci Remote Sens 2005; 43: 1351–62. [Google Scholar]
- 14. Camps‐Valls G, Gomez‐Chova L, Calpe‐Maravilla J et al. Robust support vector method for hyperspectral data classification and knowledge discovery. IEEE Trans Geosci Remote Sens 2004; 42(7): 1530–42. [Google Scholar]
- 15. Melgani F, Bruzzone L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans Geosci Remote Sens 2004; 42(8): 1778–90. [Google Scholar]
- 16. Huang C, Davis LS, Townshend JR. An assessment of support vector machines for land cover classification. Int J Remote Sens 2002; 23(4): 725–49. [Google Scholar]
- 17. Brown M, Lewis HG, Gunn SR. Linear spectral mixture models and support vector machines for remote sensing. IEEE Trans Geosci Remote Sens 2000; 38(5): 2346–60. [Google Scholar]
- 18. Akbari H, Kosugi Y, Kojima K, Tanaka N. Hyperspectral Image segmentation and its application in abdominal surgery. Int J of Functional Informatics and Personalised Medicine 2009; 2(2): 201–16. [Google Scholar]
- 19. Bao Y, Liu Z. A fast grid search method in support vector regression forecasting time series. LNCS 2006; 4224: 504–11. [Google Scholar]
- 20. Akbari H, Kosugi Y, Kojima K, Tanaka N. Detection and analysis of the intestinal ischemia using visible and invisible hyperspectral imaging. IEEE Trans Biomed Eng 2010; 57(8): 2011–7. [DOI] [PubMed] [Google Scholar]
- 21. Richardson AD, Duigan SP, Berlyn GP. An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol 2002; 153: 185–94. [Google Scholar]
- 22. Moran JA, Mitchell AK, Goodmanson G, Stockburger KA. Differentiation among effects of nitrogen fertilization treatments on conifer seedlings by foliar reflectance: a comparison of methods. Tree Physiol 2000; 20: 1113–20. [DOI] [PubMed] [Google Scholar]
- 23. Datt B, McVicar TR, Van Niel TG, Jupp DLB, Pearlman JS. Preprocessing EO‐1 hyperion hyperspectral data to support the application of agricultural indexes. IEEE Trans Geosci Remote Sens 2003; 41: 1246–59. [Google Scholar]
- 24. Poss JA, Russell WB, Grieve CM. Estimating yields of salt‐ and water‐stressed forages with remote sensing in the, visible and near infrared. J Environ Qual 2006; 35: 1060–71. [DOI] [PubMed] [Google Scholar]
- 25. Fenoglio‐Preiser C, Carneiro F, Correa P. Tumours of the stomach. In: Hamilton SR, Aaltonen LA, eds. World Health Organization Classification of Tumors: Pathology and Genetics of Tumors of the Digestive System. Lyon, France: IARC Press, 2000; 37–52. [Google Scholar]
- 26. Ahmad Z, Idrees R, Sahar Azad N, Ahmed R, Ahsan A, Asghar N. Gastric carcinoma: typing, staging, lymph node and resection margin status on gastrectomy specimens. JCPSP 2007; 17(9): 539–42. [PubMed] [Google Scholar]
- 27. Cappell MS, Friedel D. The role of esophagogastroduodenoscopy in the diagnosis and management of upper gastrointestinal disorders. Med Clin North Am 2002; 86: 1165–216. [DOI] [PubMed] [Google Scholar]
- 28. Andersson‐Engels SJ, Johansson J, Svanberg K, Svanberg S. Fluorescence imaging and point measurement of tissue: applications to the demarcation of malignant tumors and atherosclerotic lesions from normal tissue. Photochem Photobiol 1991; 53: 807–12. [DOI] [PubMed] [Google Scholar]
- 29. Sung KC, Mirabal YN, Atkinson EN et al. Combined fluorescence and reflectance spectroscopy for in vivo detection of cervical pre‐cancer. J Biomed Opt 2005; 10(2): 2–31. [DOI] [PubMed] [Google Scholar]
- 30. Layke JC, Lopez PP. Gastric cancer: diagnosis and treatment options. Am Fam Physician 2004; 69: 1133–40. [PubMed] [Google Scholar]
- 31. Tsenkova R. Aquaphotomics: dynamic spectroscopy of aqueous and biological systems describes peculiarities of water. J Near Infrared Spectrosc 2009; 17: 303–14. [Google Scholar]
- 32. El‐Rifai W, Powell SM. Molecular biology of gastric cancer. Semin Radiat Oncol 2002; 12(2): 128–40. [DOI] [PubMed] [Google Scholar]
- 33. Balabin RM, Safieva RZ, Lomakina EI. Comparison of linear and nonlinear calibration models based on near infrared (NIR) spectroscopy data for gasoline properties prediction. Chemom Intell Lab 2007; 88(2): 183–8. [Google Scholar]


